Abstract
We developed a transmission diffraction grating-based spectroscopic single-molecule localization microscopy (sSMLM) to collect the spatial and spectral information of single-molecule blinking events concurrently. We characterized the spectral heterogeneities of multiple far-red emitting dyes in a high-throughput manner using sSMLM. We also investigated the influence of spectral dispersion on the single-molecule identification performance of fluorophores with large spectral overlapping. The carefully tuning of spectral dispersion in grating-based sSMLM permitted simultaneous three-color super-resolution imaging in fixed cells with a single objective lens at relatively low photon budget. Our sSMLM has a compact optical design and can be integrated with conventional localization microscopy to provide add-on spectroscopic analysis capability.
1. Introduction
Super-resolution microscopy offers an opportunity to study biological processes at nanometer-level spatial resolution [1–8]. Single molecule localization microscopy (SMLM) requires moderate illumination power and exploits the stochastic fluorescence “blinking” nature of luminescent probes to capture sparsely distributed single-molecule blinking subsets [3, 9]. SMLM techniques can provide a spatial resolution up to 10 nm; however, the multi-color imaging capability is restricted to only limited numbers of discrete color channels [10, 11]. Indeed, conventional SMLM typically requires two dyes to have emission maxima with at least 100 nm separation to minimize spectral cross-talk [10, 11]. Exchanging ligand strategies enable multiplexing super-resolution imaging by sequentially labeling and subsequent dissociating the target-specific fluorescent DNA probes, however, they require additional sample preparation and prolonged acquisition process [12–14]. Recently developed spectroscopic SMLM (sSMLM) techniques simultaneously record the spatial locations and corresponding emission spectra of single-molecule blinking events, offering new imaging capabilities to resolve, in principal, unlimited number of fluorescent labels at single-molecule level [15–20].
In sSMLM, a dispersive element chromatically disperses the photons collected from each single-molecule stochastic blinking event to obtain the single-molecule emission spectrum. Meanwhile the corresponding spatial information of the blinking event is simultaneously collected in a separate optical path. As a result, sSMLM can capture single-molecule spectra with ultra-high throughput and resolve multiple molecular labels possess low single-molecule spectral variations where their emission maxima separation can be less than 10 nm [15]. The original spectrally-resolved Stochastic Optical Reconstruction Microscopy (SR-STORM) collects the spatial and spectral information of the blinking events via a dual-objective-lens system in two separate paths and disperses the photons in the spectral channel using a prism. Only bright single-molecule blinking events (> 10,000 detected photons) are utilized to reduce the noise uncertainty influence on the determination of the spectral signatures. A few examples have been demonstrated since then to collect the emission light only through a single objective lens and then split them into spatial and spectral channels using prisms as the dispersive elements [18–20]. These techniques permit sSMLM imaging in live cells and probing microenvironment changes in vitro. However, the implement of these configurations requires spatially modified optical system, typically with a beam splitter, a prism and multiple mirrors and lens. The complicated system setup may cause single-molecule signal attenuations and is challenging to integrate the system as a compact add-on module to be used with conventional localization microscopes.
On the o her hand, the grating element combines the function of the beam splitter and a dispersive element in grating-based sSMLM systems. They use minimal number of required optical elements and improve the compactness [16,17]. In fact, it has been demonstrated that placing only a piece of transmission type of diffraction grating between the detector and the conventional localization microscope port enables sSMLM imaging [17]. However, only half of photons are collected through the single objective lens comparing with the dual-objective-lens system, which complicates the multi-color sSMLM imaging acquisition using grating-based sSMLM setups, especially in high fluorescence background biological samples. Indeed, the high dispersions in the spectral channel (referred to as spectral dispersion, SD) used in these systems make it hard to precisely determine the spectral centroid (SC) of dimer blinking events because of splitting photons between zeroth and first order images. The SD has not been precisely adjusted to achieve the requisite spectral precision for multi-color imaging in grating-based sSMLM systems (we define spectral precision as the measured standard deviation of single-molecule SC under the collective influence of noise uncertainty and intrinsic single-molecule spectral variations, or spectral heterogeneity).
We developed sSMLM systems that utilize diffraction gratings to obtain the spatial and spectral information of the single-molecule blinking events simultaneously [16, 21–24]. Using sSMLM, we reported that Rhodamine dyes tagged microtubules assembled in vitro showed significant spectral heterogeneity. Taking advantage of such large spectral heterogeneity, we achieved sub-10 nm spatial resolution via photon regression algorithm [16]. In this article, we report the high-throughput characterization of single-molecule spectral heterogeneity; the SD influence on the identification performance of largely spectral overlapping dyes; and the multi-color super-resolutio imaging using transmission diffraction grating-based sSMLM under optimized SD.
