Abstract
Background
Diffuse intrinsic pontine gliomas (DIPGs) are highly aggressive pediatric brain tumors that are characterized by a recurrent mutation (K27M) within the histone H3 encoding genes H3F3A and HIST1H3A/B/C. These mutations have been shown to induce a global reduction in the repressive histone modification H3K27me3, which together with widespread changes in DNA methylation patterns results in an extensive transcriptional reprogramming hampering the identification of single therapeutic targets based on a molecular rationale.
Methods
We applied a large-scale gene knockdown approach using a pooled short hairpin (sh)RNA library in combination with next-generation sequencing in order to identify DIPG-specific vulnerabilities. The therapeutic potential of specific inhibitors of candidate targets was validated in a secondary drug screen.
Results
We identified fibroblast growth factor receptor (FGFR) signaling and the serine/threonine protein phosphatase 2A (PP2A) as top depleted hits in patient-derived DIPG cell cultures and validated their lethal potential by FGF ligand depletion and genetic knockdown of the PP2A structural subunit PPP2R1A. Further, pharmacological inhibition of FGFR and PP2A signaling through ponatinib and LB-100 treatment, respectively, exhibited strong tumor-specific anti-proliferative and apoptotic activity in cultured DIPG cells.
Conclusions
Our findings suggest FGFR and PP2A signaling as potential new therapeutic targets for the treatment of DIPGs.
Keywords: brain cancer, DECIPHER pooled shRNA screen, DIPG, FGFR, PP2A
Key Points.
1. DECIPHER pooled shRNA library screen reveals new lethality genes in DIPG cells.
2. Secondary drug screen confirms lethal effects of PP2A and FGFR inhibition.
3. PP2A and FGFR inhibitors exhibit anti-proliferative and apoptotic activity.
Importance of the Study.
In this study, we demonstrate that ponatinib and LB-100 exhibit potent therapeutic activity against patient-derived cultured DIPG cells with considerably lower efficacy on normal human astrocytes. Our findings suggest FGFR and PP2A signaling as potential new therapeutic targets for the treatment of these deadly tumors. The effects of inhibiting both of these pathways were robustly validated in our study by independent functional assays. The facts that ponatinib (an FGFR inhibitor) is known to cross the blood–brain barrier and that there are currently strong efforts to chemically modify the PP2A inhibitor LB-100 with the aim of also introducing blood–brain barrier permeability further enhance the possible translational impact of our findings.
Diffuse intrinsic pontine gliomas (DIPGs) are highly aggressive pediatric brain tumors that arise in the pons and occur at a mean age at diagnosis of 6 to 7 years.1 Due to the highly sensitive tumor location and infiltrative growth patterns, DIPGs are surgically inaccessible and display a dismal prognosis—with a median overall survival of 8 to 11 months after diagnosis and a 2-year survival rate of less than 10%.1,2
In the past years, an increase in the availability of autopsy material as well as, more recently, stereotactic biopsy sampling has helped to significantly improve the knowledge of the genetic and epigenetic landscapes and the understanding of the biology of DIPGs. They are characterized by a recurrent mutation of lysine 27 to methionine (K27M) in the histone H3.3 and H3.1 encoding genes H3F3A and HIST1H3A/B/C, respectively,3 leading to a functional inhibition of the Polycomb repressive complex 2 and thus a global reduction in the repressive histone modification H3K27me3.4–6 Besides this mutation, found in about 80% of DIPGs,3 a great heterogeneity of additional genetic alterations was reported, including TP53/PPM1D mutations, activating ACVR1 and PIK3CA mutations, amplifications of PDGFRA, MET, MYC, CDK4/6, and CCND1/2/3, as well as loss of PTEN and RB1.7–10 However, despite numerous clinical trials involving diverse chemotherapeutic drugs or targeted agents, no advances have been made in improving the survival of DIPG patients.2,11 The current standard of care is radiotherapy,12 which usually temporarily improves symptoms, but tumors typically progress rapidly within a few months. Thus, to date there is no effective treatment available and DIPGs are still the leading cause of brain tumor–related death in children.2,11
The poor clinical outcome and lack of curative treatment options for DIPG patients emphasize the desperate need for the development of novel targeted therapies. Global changes in H3K27 trimethylation and DNA methylation patterns lead to an extensive transcriptional reprogramming,4,5 and other single gene alterations as noted above tend to each occur in only a relatively small proportion of tumors. The identification of single therapeutic targets based on a molecular rationale is thus a significant challenge. We therefore conducted a large-scale gene knockdown study using a pooled short hairpin (sh)RNA library in combination with next-generation sequencing on patient-derived H3.3K27M-mutated DIPG cell cultures in order to identify critical nodes of tumor development and maintenance that potentially allow for the establishment of novel treatment strategies.
Materials and Methods
Cell Culture
HEK293T cells (CRL-1273, American Type Culture Collection) were cultured as previously described.13 Human astrocytes originating from fetal cerebral cortex were purchased from ScienCell (#1800) and cultured as outlined in the Supplementary Methods. Pediatric patient-derived HSJD-GBM-001 and HSJD-DIPG-007 cells were obtained from Angel M. Carcaboso, and SU-pcGBM2, SU-DIPG-XIII, -XVII, and -XXV cells were obtained from Michelle Monje. They were cultivated as floating neurospheres as previously described.14 Briefly, they were seeded in serum-free tumor stem medium composed of equal volumes of Dulbecco’s modified Eagle’s medium/F-12 and Neurobasal-A medium, 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid) buffer solution, 1 mM sodium pyruvate, 1× Minimum Essential Medium non-essential amino acids solution, 1× GlutaMax-I, 1× antibiotic-antimycotic (all Life Technologies) and supplemented with 2% (v/v) B-27 (Life Technologies), 2 µg/mL heparin (Sigma-Aldrich), 20 ng/mL epidermal growth factor (EGF), 20 ng/mL basic fibroblast growth factor (bFGF), and 10 ng/mL platelet-derived growth factor (PDGF) AA (all Peprotech). Cells were passaged every 5–6 days using Accumax (Thermo Fisher) or TrypLE (Life Technologies). Three days after seeding, half of the medium was replaced by fresh tumor stem medium containing 2× growth factors. For all patient-derived tumor cell cultures, informed consent was obtained by the respective contributing centers in accordance with local institutional review board and ethical approval.
