Acute lymphoblastic leukemia (ALL) comprises a highly heterogeneous set of diseases that are defined by their cells of origin, stage of maturation arrest, and the underlying oncogenic driver pathways. Subclassification of lymphoid neoplasms is often based on the presumed cell of origin based on T and B progenitor gene expression. In general, T-ALL has a worse prognosis1, 2 and requires more intensive therapies to achieve similar remission rates as their B-ALL counterparts.3 Given the common origin of these cells from committed lymphoid progenitors, investigators have suggested that the differences in clinical presentation of T- and B-ALL are likely accounted for by the underlying differences in the oncogenic drivers of transformation. For example, T-ALLs often express oncogenic transcription factors and acquire activating NOTCH1 mutations that elevate MYC activity to drive tumor initiation and maintenance.4–6 By contrast, B-ALLs are more frequently driven by chromosomal rearrangements that create fusion oncogenes and have recurrent chromosomal aberrations.7, 8 Despite these differences, MYC is also commonly activated in human B-ALL9 and drives leukemia initiation and growth in mouse models,10 suggesting important roles for this pathway in both T- and B-ALL. It is clear that underlying oncogenic drivers exert important roles in regulating the functional characteristics of leukemia cells, including regulating the overall fraction of leukemia stem cells (LSCs) that drive continued tumor growth and relapse following therapy resistance. Yet, to date, the effect of cell lineage on influencing LSC number and self-renewal is largely unknown, accounted for in part, by lack of experimental models to address these questions.
Here, we have used a zebrafish transgenic model of Myc-induced ALL to investigate the roles of cell lineage on modulating growth, aggression, and leukemia stem cell (LSC) frequency in T, B, and biphenotypic ALL. Previously, the rag2-Myc transgenic zebrafish model has been exploited to develop robust T-ALL models when introduced into AB strain zebrafish.11, 12 Moreover, findings in this model have led to significant new insights into human T-ALL.13, 14 Using this same transgenic approach, we had previously generated leukemias in syngeneic CG1 strain zebrafish and performed large-scale cell transplantation assays to assess latency and LSC frequency differences between intra- and inter-tumoral clones.13 These experiments required implanting single LSCs into large cohorts of transplant animals, thus creating monoclonal tumors (schematic shown in Figure 1A). Each monoclonal leukemia was then assessed for latency of regrowth and LSC frequency using large-scale limiting dilution cell transplantation.13 From these previously published studies, we uncovered that clonal heterogeneity is common in T-ALL, with individual leukemia clones exhibiting striking differences in latency, aggression, and LSC frequency, and also identified the PI3-kinase/AKT pathway as a driver of elevated growth and LSC frequency in both zebrafish and human disease.13
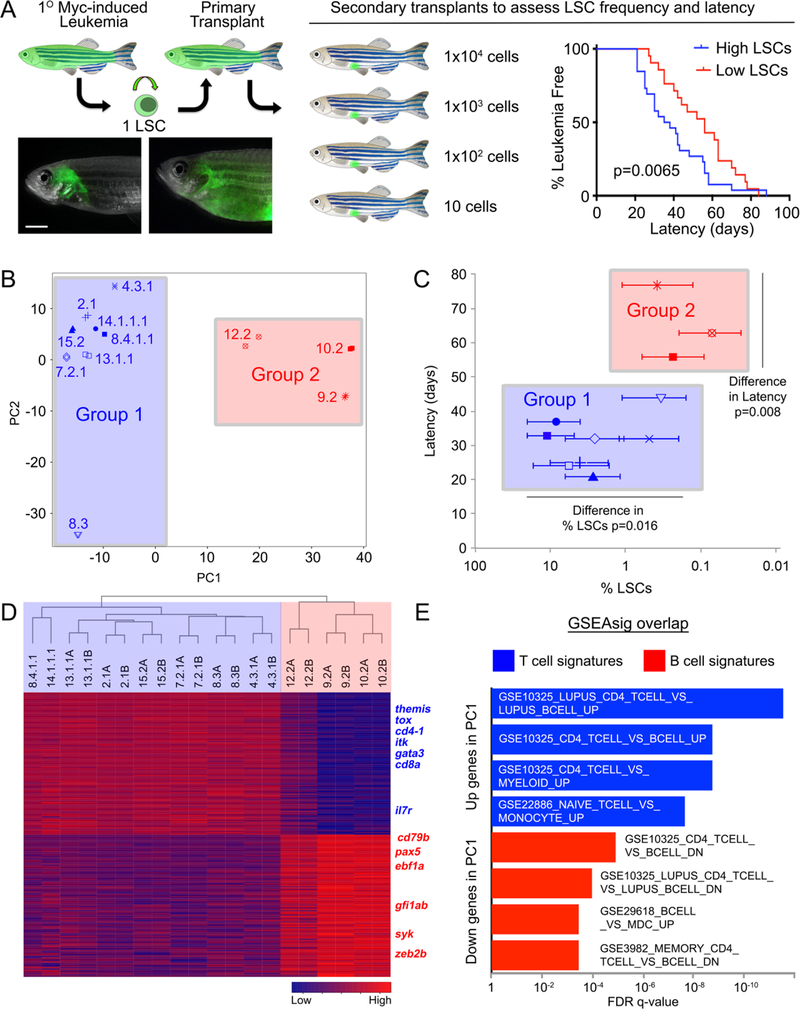
Figure 1. Myc-induced ALLs comprise two distinct molecular subtypes that differ in latency and leukemia stem cell number.
