Abstract
Radiation therapy is used in ~50% of cancer patients to reduce the risk of recurrence and in some cases improve survival. Despite these benefits, doses can be limited by toxicity in multiple organs, including the heart. The underlying causes and biomarkers of radiation-induced cardiotoxicity are currently unknown, prompting the need for experimental models with inherent differences in sensitivity and resistance to the development of radiation-induced cardiotoxicity. We have identified the parental SS (Dahl salt-sensitive/Mcwi) rat strain to be a highly-sensitized model of radiation-induced cardiotoxicity. In comparison, substitution of rat chromosome 3 from the resistant BN (Brown Norway) rat strain onto the SS background (SS-3BN consomic) significantly attenuated radiation-induced cardiotoxicity. SS-3BN rats had less radiation-induced cardiotoxicity than SS rats, as measured by survival, pleural and pericardial effusions, echocardiogram parameters, and histological damage. Mast cells, previously shown to have predominantly protective roles in radiation-induced cardiotoxicity, were increased in the more resistant SS-3BN hearts postradiation. RNA sequencing from SS and SS-3BN hearts at 1 wk postradiation revealed 5,098 differentially expressed candidate genes across the transcriptome and 350 differentially expressed genes on rat chromosome 3, which coincided with enrichment of multiple pathways, including mitochondrial dysfunction, sirtuin signaling, and ubiquitination. Upstream regulators of enriched pathways included the oxidative stress modulating transcription factor, Nrf2, which is located on rat chromosome 3. Nrf2 target genes were also differentially expressed in the SS vs. SS-3BN consomic hearts postradiation. Collectively, these data confirm the existence of heritable modifiers in radiation-induced cardiotoxicity and provide multiple biomarkers, pathways, and candidate genes for future analyses.
NEW & NOTEWORTHY This novel study reveals that heritable genetic factors have the potential to modify normal tissue sensitivity to radiation. Gene variant(s) on rat chromosome 3 can contribute to enhanced cardiotoxicity displayed in the SS rats vs. the BN and SS-3BN consomic rats. Identifying genes that lead to understanding the mechanisms of radiation-induced cardiotoxicity represents a novel method to personalize radiation treatment, as well as predict the development of radiation-induced cardiotoxicity.
Keywords: cardiotoxicity, consomic, genomics, radiation therapy, RNA sequencing
INTRODUCTION
In 2015, heart disease and cancer were the two leading causes of death in the United States, accounting for 45% of all deaths (20). The heavy burden of these two diseases is expected to grow substantially over the next generation, as the population of patients with coexisting heart disease and cancer is predicted to increase exponentially, resulting from better treatments and increased life expectancy (11). Radiation therapy (RT) is received by >50% of all cancer patients, and whereas RT can improve local control and survival, it can also cause a wide range of side effects, including damage to the heart (i.e., cardiotoxicity) (14).
The survival of cancer patients has improved over the past few decades (41), resulting in higher numbers of survivors who have received radiation to the heart and are at risk for radiation-induced cardiotoxicity, which can include coronary artery disease, pericarditis, cardiomyopathy, myocardial fibrosis, pericardial effusion, and arrhythmias (14, 55). The risk of cardiotoxicity is increased in patients who receive incidental radiation to the heart, such as breast cancer patients with left-sided tumors (48), and the risk of major coronary events increases ~7.4% per Gray (Gy) of mean dose exposed to the heart (10). While modern RT techniques can reduce radiation dose to the heart in some breast cancer patients, even low doses of radiation can lead to chronic cardiac dysfunction in a significant proportion of patients (10, 47). Moreover, the risk of radiation-induced cardiotoxicity is further exacerbated by cardiotoxic chemotherapies (e.g., anthracyclines) that are frequently prescribed to breast cancer patients.
Mounting evidence suggests that complex genetic modifiers contribute to the risk of radiation-induced toxicity in cancer patients (21, 26). However, many of the existing genetic association studies are limited by small cohort sizes and multiple confounding factors, thus posing a challenge to identifying the heritable risk factors of radiation-induced cardiotoxicity. Another strategy for identifying complex genetic modifiers of cardiovascular phenotypes is through mapping studies using chromosome substitution strains (i.e., consomics) (9, 38), which are derived from two parental strains with different susceptibilities to a trait of interest, such as radiation-induced cardiotoxicity. However, to date, experimental models of genetic susceptibility to radiation-induced cardiotoxicity have not been characterized.
Here, we identified the Brown Norway (BN) strain and the Dahl salt-sensitive/Mcwi (SS) rat strains as the first experimental models for mapping the genetic modifiers of radiation-induced cardiotoxicity. We found that the BN strain was relatively resistant to radiation-induced cardiotoxicity, whereas the SS strain developed severe cardiac dysfunction. Substitution of the BN-derived rat chromosome 3 (RNO3) into the SS background (i.e., SS-3BN consomic) completely recapitulated the protected phenotypes observed in the BN, indicating that genetic modifier(s) of radiation-induced cardiotoxicity reside on RNO3. RNA-sequencing (RNA-seq) was used for prioritizing the list of potential candidates, which nominated multiple genes and pathways that were differentially expressed in the left ventricles of SS and SS-3BN rats after radiation. Collectively, the SS-3BN consomic model, coupled with future congenic studies, is the first experimental strategy for mapping the genetic modifiers of radiation-induced cardiotoxicity, which will facilitate the discovery of novel biomarkers and therapeutics for averting radiation-induced cardiotoxicity in high-risk patients in the future.
MATERIALS AND METHODS
Rats and irradiation procedure.
SS, BN, and SS/BN consomic SS-3BN rats (Medical College of Wisconsin), aged 10 to 12 wk, were randomly allocated to different treatment groups. Local heart irradiation was performed using the high-precision image-guided X-RAD SmART irradiator (Precision X-Ray, North Branford, CT). The output of the SmART was regularly checked using a calibrated ionization chamber. Rats were anesthetized by 3% isoflurane/room temperature air inhalation for the duration of each treatment. Pilot V1.8 Imaging Software (University Health Network, Toronto, Canada) was used to create two-dimensional projections over 360° to provide computed tomography scans in transverse, sagittal, and frontal views, with each projection on the heart marked and centered to fit into the collimator. Rats were positioned in the prone position. A circular 1.5-cm-diameter collimator was used that encompassed the whole heart. The central axis of the beam (isocenter) was set in the center of the heart, with radiation dose to isocenter of 24 Gy × 1 fraction or 9 Gy × 5 fractions given once daily, with equally weighted anterior-posterior and 2 lateral beams (1:1:1, 225 kVp, 13 mA, 0.32 mm Cu, 2.69 Gy/min). Control rats received sham irradiation. Monte-Carlo-based treatment planning was used to precisely calculate irradiation doses (MAASTRO Radiotherapy Clinic). Males and females were used as described in the results. All rats were maintained in single ventilated cages under pathogen-free conditions at our Biomedical Research Center maintained at a temperature of 23°C on a 12-h:12-h light-dark cycle with access to standard diet (0.4% salt) and water. All procedures in this study were performed according to the American Guidelines for the Ethical Care of Animals and approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin.
Echocardiography.
Cardiac function was assessed using echocardiography, with M-mode readings and two-dimensional strain analysis, on rats receiving cardiac irradiation and control rats at baseline, 3 mo, and 5 or 6 mo after radiation. An echocardiograph Vivid 7 (General Electric, Wauwatosa, WI) was used with an M12L (11-MHz) linear-array transducer. Closed-chest imaging was conducted in the short-axis view at the mid-left ventricle level, verified by the presence of prominent papillary muscles, by a sonographer blinded to the study groups. Image depth was 2.5 cm, and images were acquired at 258 frames/s with electrocardiographic gating. Using the anatomical M-mode feature of the EchoPac software (General Electric, Wauwatosa WI), an M-mode display was generated from raw data two-dimensional images with the line selected passing through the anterior and inferior segments. Percent ejection fraction (EF) was measured using left ventricle end-diastolic volume (LVEDV) and left ventricle end-systolic volume (LVESV) using the formula EF = LVEDV − LVESV/LVEDV × 100. Percent fractional shortening (FS) was calculated with the formula: LVEDD − LVESD/LVEDD × 100, where LVEDD is left ventricle end-diastolic dimension and LVESD is left ventricle end systolic dimension. For every measurement, three consecutive heartbeats were measured, and the average used for analysis (40). For strain analysis, the images were processed with EchoPac Q analysis software (General Electric, Wauwatosa, WI). A cardiac cycle was defined from the peak of one R wave to the peak of the following R wave. For radial and circumferential strain, the endocardial border was traced in an end-systolic frame in the short-axis view at the midventricle, as identified by the prominent papillary muscles. The outer border was adjusted to approximate the epicardial border. The computer then provided a profile of radial (myocardial deformation toward the center) and circumferential (myocardial deformation along the curvature) strain (%) with time. Three consecutive heartbeats were measured, and the average used for analyses (40, 49). The quantification of rat pericardial effusions was based on the American Society for Echocardiography consensus statement for the quantification of human pericardial disease (30). Anatomic position, circumferential location, and size of the effusion present in the echolucent space between visceral and parietal pericardium was measured and categorized as zero, mild, moderate, or large. At end diastole, mild effusions seen measured <3 mm in size in any one-dimension, moderate effusions measured 3–6 mm, and large effusions measured >6 mm (30).
Histological analysis.
For histological examination, hearts were fixed in zinc formalin at 5 mo postradiation or time of death for those rats that died of heart failure before the 5-mo time point. Four-micrometer sections were stained with hematoxylin and eosin, according to standard methods. Histological characteristics of coagulative necrosis and cardiomyocyte nuclear pleomorphism were scored on blinded samples by a board-certified pathologist (P.E.N.), using a scale of 0–4: 0 (not present), 1 (trace amount present), 2 (mild amount present), 3 (moderate amount present), and 4 (large amount present). Histochemical analyses were performed on heart sections fixed for 48 h in 10% zinc formalin and transferred to 70% ethanol. Tissue sections were embedded in paraffin. For mast cell staining, sections were dewaxed in CitroSolv (Fisher Scientific, Hampton, NH) for 5 min and rehydrated in decreasing concentrations of ethanol. Each slide was covered in a 0.1% toluidine blue in 1% sodium chloride (pH 2.0) for 3 min. All images were acquired using a Nikon Eclipse 50i upright microscope equipped with a Nikon Digital sight DS-U3 camera and NIS Elements BR software (Nikon Instruments, Melville, NY). Staining was quantified by counting the number of cells per high-power field on blinded samples by two independent investigators.
