Abstract
The evolutionarily conserved mTOR signaling pathway plays essential roles in cell growth, proliferation, metabolism and responses to cellular stresses. Hyperactivation of the mTOR signaling is observed in virtually all solid tumors and has been an attractive drug target. In addition to changes at genetic levels, aberrant activation of the mTOR signaling is also a result from dysregulated post-translational modifications on key pathway members, such as phosphorylation that has been extensively studied. Emerging evidence also support a critical role for ubiquitin-mediated modifications in dynamically regulating the mTOR signaling pathway, while a comprehensive review for relevant studies is missing. In this review, we will summarize all characterized ubiquitination events on major mTOR signaling components, their modifying E3 ubiquitin ligases, deubiquitinases and corresponding pathophysiological functions. We will also reveal methodologies that have been used to identify E3 ligases or DUBs to facilitate the search for yet-to-be discovered ubiquitin-mediated regulatory mechanisms in mTOR signaling. We hope that our review and perspectives provide rationales and strategies to target ubiquitination for inhibiting mTOR signaling to treat human diseases.
1. Overview of the mTOR signaling pathway.
The mTOR signaling pathway is an evolutionarily conserved kinase cascade playing essential roles in regulating key cellular functions, including but not limited to cell growth, proliferation, autophagy, metabolism and DNA damage. The TOR genes were firstly identified in yeast by the Hall group in 1991. Afterwards, mTOR was purified by both Schreiber and Snyder groups and its function on cell proliferation and protein synthesis was discovered by multiple groups including Vézina, Sonenberg, Blenis, Thomas, Crabtree, Gelfand and others. The first mTOR-deleted mouse model was established by Peterson and Sabatini groups, which confirms the essential role of mTOR in cell survival and development. Afterwards, the mTOR signaling cascade was characterized by both genetic and biochemical approaches (please refer to many great mTOR review articles from Hall, Sabatini and other groups for references) (Figure 1).
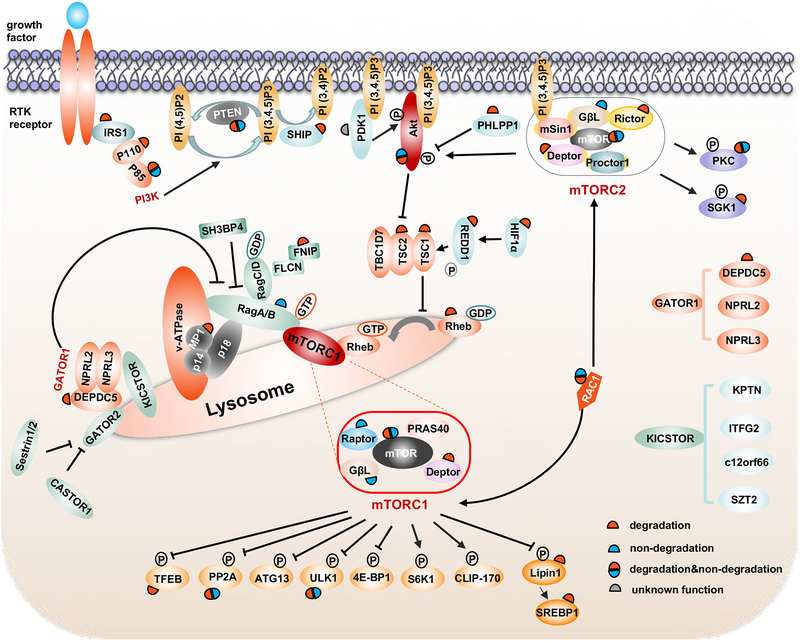
Figure 1. Overview of the ubiquitin regulations on mTOR signaling components.
This is a cartoon illustration of key mTOR signaling pathway members, their cellular localization and reported ubiquitin-mediated regulations. The red half circle indicates degradation-oriented ubiquitination, the blue half circle indicates non-degradation-oriented ubiquitination, and the gray half circle indicates ubiquitination with unknown function.
2. Overview of the ubiquitin signaling.
Ubiquitin is a small molecule composed of 76 amino acids. Ubiquitination is usually considered as a protein modification by attaching the ubiquitin moiety through one of the seven lysine residues (K6, K11, K27, K29, K33, K48 and K63), or to the amino terminus (M1) in each ubiquitin to form poly-ubiquitin chains. Recently, beyond protein-related function, we found that K63-linked poly-ubiquitin chains also bind DNA to facilitate DNA damage response1. Nonetheless, protein ubiquitination is governed by three types of enzymes as a cascade of a three-step reaction, including the E1 ubiquitin activating enzyme, the E2 ubiquitin conjugating enzyme and the E3 ubiquitin ligase. In mammalian genome, there are ~2 E1 enzymes, ~30 E2 enzymes and ~1,000 E3 ligases. There are three major types of E3 ubiquitin ligases in mammals including the Cullin-Ring (really interesting protein) (CRL) family of E3 ligases, HECT-domain containing E3 ligases and RBR (ring-between-ring) E3 ligases. E3 ligases are substrate recognition subunits, which recognize specific protein motif sequences in substrates for binding. These protein motif sequences are defined as “degron”s. CRL is the largest family of E3 ligases and usually CRL E3s are structurally composed of Rbx1 (also named Roc1, binds E2 through its Ring domain), Skp1, Cullin (including one of Cullin 1, 2, 3, 4A, 4B, 5 and 7) and a E3 ligase. F-box proteins is a major type of CRL-E3 ligases bearing a F-box motif with ~50 amino acids (termed as F-box) as a protein binding motif mediating F-box binding to Skp1. There are 69 F-box proteins in mammals2, thus forming 69 SCF (Skp1/Cullin1/F-box) E3 ligase complexes. Except several well-characterized SCFs including Fbw7, β-TRCP and Skp2, the function of the majority of F-box proteins are just began to be appreciated (please refer to3–5 for review). In addition to protein degradation, recognition of degrons by E3 ligases can also promote non-degradable ubiquitination via other linkages than degradation oriented K11 and K48 linkages, such as K6 and K63 linkages.
Previously, phosphorylation and its associated physiological functions on key mTOR pathway members have been extensively studied. Recently, ubiquitin-mediated regulation of mTOR signaling emerges as an important approach to diversely regulate functions of mTOR by either degrading the protein targets to terminate the mTOR signaling, modulating protein cellular localization, or promoting protein association with its binding partners to alter its biological functions. Importantly, the ubiquitin system has been considered as a druggable target with various effective agents available. Thus, in this review, we will summarize major ubiquitination events identified to date on major mTOR pathway components, E3 ubiquitin ligases and deubiquitinases regulating these ubiquitination processes, and function of the ubiquitin modifications, hoping to provide the first comprehensive view of ubiquitin-mediated regulation of the mTOR signaling.
3. Degradation-oriented and ubiquitin-mediated mTOR signaling regulations.
mTOR can be activated by degradation of mTOR inhibitory proteins or inactivated by degradation of mTOR activating partners, both of which are controlled by the protein ubiquitination process. In this section, we will summarize major degradation-oriented ubiquitination modifications on the mTOR signaling components to illustrate roles of these degradation-oriented protein stability control mechanisms in regulating mTOR activity and function in distinct disease and development settings (Figure 1 and Table 1).