Genomic and Epigenomic Profiling
DNA methylation profiling was performed using the Infinium MethylationEPIC (850k) BeadChip array (Illumina) according to the manufacturer’s instructions at the DKFZ Genomics & Proteomics Core Facility. Filtering and genome-wide copy number analyses were performed as previously described,15 using the “conumee” package in R (http://www.bioconductor.org).
For whole exome sequencing: exome capture, sequencing, and data analysis were performed as described in the Supplementary Methods.
RNA sequencing libraries were prepared using the TruSeq RNA Sample Preparation Kit v2 (Illumina) following the manufacturer’s instructions and sequenced as described in the Supplementary Methods.
Lentiviral Transduction
The DECIPHER human Module 1 shRNA plasmid library (DHPAC-M1-P, 5043 Signaling Pathway Targets; 27 500 shRNAs) was purchased from Cellecta. High-throughput lentiviral packaging was performed according to the manufacturer’s guidelines and as described in the Supplementary Methods. Since the transduction efficiency of different cell types can vary widely, the amount of virus required to transduce a specific percentage of cells (eg, about 40% for pooled shRNA screening) was determined for each cell line individually, as outlined in the Supplementary Methods.
Pooled shRNA Library Screen
The shRNA screening procedure was established based on Cellecta’s guidelines. For each screen, 4.5–6 × 107 cells were infected with the shRNA library virus at varying dilutions, in order to obtain equal infection rates of about 40% and thus transduce at least 400 cells per shRNA. Cells were distributed to 20–24 × 175 cm2 cell culture flasks. Eighteen hours after infection, half of the cells were harvested, snap-frozen in liquid nitrogen, and stored at −20°C (baseline sample). For remaining cells, a complete medium change was performed. Seventy-two hours after infection, spheres from all flasks were pooled and dissociated. Anticipated transduction efficiencies were confirmed by measuring the percentage of red fluorescent protein–positive cells with an LSR II flow cytometer. Residual cells were replated into 20–24 × 175 cm2 cell culture flasks in medium containing 0.25–0.7 µg/mL puromycin, with concentrations determined beforehand in order to kill about 90% of the uninfected cells within 72 hours. Twenty-four hours after addition of puromycin, two-thirds of the medium was exchanged with fresh puromycin-containing medium in order to remove toxic products produced by dying cells. After 72 hours of puromycin treatment, cells were again pooled and dissociated and the percentage of red fluorescent protein–positive cells was determined. At least 3 × 107 cells were replated in order to maintain a library representation of at least 1000-fold, with cell densities of about 3 × 104 cells/mL. Cells were further propagated for 7–20 days until 8 or 9 population doublings were reached in total, calculated from counted cells. During this time, cells were split every 3–6 days and a half medium change was conducted every 3 days in between. Endpoint samples of at least 3 × 107 cells (>1000-fold library representation) were harvested, snap-frozen in liquid nitrogen, and stored at −20°C.
Amplification, high-throughput sequencing, and analysis of shRNA barcodes were performed as outlined in the Supplementary Methods. See also Supplementary Tables 1−4 referred to therein.
Hit Validation
Individual shRNAs of the DECIPHER pooled shRNA library were cloned into the pRSI12-U6-(shRNA)-UbiC-TagRFP-2A-Puro vector backbone as described in the Supplementary Methods. Details on drug sources, growth factor depletion, western blot analysis, viability, and apoptosis readouts are outlined in the Supplementary Methods. See also Supplementary Tables 5−7 referred to therein.
Statistical Analysis
All plotted data represent mean ± SD. GraphPad Prism 6 and R were used for statistical analyses. Comparisons between different groups were made using one-way ANOVA followed by Dunnett’s post-hoc test (significances: *P < 0.05, **P < 0.01, ***P < 0.001).
Data Availability
RNA sequencing, whole-exome sequencing, and 850k methylation array data of patient-derived tumor cell cultures are available at the European Genome-phenome Archive (EGAS00001003404). Deconvoluted sequencing data of individual shRNA screening samples can be found in Supplementary Table 2.
Results and Discussion
Pooled shRNA Library Screen for DIPG-Specific Therapeutic Targets
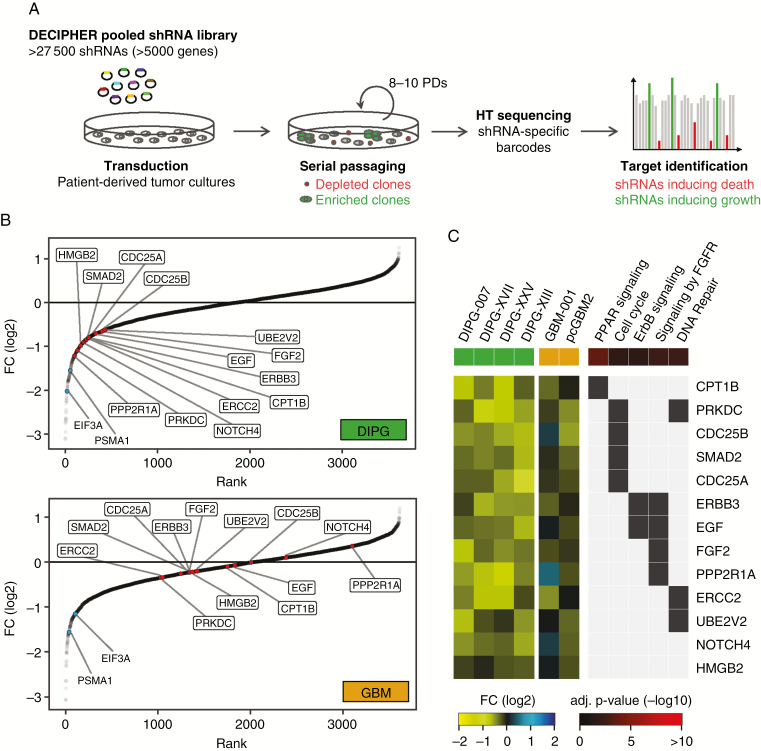