2. Methods and materials
2.1. sSMLM setup
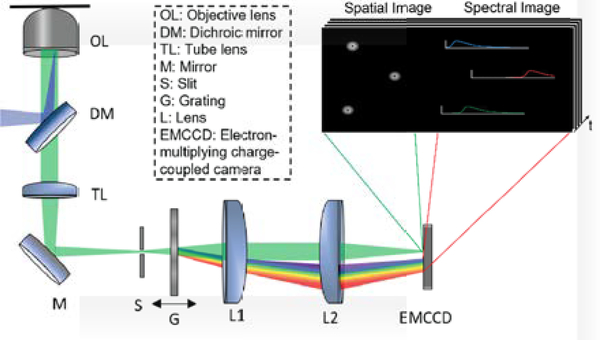
The schematic of our sSMLM system is shown in Fig. 1. A 642-nm CW laser was focused at the back focal plane of a Nikon Ti microscope body equipped with a perfect focus module. The light was illuminated onto the sample by a 100 X TIRF objective lens (CFI Apochromat 100X, NA=1.49, Nikon) with an illumination angle slightly smaller than the TIRF module requirement. The emitted photons were collected by the same objective lens. After passing through a tube lens (TL), the collected photons were routed by a mirror (M) to an entrance slit (S), which confines the field-of-view (FOV) of the spatial image, and then split into the zeroth-order and first-order images with a ratio of ~1:3 by a transmission grating (G, 100 grooves/mm, STAR100, Paton Kawskely Education Ltd.). The grating position relative to the imaging lens (L1, the focal length f = 50 mm) was adjusted to tune the SD from 6 to 9 nm/pixel while the zeroth-order images remained unmodified adapting the optics setup in Fig. 1. The SD of 3 nm/pixel was achieved by placing the grating in the collimating beam path between L1 and L2 [23]. Photons in the zeroth and first orders were respectively focused onto two different regions of an electron-multiplying charge-coupled device (EMCCD) camera (ProEM 532, Princeton Instrument) by an imaging lens (L2, f = 50 mm).
Fig 1.
Schematic of the sSMLM experimental system. The fluorophores are excited, and the emitted light is collected by an objective lens and passing through the dichroic mirror, tube lens, mirror and an entrance slit sequentially. Then the light passes through a transmission grating to be split into zeroth-order and first-order light, which provide the spatial and spectral information, respectively, and then two imaging lenses. The inset illustrates the zeroth-order (spatial) and first-order (spectral) images captured by the EMCCD.
2.2. Sample preparation
NHS-ester functionalized Alexa Fluor 647 (AF647, Thermofisher), CF660C and CF680 (Biotinum), Dyomics 634 (Dy634, Dyomics Inc.), Cy®5.5 (Sigma) were stored at −20 °C. Primary antibodies mouse anti-TOM20 (Santa Cruz), sheep anti-tubulin (Cytoskeleton), and chicken anti-ATPB (Abcam) were aliquoted and stored at −80 °C. Secondary antibodies Donkey anti-mouse, anti-sheep IgG, and anti-chicken IgY (Jackson ImmunoResearch) were aliquoted and stored at 4 °C. Fluorescent dye-antibody conjugations were performed by NHS-coupling reaction of the NHS-ester functionalized dyes with the secondary antibodies following a literature protocol [18]. Briefly, Dye (0.4 μL, 5 mM in DMSO), IgG/IgY proteins (100 μL, 1 mg/mL in PBS) and Sodium Bicarbonate (10 μL, 1 M in water) were mixed overnight at 25 °C. The mixture was purified by Nap-5 size exclusion column, concentrated with Amicon Ultra-0.5 Centrifugal Filter units, characterized by a NanoDrop Spectrophotometer to give 1–2 dyes per antibody and stored at 4 °C.
To measure spectral heterogeneity of far-red emitting fluorescent probes, the NHS-ester of Dy634, AF647, CF660C, CF680 and Cy5.5 were conjugated with anti-mouse IgG, adsorbed on the surface of No. 1 cover glass at a concentration of 2 μg mL−1 for 5 min and rinsed thoroughly with PBS. Before the experiment, a freshly prepared imaging buffer was added to support single-molecule detection (see below).
