Abstract
Background
Posterior fossa A (PFA) ependymomas are one of 9 molecular groups of ependymoma. PFA tumors are mainly diagnosed in infants and young children, show a poor prognosis, and are characterized by a lack of the repressive histone H3 lysine 27 trimethylation (H3K27me3) mark. Recently, we reported overexpression of chromosome X open reading frame 67 (CXorf67) as a hallmark of PFA ependymoma and showed that CXorf67 can interact with enhancer of zeste homolog 2 (EZH2), thereby inhibiting polycomb repressive complex 2 (PRC2), but the mechanism of action remained unclear.
Methods
We performed mass spectrometry and peptide modeling analyses to identify the functional domain of CXorf67 responsible for binding and inhibition of EZH2. Our findings were validated by immunocytochemistry, western blot, and methyltransferase assays.
Results
We find that the inhibitory mechanism of CXorf67 is similar to diffuse midline gliomas harboring H3K27M mutations. A small, highly conserved peptide sequence located in the C-terminal region of CXorf67 mimics the sequence of K27M mutated histones and binds to the SET domain (Su(var)3-9/enhancer-of-zeste/trithorax) of EZH2. This interaction blocks EZH2 methyltransferase activity and inhibits PRC2 function, causing de-repression of PRC2 target genes, including genes involved in neurodevelopment.
Conclusions
Expression of CXorf67 is an oncogenic mechanism that drives H3K27 hypomethylation in PFA tumors by mimicking K27M mutated histones. Disrupting the interaction between CXorf67 and EZH2 may serve as a novel targeted therapy for PFA tumors but also for other tumors that overexpress CXorf67. Based on its function, we have renamed CXorf67 as “EZH Inhibitory Protein” (EZHIP).
The C-terminal part of EZHIP/CXorf67 is sufficient to bind and inhibit PRC2.
The C-terminus of EZHIP sequesters EZH2 activity by mimicking the H3K27M peptide.
EZHIP overexpression in PFA ependymoma leads to upregulation of PRC2 target genes.
Importance of the Study.
PFA ependymomas remain incurable in a large proportion of pediatric patients, but molecularly targeted therapies are lacking due to the absence of a clear driver. Downregulation of the repressive histone mark H3K27me3 has been reported as a distinct feature of these tumors, which correlated with overexpression of the previously uncharacterized protein CXorf67. We have identified the exact mechanism of action of how CXorf67, renamed EZHIP, inhibits PRC2 function, thereby causing downregulation of H3K27me3. We show that a short sequence in EZHIP, mimicking K27M mutated histones, is sufficient to inhibit EZH2 methyltransferase activity. Overexpression of EZHIP and rare but mutually exclusive H3K27M mutations in PFA ependymoma thus both lead to a de-repression of PRC2 target genes and cause upregulation of genes involved in neurodevelopmental processes. Our findings have identified overexpression of EZHIP as an oncogenic mechanism that may serve as the basis for developing the first targeted therapies for PFA ependymomas.
Ependymomas are neuroepithelial tumors occurring throughout all compartments of the central nervous system (CNS) and affecting both children and adults. Pediatric ependymomas arise almost exclusively intracranially and are associated with a poor 10-year overall survival of around 60%.1,2 Using DNA methylation profiling, we previously identified 9 molecular groups of ependymoma that show clear differences in demographics, molecular characteristics, and patient outcome.3 The vast majority of pediatric infratentorial ependymomas belong to the posterior fossa A (PFA) group, characterized by a poor prognosis and distinct from the PF-subependymoma and PFB groups, which are more prevalent in adult patients and have a much better outcome.3–5 Initial sequencing studies showed an absence of recurrent mutations or gene fusions in PFA ependymomas, suggesting that these tumors might be epigenetically driven.6,7 Interestingly, immunohistochemistry revealed that all PFA tumors, but no tumors from other molecular groups of ependymoma, lack the epigenetic histone H3 lysine 27 trimethylation (H3K27me3) mark that is thought to be involved in the negative regulation of gene expression.8 It has been shown that a reduction of the H3K27me3 mark drives aberrant gene expression in histone H3 K27M (H3K27M) mutant diffuse midline gliomas,9 suggesting that this may also hold true for PFA ependymomas. Indeed, PFA tumors also exhibit increased expression of genes involved in angiogenesis, cell cycle, and receptor tyrosine kinase signaling.4,5 However, H3K27M mutations, which inhibit the catalysis of H3K27 trimethylation by polycomb repressive complex 2 (PRC2), are rare in PFA ependymoma, suggesting a distinct mechanism responsible for reduced H3K27me3 levels.10–12
Recently, we found overexpression of an uncharacterized gene called chromosome X open reading frame 67 (CXorf67) in nearly all PFA ependymomas.12 Additionally, mutations in CXorf67 were identified in 9.4% of PFA ependymomas. Interestingly, expression and mutations of CXorf67 in PFA ependymomas were mutually exclusive with H3K27M mutations present in 4.2% of PFA ependymomas. Moreover, we found that CXorf67 interacts with core components of PRC2 such as enhancer of zeste homolog 2 (EZH2) and suppressor of zeste 12 homolog (SUZ12), and ectopic expression of CXorf67 in vitro resulted in a strong downregulation of H3K27me3, suggesting a crucial role of CXorf67 in modulating PRC2 function.12 However, a detailed molecular description of CXorf67 and its functional domains is still lacking. Also, the role of the mutations and the exact mechanism of how the protein may regulate PRC2 function have not yet been identified. In this study, we thus aimed to identify and characterize putative functional domains of CXorf67 and to elucidate the mode of action of how CXorf67 interacts with EZH2 and inhibits PRC2 function.
Materials and Methods
Cell Culture
HEK293T cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum.