(A) A schematic of the cell transplantation screen previously used to identify differences in latency and leukemia stem cell number (LSCs) between ALL clones (Blackburn et al., Cancer Cell, 2014). ** denotes p=0.0065, by Wilcoxon rank order test. (B) RNA sequencing and principal component analysis of ALLs with high and low LSCs. All samples, except for 14.1.1.1 and 8.4.1.1, were assessed as technical replicates. Group 1 leukemias are demarcated by blue shading, while group 2 leukemias have been shaded in red in panels B, C, and D. (C) The two molecularly-defined subgroups differ in latency and LSC frequency when assessed by Wilcoxon Rank Sum Test. (D) Heat map and hierarchical clustering of group 1 and group 2 leukemias showing the top 100 genes positively and negatively correlated with PC1. (E) Gene set enrichment analysis using genes positively and negatively correlated with PC1. Genes positively correlated with PC1 are associated with T cells while those found to be negatively correlated with PC1 were B cell associated genes.
Using this previously generated library of well-annotated monoclonal ALLs defined by Blackburn et al. (2014), we selected leukemias that harbored high (>1%) and low numbers of LSCs (<1%, Supplemental Table 1).13 These leukemias were subjected to bulk RNA sequencing and Principal Component Analysis (PCA) was performed to identify molecular differences between clones. Principal Component 1 (PC1) and PC2 represent the top two gene expression profiles derived as dimensions from Principal Component Analysis. This analysis confirmed that replicate samples always segregated with one another, and more importantly, identified two easily discernable molecular subgroups of leukemia, labeled group 1 (in blue) and group 2 (in red, Figure 1B). Interestingly, the two clusters in the PCA corresponded to significant differences in tumor behavior: group 1 leukemias had significantly shorter latency of leukemia regrowth in transplanted fish and had more LSCs when compared with group 2 (Figure 1C, Wilcoxin Rank Order Test). Hierarchical clustering using the 100 most highly variable, positively and negatively correlated transcripts identified from PC1, affirmed the classification of two molecularly distinct ALL groups (Figure 1D, Supplemental Table 2). Unexpectedly, GSEA analysis uncovered that PC1 was defined by T and B cell lineage genes, with T cell genes being highly expressed in the fast growing and high LSC containing leukemias while B cell genes were confined to leukemias with longer latency and low LSCs (Figure 1D and E). PC2 genes comprised proliferation genes (Supplemental Table 3), identifying T-ALL clone 8.3 as having lower overall proliferative potential and likely accounting for its low percentage of LSCs.