For multi-color sSMLM imaging, COS-7 cells (ATCC) were grown in DMEM (Gibco/Life Technologies) supplemented with 2 mM L-glutamine (Gibco/Life Technologies), 10% fetal bovine serum (Gibco/Life Technologies), and 1% penicillin / streptomycin (10,000 U mL−1, Gibco/LifeTechnologies) at 37 °C with 5% CO2. The cells were plated on No.1 borosilicate bottom 8-well Lab-Tek™ Chambered Coverglass with low confluency. After 48 hours, the cells were fixed in pre-warmed 3% Paraformaldehyde and 0.1 % Glutaraldehyde in PBS for 10 min, washed with PBS twice, quenched with freshly prepared 0.1 % Sodium Borohydride in PBS for 7 min and rinsed with PBS for three times at 25 °C. The fixed samples were permeabilized with a blocking buffer (3% BSA, 0.5% Triton X-100 in PBS) for 20 min and then incubated with the primary antibodies (Sheep anti-tubulin, 10 μg mL−1 for three-color samples and 2.5 μg mL−1 for other samples), (Chicken anti-ATPB, 2 μg mL−1 ), (Mouse anti-TOM20, 5 μg mL−1) in blocking buffer for 1h at room temperature and rinsed with a washing buffer (0.2% bSa, 0.1% Triton X-100 in PBS) for three times. The fixed samples were further incubated with the corresponding Donkey secondary antibodies-dye conjugates (Anti-Sheep AF647 and Anti-Mouse CF660C for two-color imaging experiment; Anti-Chicken AF647, Anti-Sheep CF660C and Anti-Mouse CF680 for three-color imaging experiment, 2.5 μg mL−1 in blocking buffer) for 40 min, washed thoroughly with PBS for three times at 25 °C and stored at 4 °C.
2.3. Imaging acquisition and data processing
We measured the system’s SD using a fluorescent calibration lamp with an entrance slit width of 10 μm. The emission peaks of 487.7, 546.5, 611.6 and 707.0 nm were used to calculate the SD with a first polynomial fit. For the spectral heterogeneity measurement, the entrance slit width was adjusted to 50 μm to reduce the background noise in the spectral channel. For multi-color imaging in cells, the slit width was adjusted to 5 mm to provide an adequate FOV while preventing overlapping between spatial and spectral image channels.
Prior to imaging, an imaging buffer containing 50 mM Tris (pH = 8.0), 10 mM NaCl, 0.5 mg mL−1 Glucose Oxidase (Sigma, G2133), 2000 U/mL Catalase (Sigma, C30), 10% (w/v) D-Glucose, and 100 mM Cysteamine was added to the sample chamber. The imaging experiments were typically finished in one hour or replenished with fresh prepared buffers every hour. The single-molecule blinking lasts for tenths of milliseconds with a few thousand of photons, the spatial locations and the spectral information of each single-molecule blinking were collected with an exposure time of 20 ms. We respectively recorded 5,000, 20,000 and 30,000 frames for spectral heterogeneity measurement, the two-color, and three-color sSMLM imaging reconstructions.
The single-molecule localization of each blinking event was processed using ThunderSTORM [25]. The corresponding single-molecule emission spectrum of every blinking event was obtained using its spectral image with the spatial image as the reference for the calibration process. The spectral detection channel (650 – 750 nm) was selected to collect at least 90% of the dye emission while minimizing the noise uncertainty contribution on the SC of each blinking event. This detection channel corresponds to 34 ×3, 17×3 and 12×3 pixel areas for the SD of 3, 6, and 9 nm/pixel on the camera, respectively. The SC of the emission spectrum was calculated as the weighted average of each individual spectrum [15]. The single-molecule SCs were used to identify and classify the single-molecule blinking from different dye molecules.
For two-color imaging, we respectively preset the spectral windows for AF647 and CF660C to 683–689 nm and 692–698 nm based on their SCs and spectral precisions (see section 3.1). For misidentification characterizations, we imaged AF647-labeled cells and calculate the spectral precision and the number of misidentified single molecules classified into both spectral windows. We applied different photon threshold filters (500–2500) for the spectral channel to discard the blinking events dimmer than the filter thresholds. For spectral precision measurement and multi-color imaging experiments, we filtered out all the blinking events less than 500 photons. Each single-molecule event was assigned a SC value, and we collected all the events with their SCs within the predefined spectral windows for each dye. Error bar in Figure 4c refers to the standard errors of spectral precision measured across different experimental acquisitions. In each experiment, 10,000 single molecules were analyzed, and a spectral precision value was provided based on the standard deviation of the spectral centroids of the 10,000 single molecules. In total, 10 samples were analyzed for each condition to provide the standard errors.
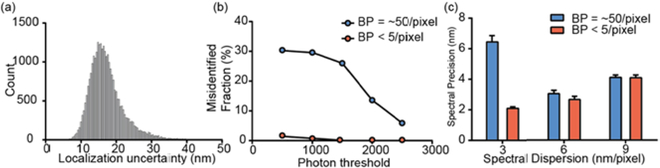
Fig. 4.
sSMLM imaging parameter characterizations. (a) Localization uncertainty of sSMLM images of COS-7 cells which the microtubule and mitochondria were labeled with AF647 and CF660C respectively; (b) Misidentified fraction of AF647 as CF660C in a pre-determined collection window of 692–698 nm based on the single-molecule SCs under different BP of 5 and 50 per pixel respectively with different photon thresholds of AF647 labeled microtubules of COS-7 cells at 3 nm/pixel SD; (c) Spectral precision measurements at different SDs of AF647 labeled microtubules in COS-7 cells. Error bars indicate the standard errors in each condition (Sample size for each condition = 10).
