Abstract
Due to importance of screening for colorectal cancer the present study was conducted and aimed at investigating the quantitative methylation of vimentin gene in stool, tumor tissue, and healthy tissue of patients with colorectal cancer (CRC) in comparison with healthy individuals. This is a case-control study in which we measures methylation of the vimentin in tumoral tissue, normal tissue and stool specimens were collected from forty-nine CRC patients as case group and stool and normal tissue specimens were collected from thirty healthy individuals as control group. There was no statistically significant difference in methylation of vimentin in normal tissue (P > 0.05) between the two groups. Moreover, the status of methylated or unmethylated vimentin gene in tumor and stool tissues in the case group was not significantly correlated with their mean age and sex (P > 0.05). This study showed that the specificity and sensitivity of vimentin methylation in stool of CRC patients are 100% and 60%, respectively. Furthermore, the methylation of vimentin in stool of CRC patients has a high-positive predictive value (100%). The results of this study suggested that methylation of the vimentin gene in the stool can be used as a specific marker for the detection and screening of CRC.
Key words: Colorectal cancer, methylation, screening, stool, vimentin
INTRODUCTION
Colorectal cancer (CRC) is considered as one of the most common malignant tumors in the gastrointestinal tract. It is the common cause of death throughout the world. Its prevalence is higher in men than that in women and is increasing significantly with aging. CRC, as a life-threatening malignant tumor, is the second leading cancer in men following lung cancer. It is also the third most common cancer among women, following breast and lung cancers.[1,2,3] The development of carcinoma from adenoma is known as adenoma-carcinoma. Most cases of CRC are adenocarcinomas, originating almost from adenomas. Adenoma is a neoplastic polyp, which varies from small and mobile tumors to large and stable lesions. Two distinct pathogenic pathways are involved in the development of CRC, and gradual accumulation of different mutations is involved in these pathways. The first pathway is Adenomatous Polyposis Coli gene (APC)/β-catenin, characterized by chromosomal instability. It leads to gradual accumulation of mutations in a set of tumor suppressor genes and enzymes. The second pathway is characterized by genetic lesions in DNA mismatch repair genes, which are sporadic in 10%–15% of cases. Among these DNA mismatch repair genes, MLH1 is one of the most well-known genes involved in carcinogenesis.[4,5]
MATERIALS AND METHODS
This research was a case–control study performed on 79 patients who admitted to the endoscopy ward of Ghaem Hospital and Imam Reza Hospital in Mashhad from May to March 2017 using EpiTect Plus FFPE Bisulfite kits. The research inclusion criteria for case group were the malignant colon tumor approved by the pathologist and observing the tumor through colonoscopy and for control group were the participants who underwent colonoscopy due to hemorrhoid or screening and lack of malignant lesion in the colonoscopy and no family history of colon cancer. The research exclusion criteria were benign tumor in pathological examination of the samples, hemodynamic instability during endoscopy (BP <90.60 and HR >110), and patient dissatisfaction. In this study, the delta-delta Ct method was used for real-time polymerase chain reaction (PCR). In relative quantitative measurement, the considered gene information is shown relative to a calibrator or an internal gene control.[6] The data were analyzed by the statistical tests using SPSS 11.5, IBM SPSS Statistics for Windows (IBM SPSS, Armonk, NY, USA).
Primary colorectal tumor samples and normal tissue samples surrounding the tumor (5–10 cm distant from the tumor) were collected from 49 patients. The tissue samples were taken from the control group. In addition, stool samples were collected from both case and control groups. Then, DNA was extracted from normal and colon cancer tissue samples. DNA was extracted from normal and cancer tissues using FavorPrep Tissue Genomic DNA Extraction Mini Kit. DNA was also extracted from stool samples of patients with CRC and healthy participants using the QIAamp DNA Stool Mini Kit. EpiTect® Fast DNA Bisulfite Kit was used to bisulfate DNA in the present study. DNA fragments extracted from tumor tissue, healthy tissue, and stool samples were amplified using quantitative methylation-specific PCR after bisulfate, and the vimentin gene methylation was measured in them. The ACTB gene was used as an internal control to normalize the conditions, and all of the steps mentioned for this gene were also performed. The primer sequence for the vimentin gene was as follows:
Methylated sense (forward), 5'TCGTTTCGAG GTTTTCGCGTTAGAGAC-3'
Methylated antisense (reverse), 5'-CGACTAAAA CTCGACCG ACTCGCGA-3'.