RNA expression analysis.
Total RNA was extracted by TRIzol (Thermo Fisher Scientific, Waltham, MA) from the left ventricles of 1-wk postradiation or mock-treated rats, treated as noted above (n = 4–5/group), per the manufacturer’s instructions. For RNA-seq, this was followed by library preparation using Illumina’s TruSeq RNA library kit per the manufacturer’s instructions and sequencing on an Illumina HiSeq2500 (Illumina, San Diego, CA). Individual libraries were prepared for each sample, indexed for multiplexing, and then sequenced on an Illumina HiSeq2500 (19). The Trim Galore program (version 0.4.1) was used to trim bases with a Phred quality score <20 [https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/] and only reads with a Phred quality score ≥20 were taken for analysis. The RSEM program function “rsem-prepare-reference” (version 1.3.0) was used to extract the transcript sequences from the rat genome (build Rnor6.0) (33) and to generate Bowtie2 indexes (Bowtie2 version 2.2.8) (32), followed by read alignment using the “rsem-calculate-expression” function. Differential expression analysis was performed using the Bioconductor package DESeq2 (version 1.12.4) (36) to compute log2 fold changes and false discovery rate-adjusted P values. For gene ontology pathway analysis, genes used in the analysis were those with a false discovery rate threshold of < 5 × 10−5. Data were analyzed for molecular and functional pathway enrichment using Ingenuity Pathway Analysis (IPA; Qiagen, Redwood City, CA). All raw sequencing data can be accessed from the Sequence Read Archive, BioProject ID PRJNA525087 (https://www.ncbi.nlm.nih.gov/bioproject/525087).
For quantitative PCR (qPCR), total RNA was extracted from the left ventricles of rats using Trizol (Thermo Fisher Scientific) and RNeasy (Qiagen, Redwood City, CA), according to the manufacturer’s instructions. Total RNA was transcribed to cDNA with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA), according to the manufacturer’s protocol. Quantification reactions were conducted using Applied Biosystem QuantStudio 6 Flex Real‐Time PCR instrument. qPCR was conducted using 1 μl cDNA, 10 μl SYBR Green PCR Master Mix (Applied Biosystems), 1 μl of 18 μM forward and reverse primer mix, and nuclease free‐water in a final volume of 20 μl. The reaction was carried out under the following conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Data were normalized to 18 s, and relative mRNA expression was calculated as the fold change using the ΔΔCt method (50). Primers for qPCR reactions are listed in Supplemental Table S1 (all supplemental material is available at https://doi.org/10.6084/m9.figshare.8079590.v1).
Statistical analysis.
Analyses of the echocardiogram data and histological scores were evaluated by a Student’s t-test. The criterion for significance was P < 0.05. Data are reported as means ± SE. For analysis of Kaplan-Meier survival curves, a log-rank (Mantel-Cox) test was used to determine significance. For analyses of scores assigned to pericardial effusions and histological features, Fisher’s exact test was used to determine significance. Histological criteria were grouped as ≤2 (not present to mild amount present) and >2 (moderate to large amount present) for Fisher’s exact tests. For histological criteria, Wilcoxon signed-rank test also gave similar results to those reported for Fisher’s exact tests. Criteria for significance was P < 0.05.
RESULTS
Phenotypic analysis of cardiac function in the SS and BN parental rat strains after radiation.
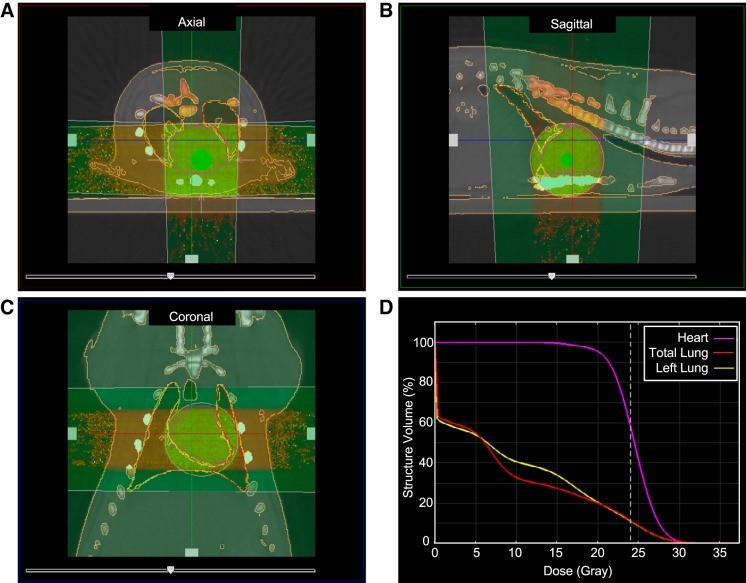
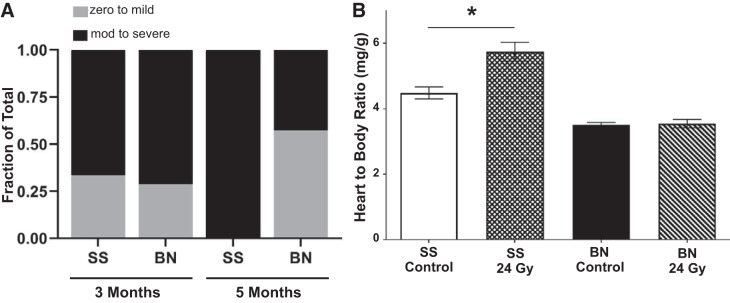
To first assess whether the parental SS and BN rat genomes harbored differential genetic modifiers of radiation-induced cardiotoxicity, female SS and BN rats at 10–12 wk of age were subjected to 24 Gy of localized imaged-guided heart irradiation or mock treatment (n = 7–11 rats per radiation group, n = 4 for control groups). Images of a representative rat with radiation plan and dose-volume histogram summary are provided in Fig. 1. At 5 mo postradiation, there was a trend for worse pericardial effusions in the SS rats, with 100% of the SS rats that survived 5 mo (6 of the 6 surviving) displaying severe pericardial effusion, compared with 43% of BN rats (3 of 7; Fig. 2A). This was not significant (P = 0.07), but in part this could be because the most severely affected SS rats had died by 5 mo postradiation, reducing the number of SS rats subjected to a 5-mo echocardiogram. However, the SS rats had significantly larger heart-to-body-weight ratios compared with SS controls at 5 mo [change (∆) 1.25 mg/g; P < 0.05], whereas no significant differences in heart weight were observed in the BN strain (∆0.04 mg/g, Fig. 2B). The SS strain also had significantly increased mortality in response to radiation (P < 0.05), with only 55% of rats (6/11) surviving to 5 mo postradiation compared with the 100% survival of BN rats (9/9). Collectively, these data demonstrate that the SS rat is significantly more sensitive to radiation-induced cardiac toxicity compared with the highly resistant BN rat strain.
Fig. 1.
Image-guided localized cardiac radiation technique. Computed tomography images of a representative female rat at 10 wk of age with a 1.5-cm-diameter collimator plan of 24 Gy to the heart isocenter in equally weighted beams (anterior-posterior and two laterals) are shown in the axial (A), sagittal (B), and coronal (C) planes. D: dose volume histogram demonstrating dose to the heart, total lungs, and the left lung.
Fig. 2.
SS (Dahl salt-sensitive/Mcwi) rats exhibit a radiation-induced cardiotoxicity permissive phenotype, compared with the BN (Brown Norway) rats. Female adult rats were treated with 24 Gy of image-guided cardiac radiation therapy (RT). A: pericardial effusions were similar at 3 mo, with 6/9 female SS rats vs. 5/7 BN rats having moderate (mod) to large amounts of fluid, but at 5 mo all SS rats had moderate to large pericardial effusions (6/6 SS vs. 3/7 BN rats), as measured via echocardiogram. B: heart-to-body ratio was measured at 5 mo in unirradiated or postradiation rats, as a marker of cardiac hypertrophy. At 5 mo, cardiac hypertrophy was evident in the SS female rats, but not in the BN female rats. Values are means ± SE; n = 4 rats in control groups and n = 7–11 rats used for radiation groups. *P < 0.05.
Phenotypic analysis of cardiac function in the SS-3BN consomic rat strain after RT.
A consomic panel derived from the SS and BN parental rat strains has been widely used to map genetic modifiers of various cardiovascular (8, 16–18, 22, 46) and cancer phenotypes (2, 15, 19), including a recent genomewide analysis of non-irradiation cardiac injury (https://rgd.mcw.edu/rgdweb/phenominer/table.html?terms=CMO:0000695,RS:0000803,RS:0000145,RS:0000811&fmt=2&species=3). Of the phenotyped consomic strains, the SS-3BN consomic was highly resistant to cardiac injury compared with the SS, suggesting that genetic modifier(s) localized to rat chromosome 3 (RNO3) might also convey resistance to the radiation-induced cardiotoxicity that was observed in the BN parental strain (Fig. 2).