Production of Lentiviral Particles and Generation of Stable Cell Lines
Lentiviral constructs were generated by replacing the Ngn2 gene of the FUW-TetO-Ngn2-T2A-puromycin construct published by Zhang et al13 with DNA sequences encoding for the CXorf67 full-length protein or CXorf67 truncates carrying a C-terminal FLAG-HA-tag. Lentiviruses were produced by co-transfecting lentiviral constructs with psPAX2 and pMD2.G into low-passage HEK293T cells using FugeneHD (Promega). For the generation of stable cell lines, supernatant containing lentiviral particles was collected and added to HEK293T cells. Protein expression was induced by addition of 1 µg/mL of doxycycline followed by selection with 1 µg/mL of puromycin.
Co-Immunoprecipitation and Mass Spectrometry Analysis
For co-immunoprecipitation (co-IP), CXorf67 full-length or truncate expressing cell lines were resuspended in lysis buffer (20 mM Tris-HCl pH 8, 200 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton-X100). The resulting cell lysates were precleared using mouse immunoglobulin G agarose beads (Sigma-Aldrich). Next, FLAG-tagged proteins were purified by incubating the precleared lysate with ANTI-FLAG-M2 affinity gel (Sigma-Aldrich). Proteins were eluted in elution buffer (50 mM NH4HCO3, 15 mM dithiothreitol, 0.1% sodium dodecyl sulfate) and further analyzed using an Orbitrap Fusion (Thermo Fisher Scientific) mass spectrometer. The mass spectrometry (MS) proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE14 partner repository with the dataset identifier PXD012318. A detailed description of the above procedures can be found in the Supplementary Material.
Western Blot Analysis
Whole cell lysates, nuclear and cytoplasmic extracts, or eluted fractions from co-IP experiments were separated on a 4–12% Bis-Tris gradient gel (Invitrogen) followed by transfer onto a 0.2 µm polyvinylidene difluoride membrane. Primary antibodies used for western blot analysis were targeted against H3K27me3, histone H3, FLAG-tag, EZH2, SUZ12, embryonic ectoderm development (EED), β-tubulin, lamin B1, or β-actin. Secondary antibodies used were goat anti-mouse horseradish peroxidase (HRP) and goat anti-rabbit HRP. Chemiluminescence was detected using the Chemostar ECL Imager device (Intas Science Imaging). A detailed protocol regarding sample preparation and antibodies utilized can be found in the Supplementary Material.
Immunocytochemistry
For immunocytochemistry, cells were fixed in 4% paraformaldehyde and stained with primary antibodies against FLAG-tag or H3K27me3. Secondary antibodies used were donkey anti-mouse Alexa Fluor 568 and donkey anti-rabbit Alexa Fluor 488. Cell nuclei were counterstained with 4′,6′-diamidino-2-phenylindole. Images were obtained using the Zeiss LSM 800 confocal microscope (Leica). A detailed description of the immunocytochemistry procedure and antibodies utilized can be found in the Supplementary Material.
EZH2 Inhibition
Inhibition of EZH2 activity was evaluated using the EZH2 Chemiluminescent Assay Kit (BPS Bioscience) as previously described.9 The K27M peptide (ATKAA RMSAPATGGVKKPHRYR), CXorf67 conserved peptide (VRMRASSPSPPGR), CXorf67 peptide (VWHAVRMRASS PSPPGRFFLP), and the corresponding M to G variant peptide (VWHAVRGRASSPSPPGRFFLP) were synthesized by GenScript. GSK126 was purchased from ApexBio (A3446). Dose-response curves were generated with GraphPad Prism 6 and based on triplicate measurements.
Multiple Sequence Alignment Analysis
CXorf67 ortholog sequences were obtained using the Identical Protein Groups resource of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/ipg/; Accessed September 15, 2018). RefSeq was selected as the source database. Protein sequences were exported as a single FASTA file which was used as the input for the Clustal Omega tool.15 Results were visualized in MVIew16 to identify consensus regions. All tools were used with the standard parameters and accessed via the EMBL-EBI webpage (https://www.ebi.ac.uk/Tools/msa/clustalo/ and https://www.ebi.ac.uk/Tools/msa/mview/; Accessed March 12, 2019).
Gene Expression Analysis
Gene expression profiles of untransduced HEK293T cells and HEK293T cells overexpressing CXorf67-N, CXorf67-M, CXorf67-C, or CXorf67-Full were generated on Affymetrix GeneChip human Genome U133 Plus 2.0 (U133v2) arrays. These profiles have been deposited in Gene Expression Omnibus under accession number GSE127941. Gene expression profiles of PFA tumors have been previously deposited in Gene Expression Omnibus under the identifier GSE64415. The corresponding CEL files were uploaded and analyzed using the R2 platform for genomic analysis and visualization (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi; Accessed March 4, 2019). Gene Ontology (GO) enrichment analysis of genes upregulated in CXorf67-Full cells was performed using the DAVID (Database for Annotation, Visualization and Integrated Discovery) tool.17 Gene Set Enrichment Analysis (GSEA) was performed using the GSEA software18 with a custom-made gene set that was based on the PRC2 target gene list from Bracken et al,19 filtered for genes occupied by SUZ12.