White blood cells of healthy blood donors and the following primers were used for promoter regions of the nonmethylated ACTB gene:
5' TGGTGATGGAGGAGGTTTAGTAAGT 3'
5' AACCAATAAAACCTACTCCTC CCTAA 3'.
It should be noted that commercial kit of QIAGEN N.V. was used when the samples were methylated. In addition, a negative control tube, containing all the PCR elements except for template, was used to examine the possibility of infection for each PCR reaction. In the delta-delta-CT formula, the number 2 represents the efficiency of the PCR reaction. This number is 2 (100%) for the best and most optimal reaction. For this purpose, a serial of dilutions (10, 100, 1000, 100,000, and 1,000,000) was prepared from the DNA of the vimentin and ACTB genes, and real-time PCR reaction was performed on them. After performing the reaction and plotting the CT chart versus initial dilution of the DNA, the line slope was obtained. Using the following formula, the efficiency of the reaction is obtained: Efficiency = 10^[–1/slope] [Charts 1–4].
Chart 1.
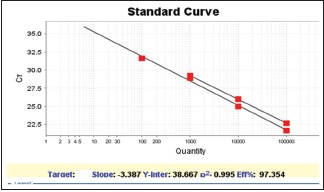
Vimentin and ACTB gene standard chart
Chart 4.
Display chart
Chart 2.
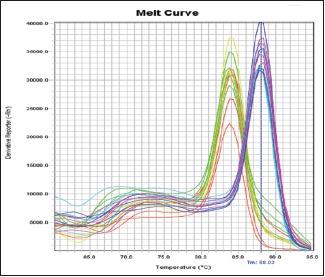
Melt-curve display of vimentin and ACTB gene expression
Chart 3.
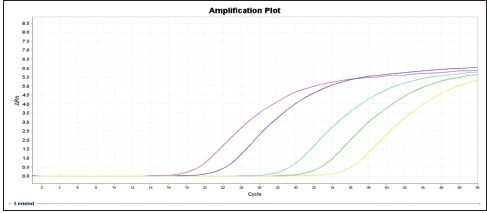
Amplification display chart in different dilutions of vimentin
In this study, the efficiency of the reaction in the vimentin gene is 97%. The value of slope is 3.3.
The methylated DNA level was obtained by calculating the ratio of vimentin gene methylation to ACTB gene methylation using delta-delta-CT method. Results were evaluated based on the relative quantitative method. Statistical data of this study were analyzed using SPSS 11.5. To compare the difference between levels of methylation and other values among the groups, t-test or ANOVA was used. Fisher's exact test was also used to analyze the classified data. Data values and results were expressed as mean ± SEM. P < 0.05 was considered as statistically significant.
RESULTS
The underlying information of patients in both case and control groups is presented in Table 1.
Table 1.
Underlying characteristics at the beginning of the study
| Characteristic | Case group (n=49) (%) | Control group (n=30) (%) | P |
|---|---|---|---|
| Age | 61.18 | 58.77 | 0.24 |
| Gender (male) | 67.3 | 56.7 | 0.33 |
| Gender (female) | 32.7 | 43.3 | 0.33 |
Chisquare t-test was used to examine the difference of means
The results of the frequency of vimentin gene methylation are shown in Table 2. Normal tissue was taken in a distance of 5–10 cm from the tumor tissue in the case group.
Table 2.