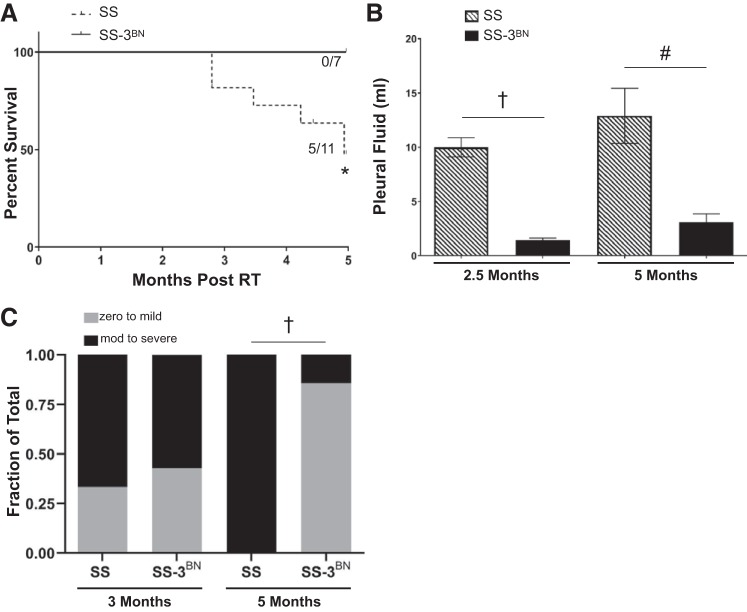
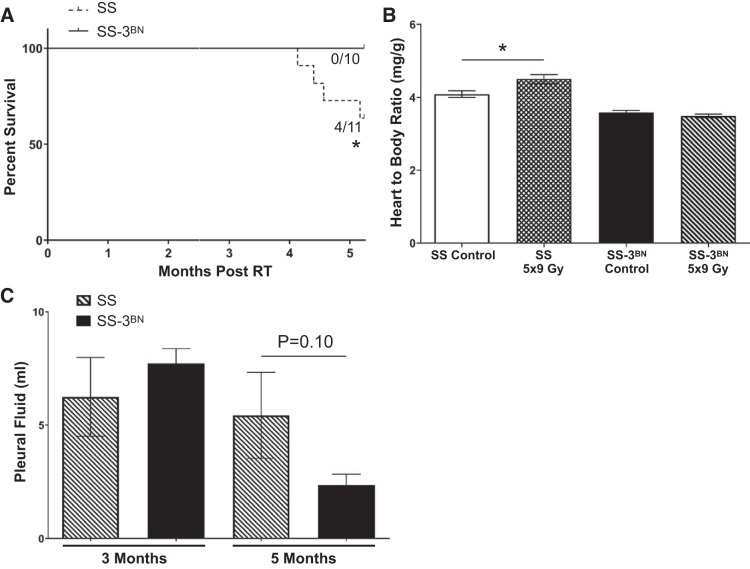
To test whether RNO3 harbors genetic modifier(s) of radiation-induced cardiotoxicity, female SS and SS-3BN consomic rats at 10–12 wk of age received 24 Gy of localized cardiac radiation or mock treatment (n = 7–11 rats per radiation group, n = 4 rats per control group), with assessment of cardiac structure and function by echocardiography at baseline, 3 mo, and 5 mo. Starting ~90 days after localized cardiac irradiation, the SS rats began experiencing mortality, with 45% (5/11) dying by 5 mo postradiation, whereas 0/7 of the SS-3BN consomic rats died by 5 mo (Fig. 3A). At 2.5 and 5 mo after radiation, the female SS rats also had significantly increased volumes of pleural effusions, compared with the SS-3BN consomic rats (Fig. 3B). At 5 mo postradiation, the SS-3BN female consomic rats also had significantly lower incidence of moderate to severe pericardial effusions compared with the SS rats, surviving to 5 mo [1/7 (15%) vs. 6/6 (100%), P < 0.01] (Fig. 3C). Collectively, these data suggest that the SS-3BN consomic recapitulates the protective phenotypes observed in the BN parental rat strain.
Fig. 3.
SS (Dahl salt-sensitive/Mcwi) rats exhibit worse mortality and increased effusions after localized cardiac irradiation compared with the SS-3BN (substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background) rats. A: increased cardiac mortality was observed after 24 Gy localized cardiac radiation therapy (RT) in SS female rats (5/11 SS vs. 0/7 SS-3BN rats; *P < 0.05). B: at 2.5 and 5 mo post-RT, SS (n = 6) female rats had increased pleural effusion volumes compared with SS-3BN (n = 7) rats, quantified at time of euthanasia. The volume of pleural effusion was larger in SS vs. SS-3BN rats at 2.5 mo postradiation, as well as at 5 mo in the surviving SS vs. SS-3BN rats: †P < 0.01; #P < 0.001. Values are means ± SE. C: at 3 mo, SS and SS-3BN had comparable number of rats with moderate (mod) to severe pericardial effusion, but the SS surviving to 5 mo postradiation had significantly more moderate to severe pericardial effusions (6/6 SS vs. 1/7 SS-3BN rats) detected via echocardiogram: †P < 0.01.
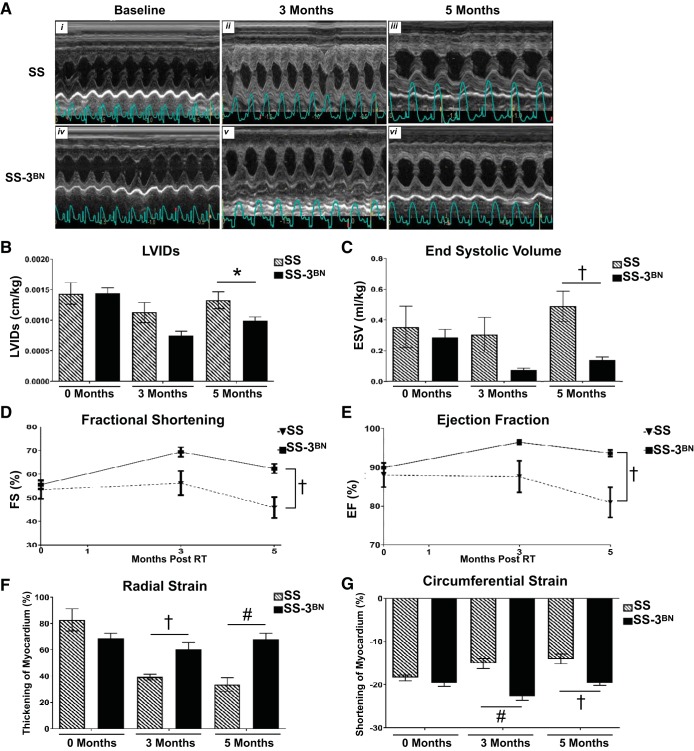
As outlined in Table 1, multiple echocardiograph parameters also indicated that the RNO3-specific genetic modifier(s) protect the SS-3BN consomic strain from radiation-induced cardiotoxicity. M-mode echocardiogram imaging demonstrated hyperdynamic systolic function in the SS-3BN consomic compared with severe dysfunction in the SS (representative images in Fig. 4A). Body weight-normalized left ventricular internal diameter at end systole (LVIDs, Fig. 4B) and end-systolic volume (ESV, Fig. 4C) were significantly increased at 5 mo postradiation in the surviving female SS rats vs. the SS-3BN consomic rats. In addition, the percent FS and EF were preserved to slightly increased after radiation in the SS-3BN consomic rats, whereas the SS rats had decreased FS and EF at 5 mo postradiation (Fig. 4, D and E). Finally, analysis of both radial and circumferential strain indicated that, compared with SS hearts at 5 mo postradiation, the SS-3BN consomic hearts had significantly reduced myocardium deformation, consistent with decreased systolic dysfunction (Fig. 4, F and G).
Table 1.
Echocardiogram data for female SS and SS-3BN rats treated with 24 Gy of localized, image-guided irradiation
| Baseline |
3 mo |
5 mo |
||||
|---|---|---|---|---|---|---|
| SS | SS-3BN | SS | SS-3BN | SS | SS-3BN | |
| IVSd, cm/kg | 0.83 ± 0.03 | 0.91 ± 0.03 | 1.08 ± 0.05 | 0.99 ± 0.06 | 0.90 ± 0.04 | 0.96 ± 0.06 |
| LVIDd, cm/kg | 3.01 ± 0.16 | 3.23 ± 0.08 | 2.42 ± 0.09 | 2.47 ± 0.14 | 2.68 ± 0.11 | 2.61 ± 0.06 |
| LVIDs, cm/kg | 1.44 ± 0.18 | 1.44 ± 0.09 | 1.08 ± 0.15 | 0.75 ± 0.07 | 1.33 ± 0.14 | 0.99 ± 0.06* |
| LVPWd, cm/kg | 0.92 ± 0.06 | 0.86 ± 0.02 | 1.03 ± 0.05 | 0.89 ± 0.03* | 0.94 ± 0.03 | 0.96 ± 0.07 |
| EDV, ml/kg | 2.72 ± 0.40 | 2.79 ± 0.17 | 2.01 ± 0.17 | 1.97 ± 0.26 | 2.58 ± 0.18 | 2.15 ± 0.12 |
| ESV, ml/kg | 0.35 ± 0.13 | 0.29 ± 0.05 | 0.29 ± 0.11 | 0.07 ± 0.01 | 0.49 ± 0.10 | 0.14 ± 0.02† |
| SV, ml/kg | 2.36 ± 0.31 | 2.49 ± 0.13 | 1.72 ± 0.11 | 1.89 ± 0.25 | 2.08 ± 0.19 | 2.01 ± 0.11 |
| LVM, g/kg | 5.71 ± 0.16 | 5.87 ± 0.10 | 6.32 ± 0.20 | 5.56 ± 0.13* | 6.04 ± 0.23 | 5.92 ± 0.30 |
| FS, % | 53.36 ± 3.82 | 55.54 ± 1.97 | 56.18 ± 5.15 | 69.35 ± 2.06 | 45.87 ± 4.41 | 62.34 ± 1.97† |
| EF, % | 87.96 ± 3.09 | 89.89 ± 1.25 | 87.59 ± 4.03 | 96.44 ± 0.56 | 80.93 ± 3.88 | 93.64 ± 0.84† |
Values are means ± SE. EDV, end-diastolic volume; EF, ejection fraction; ESV, end-systolic volume; FS, fractional shortening; IVSd, interventricular septal wall thickness during diastole; LVIDd, left ventricular internal diameter at end diastole; LVIDs, left ventricular internal diameter at end systole; LVM, left ventricular mass; LVPWd, left ventricular posterior wall thickness at diastole; SS, Dahl salt-sensitive/Mcwi; SS-3BN, substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background; SV, stroke volume.