Modeling Peptide Binding to EZH2
For modeling the binding of CXorf67 to EZH2, the binding of the CXorf67 peptide to EZH2 (Protein Data Bank [PDB]: 6c23,20 chain K) was probed using PepSite,21 which gave a highly significant (P < 0.005) match at the H3K27M binding site. The structure of EZH2 in complex with H3K27M (PDB: 5hyn22) was then used to construct a model of the peptide WHAVRMRAS (401–409 from CXorf67) on the H3 peptide using Modeller.23 Finally, the peptide was superimposed onto the known structure using STAMP.24
Results
Core Components of PRC2 Interact with the C-Terminal Region of CXorf67
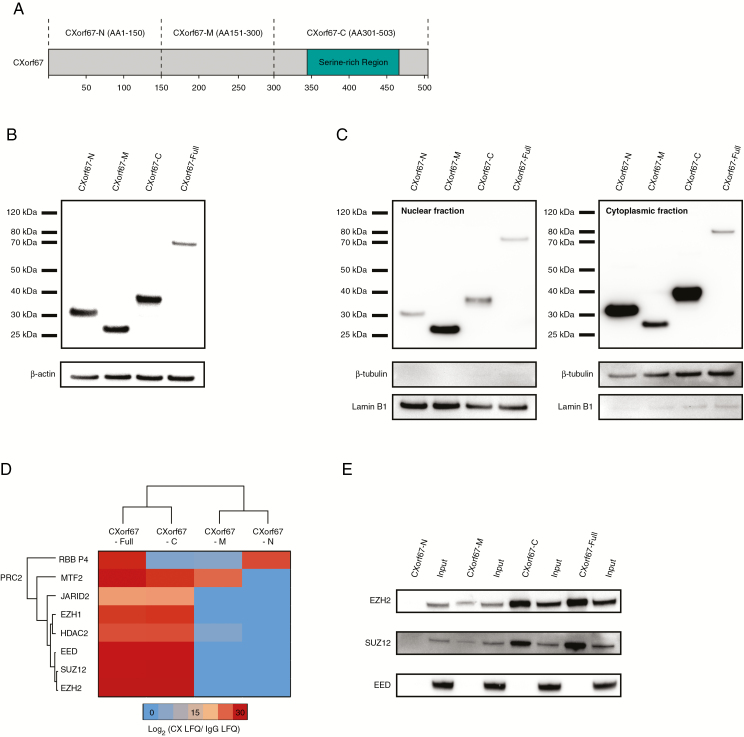
Previously, we showed that CXorf67 directly interacts with the PRC2 core components EZH2 and SUZ12, leading to inhibition of the complex and reduction of H3K27me3 in tested cell lines.12 However, the functional domain(s) of CXorf67 responsible for binding and inhibition of PRC2 are currently unknown. CXorf67 is predicted to be an intrinsically disordered protein and only a serine-rich domain has been identified in the C-terminus.12 To identify functional domains of CXorf67, we used lentiviral delivery to generate stable HEK293T cell lines expressing different CXorf67 truncates. For our initial approach, we started with 3 FLAG-HA-tagged truncates covering either the N-terminus (amino acids 1–150; CXorf67-N), the middle region (amino acids 151–300; CXorf67-M), or the serine-rich C-terminus (amino acids 301–503; CXorf67-C) of the protein (Fig. 1A). Expression of truncates was regulated by a tetO promoter allowing for a controlled induction of protein expression by the addition of doxycycline.13 Using the same inducible system, we also generated a cell line expressing the full-length protein (CXorf67-Full) for control purposes. Following selection and induction of protein expression, western blot analysis of whole cell lysates revealed single bands for each truncate and the full-length protein, confirming their successful expression (Fig. 1B). We have previously shown that the main accumulation site of CXorf67 is in the nucleus.12 To validate whether our CXorf67 protein variants could also localize to the nucleus, we isolated cytoplasmic and nuclear fractions from each transduced cell line and analyzed them for the presence of FLAG-tagged proteins (Fig. 1C). Indeed, the full-length protein and its truncated variants could be found in both fractions, making them suitable for downstream analyses.
Fig. 1.
Mass spectrometry analysis of CXorf67 truncates reveals interaction of the C-terminus of CXorf67 with PRC2 core components. (A) Overview of CXorf67 and the 3 truncates used in this study. The serine-rich region of the protein is highlighted in turquoise. (B) Detection of FLAG-tagged CXorf67 truncates and the full-length protein in lysates of HEK293T cells after transduction. (C) Western blot analysis of nuclear and cytoplasmic fractions obtained from transduced HEK293T cells. Both fractions contain CXorf67 truncates and the full-length protein. Lamin B1 was used as a nuclear loading control and β-tubulin as a loading control for the cytoplasmic fraction. (D) Heatmap depicting PRC2 components interacting with CXorf67-Full and at least 1 truncate. The scale bar depicts the ratio of the log2 signal intensities (LFQ) of PRC2 related proteins within each CXorf67 variant experiment over their respective immunoglobulin G control. (E) Validation of CXorf67-C interaction with EZH2 and SUZ12 by co-IP. A faint interaction can also be observed for CXorf67-M.
To identify which region of CXorf67 binds to PRC2 core components, we performed co-IP of the truncates or the full-length protein using an anti-FLAG antibody. Eluates were subjected to MS analysis to identify putative interaction partners. The resulting lists of interaction partners for each truncate were compared with the full-length protein to exclude proteins that were only binding to the truncated forms. We then selected for PRC2 components that interacted with CXorf67 truncates indicating the presence of a functional domain in this region (Fig. 1D). Data from the MS analysis showed that most PRC2 components, including the PRC2 core components EZH2, SUZ12, and EED, interacted exclusively with the C-terminal part of CXorf67 (CXorf67-C) and the full-length protein but not with the other truncates. Indeed, co-IP of CXorf67-C followed by western blotting resulted in the successful pull-down of the PRC2 core components EZH2 and SUZ12, thereby validating our MS results and being in line with our previous findings12 (Fig. 1E). Interestingly, we also detected a faint interaction of EZH2 and SUZ12 with CXorf67-M that was not picked up during the MS analysis. In contrast, a direct interaction with EED could not be validated for any of the truncates or the full-length protein.