For three-color sSMLM imaging, 6-nm/pixel SD and 500-photon threshold were applied. We used spatial information of all the single-molecule events after the above classification methods to render the pseudo color-coded sSMLM images. Based on the spectral precision, the preset spectral windows of 683–689 nm, 692–698 nm, and 703–709 nm for the three dyes (AF647, CF660C and CF680) were used. We firstly acquired AF647, CF660C and CF680 individually labeled cell samples. Using these single-dye stained samples, we calculated the spectral precision of every dye under the same imaging condition (Figure 5a) and the number of single molecules classified into the other two spectral windows against the total population as the identification fraction (Figure 5b). The possibilities of mis-identification (channel shift) are 5% between AF647 and CF660C and between CF660C and CF680, and 0.5% between AF647 and CF680.
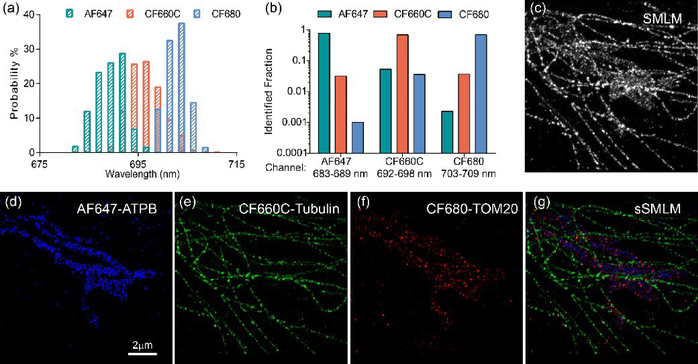
Fig. 5.
Multi-color sSMLM imaging of COS-7 cells, (a) SC distribution of AF647, CF660C and CF680; (b) Identification fraction of the three dyes in the preset channels; (c) SMLM imaging of a COS-7 cell which the ATPB, Tubulin and TOM20 proteins were labeled with AF647, CF660C and CF680 respectively; (d-f) pseudo color-coded images of the same sample in (c) classified in three spectral windows based on their spectral signature and the sSMLM image (g).
The morphology-based drift correction was performed according to reference [25] from the original spatial image reconstruction before spectral classification. Prior to sSMLM image rendering, the drift correction file obtained from the original spatial localization image was applied to each classified imaging channel.
3. Results and discussion
3.1. Single-molecule fluorescence spectral signature characterization
We discovered that Rhodamine dyes possess large single-molecule fluorescence spectral heterogeneity when labeled with proteins. Their single-molecule SCs can fluctuate more than 50 nm. Taking this advantage, we employed spectral regression algorithm to achieve sub-10 nm spatial resolution using sSMLM [16]. In contrast, certain dyes were reported to show reduced spectral heterogeneity at the single-molecule level and the identification of more than three types of dye molecules emitted at similar spectral regions based on their single-molecule SC were achieved with prism-based sSMLM system [15].
To validate our sSMLM system for simultaneous multi-color imaging, spectral heterogeneities of multiple far-red emitting fluorophores were firstly measured. We collected single-molecule blinking events with more than 500 photons and a narrow spectral detection window (650–750 nm) to characterize the spectral heterogeneity under minimal noise contribution. The width of the entrance slit was adjusted to 50 μm, which provides a background photon (BP) less than 5 per pixel for each blinking event in the corresponding spectral image. The measurement of SCs variation of red fluorescent nanosphere under the same experimental condition showed a spectral precision of 1.3 nm. This evidence suggested that the spectral variation more than 1.3 nm should be predominantly contributed by the intrinsic single-molecule fluorescence spectral heterogeneity using this particular system setup. In other words, we can measure the spectral heterogeneity of dyes unambiguously down to 1.3 nm. The FOV in the spatial image was restricted within a 5×120 pixel area. We can collect 5,000–10,000 single-molecule blinking events within 5000 frames (~100 s).