Frequency of vimentin gene methylation in both groups
| Group (n) | Gene extraction source | Methylation, n (%) | Nonmethylation, n (%) |
|---|---|---|---|
| Case (n=49) | Tumor tissue | 32 (65.3) | 17 (34.7) |
| Normal tissue | 5 (10.2) | 44 (89.8) | |
| Stool | 24 (60) | 16 (40) | |
| Control (n=30) | Normal tissue | 2 (6.7) | 28 (93.3) |
| Stool | 0 (0) | 30 (100) |
Table 3 illustrates the result of the comparison of vimentin gene methylation in the normal tissues of both case and control groups. Results of Chi-square test showed no statistically significant difference between the two groups in terms of vimentin gene methylation in normal tissue (P > 0.05).
Table 3.
Comparison of the vimentin gene methylation in normal tissues in both groups
| Group | Methylation in normal tissue | Nonmethylation in normal tissue | P |
|---|---|---|---|
| Case | 5 (10.2) | 44 (89.8) | 0.56 |
| Control | 2 (6.7) | 28 (93.3) |
Table 4 illustrates the result of the comparison of mean age in patients gene methylation in tumor and stool tissues of the case group and showed no statistically significant age difference between the the methylation in tumor and stool tissues of case group (P > 0.05).
Table 4.
Comparison of the vimentin gene methylation in tumor and stool tissues of the case group and its association with mean age
| Variable | n | Mean age | P |
|---|---|---|---|
| Tumor tissue | |||
| Methylation | 32 (65.3) | 61.60 | 0.51 |
| Nonmethylation | 17 (34.7) | 59.21 | |
| Stool tissue | |||
| Methylation | 24 (60) | 63.32 | 0.26 |
| Nonmethylation | 16 (40) | 61.29 |
Table 5 illustrates the result of Comparing the vimentin gene methylation in tumor and stool tissues of the case group and its relationship with gender and showed no statistically significant gender difference between the the methylation in tumor and stool tissues of case group (P > 0.05).
Table 5.
Comparing the vimentin gene methylation in tumor and stool tissues of the case group and its relationship with gender
| Variable | Gender (male) | Gender (female) | P |
|---|---|---|---|
| Tumor tissue | |||
| Methylation | 20 (60.6) | 12 (75) | 0.32 |
| Nonmethylation | 13 (39.4) | 4 (25) | |
| Stool tissue | |||
| Methylation | 16 (57.1) | 8 (66.7) | 0.57 |
| Nonmethylation | 12 (42.9) | 4 (33.3) |
Table 6 illustrates the Sensitivity, specificity, positive predictive value, and negative predictive value of the Valentin gene methylation.
Table 6.
Sensitivity, specificity, positive predictive value, and negative predictive value of the vimentin gene methylation
| Variable | Tumor tissue (%) | Normal tissue (%) | Stool (%) |
|---|---|---|---|
| Sensitivity | 65.30 | 10.2 | 60 |
| Specificity | - | 93.33 | 100 |
| Positive predictive value | 100 | 71.42 | 100 |
| Negative predictive value | - | 38.35 | 65.21 |
DISCUSSION
The research suggests that methylation of DNA of certain genes is associated with CRC development and seen in the stools of patients with colorectal adenomas. Thus, it is recommended to evaluate the feasibility of using DNA methylation biomarkers in diagnosing colorectal adenomas. Observations have approved that DNA methylation biomarkers are very sensitive and specific. These markers can also be used in diagnosing CRC and adenoma at an early stage.[4] The presence of TFPI2 as a biomarker in fecal DNA-based tests was used for the early diagnosis of CRC. After performing this test on the two groups of patients with nonmetastatic CRC and the control group with no cancer and moderate risk, methylated TFPI2 was diagnosed in the stool DNA of patients with CRC (Grades 1–3) with a sensitivity of 76%–89% and specificity of up to 93%. The results of this research revealed that the diagnosis of TFPI2 methylation in stool DNA may be used as a technique in screening the CRC.[7] Li et al. evaluated hypermethylation of 4010 genes using Methylated-CpG Island Recovery Assay (MIRA)-based microarrays in CRC. The findings of their research indicated that 211 genes were hypermethylated in the cancer tissue.[8]