P < 0.05;
P < 0.01.
Fig. 4.
Echocardiogram measurements indicate SS (Dahl salt-sensitive/Mcwi) female rats have poorer heart function compared with SS-3BN (substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background) rats after 24 Gy localized heart radiation. A: M-mode echocardiogram images of SS and SS-3BN rats that received 24 Gy of localized heart radiation therapy at baseline, 3 mo, and 5 mo postradiation. B: left ventricular internal diameter end systole (LVIDs) normalized to body weight is higher in SS vs. SS-3BN rats after radiation. C: end-systolic volume (ESV) normalized to body weight is higher in SS rats postradiation vs. SS-3BN rats. D: fractional shortening (FS) is reduced in SS rats at 5 mo postradiation vs. SS-3BN rats. E: ejection fraction (EF) is reduced in SS rats at 5 mo postradiation vs. SS-3BN rats. F: radial strain was lower in SS rats at 3 and 5 mo postradiation. G: circumferential strain showed SS rats had a decreased ability to contract, indicated by a smaller negative percentage. Values are means ± SE. *P < 0.05. †P < 0.01. #P < 0.001.
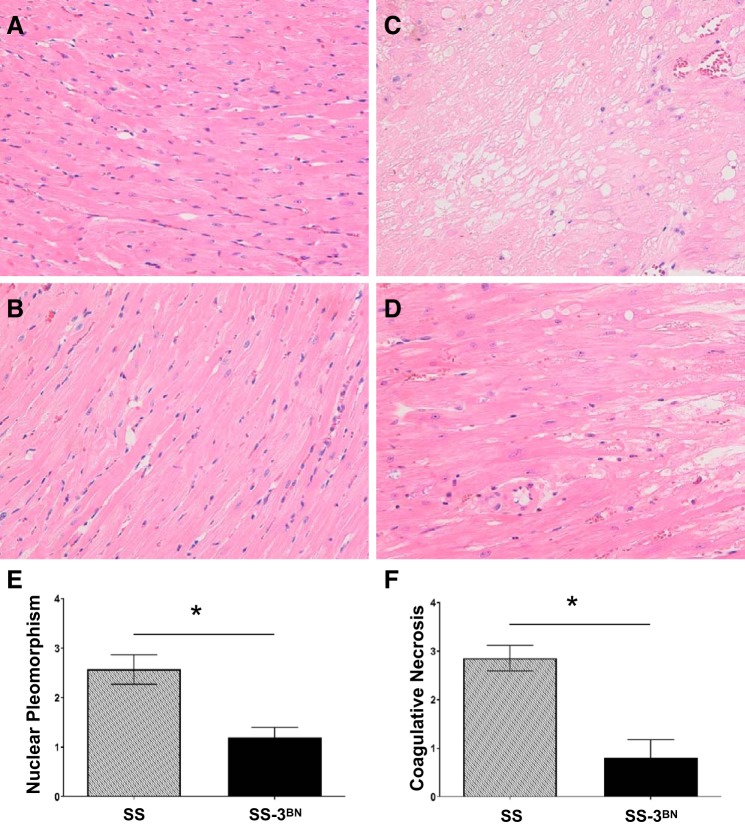
Histological analysis of blinded hematoxylin-and-eosin-stained slides from hearts of female SS and SS-3BN rats that received 24 Gy of localized radiation was performed by a board-certified pathologist, specifically quantifying the severity of coagulative necrosis and cardiomyocyte nuclear pleomorphism. Blinded analysis from female SS and SS-3BN consomic rat hearts that did not receive radiation at 8 mo of age (similar age as the 5 mo postradiation rats) revealed no coagulative necrosis or cardiomyocyte nuclear polymorphism (Fig. 5, A and B). Hearts from female rats at 5 mo postradiation or at the time of death revealed that the SS rat hearts exhibited more overall damage than SS-3BN hearts, with increased coagulative necrosis and cardiomyocyte nuclear pleomorphism (Fig. 5, C and D), and scoring of these features on a semiquantitative scale of 0–4 (see materials and methods) revealed the SS hearts had significantly more nuclear pleomorphism than the SS-3BN hearts (2.6 ± 0.30 vs. 1.2 ± 0.2, P < 0.05, Fig. 5E), as well as significantly more coagulative necrosis than the SS-3BN hearts (2.9 ± 0.26 vs. 0.8 ± 0.37, P < 0.05, Fig. 5F).
Fig. 5.
Representative histological features of unirradiated and postradiation SS (Dahl salt-sensitive/Mcwi) and SS-3BN (substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background) rat hearts. SS and SS-3BN female rat hearts were fixed and hematoxylin-and-eosin-stained 5 mo after receiving 24 Gy of localized cardiac irradiation or at time of death in the irradiated group. Age-matched unirradiated rat hearts were also examined. No coagulative necrosis or nuclear pleomorphisn were seen in the SS (A) or SS-3BN (B) unirradiated rats. C: at 5 mo post-radiation therapy, multiple relatively large foci of myocardial coagulative necrosis were typical in the SS hearts with abundant nuclear pleomorphism. D: the SS-3BN hearts exhibited less cardiomyocyte nuclear pleomorphism. Coagulative necrosis was focally present in the SS-3BN hearts, but the necrotic foci were smaller and more incipient rather than well developed compared with that seen in the SS hearts. Original magnifications ×20. Semiquantitative scoring of nuclear pleomorphism (E) and coagulative necrosis (F) in the SS and SS-3BN hearts is shown. Values are means ± SE. *P < 0.001.
Fractionated radiation (9 Gy × 5 fractions given once daily) to the heart of female rats using identical image-guided radiation techniques as above (Fig. 1) demonstrated similar trends to the 24 Gy × 1 fraction data, with significantly increased LVIDs, left ventricular volume at end diastole, ESV, and end-diastolic volume in the SS rats compared with the SS-3BN rats (Table 2). In addition, the percent FS and EF were significantly decreased in the SS vs. SS-3BN rats (Table 2). Similar to the 24 Gy × 1 fraction data, increased mortality was observed in the SS rats compared with the SS-3BN rats treated with 9 Gy × 5 fractions, with 4/11 rats dying by 5 mo postradiation, whereas 0/10 of the SS-3BN rats died before the 5-mo end point (P < 0.05, Fig. 6A). The SS rats had significantly larger heart-to-body weight ratios compared with SS controls at 5 mo (∆0.41 mg/g; P < 0.05), whereas no significant differences in heart weight were observed in the SS-3BN rats (Fig. 6B). At 3 mo postradiation, there was no difference in the volume of pleural effusions, whereas at 5 mo postradiation, the female SS rats had increased volumes of pleural effusions compared with the SS-3BN consomic rats (Fig. 6C). This was not significant (P = 0.100), but in part this could be due to the variability seen in the SS rats, as well as the fact that three SS rats died before the 5-mo end point without quantification of their effusions.
Table 2.
Echocardiogram data for female SS and SS-3BN rats treated with 9 Gy × 5 daily fractions of localized, image-guided irradiation
| Fractionated RT |
||
|---|---|---|
| SS | SS-3BN | |
| n | 8 | 9 |
| IVSd, cm/kg | 0.90 ± 0.04 | 1.07 ± 0.04* |
| LVIDd, cm/kg | 2.93 ± 0.09 | 2.55 ± 0.05† |
| LVPWd, cm/kg | 1.03 ± 0.04 | 1.06 ± 0.04 |
| LVIDs, cm/kg | 1.97 ± 0.09 | 0.89 ± 0.05† |
| EDV, ml/kg | 3.05 ± 0.14 | 1.91 ± 0.09† |
| ESV, ml/kg | 1.03 ± 0.10 | 0.09 ± 0.02† |
| SV, ml/kg | 2.04 ± 0.08 | 1.80 ± 0.08 |
| LVM, g/kg | 6.55 ± 0.23 | 6.22 ± 0.10 |
| FS, % | 32.70 ± 1.40 | 64.86 ± 1.90† |
| EF, % | 66.90 ± 1.97 | 94.82 ± 0.86† |
Values are means ± SE; n, no. of rats. EDV, end-diastolic volume; EF, ejection fraction; ESV, end-systolic volume; FS, fractional shortening; IVSd, interventricular septal wall thickness during diastole; LVIDd, left ventricular internal diameter at end diastole; LVIDs, left ventricular internal diameter at end systole; LVM, left ventricular mass; LVPWd, left ventricular posterior wall thickness at diastole; RT, radiation therapy; SS, Dahl salt-sensitive/Mcwi; SS-3BN, substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background; SV, stroke volume.
P < 0.05;
P < 0.01.
Fig. 6.
SS (Dahl salt-sensitive/Mcwi) rats that received 9 Gy × 5 localized cardiac radiation exhibit worse mortality and cardiac hypertrophy, and a trend toward increased pleural effusions, compared with the SS-3BN (substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background) rats. A: increased cardiac mortality was observed after 9 Gy × 5 Gy localized cardiac radiation therapy (RT) in SS female rats (4/11 SS vs. 0/10 SS-3BN rats expired: *P < 0.05). B: at 5 mo, cardiac hypertrophy was evident in the SS female rats, but not in the SS-3BN female rats, as measured by heart-to-body weight ratios: *P < 0.05. C: at 3 mo postradiation, there was no difference in the volume of pleural effusions between SS and SS-3BN rats, and at 5 mo postradiation the female SS rats had a trend toward increased volumes of pleural effusions compared with the SS-3BN consomic rats (P = 0.10). Values are means ± SE; n, no. of rats.
Male SS rats also exhibited increased cardiac damage compared with male SS-3BN rats. Male SS and SS-3BN rats received one fraction of 24 Gy to the heart, as described above and shown in Fig. 1. Echocardiograms were obtained at baseline, as well as 3 and 6 mo postradiation. No significant difference was seen in pericardial and pleural effusions between SS and SS-3BN males. However, the SS male rats had decreased cardiac function at 3 and 6 mo (Table 3), with weight-adjusted LVIDs and ESV increased in the SS vs. SS-3BN rats at 3 mo (LVIDs 0.94 ± 0.06 vs. 0.55 ± 0.04 cm/kg, P < 0.01; ESV 0.37 ± 0.06 vs. 0.065 ± 0.01 ml/kg, P < 0.01), and percent FS and EF decreased in the SS vs. SS-3BN rats at both 3 and 6 mo (Table 3).