Given that the rag2-Myc transgenic model had never generated B-ALL in other strains of zebrafish,11,12,15 we next sought to independently confirm cell lineage by assessing T and B cell receptor expression. As expected, all group 1 leukemias expressed tcr-beta, with most expressing a single unique variable region, independently confirming 1) derivation from a single LSC, 2) monoclonality of each ALL, and 3) correct classification as T-ALL. In contrast to what is seen in human, the tcrbc1 and c2 constant regions are not excised from the zebrafish genomic locus following receptor recombination and hence each can be used to create a functional receptor following splicing. As reported in human, a small subset of T cells can also express two TCR-beta chains,16, 17 likely accounting for the expression from both recombined alleles in T-ALL clone 15.2. Meanwhile, clones 9.2 and 10.2 failed to express tcr-beta or other T cell receptors including alpha, delta, and gamma, but rather expressed the constant region of IgHC (Figure 2A). Importantly, only the IgHC constant regions for IgM, IgD, and IgZ were expressed while the variable gene segments were not, suggesting that the IgHC locus is open but that rag1/2-mediated receptor recombination had not yet occurred in these B-ALLs (Supplemental Table 4). Additional gene expression analysis showed that T-ALLs were arrested at a CD4+/CD8+ cortical thymocyte stage as previously described,18 while B-ALLs 9.2 and 10.2 express genes pro-B marker genes including rag1/2, ebf1, pax5, dntt (tdt), and cd79a (Figure 2B). Remarkably, Myc expression levels were not significantly different between T- and B-ALLs (Supplemental Figure 1), failing to account for the observed differences in LSC frequency and latency differences between leukemias. Together, these results show that CG1 strain zebrafish develop both Myc-induced T- and B-ALL and that cell of origin likely has important roles in regulating LSC frequency and overall aggression. Given that all previous leukemia generated in this model were suggested to be T-ALLs, our refined analysis has led to the unexpected finding of B-ALL in a small fraction of CG1 strain fish.13
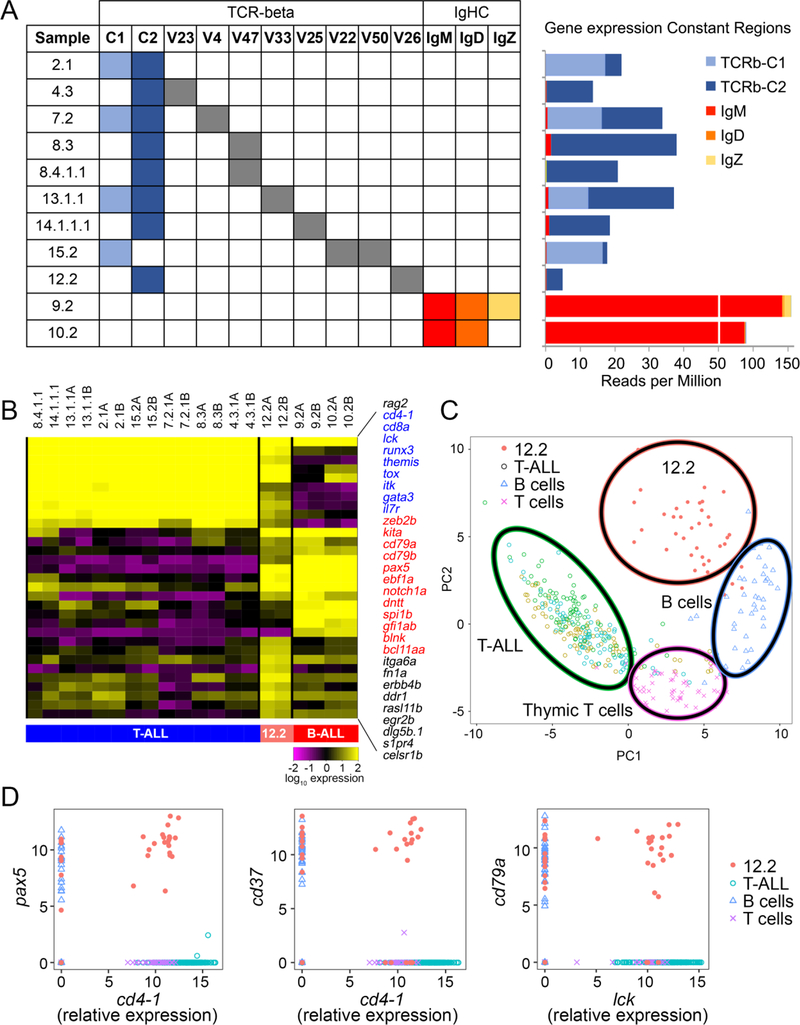
Figure 2. B cell molecular programs drive lower aggression and stem cell activity in acute lymphoblastic leukemia.