The C-Terminal Region of CXorf67 Is Sufficient to Induce Downregulation of H3K27me3
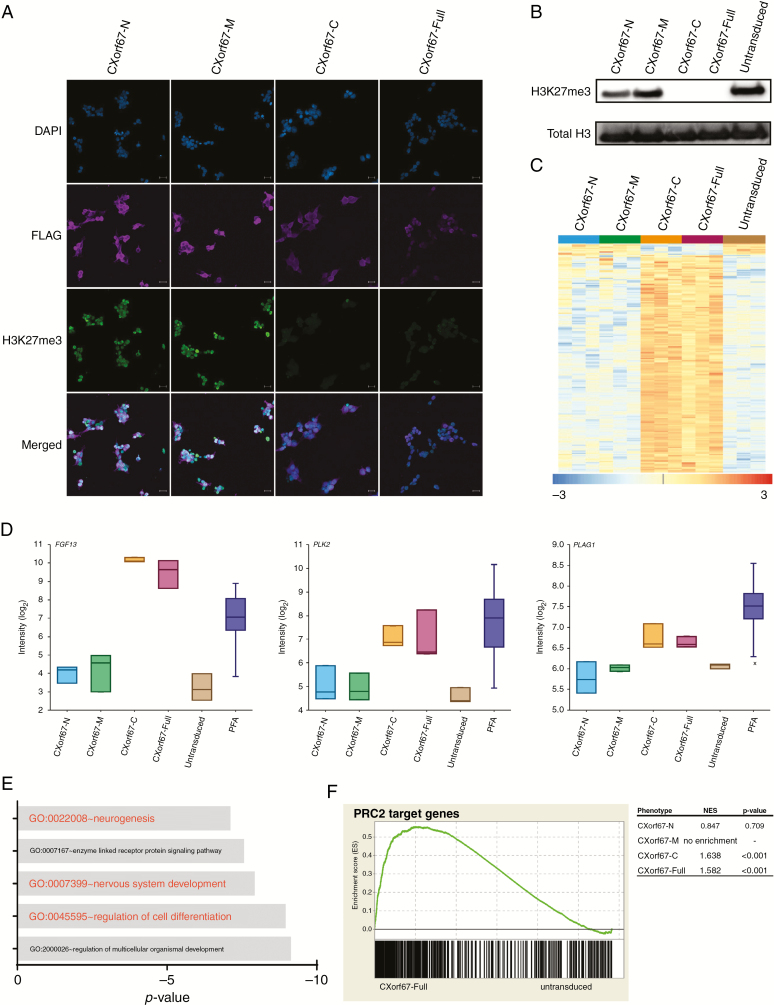
Having identified the C-terminal part of CXorf67 as the main region to interact with PRC2 core components, we wondered whether this region is also sufficient to inhibit PRC2 function. To investigate this, we stained each of our 4 transduced cell lines for the presence of FLAG-tagged proteins and H3K27me3. Indeed, CXorf67-C and CXorf67-Full expressing cell lines showed a robust reduction of H3K27me3, which was not observed in CXorf67-N or CXorf67-M expressing cells (Fig. 2A). This strong reduction of H3K27me3 levels caused by the overexpression of either CXorf67-C or the full-length protein was confirmed by western blot analyses of cell extracts (Fig. 2B). In contrast, neither CXorf67-N nor CXorf67-M had a substantial influence on H3K27me3 levels in these cells. Consequently, we concluded that the crucial domain responsible for binding and inhibition of PRC2 is located at the C-terminus of CXorf67.
Fig. 2.
The C-terminus of CXorf67 is crucial for PRC2 binding and inhibition. (A) Staining of HEK293T cells expressing CXorf67 truncates or the full-length protein using an H3K27me3 antibody. Cells expressing CXorf67-C or CXorf67-Full show a reduced H3K27me3 signal. Scale bar: 20 µm. (B) Western blot analysis of H3K27me3 levels in HEK293T cells expressing CXorf67 truncates or the full-length protein. A strong downregulation of H3K27me3 can be observed for CXorf67-C and CXorf67-Full. (C) Heatmap depicting significantly (P < 0.05) deregulated genes (n = 198) upon overexpression of CXorf67-Full in HEK293T cells. A strong deregulation of the same genes can also be observed for CXorf67-C expressing cells (n = 3 per condition). The scale bar depicts the z-scores of the gene expression levels. (D) Comparison of expression levels for FGF13, PLK2, and PLAG1 in transduced cell lines (n = 3), untransduced control cells (n = 3), and PFA tumors (n = 55). (E) GO enrichment analysis of CXorf67-Full upregulated genes (n = 188). X-axis depicts the p-value (log10). (F) GSEA showing significant enrichment of PRC2 target genes in CXorf67-C and CXorf67-Full cells compared with untransduced control cells. No significant enrichment is observed for the other truncated variants. A representative plot for CXorf67-Full is shown. NES: normalized enrichment score.
We next sought to identify transcriptional changes that may be associated with CXorf67-mediated downregulation of H3K27me3. Strikingly, transcriptional changes upon overexpression of CXorf67-Full were highly similar to those observed in CXorf67-C cells (Fig. 2C). In contrast, no deregulation of these genes was observed in any of the other truncate expressing cell lines. Interestingly, genes upregulated in CXorf67-C and CXorf67-Full expressing cells such as FGF13 and PLK2 were also found to be increased in PFA tumors (Fig. 2D). Although not part of the top differentially expressed genes, PLAG1, which has been repeatedly shown to be upregulated in diffuse midline gliomas with H3K27M mutations,25,26 was also found to be increased upon overexpression of CXorf67-C or CXorf67-Full. We next asked whether these genes could be associated with distinct functional processes. Indeed, GO enrichment analysis of upregulated genes showed a significant enrichment of GO terms associated with neurodevelopmental processes and differentiation (Fig. 2E). Furthermore, GSEA revealed that downregulation of H3K27me3 by both CXorf67-C and CXorf67-Full resulted in a de-repression of PRC2 target genes, which was not observed for the other truncates (Fig. 2F).