Four patient-derived H3.3K27M-mutated DIPGs as well as 2 pediatric H3.3 wild-type glioblastoma (GBM) cell cultures were selected for shRNA screening and extensively profiled by DNA methylation array and whole-exome and RNA sequencing (Supplementary Figure 1). DIPG and GBM cells were screened for shRNAs inducing cell death using the DECIPHER pooled shRNA library Module 1, which targets 5043 signaling pathway genes with multiple shRNAs per gene (Fig. 1A). High quality of the screening procedure was confirmed by analyzing the correlation and library representation of baseline and endpoint samples, as well as by verification of internal control shRNAs (Supplementary Figure 2). Bioinformatics analysis of screening results expectedly revealed many known essential genes among the top depleted hits for both DIPG and GBM cell cultures, including proteasome and ribosome subunits, as well as RNA transport factors (Supplementary Figure 3A, Supplementary Table 8). In total 110 DIPG-specific hits could be identified by setting cutoffs to fold change of logFC (DIPG) < −0.6 and logFC (GBM) > −0.4, with an adjusted P-value (DIPG) < 0.1 (Supplementary Figure 3B, Supplementary Table 9). Since our aim was the identification of factors that can be therapeutically targeted, we screened the list of DIPG-specific hits for the availability of target-specific inhibitors. For further validation, we identified 13 hits for which inhibitors were available that either directly target the respective screening hit (eg, LB-100 targeting serine/threonine protein phosphatase 2A [PP2A], the structural subunit of which is encoded by, among others, PPP2R1A16) or target an interaction partner or associated factor of the screening hit (eg, ponatinib targeting fibroblast growth factor receptor [FGFR] 1 as an interaction partner of FGF2) (Table 1). As shown in Fig. 1B and Supplementary Figure 4, we found the selected hits to rank high for depletion in DIPG but not in GBM reference screens, while shRNAs targeting the known essential genes EIF3A (eukaryotic translation initiation factor 3 subunit A) and PSMA1 (proteasome subunit alpha type 1) were strongly depleted in all screens performed. According to a functional enrichment analysis of DIPG-specific hits using Enrichr,17 selected druggable hits could be assigned to, among others, peroxisome proliferator-activated receptor signaling, cell cycle, ErbB signaling (erb-b receptor tyrosine kinase), FGFR signaling, and DNA repair (Fig. 1C, Supplementary Tables 10 and 11). Consequently, we also included inhibitors targeting distinct DNA repair pathways, such as homologous recombination, in our list of DIPG hit–specific inhibitors, as well as the poly-ADP ribose polymerase (PARP) 1 and 2 inhibitor niraparib and the alkylating agent temozolomide as independent reference drugs based on a literature search (Table 1).
Fig. 1.
Identification of DIPG-specific hits by DECIPHER pooled shRNA library screening. (A) Schematic representation of a pooled shRNA library screen. Cells were infected with the pooled shRNA library virus at <1 shRNA per cell and propagated for 8 to 10 population doublings. ShRNAs targeting lethal genes were depleted from the pool and can be identified by their underrepresented barcodes via high-throughput sequencing. (B) Ranking of combined fold change (FC) (endpoint/baseline) values for the top 3 shRNAs of each target gene in DIPG and GBM screens. Indicated DIPG-specific hits selected for further validation (red), based on the availability of pharmacological inhibitors, ranked high for depletion in DIPG but not in GBM cell cultures, whereas shRNAs targeting the known essential genes EIF3A and PSMA1 (blue) were depleted in both groups. (C) Heatmap of selected, druggable DIPG-specific hits illustrating the respective fold change values in individual DIPG and GBM screens as well as selected associated enriched Kyoto Encyclopedia of Genes and Genomes and Reactome signaling pathways. EIF3A = eukaryotic translation initiation factor 3 subunit A; PSMA1 = proteasome subunit alpha type 1.
Table 1.
Selected DIPG-specific screening hits with available inhibitors
| Screening Hit | Rank | Fold Change (log2) | Adj. P-value | Inhibitor | Targets |
|---|---|---|---|---|---|
| PPP2R1A | 6 | −1.220 | 0.012 | LB-100 | PP2A |
| PRKDC | 11 | −1.092 | 0.025 | NU7026 | DNA-PK |
| NU7441 | |||||
| HMGB2 | 17 | −0.982 | 0.058 | Inflachromene | HMGB2 |
| NOTCH4 | 24 | −0.903 | 0.027 | RO4929097 | γ-Secretase |
| CDC25B | 28 | −0.866 | 0.002 | NSC95397 | CDC25A, CDC25B |
| CDC25A | 33 | −0.843 | 0.027 | ||
| SMAD2 | 32 | −0.851 | 0.001 | LY2109761 | TGF-β receptor type I/II |
| ERCC2 | 39 | −0.813 | 0.078 | Spironolactone | XPB/ERCC3 |
| CPT1B | 41 | −0.803 | 0.053 | Etomoxir | CPT-1, PPARα |
| ERBB3 | 42 | −0.793 | 0.011 | Sapitinib | ERBB3 |
| EGF | 77 | −0.704 | 0.025 | Afatinib | ERBB2, EGFR |
| FGF2 | 94 | −0.656 | 0.072 | AZD4547 | FGFR1, FGFR2, FGFR3 |
| Dovitinib | FGFR1, FGFR3 | ||||
| PD173074 | FGFR1 | ||||
| Ponatinib | |||||
| UBE2V2 | 101 | −0.620 | 0.012 | NSC697923 | Ubc13/UBE2N-UEV1A |
| DNA repair | n/a | n/a | n/a | Gimeracil | DPYD/homologous recombination |
| YU238259 | Homology-dependent DNA repair | ||||
| SCR7 | LIG4/non-homologous end joining | ||||
| Reference drugs | |||||
| – | – | – | – | Niraparib | PARP1, PARP2 |
| – | – | – | – | Temozolomide | Alkylating DNA |
TGF-β = transforming growth factor beta; PPAR = peroxisome proliferator-activated receptor.