(A) T cell receptor beta and Ig heavy chain expression within individual ALL clones. Left plot shows only the tcrb variable regions that were expressed within each clone. Right plot displays expression for each tcrb and IgHC constant region for clones in the same order as presented on the left. (B) Heatmap showing gene expression of T and B cell associated genes and genes specifically expressed in biphenotypic B/T-ALL clone 12.2. (C) Single cell quantitative real-time PCR and principal component analysis visualization showing that 12.2 represents a clearly distinct molecular subtype when compared with T-ALL clones 8.4.1.1, 13.1.1.1, and 14.1.1.1, and normal rag2-dsRed+ thymic T cells and marrow-derived B cells. (D) Gene expression correlation comparing T vs B cell genes within individual ALL cells (x- versus y-axis, respectively), confirming that clone 12.2 is a bi-phenotypic B/T-ALL.
In completing our receptor gene expression studies, we also observed that leukemia 12.2 had both T and B cell characteristics. This leukemia recombined and expressed tcr-beta, failed to express IG heavy chain, and exhibited expression of both T and B lineage markers along with unique genes not found in the other leukemias (Figure 2B). These data raised the interesting possibility that leukemia 12.2 was a mixed phenotypic acute leukemia (MPAL). In fact, the World Health Organization recently published a revision for acute leukemia classification that now includes new criteria for classification and diagnosis of MPAL,19, 20 uncovering that MPALs occur in approximately 5% of pediatric and adult acute leukemia.21 Because clinical MPAL cases can be further subclassified into either bilineal, a leukemia that contains two distinct blast populations, or biphenotypic ALL, a leukemia that contains a single blast population that expresses multiple lineage markers,22 we next used single cell gene expression analysis to clarify if leukemia 12.2 was a mixture of two distinct blast types or if they were biphenotypic.18 Principal component analysis showed that tumor 12.2 was molecularly distinct and separated cleanly into its own PCA grouping when compared with non-transformed thymic T cells, marrow-derived B cells and three independent T-ALL clones (Figure 2C). Remarkably, gene expression correlation analysis revealed that individual leukemia cells from 12.2 indeed expressed both T and B cell genes, confirming assignment as biphenotypic B/T-ALL (Figure 2D). Human biphenotypic B/T-ALL is exceeding rare and to date has been characterized by CD3ε T cell marker expression in combination with CD19 and either CD79a, CD22 or CD10 B cell marker expression23. Zebrafish orthologs to many of these genes have yet to be fully described and antibodies are lacking. Moreover, detailed transcriptional analysis of human B/T-ALL has yet to be completed, obviated direct comparison of human and zebrafish disease at this time. Future experiments will assess if there are species-specific differences in this disease entity or, more importantly, if common molecular programs drive their growth.
In summary, our results have uncovered unexpected zebrafish strain differences in driving cell fate decisions in the genesis of ALL and provide new and exciting avenues for modeling B-ALL and biphenotypic B/T-ALL using zebrafish models. To date only a single report has characterized the development of B cell leukemia models in the zebrafish and no robust models of biphenotypic B/T-ALL have been reported.24 Perhaps more importantly, we have used our zebrafish model to show that cell lineage is a major determinant of oncogenic phenotypes in ALL, with T cell pathways driving elevated growth, aggression, and LSC self-renewal. By contrast, intrinsic B cell pathway activation leads to reduced growth and leukemia stem cells. These results provide new insights into the clinical differences between T- versus B-ALL and suggest that underlying differences in cell of origin likely account for superior clinical outcome in B-ALL patients, with the prediction that B cell lineage leukemias will have lower aggression and overall numbers of relapse driving LSCs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Jessica Blackburn and Finola Moore for supplying zebrafish leukemias for RNA sequencing and Fluidigm single cell PCR analysis; the MGH Flow Cytometry Core for help with single cell sorting; Na Qu and Dr. Toshi Shioda for help with NextGen sequencing; and Dr. Antony Anselmo for superior bioinformatics analysis. This work is supported by NIH grant R24OD016761 and R01CA211734 (D.M.L.) and by the Fund for Scientific Research Flanders (FWO Vlaanderen, doctoral grant S.L.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Silverman LB, Gelber RD, Dalton VK, Asselin BL, Barr RD, Clavell LA, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Dana-Farber Consortium Protocol 91–01. Blood 2001; 97(5): 1211–1218. [DOI] [PubMed] [Google Scholar]