Identification of a Short Peptide Sequence Responsible for Binding and Inhibition of EZH2
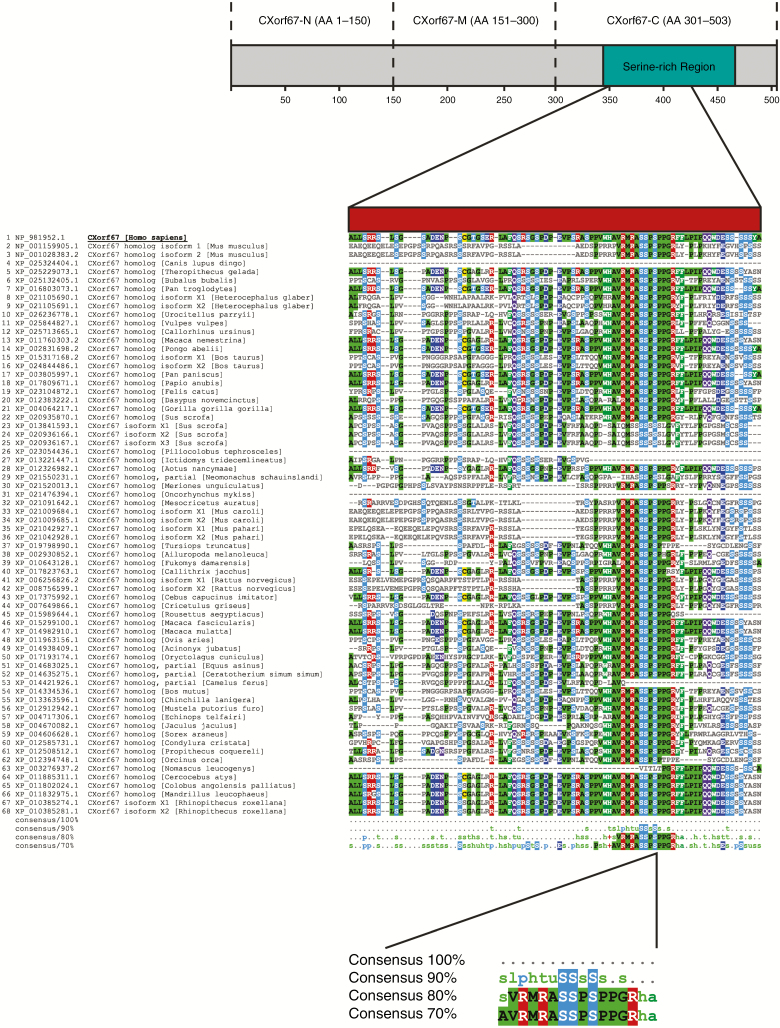
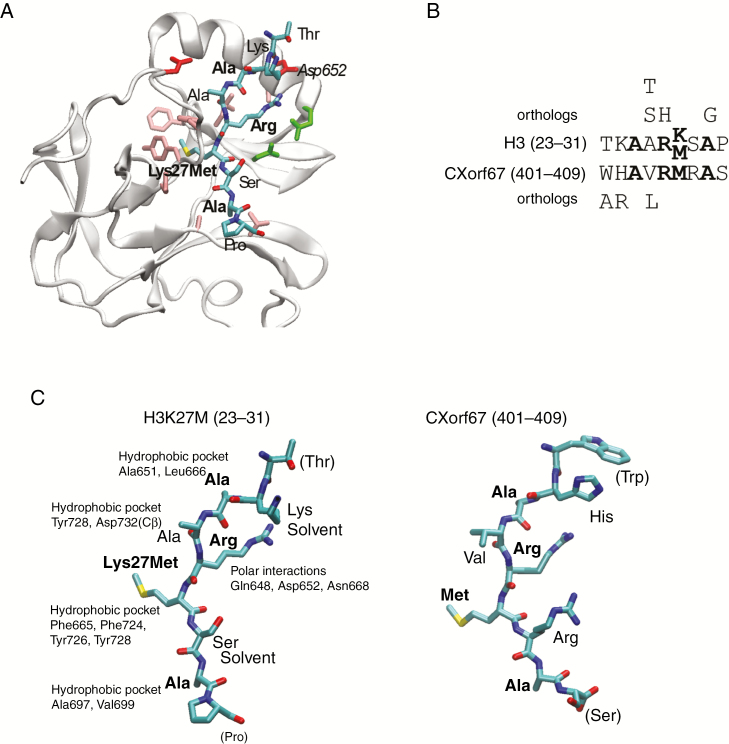
Having identified the C-terminus of CXorf67 as a presumable key factor in mediating H3K27 hypomethylation, we focused our attention on this region to elucidate the exact mechanism of PRC2 binding and inhibition. Driven by the hypothesis that the protein domain responsible for this critical function may be conserved among other species, we used the Clustal Omega tool15 to perform sequence alignments of multiple CXorf67 orthologs and visualized conserved regions corresponding to the C-terminal region using MView.16 Interestingly, a 13 amino acid long peptide sequence, matching amino acids 404 to 416 of human CXorf67, was found to be strongly conserved among 67 CXorf67 orthologs (Fig. 3). Next, we asked whether this peptide sequence is sufficient to inhibit PRC2 and whether the underlying mechanism might be similar to the one observed in H3K27M tumors, such as diffuse midline gliomas. Therefore, we relied on the study by Justin and colleagues,22 which had previously investigated the structure of EZH2 bound by an H3K27M peptide (Fig. 4A). Strikingly, part of the conserved peptide sequence of CXorf67 showed a strong similarity to the histone H3 mutant sequence where the Lys-to-Met mutation (27) matched a Met (406) of CXorf67. Additionally, both sequences contained an arginine in the (−1) position of lysine/methionine as well as 2 overlapping alanines (Fig. 4B). Furthermore, in silico analyses predicting the binding of either the H3K27M peptide or part of the CXorf67 conserved peptide to EZH2 showed that both peptides could bind to EZH2 in a similar manner (eg, with non-identical amino acids likely performing similar interactions), suggesting that CXorf67 induces H3K27 hypomethylation via a similar moiety as H3K27M mutations (Fig. 4C).
Fig. 3.
The C-terminus of human CXorf67 contains a highly conserved amino acid sequence. Multiple sequence alignment of CXorf67 ortholog protein sequences identifies a highly conserved 13 amino acid sequence in the C-terminus of human CXorf67. Alignments were performed for the entire protein but the figure shows only the indicated region in the C-terminus of CXorf67. Amino acids that match the human CXorf67 sequence are colored.
Fig. 4.