IRS-1 (insulin receptor substrate 1) is essential for transducing extra-cellular signals such as insulin or insulin-like growth factors to PI3K and mTOR. Loss of or reduced IRS-1 expression results in insulin resistance, which is a major determinant of type 2 diabetes. In early 2000s, IRS-1 protein stability was observed to be controlled in a Ser phosphorylation-dependent manner6. Afterwards, inflammation-induced SOCS1 (suppressor of cytokine signaling 1) and SOCS3 expression recruits IRS-1 to the elongin BC ubiquitin ligase for IRS-1 ubiquitination and degradation7. Another independent proteomic study found that IRS-1 is also a Fbw8CUL7 substrate and its degradation by Fbw8CUL7 is controlled by IRS-1 phosphorylation8. Furthermore, Fbxo40 was observed to control IRS-1 protein turnover in skeleton muscle9. Thus, the IRS-1 protein stability is controlled by a set of E3 ubiquitin ligases under distinct physiological settings.
PI3K (phosphoinositide 3-kinase) is firstly identified by Lewis C. Cantley and collogues as a lipid kinase for PI(3,4,5)P3 production. It is a protein kinase complex composed of p110 (α, β and γ) catalytic subunits and p85 (α and β) regulatory subunits. The catalytic subunit p110α is recognized by a plasma membrane attached E3 ligase NEDD4L (neural precursor cell expressed, developmentally down-regulated 4-like) for K29-linked ubiquitination and degradation10, resulting in compromised mTOR activation. While the complex-free p85β subunit is targeted by Fbxl2 for degradation in a p85β-Tyr655 dephosphorylation-dependent manner11. Specifically, Fbxl2 mediates the degradation of PTPL1-dephosphorylated p85β to prevent its competition with active PI3K (composed of p110/p85 heterodimers) for phospho-Tyr docking at plasma membrane11. Similarly, Fbxl2 targets IP3R3 for ubiquitination and degradation to limit calcium flux from the ER to mitochondria, mitochondrial calcium overload and apoptosis-events that are counteracted by PTEN12. Via these mechanisms, Fbxl2 ensures sufficient activation of the PI3K signaling cascade and promotes cell survival. Intriguingly, viruses (such as hepatitis C) hijack these regulatory mechanisms to inhibit virus-induced apoptosis and establish chronic infection by binding and unmasking IP3R3 degron13. In another report, p85 is recruited to HSP70/CHIP-E3 ligase via p42 for ubiquitination and degradation14. Ubiquitin-mediated p85 degradation leads to dampened PI3K activation. On the other hand, ubiquitin-mediated activation mechanism for PI3K remains unclear.
As a lipid phosphatase, PTEN antagonizes PI3K function by converting PI(3,4,5)P3 into PI(4,5)P2. The E3 ligase WWP2 was identified to negatively govern PTEN protein stability15. Interestingly, XIAP1, a pro-apoptotic protein, displays an E3 ligase activity in ubiquitinating and degrading PTEN16. Moreover, the E3 ligases DCAF13CUL4B17 and MKRN118 were also observed to ubiquitinate and destruct PTEN proteins in osteosarcoma and cervical cancer cells, respectively. Thus, PTEN expression is tightly controlled by multiple E3 ligases to ensure its proper physiological function as a dual lipid and protein phosphatase. In addition to the 26S proteasome pathway, PTEN also undergoes lysosomal degradation, which is mediated by SYNJ2BP19. Whether PTEN stability, activity or cellular localization can be regulated by atypical linkage ubiquitination events remain to be determined. Notably, PI(3,4,5)P3 can also be dephosphorylated into PI(3,4)P2 by the lipid phosphatase SHIP. The stability of SHIP is governed by the oncogenic-fusion E3 ligase BCR-ABL20,21 in hematopoietic cancer.
PDK1, the major Akt upstream kinase phosphorylating Akt-T308, has been observed to undergo mono-ubiquitination within its kinase domain22. However, the physiological function for mono-ubiquitination of PDK1 remains unclear, but it is not tightly associated with PDK1 kinase activity22.
mTOR is the essential kinase component for both mTORC2 and mTORC1 complexes. The first mTOR protein stability control mechanism was discovered in 2008 and the tumor suppressive E3 ligase Fbw7 was observed to target mTOR for ubiquitination and degradation in a GSK3-phosphorylation dependent manner23. In colorectal cancer, Fbx8, a metastasis suppressive E3 ligase partially exerts its tumor suppressor function through recognizing and degrading mTOR24.
Deptor is an endogenous mTOR inhibitor suppressing both mTORC1 and mTORC2 activation25. Thus, depletion of β-TRCP, a E3 ligase targeting Deptor for ubiquitination and degradation triggered by Deptor phosphorylation via mTOR, CKI, RSK or p38 (see Table 1 for references), led to accumulation of Deptor and subsequently reduced mTORC1 activation. In prostate cancer, the Ring type of E3 ligase SAG (sensitive to apoptosis gene, also named Rbx2) also targets Deptor for destruction, through which SAG activates the mTORC2/Akt signaling to promote prostate tumorigenesis26.
Rictor is an integral component for the mTORC2 complex and indispensable for mTORC2 kinase activity. Fbw7-mediated Rictor ubiquitination relies on GSK3-mediated Rictor-T1695 phosphorylation27, resulting in attenuated mTORC2 activity towards phosphorylating Akt, SGK and PKC.