Table 3.
Echocardiogram data for male SS and SS-3BN rats treated with 24 Gy × 1 fraction of localized, image-guided irradiation
| Baseline |
3 mo |
6 mo |
||||
|---|---|---|---|---|---|---|
| SS | SS-3BN | SS | SS-3BN | SS | SS-3BN | |
| IVSd, cm/kg | 0.60 ± 0.04 | 0.71 ± 0.03 | 0.52 ± 0.04 | 0.63 ± 0.03 | 0.63 ± 0.03 | 0.56 ± 0.02 |
| LVIDd, cm/kg | 2.48 ± 0.14 | 2.80 ± 0.11 | 1.81 ± 0.04 | 1.89 ± 0.05 | 1.79 ± 0.07 | 1.97 ± 0.04* |
| LVIDs, cm/kg | 1.26 ± 0.11 | 1.44 ± 0.10 | 0.94 ± 0.06 | 0.55 ± 0.04† | 0.96 ± 0.07 | 0.90 ± 0.05 |
| LVPWd, cm/kg | 0.62 ± 0.03 | 0.70 ± 0.04 | 0.61 ± 0.03 | 0.65 ± 0.03 | 0.64 ± 0.04 | 0.61 ± 0.02 |
| EDV, ml/kg | 3.19 ± 0.37 | 3.15 ± 0.29 | 2.09 ± 0.11 | 1.84 ± 0.13 | 2.00 ± 0.17 | 2.20 ± 0.10 |
| ESV, ml/kg | 0.50 ± 0.09 | 0.50 ± 0.10 | 0.37 ± 0.06 | 0.07 ± 0.01† | 0.37 ± 0.07 | 0.26 ± 0.04 |
| SV, ml/kg | 2.70 ± 0.28 | 2.64 ± 0.21 | 1.72 ± 0.06 | 1.77 ± 0.12 | 1.63 ± 0.14 | 1.94 ± 0.08 |
| LVM, g/kg | 4.75 ± 0.25 | 5.15 ± 0.21 | 4.19 ± 0.14 | 4.19 ± 0.11 | 4.46 ± 0.25 | 4.13 ± 0.08 |
| FS, % | 49.27 ± 1.50 | 48.91 ± 2.37 | 48.73 ± 2.47 | 70.95 ± 1.82† | 46.87 ± 2.51 | 54.27 ± 1.97* |
| EF, % | 85.05 ± 1.27 | 84.66 ± 2.00 | 83.45 ± 1.99 | 96.77 ± 0.53† | 81.92 ± 2.67 | 88.50 ± 1.37* |
Values are means ± SE. EDV, end-diastolic volume; EF, ejection fraction; ESV, end-systolic volume; FS, fractional shortening; IVSd, interventricular septal wall thickness during diastole; LVIDd, left ventricular internal diameter at end diastole; LVIDs, left ventricular internal diameter at end systole; LVM, left ventricular mass; LVPWd, left ventricular posterior wall thickness at diastole; SS, Dahl salt-sensitive/Mcwi; SS-3BN, substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background; SV, stroke volume.
P < 0.05;
P < 0.01.
Assessment of mast cell infiltration in SS and SS-3BN hearts after RT.
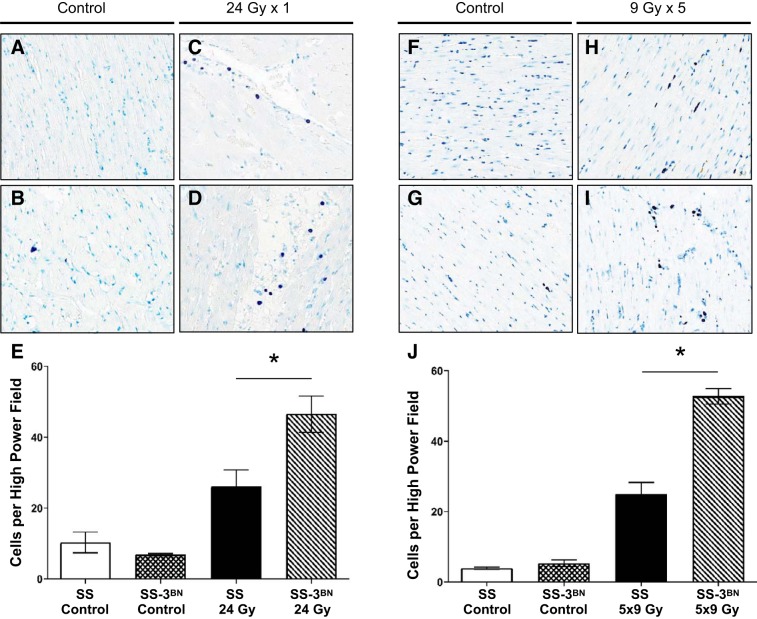
Previous work has demonstrated that mast cells can play a protective role in the development of radiation-induced cardiotoxicity (5). We examined the potential role of mast cell infiltration in our radiation-induced cardiotoxicity by staining for mast cells in the SS and SS-3BN hearts 10 wk after 24 Gy × 1 or 9 Gy × 5. This demonstrated that the SS-3BN rats had significantly increased levels of mast cells in the left ventricle after both the single fraction (Fig. 7, A–E), as well as the five fraction radiation regimens (Fig. 7, F–J). There were no differences in the levels of mast cells between the unirradiated SS and SS-3BN hearts (Fig. 7, E and J).
Fig. 7.
SS (Dahl salt-sensitive/Mcwi) rat hearts have decreased mast cell infiltration compared with SS-3BN (substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background) rat hearts at 10–12 wk following 24 Gy × 1 or 9 Gy × 5 localized heart radiation. Mast cells were stained using toluidine blue in left ventricle sections from female rats 10 wk after localized cardiac treatment with 24 Gy × 1 (A–E) or 12 wk after 9 Gy × 5 (F–J). There was no difference between the mast cell infiltrates in untreated SS vs. SS-3BN (A vs. B and F vs. G). However, after radiation, mast cell infiltrates increased, and the number of mast cells was significantly higher in the SS-3BN rats after both 24 × 1 (SS vs. SS-3BN; C and D) and 9 Gy × 5 (SS vs. SS-3BN; H and I). This is summarized for 24 Gy × 1 (E) and 9 Gy × 5 (J). *P < 0.05. Values are means ± SE.
RNA-seq analysis of SS and SS-3BN hearts following radiation.
To begin identifying the molecular gene networks that are altered in response to radiation by the genetic modifier(s) on RNO3, left ventricles of SS and SS-3BN hearts at 1 wk post-RT (n = 4–5/group) were analyzed by RNA-seq. A total of 5,098 genes were differentially expressed (FDR < 5 × 10−5) in SS-3BN-irradiated left ventricles compared with SS (Supplemental Table S2), of which 350 differentially expressed genes were localized to RNO3 and can, therefore, be considered as candidates (Supplemental Table S3). qPCR was used to validate the differential expression of genes differentially expressed between the SS and SS-3BN rats 1 wk after radiation. These results demonstrated that 23 of 30 genes tested were differentially expressed in SS vs. SS-3BN similarly to the RNA-seq results (Supplemental Fig. S1).
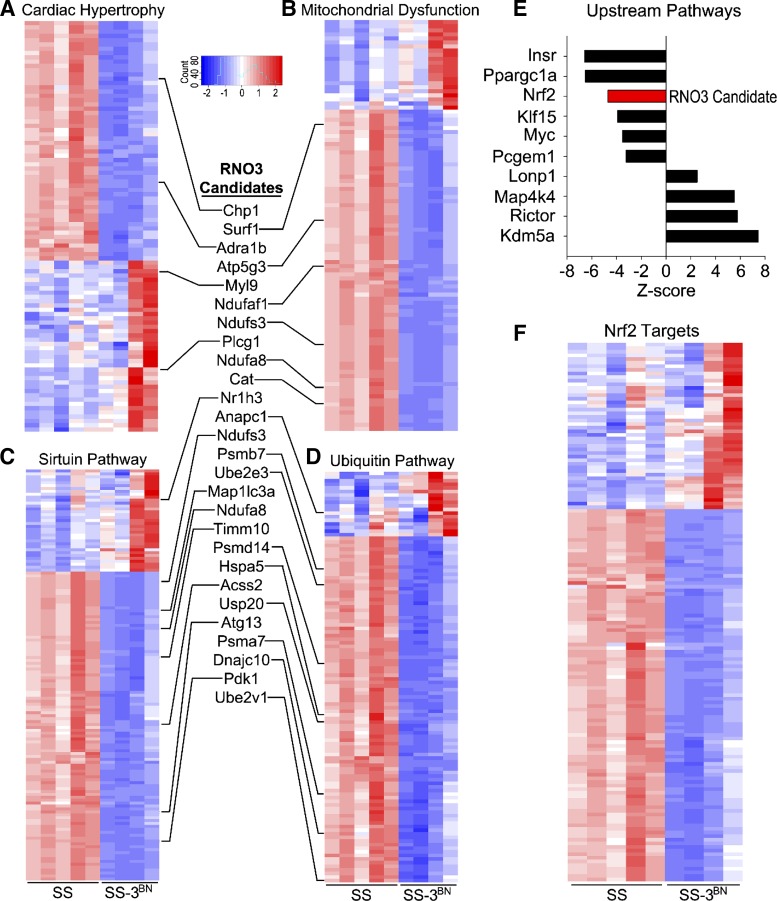
Gene ontology (GO) enrichment analysis using IPA revealed that the most enriched GOs included cardiac hypertrophy signaling (P = 7.8 × 10−12, Fig. 8A, Supplemental Table S4), mitochondrial dysfunction (P = 3.9 × 10−27, Fig. 8B, Supplemental Table S5), sirtuin signaling (P = 4.4 × 10−25, Fig. 8C, Supplemental Table S6), protein ubiquitination (P = 7.9 × 10−16, Fig. 8D, Supplemental Table S7), and oxidative phosphorylation (P = 8.2 × 10−17, Supplemental Table S8). Multiple candidate genes on RNO3 fall within these pathways (listed in Fig. 8, inset) and, therefore, can be preliminarily prioritized as the top candidates for follow-up studies. Analysis of the upstream molecular regulators of enriched pathways in IPA were identified, including the RNO3 candidate gene, Nrf2 (also known as nuclear factor erythroid 2-like 2, Nfe2l2). Nrf2 target genes were also significantly differentially expressed in the SS vs. SS-3BN hearts following irradiation (Fig. 8F, Supplemental Table S9). Collectively, these RNA-seq data confirm the cardiac dysfunction that phenotypically differentiated SS and SS-3BN consomic rats after cardiac irradiation (Figs. 3–6) and provide multiple candidate genes, notably including Nrf2, sirtuin signaling-related genes, and mitochondrial dysfunction-related genes on RNO3 that can be pursued further as potential genetic modifiers of radiation-induced cardiotoxicity.