CXorf67 inhibits PRC2 via a small, highly conserved peptide sequence located in its C-terminus. (A) Structure of EZH2 bound to H3K27M22 where the bound peptide is shown as sticks and colored according to atom type. Residues in EZH2 binding to the peptide are shown as sticks and colored according to residue type: hydrophobic, pink; polar, green; negative, red. (B) Alignment of the peptide regions from H3 and CXorf67, where conserved residues are in bold. Residues above and below the alignment are those identified as allowed substitutions by perusing all orthologs of the 2 proteins in eggNOG.27 (C) The 2 peptides in the same orientation, where part of the CXorf67 peptide was modeled into the EZH2 structure using Modeller.23 The relevant binding pocket residues in EZH2 are labeled for each residue in H3K27M and conserved residues are shown in boldface. Structures were created using visual molecular dynamics.44
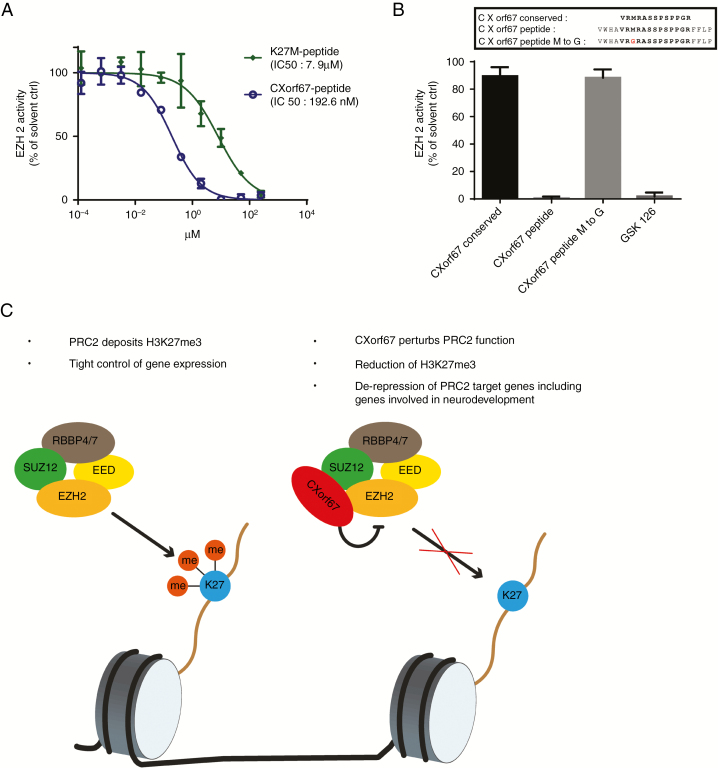
To validate these in silico predictions, we synthesized a short CXorf67 peptide based on the sequence of the conserved region to test its inhibitory properties in an EZH2 methyltransferase assay.9 We additionally incorporated 4 of the less conserved amino acids located at each end of the conserved region, since previous studies on H3K27M peptide variants had shown that excessive truncation may abolish their inhibitory properties.28 When evaluating the inhibitory capability of this CXorf67 peptide on EZH2 methyltransferase activity, we observed an IC50 (half maximal inhibitory concentration) of 192.6 nM, making this peptide even more potent than a synthesized H3K27M peptide tested in the same assay (Fig. 5A). In line with published data, even at a very high concentration (10 µM), a peptide consisting of solely the conserved region did not show inhibition of EZH2 methyltransferase activity, indicating that indeed the surrounding amino acids were aiding in the inhibitory process (Fig. 5B). It has been previously shown that methionine plays a crucial role in binding to the active site of EZH2, thereby sequestering PRC2 function in H3K27M tumors.22,29 To test this, we synthesized a variant of our CXorf67 peptide in which the Met (406) of the conserved sequence had been replaced with a glycine. As expected, this replacement abolished the inhibitory potential of the peptide, confirming that CXorf67 inhibits PRC2 function in a manner that is comparable to the one caused by H3K27M mutations (Fig. 5B).
Fig. 5.
A CXorf67 peptide incorporating the conserved region potently inhibits EZH2. (A) Methyltransferase assay to evaluate the inhibitory capabilities of the CXorf67 and the K27M peptides. (B) Evaluation of EZH2 inhibition by CXorf67 peptide variants at a single concentration of 10 µM. Peptide variants encompass the conserved region alone (CXorf67 conserved), the longer variant incorporating surrounding amino acids (CXorf67 peptide) and the longer peptide variant in which the methionine has been replaced by a glycine (CXorf67 peptide M to G). GSK126 was used as a positive control. (C) Proposed model of CXorf67-mediated H3K27 hypomethylation. When CXorf67 is absent, PRC2 mediates trimethylation of H3K27, resulting in a tight regulation of gene expression. In contrast, the presence of CXorf67 results in the inhibition of PRC2. In detail, the conserved domain of CXorf67 sequesters EZH2, thereby abolishing its methyltransferase activity. The resulting decrease in H3K27me3 levels allows the de-repression of PRC2 target genes, including genes involved in neurodevelopment which contributes to the tumorigenesis of PFA ependymoma.
Discussion
Downregulation of H3K27me3 levels has been described as a hallmark of diffuse midline gliomas, where it is the main driving force of aberrant gene expression.9 So far, the main culprits responsible for this observation are H3K27M mutations, which have been shown in numerous studies to trap and inhibit EZH2, thereby preventing H3K27 trimethylation.30–32 Only recently, expression of CXorf67, a hallmark of PFA ependymoma, has been proposed as an alternative mechanism for downregulation of H3K27me3.12 However, the exact mechanism of how CXorf67 inhibits PRC2 function was unknown.