Akt, SGK and PKC are three well-characterized mTORC2 kinase substrates. Akt, also known as PKB (protein kinase B), is the major downstream protein kinase mediating the oncogenic feature of mTORC2. Catalytic-active Akt being phosphorylated on both T308 and S473 is thought to be targeted by multiple E3 ligases for ubiquitination and degradation. For example, TTC3 (tetratricopeptide repeat domain 3) promotes K48-linked poly-ubiquitination and destruction of doubly phosphorylated nuclear Akt28. Interestingly, Akt-mediated phosphorylation of TTC3 at Ser378 is necessary to activate TTC3 E3 ligase28, revealing a feedback mechanism for Akt to restrain its own activity. Another mitochondrial E3 ligase MULAN (mitochondrial ubiquitin ligase activator of NF-κB) targets Akt1 and Akt2, but not Akt3, for ubiquitination and degradation. Specifically, MULAN recognizes phospho-Akt to add K48-linked polyubiquitin chains on Lys284 in Akt kinase domain for Akt destruction29. Interestingly, in head-and-neck cancer, NTP (non-thermal plasma, an ionized gas) induces MULAN expression by elevating cellular ROS levels to facilitate Akt degradation, through which NTP exerts is anti-cancer ability30. Moreover, RFP2 (ret finger protein 2), a RBCC (RING finger, B-box, and coiled-coil domain) family of E3 ligases frequently deleted in various tumor types, promotes cell apoptosis in part by degrading both Akt and Mdm231. In addition, BRCA1 (breast cancer susceptibility gene 1) also negatively regulates Akt activation by serving as a E3 ligase that targets phospho-Akt for ubiquitination and degradation32. Given that BRCA1 is tightly connected with genome instability and familial breast tumorigenesis, whether DNA damage triggers BRCA1-medaited Akt degradation remains to be explored. Similarly, the U-box E3 ligase CHIP (C-terminal Hsp70 -interacting protein) targets phospho-Akt for degradation33, through which CHIP exerts a tumor suppressor function. However, a recent report suggests something opposite-that overexpressed CHIP in prostate cancer leads to increased Akt activation34 to facilitate prostate cancer growth34. This discrepancy warrants further investigations to determine whether CHIP differentially regulates Akt ubiquitination and activity in a cellular context-dependent manner. In addition to cancer, Akt stability control also plays a critical role in neurodegeneration diseases. To this end, CHIP degrades Akt in Alzheimer’s disease, leading to reduced Tau degradation by CHIP35. Thus, CHIP and Akt co-regulate Tau protein stability. A CUL1-based F-box E3 ligase Slimb degrades Akt to facilitate ddaC dendrite pruning by suppressing the Akt signaling36. Moreover, ZNRF1 (zinc and ring finger 1) promotes Wallerian degeneration through activating the GSK3/CRMP2 signaling achieved by terminating Akt signaling by promoting Akt ubiquitination and degradation37. In addition, the TTC3/Akt/eNOS signaling (TTC3 targets Akt for degradation) also plays a critical role in Down Syndrome28. Taken together, most of identified E3 ligases largely target phospho-Akt (pT308 and pS473) for ubiquitination and degradation. In addition to canonical phosphorylation events on S473 and T308, we identified a novel Akt-tail phosphorylation event that may function independent of these doubly phosphorylated Akt to promote Akt activation38. Whether and how any E3 ubiquitin ligases recognize tail-phosphorylated Akt or non-modified Akt, which accounts for the majority of Akt species under physiological conditions remains elusive.
In addition to proteasome-mediated protein stability control, Akt has also been demonstrated to be degraded through the autophagy process triggered by AMPK activation upon treatment with curcumin, a yellow bioactive compound from plant turmeric39. Moreover, Wnt5a/Wnt11-mediated cell differentiation results in reduced Akt activation, as well as a decrease of total Akt proteins due to caspase-dependent Akt degradation40. Interestingly, caspase 3 cleavages Akt at Asp108 and Asp119, which inactivates Akt under detachment-induced cell apoptosis by death receptor signaling in epithelial cells41. In Alzheimer’s disease, caspase3-meidated Akt cleavage leads to reduced phosphorylation and activation of an Akt substrate GSK3, where GSK3 induces Tau phosphorylation to promote disease development42. Taken together, these studies demonstrate that Akt activity/stability is negatively regulated by proteasome-mediated degradation, autophagy and caspase cleavage under distinct pathophysiological conditions.
In addition to Akt degradation, dephosphorylation of Akt also contributes to Akt inactivation, facilitated by phosphatases PP2A (protein phosphatase 2A) and PHLPP1/2 (PH Domain And Leucine Rich Repeat Protein Phosphatase 2), on Thr308 and Ser473, respectively. PHLPP1 is degraded by β-TRCP43 in a GSK3 and CKI-phosphorylation-dependent manner or by SAG26, leading to increased Akt activity. The PP2A heterotrimeric phosphatase is composed of a structural A, a catalytic C and a regulatory B subunit. DCAF1CUL4 targets PP2A-A for destruction to control oocyte meiotic maturation44. KLHL15CUL3 recognizes and degrades PP2A-B, thus promoting its exchange with other PP2A regulatory subunits45. The E3 ligase NOSIP promotes PP2A-C degradation to regulate craniofacial development46, and the inability of the E3 ligase MID1 to degrade PP2A-C contributes to Opitz Syndrome47. Notably, MID1-mediated PP2A-C destruction depends on the Bbox1 domain in MID1 and an MID1 binding protein Alpha4.
There are three isoforms of SGK (serum and glucocorticoid-induced protein kinase) in mammals, namely SGK1, SGK2 and SGK3. SGK1 regulates ion channels, membrane trafficking and cell growth to play critical roles in hypertension, diabetic neuropathy, trauma and neurodegenerative diseases48. Basal SGK1 protein levels are low in mammalian cells cultured in vitro due to active proteasomal degradation. The E3 ligase NEDD4–2 ubiquitinates and degrades SGK1, while SGK1-mediated NEDD4–2 phosphorylation antagonizes this process49. In addition, CHIP co-localizes with SGK1 at ER (endoplasmic reticulum) to promote SGK1 protein turnover under stress conditions50. Interestingly, Rictor, an essential mTORC2 component governing SGK phosphorylation and activation, exerts an E3 ligase function by complexing with CUL1 to promote SGK1 degradation51. Thus, on one hand, Rictor facilitates SGK1 activation by facilitating SGK1 phosphorylation in a mTORC2 kinase-dependent manner, while on the other hand, Rictor destabilizes SGK1 proteins in a E3 ligase-dependent manner.
PKC is a large family of protein kinases with 15 members. Specifically, isoforms α, βI, βII and γ belong to classical PKCs; δ, ε, η and θ are defined as novel PKCs and Mζ and ι / λ are atypical PKCs. PKCs usually phosphorylate potent activators for transcription and promote tumorigenesis. Interestingly, PMA and insulin induce ubiquitination and degradation of PKCδ, but not PKCα nor PKCε in a PKCδ-Y311 phosphorylation-dependent manner52 in muscle. Under nomoxia conditions, atypical PKCζII is recognized and degraded by the tumor suppressive E3 ligase pVHL through its N-terminal PB1 domain53. While under hypoxia, PKCζ is rapidly ubiquitinated and degraded by HOIL-1L (heme-oxidized IRP2 ubiquitin ligase 1L) through K48-linked polyubiquitination (see Table 1 for references).
Akt-mediated TSC2 phosphorylation is the critical signaling node for connecting mTORC2 to mTORC1. The TSC complex is composed of TSC2, TSC1 and TBC1D7 and considered tumor suppressive given to its function in suppressing the Rheb/mTORC1 signaling. Loss of TSC2 or TSC1 is observed as the cause of TSC (tuberous sclerosis complex) disease with formation of benign tumors in kidney, lung, heart and skin. Thus, understanding regulatory mechanisms governing TSC complex protein stability control may provide new insights into TSC treatments. TSC2 protein stability is controlled by multiple E3 ligases, including Fbw5CUL4/DDB1, Pam54, E6AP (independent of HPV E6) and HERC1 (see Table 1 for references). Interestingly, TSC1 binding to TSC2 protects TSC2 from being recognized and degraded by Fbw5, Pam and HERC1. RB1CC1 (RB1-inducible coiled-coil 1) binds TSC1 and induces TSC1 ubiquitination and degradation with help from unknown E3 ligase(s)55. In hepatocellular carcinoma, TRIM31 exerts an oncogenic function by targeting both TSC1 and TSC2 for degradation56. The identities of E3 ubiquitin ligases governing TBC1D7 protein turnovers remain completely unknown.
mTORC1 is activated by GTPases including Rheb and Rags upon growth factor and amino acid stimulation, respectively. The first evidence indicating that Rheb protein stability is regulated by proteasomes comes from the observation that hydrogen peroxide reduces Rheb protein abundance in RAW264.7 cells57. To date, only one report suggests that nitrosylated GAPDH complexes with the E3 ligase Siah1 to target Rheb for K48-linked ubiquitination and degradation58. More detailed investigations are necessary to provide more insights into whether growth signaling plays a role in this Rheb degradation process and how Rag protein stability is controlled.