- 2.Cortelazzo S, Ponzoni M, Ferreri AJ, Hoelzer D. Lymphoblastic lymphoma. Crit Rev Oncol Hematol 2011. September; 79(3): 330–343. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol 2003. October 1; 21(19): 3616–3622. [DOI] [PubMed] [Google Scholar]
- 4.Chiang MY, Wang Q, Gormley AC, Stein SJ, Xu L, Shestova O, et al. High selective pressure for Notch1 mutations that induce Myc in T-cell acute lymphoblastic leukemia. 2016. November 3; 128(18): 2229–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 2014. October; 20(10): 1130–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood 2017. March 2; 129(9): 1124–1133. [DOI] [PubMed] [Google Scholar]
- 7.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest 2012. October; 122(10): 3407–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raimondi SC, Behm FG, Roberson PK, Williams DL, Pui CH, Crist WM, et al. Cytogenetics of pre-B-cell acute lymphoblastic leukemia with emphasis on prognostic implications of the t(1;19). Journal of Clinical Oncology 1990; 8(8): 1380–1388. [DOI] [PubMed] [Google Scholar]
- 9.Raess PW, Moore SR, Cascio MJ, Dunlap J, Fan G, Gatter K, et al. MYC immunohistochemical and cytogenetic analysis are required for identification of clinically relevant aggressive B cell lymphoma subtypes. Leuk Lymphoma 2017. September 3: 1–8. [DOI] [PubMed] [Google Scholar]
- 10.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. The Journal of Experimental Medicine 1988; 167(2): 353–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 2003. February 7; 299(5608): 887–890. [DOI] [PubMed] [Google Scholar]
- 12.Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences 2005. April 26, 2005; 102(17): 6068–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackburn JS, Liu S, Wilder JL, Dobrinski KP, Lobbardi R, Moore FE, et al. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 2014. March 17; 25(3): 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobbardi R, Pinder J, Martinez-Pastor B, Theodorou M, Blackburn JS, Abraham BJ, et al. TOX Regulates Growth, DNA Repair, and Genomic Instability in T-cell Acute Lymphoblastic Leukemia. Cancer Discov 2017. November; 7(11): 1336–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez A, Feng H, Stevenson K, Neuberg DS, Calzada O, Zhou Y, et al. Loss of function tp53 mutations do not accelerate the onset of myc-induced T-cell acute lymphoblastic leukaemia in the zebrafish. Br J Haematol 2014. July; 166(1): 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davodeau F, Peyrat MA, Romagne F, Necker A, Hallet MM, Vie H, et al. Dual T cell receptor beta chain expression on human T lymphocytes. J Exp Med 1995. April 1; 181(4): 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padovan E, Giachino C, Cella M, Valitutti S, Acuto O, Lanzavecchia A. Normal T lymphocytes can express two different T cell receptor beta chains: implications for the mechanism of allelic exclusion. J Exp Med 1995. April 1; 181(4): 1587–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore FE, Garcia EG, Lobbardi R, Jain E, Tang Q, Moore JC, et al. Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J Exp Med 2016. May 30; 213(6): 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott G Aggressive B-cell lymphomas in the update of the 4th edition of the World Health Organization classification of haematopoietic and lymphatic tissues: refinements of the classification, new entities and genetic findings. Br J Haematol 2017. September; 178(6): 871–887. [DOI] [PubMed] [Google Scholar]
- 20.Brunning RD. Classification of acute leukemias. Semin Diagn Pathol 2003. August; 20(3): 142–153. [DOI] [PubMed] [Google Scholar]
- 21.Hrusak O, De Haas V, Luks A, Janotova I, Mejstrikova E, Bleckmann K, et al. Acute Leukemia of Ambiguous Lineage: A Comprehensive Survival Analysis Enables Designing New Treatment Strategies. Blood 2016; 128(22): 584.27317792 [Google Scholar]
- 22.Weinberg OK, Arber DA. Mixed-phenotype acute leukemia: historical overview and a new definition. Leukemia 2010. November; 24(11): 1844–1851. [DOI] [PubMed] [Google Scholar]
- 23.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016. May 19; 127(20): 2391–2405. [DOI] [PubMed] [Google Scholar]
- 24.Sabaawy HE, Azuma M, Embree LJ, Tsai H-J, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences 2006. October 10, 2006; 103(41): 15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.