Fig. 8.
RNA-sequencing analysis of irradiated SS (Dahl salt-sensitive/Mcwi) and SS-3BN (substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background) hearts. Total RNA was extracted, and RNA-sequencing was performed on RNA from the left ventricle of female SS and SS-3BN rats harvested 1 wk after 24 Gy of localized heart radiation or mock treatment (n = 4–5/group). Differential expression analysis was performed, and Ingenuity Pathway Analysis (IPA) gene network analysis identified the most significantly enriched gene ontologies to be cardiac hypertrophy (A), mitochondrial dysfunction (B), sirtuin signaling (C), and ubiquitin signaling (D), which included multiple candidate genes that reside on BN-derived rat chromosome 3 (RNO3; inset). E: analysis of the upstream molecular regulators of enriched pathways was also assessed by IPA, including the RNO3 candidate gene, Nrf2. F: differentially expressed Nrf2-target genes are significantly enriched following irradiation of SS and SS-3BN hearts.
DISCUSSION
In the present study, we demonstrated that heritable genetic factors on RNO3 have the potential to modify cardiac sensitivity to radiation. To our knowledge, this is the first mapping study of genetic modifiers of radiation-induced cardiotoxicity. The SS rats displayed increased cardiac damage, compared with the BN rats (Fig. 2). In fact, a large proportion of SS female rats died of heart failure starting at ~3 mo after radiation treatment (Fig. 3A). To begin localizing the genetic modifier(s) of radiation-induced cardiotoxicity, we made use of a consomic panel derived from the SS and BN parental rat strains to map genetic modifiers of radiation-induced cardiotoxicity. These data reveal that the SS-3BN consomic strain was highly resistant to cardiac injury compared with the SS strain, as measured by survival (Fig. 3A), pleural and pericardial effusions (Fig. 3, B and C), and echocardiogram parameters (Fig. 4 and Table 1). Echocardiogram results indicate that the irradiated SS rat hearts cannot contract as effectively after localized cardiac irradiation, and the SS hearts exhibit ventricular dilation and poorer systolic function, compared with SS-3BN consomic rat hearts (Fig. 4). In addition, histological damage was more severe in the SS vs. SS-3BN hearts after radiation (Fig. 5). Taken together, our data suggest that genetic modifier(s) localized to RNO3 are important for modulation of radiation-induced cardiotoxicity.
The enhanced sensitivity of SS rats vs. SS-3BN consomic rats to radiation-induced cardiotoxicity was reproduced in our study with female rats that received five daily treatments of 9 Gy of localized cardiac radiation (Table 2 and Fig. 6). Clinically, cardiac radiation exposure typically occurs in a fractionated manner. Our data demonstrate that SS rats have increased cardiac damage compared with SS-3BN rats, independent of a single or fractionated radiation dose. In addition, the genetic modifier(s) localized to RNO3 that convey resistance to radiation-induced cardiotoxicity are not only limited to female rats. The SS male rats also displayed increased cardiac damage, compared with the SS-3BN male consomic rats (Table 3). In our model, female SS rats overall had a more severe phenotype than age-matched SS male rats after receiving 24 Gy of localized cardiac radiation, as 94% (15/16) of male rats were alive 5 mo after 24 Gy of heart radiation, vs. 55% (6/11) of females (P < 0.01, data not shown). This could be in part due to sex-specific modifiers of radiation-induced cardiotoxicity, but other factors could also be contributing to this difference. For instance, the adult female rats were smaller than age-matched adult male rats, with corresponding smaller total lung volumes, and thus a higher percentage of the total lung volume would likely receive radiation using the image-guided radiation setup (Fig. 1), which could contribute to enhanced cardiac changes (25). In any case, both male and female SS rats had worse radiation-induced cardiotoxicity compared with their age-matched SS-3BN consomic rat counterparts.
Evidence supporting the existence of genetic modifiers of RT-induced cardiotoxicity.
Our study reveals novel findings that show heritable genetic factors have the potential to modify normal tissue sensitivity to radiation. Scientists in the radiogenomics field have identified a number of single nucleotide polymorphisms (SNPs) in recent years that have been confirmed in replicated studies to be associated with a number of long-term normal tissue radiation toxicities, but the toxicities were in the skin, lung, esophagus, and bowel [reviewed by Kerns et al. (26)]. Thus far, no SNPs have been shown to definitely be associated with radiation-induced cardiotoxicity. Genetic predispositions to radiation-induced cardiotoxicity would be helpful in personalizing radiation treatments to enhance the therapeutic ratio, and the etiology of such predispositions may also lead to therapies to prevent or mitigate radiation-induced cardiac damage.
Candidate modifiers of RT-induced cardiotoxicity.
Our study indicated the protective phenotype observed in the BN and consomic SS-3BN rat strains may be associated with a number of genes involved in pathways other than DNA-damage, including cardiac hypertrophy, sirtuin signaling, mitochondrial dysfunction, and the ubiquitin pathway, all identified via RNA-seq (Fig. 8). Over 300 differentially expressed genes in the SS and SS-3BN rat hearts 1 wk after radiation were on chromosome 3 and thus potential candidates for the causative loci for enhanced radiation-induced cardiotoxicity. Gene network analysis revealed that one of the upstream molecular regulators of the enriched pathways on RNO3 was Nrf2. Nrf2 is a redox-sensitive transcription factor that can regulate oxidative and cytoprotective cellular responses (23, 24). Nrf2 has been shown to play a role in a number of human diseases, and it has been shown to be important for endothelial function (35, 45), microvascular rarefaction (45), and cardioprotection (6). Nrf2 has also been shown to promote against irradiation a number of nonmalignant cell types, including mouse embryonic fibroblasts (39, 52) and breast and lung epithelial cells (12). In addition, Nrf2 signaling can reduce the severity of hematopoietic acute radiation syndrome and acute gastrointestinal radiation toxicity (28, 29), as well as reduce the severity of radiation-induced pulmonary fibrosis (59). While the role of Nrf2 in radiation-induced cardiotoxicity has not been directly determined, the effect of the knockdown of glutathione S-transferase-α4 (Gsta4) on cardiotoxicity was examined recently (4). Gsta4 is responsible for removing 4-hydroxynonenal, an electrophilic product of lipid peroxidation during oxidative stress that is a direct activator of Nrf2. Gsta4−/− mice were found to have less radiation-induced cardiac damage than wild-type controls, which was hypothesized to be due to activation of Nrf2 (4).
In the human Nrf2 locus, a number of SNPs have been shown to be associated with cardiovascular-related phenotypes, including cerebrovascular disease (31), hypertension (31, 51), triglyceride levels (13), and cardiovascular mortality (13, 51). To date, no studies have examined the associated with SNPs in Nrf2 and the incidence of radiation-induced cardiotoxicity. Examination of variants in the Nrf2 gene between SS and BN strains (https://rgd.mcw.edu/) revealed no nonsynonymous variants. However, there were a number of SNPs in noncoding regions of Nrf2 that differed in the SS and BN strains. The significance of these variants is currently unknown.
In addition to Nrf2, there are number of additional candidates on RNO3 that could be mediating differences in radiation-induced cardiotoxicity. The SS and SS-3BN rats have dramatic differences in expression of genes in mitochondrial dysfunction and sirtuin signaling pathways (Fig. 8, B and C), indicating altered mitochondrial function may be a driving mechanism for enhanced cardiotoxicity in the SS vs. SS-3BN rats.
A number of genes in these pathways reside on RNO3 (see Fig. 8, inset). For example, the PDK1 gene resides on RNO3. PDK1 can activate Akt (56), and Akt signaling has been implicated in cardiac hypertrophy after a variety of stimuli (1, 43). Sirtuins have also been shown to modulate Akt and cardiac hypertrophic responses (43, 44, 57). The sirtuin, Sirt3, has been shown to promote decreased Akt activation and decreased cardiac hypertrophic responses (57). Sirt3 is the primary deacetylase of mitochondrial proteins (34, 58), and its deacetylase activity plays a key role in maintaining mitochondrial energy metabolism equilibrium and defending against cellular damage that occurs with aging. Sirt3−/− mice demonstrate enhanced mitochondrial dysfunction (27, 58, 60), and Sirt3−/− mice exhibit enhanced doxorubicin- (42) and aortic constriction-induced cardiac hypertrophy (57). In addition, Sirt3−/− mouse embryo fibroblasts and livers are more sensitive to radiation and exhibit increased mitochondrial superoxide levels (7, 27). However, the role of Sirt3 in radiation-induced cardiac injury has not been examined.
Finally, our studies demonstrate differences in mast cell infiltration in the left ventricles of the hearts from SS vs. SS-3BN rats, with the SS-3BN rats having increased mast cells at 10–12 wk after radiation treatment with 24 Gy × 1 (Fig. 7, A–E) or 9 Gy × 5 (Fig. 7, F–J). Previous studies of radiation-induced heart disease have specifically implicated mast cells as predominantly protective against radiation-induced heart disease, with mast-cell-deficient rats exhibiting more severe cardiac changes after localized high-dose cardiac irradiation (5). While mast cells have roles in allergic reactions and immune modulation, they also have roles in tissue remodeling (37). The increase in mast cell infiltration in SS-3BN vs. SS rats is consistent with the more cardiac radiation-resistant phenotypes seen in the SS-3BN rats, but it is not yet clear whether increased mast cells are responsible for the cardiac protection in the SS-3BN rats. This needs to be examined in future studies.