The Ragulator complex (composed of LAMTOR3 (MAPKSP1 or MP1), LAMTOR2 (ROBLD3 or p14), LAMTOR1 (p18), LAMTOR4 (c11orf59) and LAMTOR5 (HBXIP)) recruits Rag GTPases to lysosome for mTORC1 activation in response to amino acids. Thus, proteasomal degradation of Ragulator negatively regulates mTORC1 activity, retards cell growth but promotes autophagy. Notably, similar to the TSC2/TSC1 complex, loss of either of the Ragulator components destabilizes other subunits in a proteasome-dependent manner59. The exact identities for E3 ligases governing Ragulator protein stability are elusive.
FLCN, together with its binding partner FNIP1/2, serve as a GAP for RagC and RagD60 to suppresses mTORC1 activation. The E3 ligase β-TRCP targets FNIP1/2 for ubiquitination and degradation in a CKI phosphorylation-dependent manner, leading to elevated mTORC1 activity and increased renal cancer growth61. GATOR1 (composed of DEPDC5, NPRL2 and NPRL3) exerts a GAP activity towards RagA and RagB to suppress mTORC1 activation; while GATOR2 (consists Mios, WDR24, WDR59, Seh1L and sec13) promotes mTORC1 activation through negatively regulating DEPDC5 in GATOR162. Upon amino acid addition, release of GATOR1-mediated suppression is necessary to facilitate Rag/mTORC1 activation, which is achieved by KLHL22-mediated DEPDC5 degradationCUL363. KICSTORs (GATSL3/GATSL1/GATSL2) recruit GATOR1 to lysosome to suppress mTORC1 activation. mTORC1 senses distinct amino acids through various amino acid sensors, such that CASTORs are arginine sensors for mTORC1 activation and Sestrin-2 is a leucine sensor. In addition to amino acids, SAMTOR is the S-adenosylmethionine sensor for mTORC1 activation, while lysosomal cholesterol is sensed by the SLC38A9/NPC1 complex that promotes mTORC1 activation. Given that these mTORC1 regulators were identified recently, the protein stability control mechanisms for these proteins remain to be further determined.
mTORC1 promotes protein synthesis by phosphorylating S6K and 4EBP1, inhibits autophagy through phosphorylating ULK1, facilitates nucleotide metabolism by phosphorylating CAD and regulates SREBP1 expression and lipid metabolism through phosphorylating Lipin1. mTORC1 and CKI-mediated phosphorylation of Lipin1 primes Lipin1 for recognition, ubiquitination and degradation by β-TRCP64. SREBP1, the target of Lipin 1, is recognized and degraded by Fbw7 upon phosphorylation by GSK3, or by RNF20 upon phosphorylation by PKA (see Table 1 for references). How the protein stability of other major mTORC1 substrates in addition to Lipin1 is controlled and whether deregulation of these ubiquitin signaling contributes to human diseases warrants further in-depth investigations.
4. Non-degradation-oriented ubiquitin-mediated mTOR signaling regulations
In addition to K48-linked poly-ubiquitination on mTOR signaling components that promotes 26S-proteasome-mediated protein degradation, K63-linked poly-ubiquitination has also been observed on mTOR components to dynamically modulate their functions in a non-degradation oriented manner. In this section, we will summarize these identified protein ubiquitination events, their modifying E3 ligases and biological consequences (Figure 1 and Table 1).
The E3 ligase Cbl-b negatively regulates p85 in a degradation-independent manner, where Cbl-b binding to p85 inhibits the recruitment of p85 to CD28 and TCRζ in T cells (see Table 1 for references). Moreover, TRAF6 promotes K63-linked poly-ubiquitination of p85α to enhance p85α binding with TβRI (TGF-β type I receptor), which subsequently activates PI3K signaling65. Interestingly, atypical-linkage ubiquitination of PTEN by RFP (Ret finger protein) inhibits PTEN phosphatase activity19 but the underlying molecular mechanism(s) remains to be determined.
The E3 ligase TRAF2 (TNF Receptor Associated Factor 2) governs K63-linked GβL poly-ubiquitination at Lys305 and Lys313 under serum-starvation conditions, which promotes Raptor binding to favor mTORC1 formation, while repelling Sin1 binding to dissociate mTORC266. Therefore, K63-linked polyubiquitination of GβL serves as a major mechanism to maintain mTOR complex homeostasis. However, the upstream signaling governing TRAF2 activity warrants further investigations. On the other hand, TRAF6, with help from p62, poly-ubiquitinates mTOR through a K63-linkage to facilitate mTORC1 activation by promoting mTOR recruitment to lysosome in response to amino acid stimulation67. In addition, K63-linked ubiquitination of Raptor by DDB1/CUL4 is also necessary for mTORC1 activation at lysosome68, and this process can be antagonized by the ubiquitin hydrolase UCHL-1-mediated Raptor deubiquitination69. Under mitochondrial stress, the E3 ligase PARKIN interacts with and ubiquitinates mTOR at Lys2066 and Lys2306 through unknown ubiquitin linkage(s) to maintain mTORC1 activity70. Notably, the detailed molecular mechanisms underlying poly-ubiquitination-mediated mTOR activation remain to be determined.
In addition to mTOR, TRAF6 also promotes K63-linked poly-ubiquitination of Akt at Lys8 and Lys14 in the Akt-PH domain to facilitate its plasma membrane attachment and activation by IGF-171. In addition, NEDD4 induces K63-linked poly-ubiquitination of phosphor-Akt, which facilitates nuclear retention of active Akt72. Interestingly, Skp2, but not TRAF6, mediates Akt poly-ubiquitination and membrane recruitment in response to activation of HER2 receptors by EGF in breast cancer to promote glycolysis and resistance to HER2 inhibition73. While in lung cancer, TRAF4, but not Skp2, promotes Akt poly-ubiquitination and activation in response to EGF74. In glioma, the Fbxl18 E3 ligase is significantly amplified to promote glioma growth through catalyzing K63-linked Akt ubiquitination to facilitate Akt activation75. It remains to be determined how distinct upstream stimuli, such as IGF-1 and EGF, triggers differential E3 ligases binding to Akt to induce Akt poly-ubiquitination, as well as why Akt poly-ubiquitination is governed by different E3 ligases responding to the same physiological cue in different cancer settings.