In another study, that vimentin gene methylation was examined in the primary carcinoma, and normal tissues of 43 patients with hepatocellular carcinoma revealed the vimentin gene methylation in 24 patients out of 43 patients (56%).[9] One study examined the status of methylation of the vimentin gene in healthy tissue and primary cancer tissue of 48 patients using a specific PCR technique to measure the methylation and to evaluate the relationship between methylation and clinical findings. The results of this research showed altered methylation of vimentin in 31 patients out of 48 patients with primary CRC, indicating the high multiplicity of methylation of the vimentin genes in advanced CRC.[10]
The presence of CRC was examined using the vimentin serum level test in a study conducted by Shirahata et al. Its results showed that of 44 colorectal patients, 4 (9%) of them showed methylation of vimentin in serum DNA. In addition, methylation of serum vimentin was seen in patients with liver cancer carcinoma caused by colon metastases, peritoneal diffusion, and distant metastases significantly, indicating that methylation of serum vimentin could be observed more in advanced CRC.[11] Another study was conducted by the same researchers to examine the status of vimentin gene methylation in serum of 242 patients with CRC.[12]
The results of the studies of Li et al. showed that the methylation of vimentin in stools of patients could diagnose the CRC by 45% accuracy.[13]
In addition, the results of several meta-analysis studies showed that the methylation of the vimentin gene might be used as a reliable biomarker with high sensitivity and specificity in diagnosing and screening of CRC and can be considered as a therapeutic target.[14,15,16,17,18,19,20,21] In addition, in a study conducted by Pan et al. in 2017 in China, the cutoff was obtained 2 for quantitative values of methylated copies of vimentin gene and for the differentiation of pathological values from the normal values found in the intestinal tissue of healthy individuals and it was accepted.[20] There are several options for measuring the status of methylation of related gene promotor, and their sensitivity and specificity are different. The selection of an appropriate method for measuring or analyzing results with various methods might be useful in improving the reliability of noninvasive methylation biomarkers in CRC.[14,21,22]
CONCLUSION
The results of this study showed that the vimentin gene methylation in the stools of patients with CRC can be considered as a marker for the diagnosis of CRC. In addition, the research results showed that the evaluation of the vimentin gene methylation in the stool of patients with CRC has high specificity and sensitivity in diagnosing and screening the CRC, increasing the hopes for clinical applications of this important diagnostic marker as soon as possible.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Wang Y, Chen PM, Liu RB. Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection. World J Gastrointest Oncol. 2018;10:15–22. doi: 10.4251/wjgo.v10.i1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tokarz P, Pawlowska E, Bialkowska-Warzecha J, Blasiak J. The significance of DNA methylation profile in metastasis-related genes for the progression of colorectal cancer. Cell Mol Biol (Noisy-le-grand) 2017;63:79–87. doi: 10.14715/cmb/2017.63.2.12. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Chen JJ, Wang AQ, Chen QQ. DNA methylation assay for colorectal carcinoma. Cancer Biol Med. 2017;14:42–9. doi: 10.20892/j.issn.2095-3941.2016.