Study limitations.
There are limitations from our study that should be acknowledged. In rat echocardiogram data, the variability between each rat in a group can be relatively large, limiting our knowledge of the average data trends. However, because the phenotypic differences in radiation-induced cardiotoxicity were large between the SS and SS-3BN rats, a number of significant differences were seen in the echocardiogram parameters between the SS and SS-3BN consomic rats. In addition, a significant portion of SS rats died before the 5-mo echocardiogram was obtained, leading to echocardiogram data for the 5-mo time points that do not include the SS rats with the most severe cardiac damage. We examined both a single large fraction of localized heart irradiation, and we also used a fractionation scheme of 9 Gy × 5, based on previous studies using this regimen for cardiac irradiation in rats (3, 53, 54). While data on a fractionated radiation regimen of 9 Gy × 5 had similar trends to the data from single fraction localized cardiac radiation (Table 2 and Fig. 6), future studies are needed with both partial heart radiation and increased fractions of smaller daily radiation doses that more closely mimic the radiation exposure seen by many cancer patients being treated with thoracic RT.
Translational impact and future considerations.
This study demonstrates that genetic variants can play a role in cardiac normal tissue radiation sensitivity. The SS rats are more sensitive to cardiac radiation than SS-3BN rats, demonstrating heritable factors on rat chromosome 3 modify radiation-induced cardiosensitivity. Gene expression analysis identified numerous potential targets on chromosome 3, including genes involved in mitochondrial dysfunction, sirtuin signaling, cardiac hypertrophy, and ubiquitin signaling.
Congenic mapping is one method to begin defining the region(s) of chromosome 3 that are critical for responses to radiation-induced cardiotoxicity to help to further limit candidates for testing. In addition, Nrf2 knockout rats (45) could be used to examine whether Nrf2 plays a critical role in protection against radiation-induced cardiotoxicity. The use of pharmacological sirtuin pathway and mitochondrial function modulators and transgenic models could be also pursued to further define the role of sirtuin signaling and mitochondrial protein acetylation and/or function in radiation-induced cardiotoxicity. This is the first study using consomic models to demonstrate heritable genetic modifiers of radiation-induced cardiotoxicity. Once identified, these modifiers will need to be validated in human studies. In the future, the results from these studies could be used to better stratify patients and develop novel therapeutics to avert or treat radiation-induced cardiotoxicity.
GRANTS
This work was supported by the Mary Kay Foundation Award Grant no. 017–29 (C. Bergom); Susan G. Komen Grant CCR17483233; the Nancy Laning Sobczak, PhD, Breast Cancer Research Award (C. Bergom); the Michael H. Keelan, Jr., MD, Research Foundation Grant (C. Bergom); the Cardiovascular Center (C. Bergom, M. Medhora, J. L. Strande) and Cancer Center (C. Bergom, M. J. Flister) at the Medical College of Wisconsin; the Institutional Research Grant 86–004–26 from the American Cancer Society (C. Bergom); the National Center for Research Resources, the National Center for Advancing Translational Sciences, and the Office of the Director of the National Institutes of Health through Grant 8KL2TR000056 (C. Bergom); National Cancer Institute Grant R01CA193343 (M. J. Flister) and Grant R01CA101841 (H. Rui); and National Institute of Allergy and Infectious Diseases Grant U01AI133594 (M. Medhora) and Grant U01AI107305 (M. Medhora).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
R.A.S., M.J.F., and C.B. conceived and designed research; R.A.S., A.F., B.L.F., L.H., T.G., P.E.N., J.L.S., and M.J.F. performed experiments; R.A.S., A.F., A.M.S., S.-W.T., L.H., Q.L., P.E.N., J.L.S., Y.S., H.R., M.J.F., and C.B. analyzed data; R.A.S., A.F., A.M.S., S.-W.T., B.L.F., L.H., M.M., P.E.N., J.L.S., M.J.F., and C.B. interpreted results of experiments; R.A.S., A.F., A.M.S., S.-W.T., T.G., P.E.N., M.J.F., and C.B. prepared figures; R.A.S., M.J.F., and C.B. drafted manuscript; R.A.S., B.L.F., L.H., T.G., M.M., P.E.N., J.L.S., M.J.F., and C.B. edited and revised manuscript; R.A.S., A.F., A.M.S., S.-W.T., B.L.F., L.H., Q.L., T.G., M.M., P.E.N., J.L.S., Y.S., H.R., M.J.F., and C.B. approved final version of manuscript.
Supplemental Data
REFERENCES
- 1.Abeyrathna P, Su Y. The critical role of Akt in cardiovascular function. Vascul Pharmacol 74: 38–48, 2015. doi: 10.1016/j.vph.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamovic T, McAllister D, Wang T, Adamovic D, Rowe JJ, Moreno C, Lazar J, Jacob HJ, Sugg SL. Identification of novel carcinogen-mediated mammary tumor susceptibility loci in the rat using the chromosome substitution technique. Genes Chromosomes Cancer 49: 1035–1045, 2010. doi: 10.1002/gcc.20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerma M, Roberto KA, Hauer-Jensen M. Prevention and treatment of functional and structural radiation injury in the rat heart by pentoxifylline and alpha-tocopherol. Int J Radiat Oncol Biol Phys 72: 170–177, 2008. doi: 10.1016/j.ijrobp.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerma M, Singh P, Sridharan V, Tripathi P, Sharma S, Singh SP. Effects of local heart irradiation in a glutathione S-transferase alpha 4-null mouse model. Radiat Res 183: 610–619, 2015. doi: 10.1667/RR13979.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerma M, Wang J, Wondergem J, Joseph J, Qiu X, Kennedy RH, Hauer-Jensen M. Influence of mast cells on structural and functional manifestations of radiation-induced heart disease. Cancer Res 65: 3100–3107, 2005. doi: 10.1158/0008-5472.CAN-04-4333. [DOI] [PubMed] [Google Scholar]
- 6.Chen QM, Maltagliati AJ. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol Genomics 50: 77–97, 2018. doi: 10.1152/physiolgenomics.00041.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman MC, Olivier AK, Jacobus JA, Mapuskar KA, Mao G, Martin SM, Riley DP, Gius D, Spitz DR. Superoxide mediates acute liver injury in irradiated mice lacking sirtuin 3. Antioxid Redox Signal 20: 1423–1435, 2014. doi: 10.1089/ars.2012.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley AW Jr, Liang M, Roman RJ, Greene AS, Jacob HJ. Consomic rat model systems for physiological genomics. Acta Physiol Scand 181: 585–592, 2004. doi: 10.1111/j.1365-201X.2004.01334.x. [DOI] [PubMed] [Google Scholar]
- 9.Cowley AW Jr, Roman RJ, Kaldunski ML, Dumas P, Dickhout JG, Greene AS, Jacob HJ. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension 37: 456–461, 2001. doi: 10.1161/01.HYP.37.2.456. [DOI] [PubMed] [Google Scholar]
- 10.Darby SC, Ewertz M, McGale P, Bennet AM, Blom-Goldman U, Brønnum D, Correa C, Cutter D, Gagliardi G, Gigante B, Jensen M-B, Nisbet A, Peto R, Rahimi K, Taylor C, Hall P. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N Engl J Med 368: 987–998, 2013. doi: 10.1056/NEJMoa1209825. [DOI] [PubMed] [Google Scholar]
- 11.Driver JA, Djoussé L, Logroscino G, Gaziano JM, Kurth T. Incidence of cardiovascular disease and cancer in advanced age: prospective cohort study. BMJ 337: a2467, 2008. doi: 10.1136/bmj.a2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Ashmawy M, Delgado O, Cardentey A, Wright WE, Shay JW. CDDO-Me protects normal lung and breast epithelial cells but not cancer cells from radiation. PLoS One 9: e115600, 2014. doi: 10.1371/journal.pone.0115600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figarska SM, Vonk JM, Boezen HM. NFE2L2 polymorphisms, mortality, and metabolism in the general population. Physiol Genomics 46: 411–417, 2014. doi: 10.1152/physiolgenomics.00178.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filopei J, Frishman W. Radiation-induced heart disease. Cardiol Rev 20: 184–188, 2012. doi: 10.1097/CRD.0b013e3182431c23. [DOI] [PubMed] [Google Scholar]
- 15.Flister MJ, Endres BT, Rudemiller N, Sarkis AB, Santarriaga S, Roy I, Lemke A, Geurts AM, Moreno C, Ran S, Tsaih S-W, De Pons J, Carlson DF, Tan W, Fahrenkrug SC, Lazarova Z, Lazar J, North PE, LaViolette PS, Dwinell MB, Shull JD, Jacob HJ. CXM: a new tool for mapping breast cancer risk in the tumor microenvironment. Cancer Res 74: 6419–6429, 2014. doi: 10.1158/0008-5472.CAN-13-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flister MJ, Hoffman MJ, Reddy P, Jacob HJ, Moreno C. Congenic mapping and sequence analysis of the Renin locus. Hypertension 61: 850–856, 2013. doi: 10.1161/HYPERTENSIONAHA.111.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flister MJ, Prisco SZ, Sarkis AB, O’Meara CC, Hoffman M, Wendt-Andrae J, Moreno C, Lazar J, Jacob HJ. Identification of hypertension susceptibility loci on rat chromosome 12. Hypertension 60: 942–948, 2012. doi: 10.1161/HYPERTENSIONAHA.112.198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flister MJ, Prokop JW, Lazar J, Shimoyama M, Dwinell M, Geurts A; International Committee on Standardized Genetic Nomenclature for Mice; Rat Genome and Nomenclature Committee . 2015 Guidelines for establishing genetically modified rat models for cardiovascular research. J Cardiovasc Transl Res 8: 269–277, 2015. doi: 10.1007/s12265-015-9626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flister MJ, Tsaih S-W, Stoddard A, Plasterer C, Jagtap J, Parchur AK, Sharma G, Prisco AR, Lemke A, Murphy D, Al-Gizawiy M, Straza M, Ran S, Geurts AM, Dwinell MR, Greene AS, Bergom C, LaViolette PS, Joshi A. Host genetic modifiers of nonproductive angiogenesis inhibit breast cancer. Breast Cancer Res Treat 165: 53–64, 2017. doi: 10.1007/s10549-017-4311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heron M. Deaths: leading causes for 2015. Natl Vital Stat Rep 66: 1–76, 2017. [PubMed] [Google Scholar]