In this study, we show that the mechanism of CXorf67-mediated inhibition of EZH2 is very similar to the H3K27M-mediated inhibition of EZH2. Our data show that the C-terminal region of CXorf67 is sufficient and responsible for binding and inhibition of EZH2, thus lowering H3K27me3 levels when overexpressed. Similar to the full-length protein, loss of H3K27me3 induced by the C-terminal region of CXorf67 led to a de-repression of PRC2 target genes and upregulated genes involved in neurodevelopmental processes—features that have also been reported in H3K27M diffuse midline gliomas.25 In contrast, expression of the N-terminus or the middle part of CXorf67 had no influence on H3K27me3 levels and did not upregulate the same genes as the full-length protein. Interestingly, a recent study in endometrial stromal sarcoma—a tumor type in which genomic alterations resulting in the perturbation of PRC2 function are a common finding—has identified a fusion protein consisting of MBTD1 (malignant brain tumor domain containing 1) and CXorf67.33 In this fusion protein, only the C-terminal region of CXorf67 is retained, further underscoring its importance in mediating PRC2 function and supporting our findings.
We have also shown that the actual C-terminal peptide sequence required for binding and inhibiting the SET domain of EZH2 is very short and is strongly conserved among other organisms. The interaction of this sequence with the SET domain follows the same mechanism as observed for the H3K27M peptide. In detail, the arginine which can be found in the (−1) position of Met (27) of H3K27M establishes interaction with the EZH2 SET domain via a series of hydrogen bonds. This allows the Met (27) residue to be placed in the lysine access channel of the EZH2 SET domain, thereby sequestering PRC2 function.22 Both the Arg (405) and Met (406) of CXorf67 match this specific pattern, while an additional overlap with histone H3 can be seen for Ala (403) and Ala (408). Whereas the His (402) and Arg (407) of CXorf67 do not match the Lys (23) and Ser (28) of histone H3, they—just like their counterparts—also point into the solvent when interacting with EZH2. This very specific combination of residues allows CXorf67 to partly “mimic” the H3K27M histone tail and to induce H3K27 hypomethylation. This is further supported by the fact that, although rare, H3K27M mutations do exist in PFA ependymoma but are 100% mutually exclusive with overexpression of CXorf67 and that replacing the methionine of our CXorf67 peptide abolished its inhibitory properties.
While our study provides a comprehensive characterization of the C-terminal function of CXorf67, the function of the N-terminal and middle region is less clear and needs to be addressed in further studies. Based on our co-IP results, the middle region of CXorf67 may aid in the recognition of EZH2 and SUZ12 but is not essential for inhibition of EZH2 activity. There is currently no evidence that the N-terminal region plays a role in recognizing the PRC2 core components EZH2 and SUZ12. However, CXorf67 is mutated in ~10% of PFA ependymoma, and mutations are almost exclusively localized to the N-terminus, where they cluster in a hotspot region of 51 amino acids, indicating that this region may contain a functional domain.12 Additionally, our MS results identified an interaction of CXorf67-N with the PRC2 component RBBP4 (retinoblastoma binding protein 4). We therefore speculate that this region may either serve as a regulatory domain that controls activity of CXorf67 or aid in the recognition of PRC2. Similar to CXorf67-M, however, this interaction does not seem to be crucial for inhibition of PRC2 function, also because both wild-type and mutant CXorf67 are able to inhibit PRC2.12
Unlike the H3K27M mutations in tumors, CXorf67 is present under physiological conditions, but it is still unclear how the expression of CXorf67 itself is regulated. In PFA ependymomas, we found that increased expression of CXorf67 is associated with promoter region hypomethylation,12 but we have not identified other genetic or epigenetic aberrations that could explain why CXorf67 is expressed in these tumors, suggesting that it is more of a lineage marker. Apart from testis, ovary, and placenta, CXorf67 is also highly expressed during pre-implantation embryo development.34,35 Previous studies have shown excessive epigenetic reprogramming during the pre-implantation phase, including the eradication of H3K27me3 from promoters of canonical PRC2 targets.36,37 One may speculate that CXorf67 aids in this eradication process by preventing the immediate reconstitution of H3K27me3 by PRC2. Alternatively, CXorf67 may only be expressed during CNS development in a small subpopulation of cells, which are then susceptible to transformation. Indeed, Bayliss and colleagues8 found that radial glia cells—the putative cell of origin of ependymoma38—displayed reduced H3K27me3 during prenatal development. Given the recent progress in the field of single-cell sequencing, transcriptomic analysis of cell populations from different stages of the developing brain may reveal not only the potential cellular origin of PFA ependymoma but also whether these cells already express CXorf67 or not.
While we have shown that CXorf67 is capable of inducing H3K27 hypomethylation, we have thus far not been able to generate PFA tumors via in utero electroporation of CXorf67-overexpressing constructs or injection of CXorf67-overexpressing neural stem cells into mice. This is in accordance with findings in the mouse modeling of diffuse midline gliomas where in addition to the H3K27M mutation, other alterations, like platelet derived growth factor B overexpression, platelet derived growth factor receptor A activation, or tumor protein 53 deficiency, are required to generate tumors.26,39 However, apart from mutations found in histone family H3 genes or CXorf67, no other recurrent mutations or amplifications have been detected in PFA ependymoma thus far, making the identification of a second hit challenging. If no second hit is necessary, cells may have to be in a specific developmental state to be susceptible to transformation by overexpression of CXorf67. Further studies are required to fully elucidate the oncogenic potential of CXorf67 overexpression on its own or in combination with other genes. Nevertheless, our current findings propose a model in which CXorf67 expression results in decreased H3K27me3 levels accompanied by a de-repression of PRC2 target genes, including genes involved in neurodevelopmental processes that may have a potential role in PFA tumorigenesis (Fig. 5C).
Our findings also have direct implications for the clinical treatment of PFA ependymoma. So far, chemotherapy has not shown any success in ependymoma, and gross total resection followed by focal irradiation is still considered to be the gold standard of treatment.40–42 We have previously shown that knockout of CXorf67 reduces cell growth in vitro.12 With the exact mechanism of EZH2 inhibition by CXorf67 uncovered, it may be possible to target and disrupt this interaction, resulting in the restoration of H3K27me3 and inhibition of tumor growth. This may be especially crucial for cases in which only partial resection can be achieved or for radiotherapy deferral strategies to spare infant patients from long-term sequelae.