Moreover, the lysosome-anchored E3 ligase RNF152 promotes RagA poly-ubiquitination through a K63-linkage upon amino acid stimulation to promote mTORC1 activation76 by engaging RagA to lysosome. Subsequently, the E3 ligase Skp2-mediated K63-linked ubiquitination of RagA at Lys15 recruits GATOR1 to turn off mTORC177. In addition, TRAF6-mediated K63-linked poly-ubiquitination of Rac1 at Lys16 promotes Rac1 activation and subsequent activation of both mTORC2 and mTORC178, while the detailed molecular mechanism(s) governing Rac1-mediated mTOR activity control remains unknown.
5. Deubiquitination in mTOR signaling.
Given that protein ubiquitination is a reversible reaction that the ubiquitin moiety can be removed by deubiquitinases (DUBs, a type of proteases), many deubiquitinases have been identified to antagonize the identified ubiquitin signaling mentioned above (Table 2). For example, USP7 stabilizes IRS-1 by removing K48-linked ubiquitin chains, and IGF-1 stimulation dissociates USP7 from binding to ubiquitinated IRS-1, thus leading to enhanced IRS-1 degradation79. PTEN can be stabilized by DUBs including USP10 in lung cancer and hepatocellular carcinoma, USP13 and OTUD3 in breast cancer, and Ataxin-3 in lung cancer through removing K48-linked ubiquitin moieties (see Table 2 for references). In addition to stability control, USP7 (also called HAUSP, herpesvirus-associated ubiquitin-specific protease) deubiquitinates PTEN to promote PTEN nuclear export, and this process can be antagonized by binding and sequesting USP7 from PTEN by PTEN phosphorylation at Ser380 by S6K80. Deptor is stabilized by OTUB181 by removing K48-linked ubiquitin chains. Although no E3 ligase(s) has been identified to promote Raptor destruction, Raptor is stabilized by USP9X in mouse embryonic brains82 to facilitate mTORC1 activation. PHLPP1, an Akt phosphatase, is stabilized by various DUBs including USP46 and WRD48/USP12 in colon cancer, USP1 in lung cancer, and Uaf1/WDR20/USP12 in prostate cancer (see Table 2 for references). Similarly, the kinase ULK1 is stabilized by USP20, which is necessary to initiate autophagy83.
Mono-ubiquitination of PDK1 is removed by USP422, which occurs at the plasma membrane where PDK1 is activated. CYLD (cylindromatosis), originally identified as a DUB inactivating the NF-κB signaling, removes K63-linked poly-ubiquitin chains from Akt to retard Akt membrane recruitment in prostate cancer84. In addition, OTUD7B is the deubiquitinase antagonizing TRAF2-goverend K63-linked GβL ubiquitination to promote GβL incorporation into mTORC2 complexes66. UCHL-1, a protease-like deubiquitinase, promotes the removal of K63-linked poly-ubiquitin chains on Raptor and GβL to modulate mTOR complex formation69. Moreover, K63-linked mTOR poly-ubiquitination is removed by USP9X85. Notably, only a handful of DUBs have been well-characterized and assigned with certain physiological functions. Thus, the physiological function and regulations on many orphan DUBs functioning in mTOR signaling need to be further determined by both mouse genetic studies and biochemical analyses. More importantly, unlike E3 ubiquitin ligases (which are not REAL enzymes but rather help E2 to transfer ubiquitin), DUBs exert enzymatic activities. Thus, DUB inhibitors might be a new line of therapeutic choices for human diseases.
6. Evolution of E3 ubiquitin ligases, DUBs and mTOR signaling components.
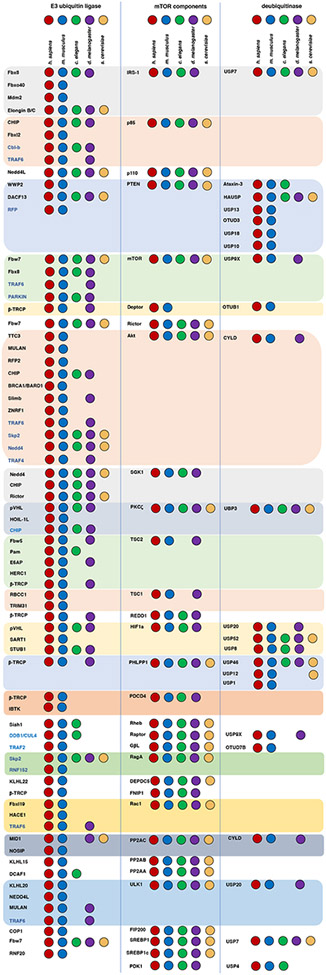
Most of the mTOR signaling components are highly conserved from yeast to mammals, with only Rheb and TSC2/TSC1 missing in certain metazoan and fungi (refer to86 for review). Phosphorylation events that play important roles in regulating mTOR signaling, is also quite conserved through evolution, given that a co-evolution pattern of both mTOR components and their modifying kinases is observed86. On the other hand, compared with conserved phosphorylation events, most E3 ligases and deubiquitinases identified are only conserved in mouse and human, but not in lower eukaryotes such as yeast and drosophila (Figure 2). These observations suggest that ubiquitin-mediated modifications on mTOR signaling components may be a late evolutionary event and ubiquitin modifications may provide new layers of regulations in higher eukaryotes in order to adapt to more sophisticated cellular environments, and may also be specifically associated with human diseases. However, more thorough studies are warranted towards these directions.
Figure 2. Evolutionary conservation of E3 ligases and DUBs regulating mTOR signaling.
The presence of mTOR signaling components (middle), identified regulating E3 ligases (left) and DUBs (right) in indicated four species (human, mouse, c. elegans and fruit fly) are indicated by dots with distinct colors. Conservation information is obtained from NCBI-homologene, genecards, KEGG pathway, uniport and literature search. Absence of any dot means either not conserved or never examined and reported.
7. Approaches used to identify E3 ligase/substrate and DUB/substrate pairs for mTOR signaling components.
In the following section, we will summarize the major experimental approaches used in previous studies to identify ubiquitin-mediated mTOR regulations. We hope these methods will inspire the characterization of unknown ubiquitin modifying enzymes for mTOR signaling components (Table 1), as well as more intensive studies for ubiquitin modifications in other major signal transduction pathways.
(1). Proteomics-mediated protein interactome approach.
In this method, either a substrate or an E3 ligase is used as the bait to pulldown its interacting proteins for further mass spectrometry analyses. This method is based on the principle that E3 ligases or DUBs form relatively stable interactions with substrates. For example, to understand the function of the orphan F-box protein Fbxl2, Fbxl2 is expressed in HeLa or HEK293T cells and immunoprecipitated for MudPIT analyses11. By this approach, p110-free p85 is identified as a bona fide Fbxl2 substrate, through which Fbxl2 negatively controls the PI3K signaling. On the other hand, TAP tagged-mTOR (as a substrate) is used as a bait in HEK293T cells and mTOR interactome was established by TAP-tag pulldown coupled mass spectrometry analyses, where USP9X was identified as a novel mTOR binding partner and a DUB to deubiquitinate mTOR for suppressing mTOR85. Similarly, triply-tagged PTEN (by S-protein, Flag and streptavidin-binding peptide, SBP) is used as the bait in a tandem purification using lysates from HEK293T cells followed by mass spectrometry analyses, and WWP2 is identified as a PTEN binding protein that targets PTEN for ubiquitination and degradation15. To further stabilize the interactions between E3/DUB and substrate, MG132, a 26S-proteasome inhibitor, is commonly used before cell collection to preserve more interactions to increase the detection sensitivity.