Additional targets of each inhibitor are specified in Supplementary Table 6.
FGFR Inhibitors and LB-100 Are Potent Drug Candidates for DIPGs
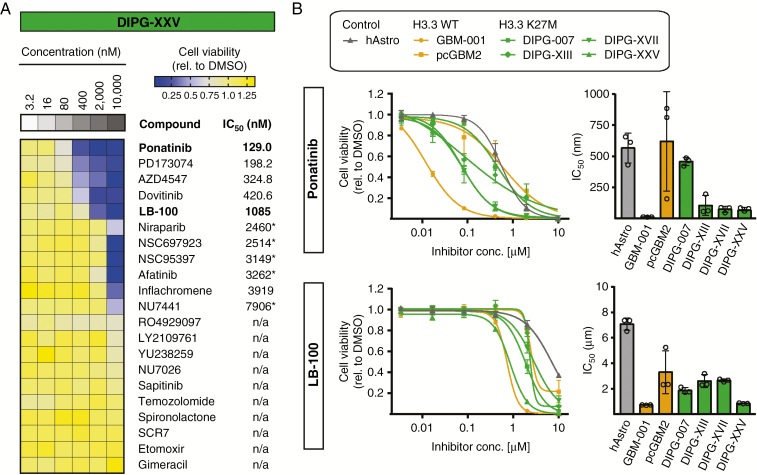
Selected inhibitors were tested at concentrations from 3.2 to 10 000 nM for 72 hours in the DIPG and GBM cell cultures and ranked according to their respective half-maximal inhibitory concentration (IC50) values (Fig. 2A, Supplementary Figs. 5 and 6). In line with a previous publication by Chornenkyy et al,18 suggesting niraparib as a therapeutic target for DIPG and pediatric high-grade astrocytoma, niraparib (as a positive control) showed some efficacy in 3 out of 4 DIPG cell cultures, with IC50 values ranging from 1.59 to 2.46 µM, as well as in GBM-001 (IC50 = 2.83 µM) (Fig. 2A, Supplementary Figures 5 and 6), thus showing consistency of our model systems with previously published findings. On the other hand, temozolomide, which is used as a first-line chemotherapeutic treatment for GBM19 but did not show any benefit in the survival of DIPG patients,20 did not affect the viability of any of the DIPG and GBM cell cultures tested, even at the highest concentration applied (Fig. 2A, Supplementary Figures 5 and 6).
Fig. 2.
Testing of shRNA screening hit–specific drugs reveals LB-100 and FGFR inhibitors as potent drug candidates for DIPGs. (A) Heatmap illustrating cell viability after treatment for 72 hours with increasing concentrations of 21 different inhibitors specified in Table 1, as exemplified by DIPG-XXV. Inhibitors are sorted according to their IC50 values, whereas n/a indicates that the IC50 could not be determined. Asterisks (*) indicate that drug responses displayed high variability. FGFR inhibitors, including ponatinib and the PP2A inhibitor LB-100 showed highest efficacy, with lowest IC50 values. (B) Dose-response curves and corresponding mean IC50 values of 72 hours ponatinib or LB-100 treatment of DIPG (green) and GBM (orange) cell cultures as well as human astrocyte control cells (gray). Both drugs showed highest efficacy in GBM-001 and DIPG cell cultures, whereas human astrocytes and pcGBM2 cells were less affected. IC50 curves and bars represent mean ± SD of 3 independent experiments.
About half of the tested inhibitors showed no effects on the viability of either DIPG or GBM cells, while some inhibitors only induced a reduction in cell viability at the highest concentration applied. Among the most effective inhibitors for all 4 DIPG cell cultures were the FGFR inhibitors AZD4547, dovitinib, PD173074, and ponatinib. The FGFR signaling pathway plays an important role in the maintenance of the self-renewing potential of a variety of physiological stem cell types, as well as in the growth of cancer stem cells isolated from human tumors (eg, breast, brain).21–23 Accordingly, bFGF/FGF2 in combination with EGF are commonly used to cultivate neural stem cells as well as patient-derived GBM and DIPG cells in vitro.14,23,24 However, while several groups reported a reduction of GBM cell growth upon inhibition of FGFR signaling,25–27 comparable studies in DIPGs are still scarce. Among all FGFR inhibitors tested, ponatinib consistently showed the highest efficacy in all 4 DIPG cell cultures (Fig. 2A, Supplementary Figures 5 and 6; the low relative IC50 values of PD173074 and AZD4547 in DIPG-XIII cells are due to the induction of stasis above 50% viability). Surprisingly, GBM-001 showed similar drug efficacy profiles to DIPGs, with even higher sensitivity to ponatinib treatment (IC50 = 39.86 nM), whereas pediatric cortical (pc)GBM2 displayed considerably lower sensitivity to ponatinib treatment, with IC50 = 2.25 µM and a generally lower sensitivity to all inhibitors tested (Supplementary Figures 5 and 6). Since both GBM cell cultures originate from frontal lobe tumors, these differences are less likely to be explained by regional differences of tumor development as they are by different genetic profiles of the cells. Interestingly, GBM-001 cells show a PDGFRA mutation, as well as overexpression and amplification of CCND1 and CCND2, respectively (Supplementary Figure 1), all of which are also frequently found in DIPGs.1,8,10 In contrast, pcGBM2 cells harbor an amplification of EGFR, which is rarely seen in DIPGs.1,10,15 Thus, it may be conceivable that the observed similarity of GBM-001 and DIPG drug sensitivities originates from related genetic profiles independent of H3.3 mutational status (Supplementary Figure 1).
Besides the indicated FGFR inhibitors, we also identified the PP2A inhibitor LB-100 as a potent inhibitor of DIPG cell viability, with highest efficacy in DIPG-007 (IC50 = 1.37 µM) and DIPG-XXV (IC50 = 1.09 µM), as well as of GBM-001 (IC50 = 918.2 nM), while pcGBM2 cells were considerably less sensitive (IC50 = 9.38 µM) (Fig. 2A, Supplementary Figures 5 and 6). PP2A is one of the major protein phosphatases in eukaryotic cells,28 and inhibition was reported to display anti-proliferative activity in cultured GBM cells either alone or in combination with radiation.29–31 We therefore selected LB-100 and ponatinib for further studies using primary human astrocytes as control cells in order to assess tumor-specific efficacy. After confirmation of astrocytic marker expression (Supplementary Figure 7), we found that ponatinib and LB-100 both displayed considerably lower efficacy in human astrocytes (Fig. 2B). Cells of pcGBM2 displayed a high variability of IC50 values between experiments, which might be associated with altered proliferation rates or subclonal evolution of the cells between different passages.