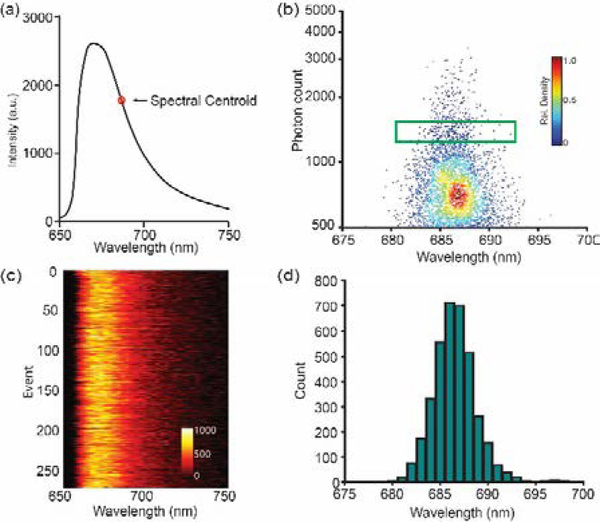
The averaged emission maximum (670 nm) and emission SC (686 nm) of AF647-IgG conjugates adsorbed on glass slide measured by the sSMLM system (Fig 2a) agreed well with literature reports [26]. The scatterplot of the emitted photon number against single-molecule SC of 6,132 blinking events revealed a relatively small SC variation primarily from 680 nm to 692 nm (Fig. 2b). Fig. 2c shows that the 260 single-molecule emission spectra with 1200–1500 detected photons (highlighted by the green box in Fig. 2b) have negligible spectral changes among each other. The histogram of SC distribution from all the 6,132 detected blinking events from AF647 depicted a relatively low spectral heterogeneity with a standard deviation (osh) of 2.1 nm as shown in Fig. 2d. The measured spectral heterogeneities of Dy634, CF660C, CF680 and Cy5,5, which emit within similar far-red spectral regions, showed slightly different values of 1.9, 3.1, 3.5 and 2.2 nm, respectively. The measured spectral heterogeneities are stable for at least 1 hours after adding fresh imaging buffer with relatively stable pH. Their measured averaged emission spectral maxima are consistent with literature values as shown in Table 1 [15, 26]. These spectral heterogeneities provide the possibility to distinguish dye labels, where their SCs are only 10 nm apart relying on the single-molecule SCs, and the simultaneous multi-color super-resolution imaging using a single objective lens.
Fig 2.
Spectral heterogeneity characterization of AF647. (a) Averaged emission spectrum of AF647 and its calculated spectral centroid (SC) position; (b) Scatterplot of photon count vs. single-molecule SC position of 6,132 AF647 single-molecule blinking events with the rectangular region highlights 260 single-molecule events with photon count from 1200–1500 and their emission spectra shown in (c); (d) Histogram of SC distribution of all the 6,132 single-molecule blinking events. Scale bar refers to the relative density of singlemolecule distribution in (b) and relative intensity in (c).
Table 1.
Single-molecule fluorescence emission signature of dye-protein conjugates.
| Dye | λEm (nm) [a] | λSC (nm) [b] | σSH (nm) [c] |
|---|---|---|---|
| Dy634 | 664 | 682 | 1.9 |
| AF647 | 670 | 686 | 2.1 |
| CF660C | 688 | 695 | 3.1 |
| Cy5.5 | 697 | 701 | 3.5 |
| CF680 | 704 | 706 | 2.2 |
λEm: averaged emission maximum
λSC: averaged emission spectral centroid
σSH: standard deviation of single-molecule spectral heterogeneity.
3.2. Simultaneous two-color sSMLM imaging in cells under different spectral dispersions
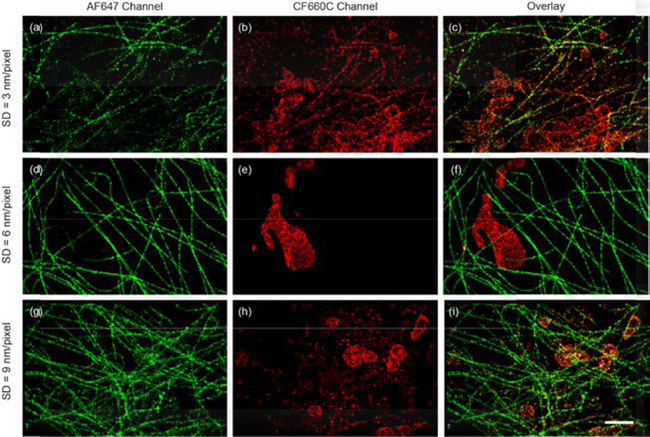
Both intrinsic single-molecule fluorescence spectral heterogeneity and background noise affect the spectral precision and determine the number of dyes or imaging channels that can be simultaneously resolved in sSMLM using SC classification method. According to the measurement in Section 3.1, the spectral heterogeneities of these far-red emitting dyes were remarkably low to permit the multi-color sSMLM imaging. As an example, the misidentification between AF647 and CF660C in the pre-defined spectral channels is only 2.0%. However, because of the limited photon budget and relatively high intracellular background, the spectral precision might significantly decrease, especially when the SD is relatively high (SD < 4 nm). Instead, lower SD of 5–7 nm/pixel could provide relatively stable spectral precision over high background conditions according to our theoretical predictions [27]. To further investigate the SD effect on multi-color sSMLM imaging experimentally, we performed two-color sSMLM imaging of COS-7 cells under different SDs where microtubule and mitochondria were labeled with AF647 and CF660C respectively.