(2). Loss-of-function genetic screens.
Another large-scale and non-biased screening approach is to utilize the loss-of-function screens facilitated by siRNAs, shRNAs or CRISPR. To this end, a siRNA screen against a DUB library identifies DUBs to remove PDK1 mono-ub22; a loss of function screen using siRNAs targeting 99 DUBs finds candidates regulating ULK1 stability83, and a siRNA library against 92 DUBs is employed to identify PTEN DUBs87. Notably, this approach is more appropriate for substrates with relatively higher levels of endogenous ubiquitination. Due to off-target effects from knockout/knockdown constructs, as well as influences from phenotypes derived from loss of essential genes, necessary controls and cautions need to be taken into consideration for this approach.
(3). Yeast-two-hybrid (Y2H) assays.
This approach is to use yeasts to perform interaction studies. For example, using a Y2H assay with a full-length Akt2 as a bait, TTC3 is found as a direct Akt binding partner that promotes Akt ubiquitination and degradation28. In addition, a direct interaction between PTEN and its E3 ligase DCAF13 is also confirmed by Y2H17. The same approach has also been used to identify Fbw5 as an E3 ligase controlling TSC2 protein stability88. Given to the presence of homologous genes in yeast, Y2H studies may generate false positive results. In addition, due to the possibility that certain post-translational modifying enzymes are not conserved in yeast, if the targeted protein-protein interactions depend on certain post-translational modifications, the Y2H assay may not faithful recapitulate the physiological conditions in mammals.
(4). Bioinformatic approaches to search for E3 ligase degrons.
With the identification of more substrates for a given E3 ligase and availability of crystal structures for E3/substrate complexes, a handful of well-defined protein motifs in substrates for specific E3 ligases have been deciphered. For example, Fbw7 prefers a consensus CDC phospho-degron (CPD) sequence I/L-I/L/P-T-P-XXXX for recognition, where a priming phosphorylation at the TP site is critical to trigger CPD binding. The E3 ligase Keap1 recognizes “ETGE” and “DLG” degrons, β-TRCP prefers a “DSGxxS” degron, and Cop1 targets a “[D/E][D/E]xxVP[D/E]” degron for binding (see89 for review). This approach has led to the successful identification of quite some substrates for E3 ligases. For example, through a genome-wide search for Fbw7 degron-containing proteins, mTOR is identified as a bona fide Fbw7 substrate through which Fbw7 negatively regulates mTOR signaling23. In addition to searching for known degrons, a bioinformatic search using Biocarta and KEGG databases from an RNA microarray analyses links Fbx8 to mTOR, and biochemical assays demonstrates that Fbx8 is an E3 ligase facilitating mTOR degradation24. Notably, the presence of a degron sequence is not necessarily indicate the candidate as a substrate, as the structure constrains around the degron, and whether degron recognition requires priming modifications also need to be taken into consideration.
(5). In-cell ubiquitination screen.
Ubiquitination assays are the most direct evidence to demonstrate whether a candidate protein exerts any E3 ligase or DUB function. For example, to identify E3 ligase(s) responsible for K63-linked Akt ubiquitination, a panel of selected E3 ubiquitin ligases are expressed in HEK293T cells in the presence of Akt1 and His-tagged ubiquitin. Using in-cell ubiquitination assays, the authors narrowed down a handful of candidates as E3(s), among which TRAF6 is further characterized as the major E3 ligase ubiquitinating and activating Akt71. Similarly, in-cell deubiquititination assays are used to identify DUBs removing K63-linked ubiquitin chains on Akt, and CYLD is found to be a major hit90. The caveats for in-cell ubiquitination assays at least include that this assay may not be able to distinguish direct or indirect effects of a given E3/DUB on ubiquitination status of a given substrate.
(6). Targeted binding screen.
Compared with proteomics approach, a selected list of E3 or DUB candidates are tested for their ability to bind a substrate. For example, using a bioinformatic motif search approach, GβL is demonstrated as a TRAF2 substrate66. In a search for DUBs antagonizing this process, all OUT family of DUBs are co-expressed with GβL and OTUD7B is identified as a major GβL binding partner and the DUB deubiquitinating GβL66. Similarly, the same panel of DUBs are ectopically expressed in HCT116 cells to examine their effects on endogenous PTEN abundance and OTUD3 is found as a major DUB negatively regulating PTEN protein stability91. In addition, through co-expression of 30 DUB ORFs and PTEN in HEK293T cells, USP13 is identified as a DUB stabilizing PTEN through deubiquitinating PTEN92. Again, this is a correlative approach that does not address a causal relationship between E3s/DUBs and substrates, thus further biochemical analyses are needed for in-depth characterization.
(7). Biochemical Arrays.
The E3/substrate pair can also be established by biochemical arrays. For example, to understand the oncogenic property of MKNR1, lysates from MKNR1-depleted HeLa cells were analyzed by a phospho-kinase array, where MKNR1 is found to target phospho-Akt for ubiquitination and degradation18. This is a more targeted approach and usually requires some hints for potential hits.
(8). Biochemical enzyme purification and LC-MS/MS identification.
This is a mixed approach to partially purify the experimental materials to facilitate mass spectrometry detection. For examine, to identify E3 ligases for p110, MEFs were fractionated by a S100 ion exchange column and each fraction was used in in vitro p110 ubiquitination assays to determine the major fractions containing the E3 ligase activity. After further purification steps the resulting proteins were analyzed by mass spectrometry for protein identifications. As a result, NEDD4L was identified as the major E3 ligase in the active fractions promoting p110 ubiquitination and degradation10.
(9). Hypothesis-driven research.
In some cases, previous literature may suggest some connections between a E3/substrate pair. For example, in a study to elucidate how mitochondrial status affects mTORC1 activity, the authors found that mTORC1 protects cells from death under mitochondria stress. Given that Parkin has a similar function with mTORC1 in this regard, the authors hypothesized and investigated whether Parkin is involved in mTORC1 regulation. As a result, Parkin indeed interacts with and ubiquitinates mTOR to maintain mTORC1 activity under mitochondria stress conditions70.
In addition to the above mentioned methods, the identification of a di-glycine signature for ubiquitin linkages enables the development of a GPS (global protein stability) profiling system based on a fluorescence-mediated genetic screen in combination with a QUINT (quantitative ubiquitination interrogation) proteomic approach created by the Elledge lab93. This non-biased system has been widely used for genome-wide measurement of protein stability that can identify either substrates for a given E3 ligase/DUB, or possible E3(s)/DUB(s) for a given substrate. In addition, multiple commercially available assay kits and services are provided to facilitate research towards these directions.