Taken together, we could identify FGFR and PP2A inhibitors as potent drug candidates for DIPGs, and the observed differential sensitivity to human astrocytes indicates a therapeutic window to potentially allow effective treatment of DIPG tumor cells with minimal effects on normal brain cells like astrocytes.
Inhibition of FGFR Signaling Exhibits Anti-Proliferative Activity in Cultured DIPG Cells
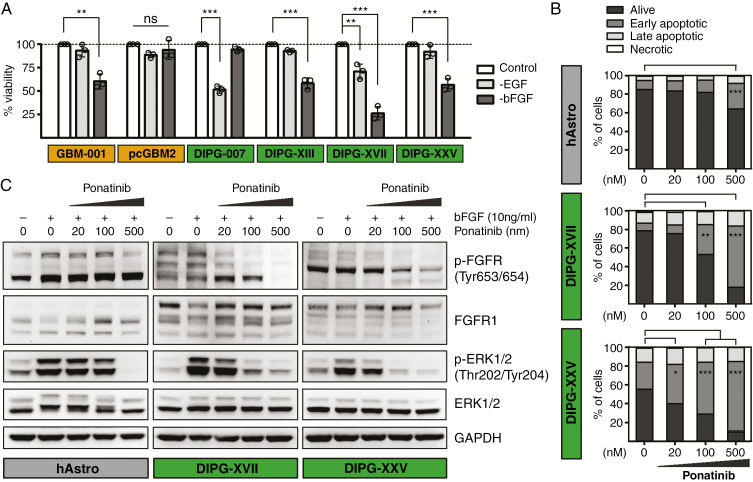
Ponatinib is a multi-kinase inhibitor that, besides FGFR1, targets several additional tyrosine kinases (eg, PDGF receptor A; Supplementary Table 6). We tested whether its therapeutic efficacy in cultured DIPG cells is FGFR dependent. We therefore depleted cells of basic FGF (bFGF/FGF2), which can bind to all 4 FGF receptor family members (FGFR1–4)32 for 72 hours and found a significant reduction in cell viability of DIPG-XIII, -XVII, and -XXV, as well as of GBM-001 (Fig. 3A). On the other hand, DIPG-007 cells were not affected by bFGF depletion but showed a strong reduction in viability upon depletion of EGF (Fig. 3A). This is in line with the previous observation that DIPG-007 cells displayed the lowest sensitivity to ponatinib treatment, but showed high sensitivity to the EGF receptor inhibitors sapitinib and afatinib, with IC50 values of 0.33 and 1.5 µM, respectively (Supplementary Figures 5 and 6). Interestingly, pcGBM2 cells did not show sensitivity to either EGF or bFGF depletion (Fig. 3A). These findings demonstrate similar patterns of sensitivity to bFGF depletion and ponatinib treatment, suggesting an FGFR-dependent mechanism of action.
Fig. 3.
Inhibition of FGFR signaling through bFGF depletion or ponatinib treatment decreases viability and induces apoptosis of cultured DIPG cells. (A) Depletion of bFGF for 72 hours significantly reduced cell viability of GBM-001 and DIPG-XIII, -XVII, and -XXV cells, while DIPG-007 cells showed reduced viability upon EGF depletion. PcGBM2 cells were not affected by EGF or bFGF depletion. (B) Ponatinib treatment for 50 hours induced apoptosis of DIPG-XVII and -XXV cells in a dose-dependent manner, while human astrocytes were affected at only the highest drug concentration. (C) Ponatinib treatment for 3 hours dose-dependently inhibited the phosphorylation of FGFR and bFGF-dependent downstream phosphorylation of ERK1/2 in DIPG-XVII and -XXV cells and, to a lesser extent, in human astrocytes. Bars represent mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ns, not significant.
To further confirm apoptotic activity of ponatinib, we performed an annexin V and propidium iodide apoptosis assay on the 2 most sensitive DIPG cell cultures, DIPG-XVII and DIPG-XXV, as well as on human astrocyte control cells using 3 drug concentrations centered around the approximate IC50 value of the 2 DIPG cell cultures. We found a dose-dependent increase in early apoptotic cells after 50 hours of ponatinib treatment, which for DIPG-XXV and DIPG-XVII reached statistical significance already at 20 and 100 nM, respectively, whereas human astrocytes only displayed significant induction of apoptosis for the highest drug concentration applied (Fig. 3B, Supplementary Figure 9A). These findings demonstrate apoptotic effects of ponatinib and again confirm DIPG-specific efficacy of the drug.
Ponatinib Inhibits FGFR Phosphorylation and Downstream Signaling in Cultured DIPG Cells
In order to confirm the activity of ponatinib on the protein level, we examined FGFR and downstream extracellular signal-regulated kinase (ERK) phosphorylation by western blot analysis. After 3 hours of treatment, ponatinib potently inhibited FGFR phosphorylation in a dose-dependent manner in DIPG-XVII and DIPG-XXV cells, whereas almost no inhibition of FGFR phosphorylation was evident in human astrocytes even at the highest drug concentration (Fig. 3C). Further, ponatinib strongly blocked the bFGF-induced downstream phosphorylation of ERK1 and 2 in DIPGs and, to a lesser extent, in human astrocytes (Fig. 3C). These findings support the observed differential sensitivity of human astrocytes and DIPG cells to ponatinib treatment. Further, analysis of mRNA expression in human astrocytes indicated absence or very low levels of FGFR2–4, while FGFR1 displayed expression levels comparable to those of GBM and DIPG cells (Supplementary Figure 8). This suggests that FGFR1 represents the main transducer of FGFR signaling in human astrocytes, and the lack of inhibitor efficacy is not due to bFGF-induced stimulation of other FGFR family members (FGFR2–4). Interestingly, differential patterns of FGFR1 isoforms were observed between human astrocytes and DIPG-XVII and DIPG-XXV cells (Fig. 3C), which most likely originate from alternative RNA splicing and/or cell type–specific alterations in FGFR glycosylation. Both mechanisms are known to significantly modulate the ligand binding specificity of FGFRs and downstream signaling33,34 which might contribute to the differential sensitivity of human astrocytes and DIPG cells to ponatinib treatment.