At the SD of 3 nm/pixel, the microtubule and mitochondria structures appeared in both AF647 and CF660C channels (Fig. 3a–c). The coordinate-based co-localization (CBC) analysis [28] indicated a CBC value of 0.19, suggesting significant overlapping between the two channels as a result of high misidentification rate and low spectral precision. These experimental evidences indicated that the relatively high intracellular background (BP = ~50/pixel) restricts the system performance to unambiguously distinguish dyes that emission spectra are only 9 nm apart at the SD of 3 nm/pixel. Presumably, limited number of photons collected in the spectral channel was dispersed to a relatively large area on the camera and the spectral precision was affected by noise uncertainty at such high BP.
Fig. 3.
Two-color sSMLM images of AF647 labeled microtubule and CF660C labeled mitochondria in COS-7 cells at different SDs (a-c, 3 nm/pixel), (d-f, 6 nm/pixel) and (g-i, 9 nm/pixel) respectively. The AF647 (a, d and g) and CF660C (b, e, and h) channels and the overlay images (c, f and i) were collected with single-molecule SC of 683–689 nm and 692–698 nm respectively. (Scale bar = 2 μm)
Then we conducted two-color sSMLM imaging under 6-nm/pixel SD. Comparing to the 3-nm/pixel case, the microtubule and mitochondria structures respectively showed only in the AF647 and CF660C channels with a CBC value of 0.01, which suggested negligible overlapping between the two channels (Fig. 3d–f). Furthermore, the two-color imaging at the SD of 9 nm/pixel showed increased overlapping between the two channels with a CBC value of 0.08 (Figs. 3g–i). In the overlaid image (Figure 3i), yellow-orangish color was observed on mitochondria region, which indicated that misidentified single molecules from the tubulin channel. Therefore, the image quality degraded in SD of 9 nm/pixel comparing to 6 nm/pixel.
3.3. Two-color sSMLM imaging performance characterization
Our sSMLM simultaneously captures the spatial and spectral information of single-molecule blinking events. For achieving high spectral precision, 75 % of the total photons were allocated to the first order image while the rest 25% of total photons were allocated into the zeroth order for spatial localization. We achieved localization uncertainty of 17–23 nm (Fig. 4a) which were comparable with literature values in the conventional SMLM under similar photon budget [26]. Notably, the spatial resolution was independent of SD since the blinking signals in the zeroth order remained unmodified. In addition, this sSMLM setup provides the opportunity to systematically tune and optimize SD for multi-color imaging with simply adjusting the grating position relative to the imaging lens (L1).
Using the same sSMLM setup, we quantified the misidentification under different SDs by imaging AF647 labeled microtubules in COS-7 cells as examples at different background levels. In the aforementioned spectral heterogeneity measurement with SD of 3 nm/pixel, low BP < 5/pixel were detected. Within 6,132 detected AF647 single molecules, 122 single-molecule SCs locate in the spectral region of 692–698 nm which was the spectral window we preset for CF660C. These values suggested a spectral precision of 2.1 nm and a misidentification rate of 2.0% between AF647 and CF660C that SCs are 9 nm apart (Fig. 4b–c). Then we adjusted the entrance slit to 5 mm to increase the FOV to 120×120 pixels (19.2×19.2 μm2) for imaging cell samples. The BP increased to ~ 50/pixel when we imaged the COS-7 cells labeled with AF647 with a concomitant decrease of spectral precision down to 6.4 nm (Fig. 4c). Under this condition, 29,862 from 100,546 detected single-molecule SCs located in the CF660C spectral window (Fig. 4b). In such high intracellular background conditions, the high misidentification rate of 29.7% restricts the imaging capability of our multi-color sSMLM system (Fig. 4b). Although applying higher photon threshold filter (2500) can decrease the misidentification rates to 5.4 % when BP was ~ 50/pixel (Fig. 4b), the number of collected localization events were not enough to render a good-quality super-resolution image with continuous microtubules until dyes photobleaching.