8. Concluding remarks
In addition to well-characterized phosphorylation-mediated mTOR regulatory mechanisms, mTOR signaling components also undergo modifications by mono-ubiquitination (such as PDK122) or poly-ubiquitination in a variety of linkages (such as K29-linked ubiquitination and degradation of p110 by NEDD4L, K48-linked ubiquitination and degradation of Akt by TTC3, K63-linked ubiquitination and activation of Akt, and multiple-linkage ubiquitination and degradation of ULK1 by NEDD4L, see Table 1 for references) that determine distinct protein fates with different effects on protein functions. In addition to the ubiquitination modification on lysine residues, recently mTOR-Lys1218 malonylation has also been reported to negatively regulate mTORC1 activity94 with unknown mechanisms, which may be due to reduced K63-linked mTOR ubiquitination.
Moreover, it is common that one E3 ligase regulates multiple substrates in the mTOR signaling. For example, CHIP promotes degradation of multiple mTOR signaling components including p8514, Akt33 and SGK150 through K48-linked poly-ubiquitination, while CHIP also facilitates K63-linked poly-ubiquitination of PKCζ for its kinase activation95. TRAF6 usually promotes K63-linked poly-ubiquitination to facilitate activation of p85, mTOR, Akt, Rac1 and ULK1 (see Table 1 for references). Other TRAF family of E3 ligases such as TRAF2 and TRAF4 have been shown to aid K63-linked ubiquitination of GβL66 and Akt74, respectively. The E3 ligase SCFβ-TRCP targets multiple proteins regulating mTOR signaling for ubiquitination and degradation including Deptor, PHLPP1, PDCD4, FNIP2 and Lipin1 (see Table 1 for references). Thus, β-TRCP functions as a hub that is utilized for the regulation of multiple aspects of the mTOR signaling following pro-growth stimuli, coordinating increased protein synthesis with pro-survival signaling. Specifically, (1) upon mitogenic stimulation, the tumor suppressor PDCD4, which inhibits the translation initiation factor eIF4A, is degraded in a β-TRCP- and S6K1 phosphorylation-dependent manner, therefore allowing efficient protein translation and cell growth96; (2) mTOR cooperates with CK1α and β-TRCP to induce the degradation of the mTOR inhibitor, DEPTOR, generating an auto-amplification loop that promotes the further activation of mTOR97; (3) alternatively, β-TRCP promotes mTOR activation through degrading FNIP2 (component of the FNIP1/FNIP2/FLCN tumor suppressive complex upstream of mTORC1) in a CKI and nutrient sensitive manner61; (4) β-TRCP regulates cell survival in cooperation with the ERK-RSK pathway by targeting BimEL for degradation98; and (5) β-TRCP indirectly promotes Akt activation by targeting PHLPP1, a phosphatase dephosphorylating and inactivating Akt, for ubiquitination and degradation in a CKI and GSK3-dependent manner43. On the other hand, NEDD4L promotes a unique K29-linked polyubiquitination of p110 to mediate p110 protein degradation, while NEDD4L produces K48-linked ubiquitin chains on ULK1 for ULK1 destruction (see Table 1 for references). Similarly, NEDD4 facilitates both K48-linked ubiquitination of SGK1 for SGK1 destruction49 and K63-linked ubiquitination of phospho-Akt to facilitate its nuclear transportation and activation72.
Furthermore, one mTOR signaling component can be targeted by different E3 ligases or DUBs for regulation in a cancer type- or cellular context-dependent manner. For example, PTEN deubiquitination is regulated by USP10 in lung cancer99 and hepatocellular carcinoma100, while the same process is regulated by USP1392 or OTUD391 in breast cancer. It remains to be determined whether cellular context-dependent upstream signaling events may determine the function of differentially identified DUBs in modulating the function of the same protein.
In summary, ubiquitin modifications play important roles in regulating the mTOR signaling by either degrading the protein targets to terminate the mTOR signaling, modulating protein cellular localization for enzyme activation/inactivation, triggering protein binding to other partners and through other unknown mechanisms. Post-translational modifications of the “degron” add an additional layer of regulation to the ubiquitination process and this might in part explain why in some cases, no inverse correlation/mutually exclusivity is observed for E3 ligases and substrates in cancer. Interestingly, it seems that ubiquitin-mediated mTOR regulation is a late evolutionary event as a gained modification and function for higher eukaryotes, which may facilitate the adaption of mammals to a more complexed and sophisticated biological system. Compared with well-studied phosphorylation-mediated regulations of the mTOR signaling, there are much to be investigated for roles of ubiquitin on many mTOR signaling components. Identification of the E3 ligases, DUBs and function of the unknown ubiquitination events in mTOR signaling will greatly advance mTOR biology and shed new lights into clinic therapeutics targeting E3 ligases or DUBs for human disease treatments by modulating mTOR activity.
Supplementary Material
Acknowledgments
The authors sincerely apologize to all those colleagues whose important work was not cited in this paper owing to space limitations, especially many prominent and pioneer work in the mTOR filed. We thank other Liu members for critical reading of the manuscript and helpful discussions. This work was supported by the NIH grants (P.L. R00CA181342), a V Foundation Research Scholar Grant (P.L. V2018–009), a UNC IBM Junior Faculty Development Award (P.L) and the UNC University Cancer Research Fund (P.L.).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Liu P, Gan W, Su S, Hauenstein AV, Fu TM, Brasher B, Schwerdtfeger C, Liang AC, Xu M & Wei W K63-linked polyubiquitin chains bind to DNA to facilitate DNA damage repair Sci Signal 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M & Harper JW Systematic analysis and nomenclature of mammalian F-box proteins Genes Dev 18, 2573–2580, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skaar JR, Pagan JK & Pagano M Mechanisms and function of substrate recruitment by F-box proteins Nat Rev Mol Cell Biol 14, 369–381, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardozo T & Pagano M The SCF ubiquitin ligase: insights into a molecular machine Nat Rev Mol Cell Biol 5, 739–751, (2004). [DOI] [PubMed] [Google Scholar]