Genetic Knockdown and Pharmacological Inhibition of PP2A Decrease Viability and Induce Apoptosis of Cultured DIPG Cells
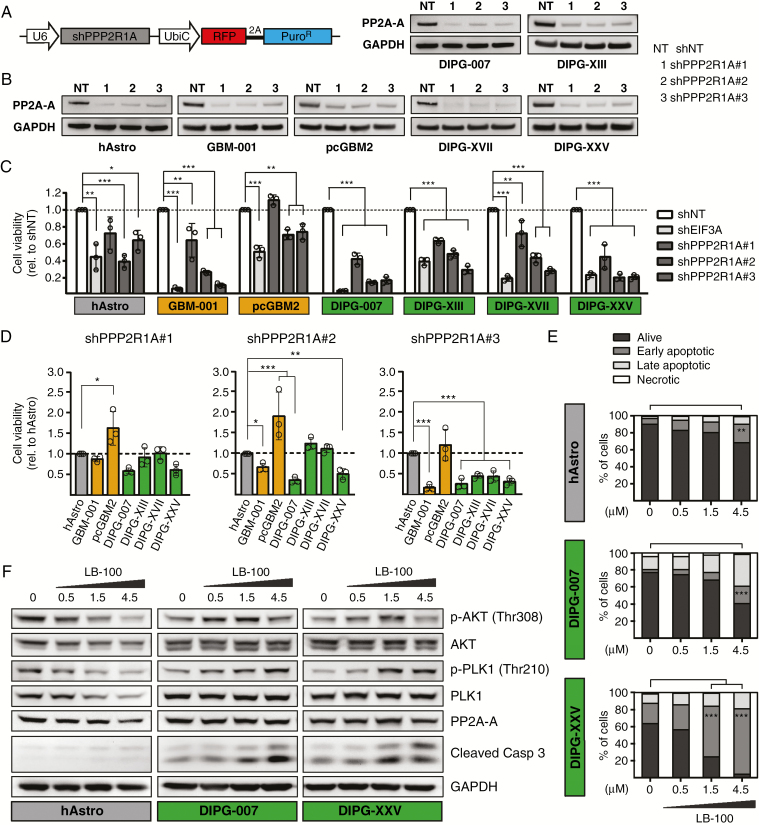
We next validated PP2A as a potential therapeutic target in DIPGs. PPP2R1A, which encodes the major isoform of the structural subunit A of PP2A (PP2A-A),16 was one of the top depleted hits for DIPGs in the shRNA screen (rank 6; Fig. 1B, C, Supplementary Figure 4, Supplementary Table 9). Out of 6 different library shRNAs targeting PPP2R1A, we selected the 3 most strongly depleted shRNAs for subcloning into the pRSI12 backbone (Fig. 4A). Corresponding shRNA viruses were titrated in the different target cells in order to obtain identical transduction efficiencies for comparative analyses. All 3 shRNAs (shPPP2R1A#1, #2, and #3) were confirmed to efficiently reduce PP2A-A protein levels in all cells (Fig. 4B). Analysis of the effects of PPP2R1A knockdown 6 days after infection revealed a potent reduction of cell viability in all DIPG cell cultures (Fig. 4C). Although human astrocyte control cells were also affected by expression of PPP2R1A shRNAs, comparative analysis revealed significant differential toxicity of shPPP2R1A#2 in DIPG-007 and DIPG-XXV cells (Fig. 4D), which were also most sensitive to pharmacological inhibition of PP2A through LB-100 treatment (Fig. 2, Supplementary Figures 5 and 6). Compared with human astrocytes, shPPP2R1A#3 showed strong differential effects in all DIPG cell cultures (Fig. 4D). This validation also revealed that GBM-001 cells displayed similar sensitivity to PPP2R1A knockdown as DIPG cells, while pcGBM2 was considerably less affected (Fig. 4C, D).
Fig. 4.
PP2A inhibition through PPP2R1A knockdown or LB-100 treatment decreases viability and induces apoptosis of cultured DIPG cells. (A) Schematic representation of the DECIPHER pRSI12 expression vector used for subcloning of shRNAs targeting PPP2R1A. (B) Knockdown of PPP2R1A with 3 independent shRNAs showed efficient reduction of PP2A-A protein levels in all cells compared with a nontargeting shRNA (shNT). (C, D) Assessment of cell viability 6 days after transduction with the indicated shRNAs. All 3 PPP2R1A shRNAs significantly reduced the viability of GBM-001 and DIPG cells compared with shNT control; shPPP2R1A#2 and #3 displayed significant differential toxicity in 2 or all 4 DIPG cell cultures, respectively, as well as in GBM-001 cells, compared with human astrocyte control cells. ShEIF3A was used as a positive control and strongly reduced the viability of all cells. (E) LB-100 treatment for 50 hours induced apoptosis of DIPG-007 and -XXV cells in a dose-dependent manner, while human astrocytes were less affected. (F) LB-100 treatment for 2.5 hours dose-dependently induced phosphorylation of Akt and PLK1 in DIPG-007 and -XXV cells, whereas in human astrocytes a decrease of pAkt and pPLK1 but also respective total protein and PP2A-A levels was observed. A dose-dependent increase in cleaved caspase-3 levels was only observed in DIPG cells. Bars represent mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. EIF3A = eukaryotic translation initiation factor 3 subunit A; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; PPP2R1A = protein phosphatase 2 subunit A alpha.