- 21.Herskind C, Talbot CJ, Kerns SL, Veldwijk MR, Rosenstein BS, West CML. Radiogenomics: a systems biology approach to understanding genetic risk factors for radiotherapy toxicity? Cancer Lett 382: 95–109, 2016. doi: 10.1016/j.canlet.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman MJ, Flister MJ, Nunez L, Xiao B, Greene AS, Jacob HJ, Moreno C. Female-specific hypertension loci on rat chromosome 13. Hypertension 62: 557–563, 2013. doi: 10.1161/HYPERTENSIONAHA.113.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hybertson BM, Gao B, Bose SK, McCord JM. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol Aspects Med 32: 234–246, 2011. doi: 10.1016/j.mam.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322, 1997. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs ER, Narayanan J, Fish BL, Gao F, Harmann LM, Bergom C, Gasperetti T, Strande JL, Medhora M. Cardiac remodeling and reversible pulmonary hypertension during pneumonitis in rats after 13-Gy partial-body irradiation with minimal bone marrow sparing: effect of Lisinopril. Health Phys 116: 558–565, 2019. doi: 10.1097/HP.0000000000000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerns SL, Kundu S, Oh JH, Singhal SK, Janelsins M, Travis LB, Deasy JO, Janssens AC, Ostrer H, Parliament M, Usmani N, Rosenstein BS. The prediction of radiotherapy toxicity using single nucleotide polymorphism-based models: a step toward prevention. Semin Radiat Oncol 25: 281–291, 2015. doi: 10.1016/j.semradonc.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17: 41–52, 2010. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J-H, Thimmulappa RK, Kumar V, Cui W, Kumar S, Kombairaju P, Zhang H, Margolick J, Matsui W, Macvittie T, Malhotra SV, Biswal S. NRF2-mediated Notch pathway activation enhances hematopoietic reconstitution following myelosuppressive radiation. J Clin Invest 124: 730–741, 2014. doi: 10.1172/JCI70812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, Cornelius C, Wright WE, Pandita TK, Shay JW. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci USA 109: E2949–E2955, 2012. doi: 10.1073/pnas.1207718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein AL, Abbara S, Agler DA, Appleton CP, Asher CR, Hoit B, Hung J, Garcia MJ, Kronzon I, Oh JK, Rodriguez ER, Schaff HV, Schoenhagen P, Tan CD, White RD. American Society of Echocardiography clinical recommendations for multimodality cardiovascular imaging of patients with pericardial disease: endorsed by the Society for Cardiovascular Magnetic Resonance and Society of Cardiovascular Computed Tomography. J Am Soc Echocardiogr 26: 965–1012.e15, 2013. doi: 10.1016/j.echo.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Kunnas T, Määttä K, Nikkari ST. Genetic polymorphisms of transcription factor NRF2 and of its host gene sulfiredoxin (SRXN1) are associated with cerebrovascular disease in a finnish cohort, the TAMRISK Study. Int J Med Sci 13: 325–329, 2016. doi: 10.7150/ijms.14849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359, 2012. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323, 2011. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814, 2007. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopes RA, Neves KB, Tostes RC, Montezano AC, Touyz RM. Downregulation of nuclear factor erythroid 2-related factor and associated antioxidant genes contributes to redox-sensitive vascular dysfunction in hypertension. Hypertension 66: 1240–1250, 2015. doi: 10.1161/HYPERTENSIONAHA.115.06163. [DOI] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maltby S, Khazaie K, McNagny KM. Mast cells in tumor growth: angiogenesis, tissue remodelling and immune-modulation. Biochim Biophys Acta 1796: 19–26, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, Jacob HJ, Cowley AW Jr. Chromosome substitution reveals the genetic basis of Dahl salt-sensitive hypertension and renal disease. Am J Physiol Renal Physiol 295: F837–F842, 2008. doi: 10.1152/ajprenal.90341.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald JT, Kim K, Norris AJ, Vlashi E, Phillips TM, Lagadec C, Della Donna L, Ratikan J, Szelag H, Hlatky L, McBride WH. Ionizing radiation activates the Nrf2 antioxidant response. Cancer Res 70: 8886–8895, 2010. doi: 10.1158/0008-5472.CAN-10-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Migrino RQ, Zhu X, Pajewski N, Brahmbhatt T, Hoffmann R, Zhao M. Assessment of segmental myocardial viability using regional 2-dimensional strain echocardiography. J Am Soc Echocardiogr 20: 342–351, 2007. doi: 10.1016/j.echo.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 66: 271–289, 2016. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 42.Pillai VB, Bindu S, Sharp W, Fang YH, Kim G, Gupta M, Samant S, Gupta MP. Sirt3 protects mitochondrial DNA damage and blocks the development of doxorubicin-induced cardiomyopathy in mice. Am J Physiol Heart Circ Physiol 310: H962–H972, 2016. doi: 10.1152/ajpheart.00832.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pillai VB, Sundaresan NR, Gupta MP. Regulation of Akt signaling by sirtuins: its implication in cardiac hypertrophy and aging. Circ Res 114: 368–378, 2014. doi: 10.1161/CIRCRESAHA.113.300536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pillai VB, Sundaresan NR, Jeevanandam V, Gupta MP. Mitochondrial SIRT3 and heart disease. Cardiovasc Res 88: 250–256, 2010. doi: 10.1093/cvr/cvq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Priestley JRC, Kautenburg KE, Casati MC, Endres BT, Geurts AM, Lombard JH. The NRF2 knockout rat: a new animal model to study endothelial dysfunction, oxidant stress, and microvascular rarefaction. Am J Physiol Heart Circ Physiol 310: H478–H487, 2016. doi: 10.1152/ajpheart.00586.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prisco SZ, Prokop JW, Sarkis AB, Yeo NC, Hoffman MJ, Hansen CC, Jacob HJ, Flister MJ, Lazar J. Refined mapping of a hypertension susceptibility locus on rat chromosome 12. Hypertension 64: 883–890, 2014. doi: 10.1161/HYPERTENSIONAHA.114.03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saiki H, Petersen IA, Scott CG, Bailey KR, Dunlay SM, Finley RR, Ruddy KJ, Yan E, Redfield MM. Risk of heart failure with preserved ejection fraction in older women after contemporary radiotherapy for breast cancer. Circulation 135: 1388–1396, 2017. doi: 10.1161/CIRCULATIONAHA.116.025434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sardaro A, Petruzzelli MF, D’Errico MP, Grimaldi L, Pili G, Portaluri M. Radiation-induced cardiac damage in early left breast cancer patients: risk factors, biological mechanisms, radiobiology, and dosimetric constraints. Radiother Oncol 103: 133–142, 2012. doi: 10.1016/j.radonc.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Schattke S, Xing Y, Lock J, Brechtel L, Schroeckh S, Spethmann S, Baumann G, Borges AC, Knebel F. Increased longitudinal contractility and diastolic function at rest in well-trained amateur Marathon runners: a speckle tracking echocardiography study. Cardiovasc Ultrasound 12: 11, 2014. doi: 10.1186/1476-7120-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108, 2008. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 51.Shimoyama Y, Mitsuda Y, Tsuruta Y, Hamajima N, Niwa T. Polymorphism of Nrf2, an antioxidative gene, is associated with blood pressure and cardiovascular mortality in hemodialysis patients. Int J Med Sci 11: 726–731, 2014. doi: 10.7150/ijms.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh A, Bodas M, Wakabayashi N, Bunz F, Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal 13: 1627–1637, 2010. doi: 10.1089/ars.2010.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sridharan V, Seawright JW, Antonawich FJ, Garnett M, Cao M, Singh P, Boerma M. Late administration of a palladium lipoic acid complex (POLY-MVA) modifies cardiac mitochondria but not functional or structural manifestations of radiation-induced heart disease in a rat model. Radiat Res 187: 361–366, 2017. doi: 10.1667/RR14643.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sridharan V, Thomas CJ, Cao M, Melnyk SB, Pavliv O, Joseph J, Singh SP, Sharma S, Moros EG, Boerma M. Effects of local irradiation combined with sunitinib on early remodeling, mitochondria, and oxidative stress in the rat heart. Radiother Oncol 119: 259–264, 2016. doi: 10.1016/j.radonc.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart JR, Fajardo LF, Gillette SM, Constine LS. Radiation injury to the heart. Int J Radiat Oncol Biol Phys 31: 1205–1211, 1995. doi: 10.1016/0360-3016(94)00656-6. [DOI] [PubMed] [Google Scholar]
- 56.Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, Painter GF, Holmes AB, McCormick F, Hawkins PT. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277: 567–570, 1997. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 57.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest 119: 2758–2771, 2009. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao R, Coleman MC, Pennington JD, Ozden O, Park S-H, Jiang H, Kim H-S, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40: 893–904, 2010. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Travis EL, Rachakonda G, Zhou X, Korhonen K, Sekhar KR, Biswas S, Freeman ML. NRF2 deficiency reduces life span of mice administered thoracic irradiation. Free Radic Biol Med 51: 1175–1183, 2011. doi: 10.1016/j.freeradbiomed.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou X, Santa-Maria CA, O’Brien J, Gius D, Zhu Y. Manganese superoxide dismutase acetylation and dysregulation, due to loss of SIRT3 activity, promote a luminal B-like breast carcinogenic-permissive phenotype. Antioxid Redox Signal 25: 326–336, 2016. doi: 10.1089/ars.2016.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.