Limited number of photons collected in the spectral channel were dispersed to a large area (34×3 pixel) on the camera and resulted in low spectral precision in the case of 3 nm/pixel SD. In turn, decreasing the dispersion may increase the signal to background ratio and spectral precision, especially for sSMLM imaging in cells under high background conditions [27]. Remarkably, the adjustment of SD to 6 and 9 nm/pixel resulted in higher spectral precisions of 3.2 nm and 4.1 nm for BP of 50 respectively (Fig. 4c). These results were consistent with our CBC analysis that 6 nm/pixel SD showed the optimal imaging performance and agreed well with our numerical simulation studies [27]. Presumably, a subpixel systematic error in mapping the spatial distribution of the single-molecule blinking events to the associated spectral signatures during the spectral calibration procedure occurs at the SD of 9 nm/pixel thus decreasing the spectral precisions to 4.1 nm under these background conditions [27]. Furthermore, the low spectral precisions appeared in both 3 and 9 nm/pixel resulted in incorrect classification of a large amount (~30% and 12% respectively) of single molecules in the two channels and degraded the image quality as discontinuous tubulin skeleton and scattered dots were observed (Fig 3a–c and g–i). In short, the SD of 6 nm/pixel showed the optimal spectral precision that tolerated relatively high intracellular background and was determined for acquiring multi-color sSMLM imaging.
3.4. Multi-color super-resolution imaging of COS-7 cell
We set the SD to 6 nm/pixel in simultaneous three-color sSMLM imaging experiments. The independent examination of the SC of AF647, CF660C and CF680 at the SD of 6 nm/pixel under the identical imaging condition labeled the microtubule structures of COS-7 cells demonstrated spectral precisions of 3.2, 4.3 and 3.1 nm, respectively (Fig. 5a). The relatively high spectral precision enabled the identification of the three dyes based on their SC positions. The classification of the SCs into three channels based on their maximal probability of SC at 683–689 nm, 692–698 nm and 703–709 nm resulted in more than 95% separation between AF647 and CF660C as well as between CF660C and CF680, where the separation between AF647 and CF680 was more than 99.5% (Fig. 5b).
A conventional SMLM image of a COS-7 cell immunofluorescent labeled with AF647, CF660C and CF680 to the ATP synthase (ATPB), Tubulin, and Mitochondrial Outer Membrane translocase complex (TOM20), respectively, is shown in Fig. 5c. The tubulin cytoskeleton can be partially distinguished based on its morphology. However, the large overlapping region between the three targets of interests remained unresolved in the single far-red detection channel (Fig. 5c). Based on their respective SCs, sSMLM separated the three labels with high specificity as shown in Fig. 5d–5f. The distinct microtubule structures were exclusively located in CF660C channel and the unresolved region in conventional SMLM can be clearly distinguishable in the pseudo color-coded images (Fig. 5e). Both the ATPB and TOM20 signals showed elongated patterns (Fig.s 5d and 5f) while the TOM20 were located at the periphery of ATPB as shown in the overlaid sSMLM image (Fig. 5g). This feature was consistent with the protein locations that TOM20 labels highlight the outer membrane of mitochondria and ATPB resides in the inner membrane of mitochondria [15]. As a result, we demonstrated that simultaneous multi-color sSMLM imaging can be achieved in immunostained cells using a single far-red detection channel with optimal SD at 6 nm/pixel.
4. Conclusion
We achieved simultaneous acquisitions of single-molecule positions and corresponding spectra of fluorescent probes with grating-based sSMLM using a single objective lens. We showed that both intrinsic single-molecule fluorescence spectral heterogeneity and noise uncertainty affect the system performance for identifying dyes emitting in close spectral region. Carefully tuning the spectral dispersion provides a suitable spectral precision for simultaneously multi-color imaging of multiple dye species with narrow spectral centroid shifts (~10 nm). We showed that using a SD of 6 nm/pixel the grating-based sSMLM can resolve three molecular labels in fixed cells at the same time with nanometer resolution. We anticipate that sSMLM will play an important role in investigating structural and dynamical biological processes involving interactions of multiple molecules.
Acknowledgments
Funding
This work was supported in part by NSF grants CBET-1706642, EFRI-1830969, and EEC-1530734; NIH grants R01EY026078 and R01EY029121; and Northwestern University Innovative Initiative Incubator (I3) Award. JLD was supported by the NSF Graduate Research Fellowship 1000231682.
C. Sun and H. F. Zhang have financial interests in Opticent Inc., which did not support this work. Other authors declare that there are no conflicts of interests related to this article.