To further confirm the induction of apoptosis upon pharmacological inhibition of PP2A through LB-100 treatment, we performed staining with annexin V and propidium iodide of the 2 most sensitive DIPG cell cultures, DIPG-007 and DIPG-XXV, as well as human astrocyte control cells. As shown in Fig. 4E, we found a significant increase in early apoptotic cells for DIPG-XXV and DIPG-007 at 1.5 and 4.5 µM LB-100, respectively, while induction of apoptosis was less pronounced for human astrocytes (Fig. 4E, Supplementary Figure 9B).
Taken together, these findings demonstrate potent reduction of cell viability and induction of apoptosis in cultured DIPG cells upon genetic knockdown and pharmacological inhibition of PP2A, respectively, with considerable differential effects compared with human astrocyte control cells.
LB-100 Induces Akt and PLK1 Phosphorylation in Cultured DIPG Cells
To elucidate the mechanism of action of LB-100, we performed western blot analysis of known PP2A targets. In line with previous publications,35,36 inhibition of PP2A by LB-100 treatment for 2.5 hours induced phosphorylation of Akt (pAkt; protein kinase B) and of polo-like kinase 1 (pPLK1) in DIPG-007 and DIPG-XXV cells in a dose-dependent manner, while PP2A levels remained unaffected (Fig. 4F). Interestingly, in both DIPG cell cultures the dose-dependent increase in pAkt levels was not continuous, with the highest LB-100 concentration of 4.5 µM resulting in pAkt levels similar to dimethyl sulfoxide control. This could be explained by the observed induction of apoptosis, indicated by increased levels of cleaved caspase-3 (Fig. 4F), and associated alterations in cellular signaling pathways that might lead to dephosphorylation of Akt. In contrast to the observed increase in Akt and PLK1 phosphorylation in DIPG cells, we found a dose-dependent decrease of pAkt and pPLK1, but also of total Akt and PLK1 levels in human astrocytes without detection of relevant levels of cleaved caspase-3 induction (Fig. 4F), suggesting a fundamentally different response in the tumor versus normal cells.
While Akt plays a role in multiple cellular processes, including proliferation and survival,37 PLK1 functions as a key regulator of cell cycle progression at the G2/M DNA damage checkpoint.38 Although both proteins are commonly known as classical oncogenes with frequent activation or overexpression in various cancer cells,39,40 they also have tumor suppressive effects depending on the cellular context.41,42 Lu et al showed that the LB-100–induced increase in pAkt levels negatively affected nuclear receptor co-repressor complex formation, leading to astroglial differentiation and reduced proliferation of U87 GBM cells.30 Further, LB-100 treatment was also shown to sensitize various cancer cells (including U87 and U251 GBM cells) to chemo- and radiotherapy.31,43 Underlying mechanisms were suggested to include premature entry into mitosis due to aberrant PLK1 activation, thus bypassing DNA damage repair, which may lead to chromosomal instability and mitotic catastrophic events (summarized by Hong et al43). The fact that we also identified several DNA repair genes among the hits for DIPGs in our shRNA screen (Supplementary Tables 9–11, Supplementary Figure 3B) indicates that DIPG cells may be unable to compensate for impairments in DNA repair pathways. As genomic instability of DIPGs has been reported,7 this supports the hypothesis that LB-100 may exert its toxic effects in DIPGs by forced cell cycle progression despite DNA damage checkpoint activation leading to mitotic catastrophe and cell death.
In this study, we demonstrate that ponatinib and LB-100 exhibit potent therapeutic activity against patient-derived DIPG and GBM cells with considerably lower efficacy on normal human astrocytes. Given the heterogeneous responses observed in the different pediatric high-grade glioma cultures, further studies will be required to identify potential predictive biomarkers. While the in vivo efficacy and tolerability of the 2 inhibitors still need to be determined, our findings suggest FGFR and PP2A signaling as potential new therapeutic targets for the treatment of these deadly tumors.
Funding
This work was supported by the DKTK topics “Molecular Analysis of Pediatric Neoplasms” and “Integrated Molecular Diagnostics for the Typing of Brain Tumors in Adults,” both part of the DKTK research program Molecular Diagnostics, Early Detection, and Biomarker Development (MDEB).
Conflict of interest statement. None declared.
Authorship statement. Conceived the strategy: D.T.W.J., J.G., K.S., P.L.; supervised the project: D.T.W.J., J.G., P.L., S.M.P.; performed the experiments: K.S., B.S.; performed bioinformatics analyses: M.I., M.Z.; analyzed RNA-seq and whole-exome sequencing data: N.J.; provided methodological expertise: D.H., M.S.; wrote the manuscript: D.T.W.J., K.S., P.L.
Supplementary Material
Acknowledgments
We thank Daniela Sohn, Lotte Hiddingh, and Achim Stephan for technical support; Michelle Monje (Stanford University) for kindly providing SU-pcGBM2, SU-DIPG-XIII, -XVII, and -XXV cells; and Angel Montero Carcaboso (Hospital Sant Joan de Déu, Barcelona) for providing HSJD-GBM-001 and HSJD-DIPG-007 cells. We also thank the DKFZ fluorescence activated cell sorting and Genomics & Proteomics Core Facilities.