Finally, the potent inhibitory effect of the CXorf67 peptide may also be utilized for the treatment of tumors driven by EZH2 overexpression or EZH2 activating mutations. Currently, most small-molecule inhibitors of EZH2 exert their function by competing with S-adenosyl-L-methionine, the methyl donor utilized by EZH2 for catalyzing H3K27 methylation.43 As shown in this study, the inhibitory mechanism of the CXorf67 peptide differs from this principle, making it a promising candidate for a novel class of EZH2 inhibitors capable of inhibiting EZH2 in the nanomolar range.
Taken together, our study provides a comprehensive characterization of CXorf67 function and identifies the C-terminus of the protein as a crucial component for the inhibition of PRC2. Based on these data, we suggested to the Human Genome Organisation (HUGO) Gene Nomenclature Committee that it change the name of CXorf67 to “EZH Inhibitory Protein (EZHIP),” a suggestion that has been approved.
Funding
J-M.H. is funded by a scholarship from the Studienstiftung des deutschen Volkes.
Supplementary Material
Acknowledgments
We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility for the generation of gene expression profiles.
Conflict of interest statement. None declared.
Authorship statement. J-M.H. designed and performed most experiments; T.M. and D.N.P. performed mass spectrometry and data analysis; M.M. generated the CXorf67 expressing cell lines. R.B.R. performed peptide modeling analyses; J.K., R.B.R., D.W.E., S.M.P., K.W.P., and M.K. contributed to the concept and design of the paper. All authors contributed to the writing of the paper.
References
- 1. Ostrom QT, de Blank PM, Kruchko C, et al. . Alex’s lemonade stand foundation infant and childhood primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2015;16(Suppl 10):x1–x36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kilday JP, Rahman R, Dyer S, et al. . Pediatric ependymoma: biological perspectives. Mol Cancer Res. 2009;7(6):765–786. [DOI] [PubMed] [Google Scholar]
- 3. Pajtler KW, Witt H, Sill M, et al. . Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Witt H, Mack SC, Ryzhova M, et al. . Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20(2):143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wani K, Armstrong TS, Vera-Bolanos E, et al. ; Collaborative Ependymoma Research Network A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012;123(5):727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parker M, Mohankumar KM, Punchihewa C, et al. . C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506(7489):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mack SC, Witt H, Piro RM, et al. . Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bayliss J, Mukherjee P, Lu C, et al. . Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 2016;8(366):366ra161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bender S, Tang Y, Lindroth AM, et al. . Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. [DOI] [PubMed] [Google Scholar]
- 10. Gessi M, Capper D, Sahm F, et al. . Evidence of H3 K27M mutations in posterior fossa ependymomas. Acta Neuropathol. 2016;132(4):635–637. [DOI] [PubMed] [Google Scholar]
- 11. Ryall S, Guzman M, Elbabaa SK, et al. . H3 K27M mutations are extremely rare in posterior fossa group A ependymoma. Childs Nerv Syst. 2017;33(7):1047–1051. [DOI] [PubMed] [Google Scholar]
- 12. Pajtler KW, Wen J, Sill M, et al. . Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 2018;136(2):211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y, Pak C, Han Y, et al. . Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78(5):785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vizcaíno JA, Csordas A, del-Toro N, et al. . 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14(4):380–381. [DOI] [PubMed] [Google Scholar]
- 17. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subramanian A, Tamayo P, Mootha VK, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20(9):1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kasinath V, Faini M, Poepsel S, et al. . Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018;359(6378):940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trabuco LG, Lise S, Petsalaki E, Russell RB. PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012;40(Web Server issue):W423–W427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Justin N, Zhang Y, Tarricone C, et al. . Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2017;1654:39–54. [DOI] [PubMed] [Google Scholar]
- 24. Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14(2):309–323. [DOI] [PubMed] [Google Scholar]
- 25. Larson JD, Kasper LH, Paugh BS, et al. . Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019;35(1):140–155.e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346(6216):1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huerta-Cepas J, Szklarczyk D, Forslund K, et al. . eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44(D1):D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown ZZ, Müller MM, Jain SU, Allis CD, Lewis PW, Muir TW. Strategy for “detoxification” of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2. J Am Chem Soc. 2014;136(39):13498–13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewis PW, Müller MM, Koletsky MS, et al. . Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwartzentruber J, Korshunov A, Liu XY, et al. . Driver mutations in histone H3.3 and chromatin remodeling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 31. Wu G, Broniscer A, McEachron TA, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mackay A, Burford A, Carvalho D, et al. . Integrated molecular meta-analysis of 1000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dewaele B, Przybyl J, Quattrone A, et al. . Identification of a novel, recurrent MBTD1-CXorf67 fusion in low-grade endometrial stromal sarcoma. Int J Cancer. 2014;134(5):1112–1122. [DOI] [PubMed] [Google Scholar]
- 34. Xie D, Chen CC, Ptaszek LM, et al. . Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20(6):804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uhlén M, Fagerberg L, Hallström BM, et al. . Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 36. Liu X, Wang C, Liu W, et al. . Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558–562. [DOI] [PubMed] [Google Scholar]
- 37. Zheng H, Huang B, Zhang B, et al. . Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell. 2016;63(6):1066–1079. [DOI] [PubMed] [Google Scholar]
- 38. Taylor MD, Poppleton H, Fuller C, et al. . Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8(4):323–335. [DOI] [PubMed] [Google Scholar]
- 39. Misuraca KL, Hu G, Barton KL, Chung A, Becher OJ. A novel mouse model of diffuse intrinsic pontine glioma initiated in Pax3-expressing cells. Neoplasia. 2016;18(1):60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khatua S, Ramaswamy V, Bouffet E. Current therapy and the evolving molecular landscape of paediatric ependymoma. Eur J Cancer. 2017;70:34–41. [DOI] [PubMed] [Google Scholar]
- 41. Merchant TE, Li C, Xiong X, Kun LE, Boop FA, Sanford RA. Conformal radiotherapy after surgery for paediatric ependymoma: a prospective study. Lancet Oncol. 2009;10(3):258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramaswamy V, Hielscher T, Mack SC, et al. . Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol. 2016;34(21):2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.