References
- 1.Rust MJ, Bates M, and Zhuang X, “Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM),” Nat. Methods 3, 793–796 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gustafsson MGL, “Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution,” Proc. Natl. Acad. Sci. USA 102, 13081–13086 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, and Hess HF, “Imaging Intracellular Fluorescent Proteins at Nanometer Resolution,” Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Sharonov A and Hochstrasser RM, “Wide-field subdiffraction imaging by accumulated binding of diffusing probes,” Proc. Natl. Acad. Sci. USA 103, 18911–18916 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Diezmann A, Shechtman Y, and Moerner WE, “Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking,” Chem. Rev 117, 7244–7275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hell SW and Wichmann J, “Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy,” Opt. Lett 19, 780–782 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Hell SW, “Nanoscopy with Focused Light (Nobel Lecture),” Angew. Chem. Int. Ed 54, 8054–8066 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Olivier N, Keller D, Rajan VS, Gonczy P, and Manley S, “Simple buffers for 3D STORM microscopy,” Biomed. Opt. Express 4, 885–899 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang B, Babcock H, and Zhuang X, “Breaking the diffraction barrier: super-resolution imaging of cells,” Cell 143, 1047–1058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates M, Huang B, Dempsey GT, and Zhuang X, “Multicolor Super-Resolution Imaging with Photo-Switchable Fluorescent Probes,” Science 317, 1749–1753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, and Betzig E, “Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes,” Proc. Natl. Acad. Sci. USA 104, 20308 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jungmann R, Avendano MS, Woehrstein JB, Dai M, Shih WM, and Yin P, “Multiplexed 3D Cellular Super-Resolution Imaging with DNA-PAINT and Exchange-PAINT,” Nat. Methods 11, 313–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai M, Jungmann R, and Yin P, “Optical imaging of individual biomolecules in densely packed clusters,” Nat. Nanotechnol 11, 798–807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schueder F, Lara-Gutierrez J, Beliveau BJ, Saka SK, Sasaki HM, Woehrstein JB, Strauss MT, Grabmayr H, Yin P, and Jungmann R, “Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT,” Nat. Commun 8, 2090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Kenny SJ, Hauser M, Li W, and Xu K, “Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy,” Nat. Methods 12, 935–938 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Dong B, Almassalha L, Urban BE, Nguyen T-Q, Khuon S, Chew T-L, Backman V, Sun C, and Zhang HF, “Super-resolution spectroscopic microscopy via photon localization,” Nat. Commun 7, 12290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bongiovanni MN, Godet J, Horrocks MH, Tosatto L, Carr AR, Wirthensohn DC, Ranasinghe RT, Lee J-E, Ponjavic A, Fritz JV, Dobson CM, Klenerman D, and Lee SF, “Multi-dimensional super-resolution imaging enables surface hydrophobicity mapping,” Nat. Commun 7, 13544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mlodzianoski MJ, Curthoys NM, Gunewardene MS, Carter S, and Hess ST, “Super-Resolution Imaging of Molecular Emission Spectra and Single Molecule Spectral Fluctuations,” PLoS One 11, e0147506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon S, Yan R, Kenny SJ, Shyu Y, Xiang L, Li W, and Xu K, “Spectrally Resolved, Functional Super-Resolution Microscopy Reveals Nanoscale Compositional Heterogeneity in Live-Cell Membranes,” J Am Chem Soc 139, 10944–10947 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Huang T, Phelps C, Wang J, Lin LJ, Bittel A, Scott Z, Jacques S, Gibbs SL, Gray JW, and Nan X, “Simultaneous Multicolor Single-Molecule Tracking with Single-Laser Excitation via Spectral Imaging,” Biophys J 114, 301–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong B, Almassalha LM, Stypula-Cyrus Y, Urban BE, Chandler JE, Nguyen T-Q, Sun C, Zhang HF, and Backman V, “Superresolution intrinsic fluorescence imaging of chromatin utilizing native, unmodified nucleic acids for contrast,” Proc. Natl. Acad. Sci. USA 113, 9716–9721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban BE, Dong B, Nguyen TQ, Backman V, Sun C, and Zhang HF, “Subsurface Super-resolution Imaging of Unstained Polymer Nanostructures,” Sci. Rep 6, 28156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong B, Soetikno BT, Chen X, Backman V, Sun C, and Zhang HF, “Parallel Three-Dimensional Tracking of Quantum Rods Using Polarization-Sensitive Spectroscopic Photon Localization Microscopy,” ACS Photonics 4, 1747–1752 (2017). [Google Scholar]
- 24.Davis JL, Dong B, Sun C, and Zhang HF, “Method to identify and minimize artifacts induced by fluorescent impurities in single-molecule localization microscopy,” J. of Biomedical Optics, 23, 106501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ovesny M, Krizek P, Borkovec J, Svindrych Z, and Hagen GM, “ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging,” Bioinformatics 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dempsey GT, Vaughan JC, Chen KH, Bates M, and Zhuang X, “Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging,” Nat. Methods 8, 1027–1036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song K, Dong B, Sun C, and Zhang HF, “Theoretical analysis of spectral precision in spectroscopic single-molecule localization microscopy,” Rev. of Sci. Instrum 89, 123703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malkusch S, Endesfelder U, Mondry J, Gelleri M, Verveer PJ, and Heilemann M, “Coordinate-based colocalization analysis of single-molecule localization microscopy data,” Histochem. Cell Biol 137, 1–10 (2012). [DOI] [PubMed] [Google Scholar]