References
- 1. Mackay A, Burford A, Carvalho D, et al. . Integrated molecular meta-analysis of 1000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol. 2006;7(3):241–248. [DOI] [PubMed] [Google Scholar]
- 3. Wu G, Broniscer A, McEachron TA, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bender S, Tang Y, Lindroth AM, et al. . Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. [DOI] [PubMed] [Google Scholar]
- 5. Chan KM, Fang D, Gan H, et al. . The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lewis PW, Müller MM, Koletsky MS, et al. . Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buczkowicz P, Hoeman C, Rakopoulos P, et al. . Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paugh BS, Broniscer A, Qu C, et al. . Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29(30):3999–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grill J, Puget S, Andreiuolo F, Philippe C, MacConaill L, Kieran MW. Critical oncogenic mutations in newly diagnosed pediatric diffuse intrinsic pontine glioma. Pediatr Blood Cancer. 2012;58(4):489–491. [DOI] [PubMed] [Google Scholar]
- 10. Wu G, Diaz AK, Paugh BS, et al. . The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jansen MH, van Vuurden DG, Vandertop WP, Kaspers GJ. Diffuse intrinsic pontine gliomas: a systematic update on clinical trials and biology. Cancer Treat Rev. 2012;38(1):27–35. [DOI] [PubMed] [Google Scholar]
- 12. Cohen KJ, Jabado N, Grill J. Diffuse intrinsic pontine gliomas—current management and new biologic insights. Is there a glimmer of hope? Neuro Oncol. 2017;19(8):1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tönjes M, Barbus S, Park YJ, et al. . BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19(7):901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin GL, Monje M. A protocol for rapid post-mortem cell culture of diffuse intrinsic pontine glioma (DIPG). J Vis Exp. 2017;( 121). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sturm D, Witt H, Hovestadt V, et al. . Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 16. Seshacharyulu P, Pandey P, Datta K, Batra SK. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 2013;335(1):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuleshov MV, Jones MR, Rouillard AD, et al. . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chornenkyy Y, Agnihotri S, Yu M, et al. . Poly-ADP-ribose polymerase as a therapeutic target in pediatric diffuse intrinsic pontine glioma and pediatric high-grade astrocytoma. Mol Cancer Ther. 2015;14(11):2560–2568. [DOI] [PubMed] [Google Scholar]
- 19. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 20. Cohen KJ, Heideman RL, Zhou T, et al. . Temozolomide in the treatment of children with newly diagnosed diffuse intrinsic pontine gliomas: a report from the Children’s Oncology Group. Neuro Oncol. 2011;13(4):410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ponti D, Costa A, Zaffaroni N, et al. . Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. [DOI] [PubMed] [Google Scholar]
- 22. Dvorak P, Dvorakova D, Hampl A. Fibroblast growth factor signaling in embryonic and cancer stem cells. FEBS Lett. 2006;580(12):2869–2874. [DOI] [PubMed] [Google Scholar]
- 23. Haley EM, Kim Y. The role of basic fibroblast growth factor in glioblastoma multiforme and glioblastoma stem cells and in their in vitro culture. Cancer Lett. 2014;346(1):1–5. [DOI] [PubMed] [Google Scholar]
- 24. Marchenko S, Flanagan L. Passaging human neural stem cells. J Vis Exp. 2007(7):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loilome W, Joshi AD, ap Rhys CM, et al. . Glioblastoma cell growth is suppressed by disruption of fibroblast growth factor pathway signaling. J Neurooncol. 2009;94(3):359–366. [DOI] [PubMed] [Google Scholar]
- 26. Zhang J, Zhou Q, Gao G, et al. . The effects of ponatinib, a multi-targeted tyrosine kinase inhibitor, against human U87 malignant glioblastoma cells. Onco Targets Ther. 2014;7:2013–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng X, Zhang B, Wang J, Xu X, Lin N, Liu H. Adenovirus-mediated transfer of siRNA against basic fibroblast growth factor mRNA enhances the sensitivity of glioblastoma cells to chemotherapy. Med Oncol. 2011;28(1):24–30. [DOI] [PubMed] [Google Scholar]
- 28. Wera S, Hemmings BA. Serine/threonine protein phosphatases. Biochem J. 1995;311(Pt 1):17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu J, Kovach JS, Johnson F, et al. . Inhibition of serine/threonine phosphatase PP2A enhances cancer chemotherapy by blocking DNA damage induced defense mechanisms. Proc Natl Acad Sci U S A. 2009;106(28):11697–11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu J, Zhuang Z, Song DK, et al. . The effect of a PP2A inhibitor on the nuclear receptor corepressor pathway in glioma. J Neurosurg. 2010;113(2):225–233. [DOI] [PubMed] [Google Scholar]
- 31. Gordon IK, Lu J, Graves CA, et al. . Protein phosphatase 2A inhibition with LB100 enhances radiation-induced mitotic catastrophe and tumor growth delay in glioblastoma. Mol Cancer Ther. 2015;14(7):1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duchesne L, Tissot B, Rudd TR, Dell A, Fernig DG. N-glycosylation of fibroblast growth factor receptor 1 regulates ligand and heparan sulfate co-receptor binding. J Biol Chem. 2006;281(37):27178–27189. [DOI] [PubMed] [Google Scholar]
- 34. Gong SG. Isoforms of receptors of fibroblast growth factors. J Cell Physiol. 2014;229(12):1887–1895. [DOI] [PubMed] [Google Scholar]
- 35. Wei D, Parsels LA, Karnak D, et al. . Inhibition of protein phosphatase 2A radiosensitizes pancreatic cancers by modulating CDC25C/CDK1 and homologous recombination repair. Clin Cancer Res. 2013;19(16):4422–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu C, Yu M, Ren Y, et al. . PP2A inhibition from LB100 therapy enhances daunorubicin cytotoxicity in secondary acute myeloid leukemia via miR-181b-1 upregulation. Sci Rep. 2017;7(1):2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9(1):59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Vugt MA, Brás A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell. 2004;15(5):799–811. [DOI] [PubMed] [Google Scholar]
- 39. Bellacosa A, Kumar CC, Di Cristofano A, Testa JR. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv Cancer Res. 2005;94:29–86. [DOI] [PubMed] [Google Scholar]
- 40. Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24(2):287–291. [DOI] [PubMed] [Google Scholar]
- 41. Lu LY, Yu X. The balance of polo-like kinase 1 in tumorigenesis. Cell Div. 2009;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Andrabi S, Gjoerup OV, Kean JA, Roberts TM, Schaffhausen B. Protein phosphatase 2A regulates life and death decisions via Akt in a context-dependent manner. Proc Natl Acad Sci U S A. 2007;104(48):19011–19016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hong CS, Ho W, Zhang C, Yang C, Elder JB, Zhuang Z. LB100, a small molecule inhibitor of PP2A with potent chemo- and radio-sensitizing potential. Cancer Biol Ther. 2015;16(6):821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing, whole-exome sequencing, and 850k methylation array data of patient-derived tumor cell cultures are available at the European Genome-phenome Archive (EGAS00001003404). Deconvoluted sequencing data of individual shRNA screening samples can be found in Supplementary Table 2.