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae JM, Rhee YY, Kim KJ, Wen X, Song YS, Cho NY, et al. Are clinicopathological features of colorectal cancers with methylation in half of CpG island methylator phenotype panel markers different from those of CpG island methylator phenotype-high colorectal cancers? Hum Pathol. 2016;47:85–94. doi: 10.1016/j.humpath.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 7.Glöckner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, et al. Methylation of TFPI2 in stool DNA: A potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–9. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Du Y, Zhang D, Wang LN, Yang C, Liu B, et al. Identification of novel DNA methylation markers in colorectal cancer using MIRA-based microarrays. Oncol Rep. 2012;28:99–104. doi: 10.3892/or.2012.1779. [DOI] [PubMed] [Google Scholar]
- 9.Kitamura Y, Shirahata A, Sakuraba K, Goto T, Mizukami H, Saito M, et al. Aberrant methylation of the vimentin gene in hepatocellular carcinoma. Anticancer Res. 2011;31:1289–91. [PubMed] [Google Scholar]
- 10.Shirahata A, Sakata M, Sakuraba K, Goto T, Mizukami H, Saito M, et al. Vimentin methylation as a marker for advanced colorectal carcinoma. Anticancer Res. 2009;29:279–81. [PubMed] [Google Scholar]
- 11.Shirahata A, Sakuraba K, Goto T, Saito M, Ishibashi K, Kigawa G, et al. Detection of vimentin (VIM) methylation in the serum of colorectal cancer patients. Anticancer Res. 2010;30:5015–8. [PubMed] [Google Scholar]
- 12.Shirahata A, Hibi K. Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer Res. 2014;34:4121–5. [PubMed] [Google Scholar]
- 13.Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–63. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo YX, Chen DK, Song SX, Wang L, Wang JP. Aberrant methylation of genes in stool samples as diagnostic biomarkers for colorectal cancer or adenomas: A meta-analysis. Int J Clin Pract. 2011;65:1313–20. doi: 10.1111/j.1742-1241.2011.02800.x. [DOI] [PubMed] [Google Scholar]
- 15.Li YW, Kong FM, Zhou JP, Dong M. Aberrant promoter methylation of the vimentin gene may contribute to colorectal carcinogenesis: A meta-analysis. Tumour Biol. 2014;35:6783–90. doi: 10.1007/s13277-014-1905-1. [DOI] [PubMed] [Google Scholar]
- 16.Du L, Li J, Lei L, He H, Chen E, Dong J, et al. High vimentin expression predicts a poor prognosis and progression in colorectal cancer: A study with meta-analysis and TCGA database. Biomed Res Int. 2018;2018:6387810. doi: 10.1155/2018/6387810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Gan A, Chen X, Wang X, He W, Zhang X, et al. Diagnostic performance of DNA hypermethylation markers in peripheral blood for the detection of colorectal cancer: A meta-analysis and systematic review. PLoS One. 2016;11:e0155095. doi: 10.1371/journal.pone.0155095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma P, Dai S, Xie M, Zou C, Liu L, Zhu Y, et al. Vimentin gene promoter hypermethylation in stool as a biomarker for colorectal neoplasm diagnosis: A meta-analysis. Int J Clin Exp Med. 2017;10:13081–8. [Google Scholar]
- 19.Yuan Y, Qu B, Yan J, Wang H, Yin L, Han Q. Diagnostic value of aberrant gene methylation in stool samples for colorectal cancer or adenomas: A meta-analysis. Panminerva Med. 2015;57:55–64. [PubMed] [Google Scholar]
- 20.Pan DY, Zeng XQ, Ma GF, Gao J, Li N, Miao Q. Label-free quantitative proteomic analysis identifies CTNNB1 as a direct target of FOXP3 in gastric cancer cells. Oncol Lett. 2018;15:7655–60. doi: 10.3892/ol.2018.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Zhu YQ, Wu YQ, Guo Y, Qi J. Diagnostic value of aberrant methylation of genes in stool for colorectal tumor: A meta-analysis. Chin J Evid Based Med. 2013;13:149–59. [Google Scholar]
- 22.Zhang H, Qi J, Wu YQ, Zhang P, Jiang J, Wang QX, et al. Accuracy of early detection of colorectal tumours by stool methylation markers: A meta-analysis. World J Gastroenterol. 2014;20:14040–50. doi: 10.3748/wjg.v20.i38.14040. [DOI] [PMC free article] [PubMed] [Google Scholar]