- 5.Davis RJ, Welcker M & Clurman BE Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities Cancer Cell 26, 455–464, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng S, Zhang W, Zheng Y, Liberman Z, Rhodes CJ, Eldar-Finkelman H & Sun XJ Glycogen synthase kinase 3 beta mediates high glucose-induced ubiquitination and proteasome degradation of insulin receptor substrate 1 J Endocrinol 206, 171–181, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rui L, Yuan M, Frantz D, Shoelson S & White MF SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2 J Biol Chem 277, 42394–42398, (2002). [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Sarikas A, Dias-Santagata DC, Dolios G, Lafontant PJ, Tsai SC, Zhu W, Nakajima H, Nakajima HO, Field LJ, Wang R & Pan ZQ The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation Mol Cell 30, 403–414, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J, Luo L, Eash J, Ibebunjo C & Glass DJ The SCF-Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling Dev Cell 21, 835–847, (2011). [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Dang T, Liu T, Chen S, Li L, Huang S & Fang M NEDD4L Protein Catalyzes Ubiquitination of PIK3CA Protein and Regulates PI3K-AKT Signaling J Biol Chem 291, 17467–17477, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L, Washburn MP & Pagano M FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade Nat Cell Biol 15, 472–480, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, Florens L, Washburn MP, Collazo-Lorduy A, Castillo-Martin M, Cordon-Cardo C, Sebti SM, Pinton P & Pagano M PTEN counteracts FBXL2 to promote IP3R3- and Ca(2+)-mediated apoptosis limiting tumour growth Nature 546, 554–558, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuchay S, Saeed M, Giorgi C, Li J, Hoffmann HH, Pinton P, Rice CM & Pagano M NS5A Promotes Constitutive Degradation of IP3R3 to Counteract Apoptosis Induced by Hepatitis C Virus Cell Rep 25, 833–840 e833, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko HR, Kim CK, Lee SB, Song J, Lee KH, Kim KK, Park KW, Cho SW & Ahn JY P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP Cell Death Dis 5, e1131, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN & Chen J WWP2 is an E3 ubiquitin ligase for PTEN Nat Cell Biol 13, 728–733, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Themsche C, Leblanc V, Parent S & Asselin E X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization J Biol Chem 284, 20462–20466, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Zhang W, Jiang K, Chen B, Wang K, Lao L, Hou C, Wang F, Zhang C & Shen H MicroRNA-300 Regulates the Ubiquitination of PTEN through the CRL4B(DCAF13) E3 Ligase in Osteosarcoma Cells Mol Ther Nucleic Acids 10, 254–268, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MS, Jeong MH, Lee HW, Han HJ, Ko A, Hewitt SM, Kim JH, Chun KH, Chung JY, Lee C, Cho H & Song J PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis Nat Commun 6, 7769, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Wu H, Li S, Xu Z, Li X, Yang Y, Li B, Li Y, Guo J & Chen H SYNJ2BP promotes the degradation of PTEN through the lysosome-pathway and enhances breast tumor metastasis via PI3K/AKT/SNAI1 signaling Oncotarget 8, 89692–89706, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruschmann J, Ho V, Antignano F, Kuroda E, Lam V, Ibaraki M, Snyder K, Kim C, Flavell RA, Kawakami T, Sly L, Turhan AG & Krystal G Tyrosine phosphorylation of SHIP promotes its proteasomal degradation Exp Hematol 38, 392–402, 402 e391, (2010). [DOI] [PubMed] [Google Scholar]
- 21.Sattler M, Salgia R, Shrikhande G, Verma S, Choi JL, Rohrschneider LR & Griffin JD The phosphatidylinositol polyphosphate 5-phosphatase SHIP and the protein tyrosine phosphatase SHP-2 form a complex in hematopoietic cells which can be regulated by BCR/ABL and growth factors Oncogene 15, 2379–2384, (1997). [DOI] [PubMed] [Google Scholar]
- 22.Uras IZ, List T & Nijman SM Ubiquitin-specific protease 4 inhibits mono-ubiquitination of the master growth factor signaling kinase PDK1 PLoS One 7, e31003, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R & Balmain A FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression Science 321, 1499–1502, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang FF, Zhang XJ, Yan YR, Zhu XH, Yu J, Ding Y, Hu JL, Zhou WJ, Zeng ZC, Liao WT, Ding YQ & Liang L FBX8 is a metastasis suppressor downstream of miR-223 and targeting mTOR for degradation in colorectal carcinoma Cancer Lett 388, 85–95, (2017). [DOI] [PubMed] [Google Scholar]
- 25.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS & Sabatini DM DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival Cell 137, 873–886, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan M, Xu J, Siddiqui J, Feng F & Sun Y Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis Mol Cancer 15, 81, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo J, Wu X, Mao Z, Khuri FR & Sun SY Rictor Undergoes Glycogen Synthase Kinase 3 (GSK3)-dependent, FBXW7-mediated Ubiquitination and Proteasomal Degradation J Biol Chem 290, 14120–14129, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suizu F, Hiramuki Y, Okumura F, Matsuda M, Okumura AJ, Hirata N, Narita M, Kohno T, Yokota J, Bohgaki M, Obuse C, Hatakeyama S, Obata T & Noguchi M The E3 ligase TTC3 facilitates ubiquitination and degradation of phosphorylated Akt Dev Cell 17, 800–810, (2009). [DOI] [PubMed] [Google Scholar]
- 29.Bae S, Kim SY, Jung JH, Yoon Y, Cha HJ, Lee H, Kim K, Kim J, An IS, Kim J, Um HD, Park IC, Lee SJ, Nam SY, Jin YW, Lee JH & An S Akt is negatively regulated by the MULAN E3 ligase Cell Res 22, 873–885, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SY, Kim HJ, Kang SU, Kim YE, Park JK, Shin YS, Kim YS, Lee K & Kim CH Non-thermal plasma induces AKT degradation through turn-on the MUL1 E3 ligase in head and neck cancer Oncotarget 6, 33382–33396, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joo HM, Kim JY, Jeong JB, Seong KM, Nam SY, Yang KH, Kim CS, Kim HS, Jeong M, An S & Jin YW Ret finger protein 2 enhances ionizing radiation-induced apoptosis via degradation of AKT and MDM2 Eur J Cell Biol 90, 420–431, (2011). [DOI] [PubMed] [Google Scholar]
- 32.Xiang T, Ohashi A, Huang Y, Pandita TK, Ludwig T, Powell SN & Yang Q Negative Regulation of AKT Activation by BRCA1 Cancer Res 68, 10040–10044, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su CH, Wang CY, Lan KH, Li CP, Chao Y, Lin HC, Lee SD & Lee WP Akt phosphorylation at Thr308 and Ser473 is required for CHIP-mediated ubiquitination of the kinase Cell Signal 23, 1824–1830, (2011). [DOI] [PubMed] [Google Scholar]
- 34.Cheng L, Zang J, Dai HJ, Li F & Guo F Ubiquitin ligase CHIP functions as an oncogene and activates the AKT signaling pathway in prostate cancer Int J Oncol 53, 203–214, (2018). [DOI] [PubMed] [Google Scholar]
- 35.Dickey CA, Koren J, Zhang YJ, Xu YF, Jinwal UK, Birnbaum MJ, Monks B, Sun M, Cheng JQ, Patterson C, Bailey RM, Dunmore J, Soresh S, Leon C, Morgan D & Petrucelli L Akt and CHIP coregulate tau degradation through coordinated interactions Proc Natl Acad Sci U S A 105, 3622–3627, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong JJ, Li S, Lim EK, Wang Y, Wang C, Zhang H, Kirilly D, Wu C, Liou YC, Wang H & Yu F A Cullin1-based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning PLoS Biol 11, e1001657, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakatsuki S, Saitoh F & Araki T ZNRF1 promotes Wallerian degeneration by degrading AKT to induce GSK3B-dependent CRMP2 phosphorylation Nat Cell Biol 13, 1415–1423, (2011). [DOI] [PubMed] [Google Scholar]
- 38.Liu P, Begley M, Michowski W, Inuzuka H, Ginzberg M, Gao D, Tsou P, Gan W, Papa A, Kim BM, Wan L, Singh A, Zhai B, Yuan M, Wang Z, Gygi SP, Lee TH, Lu KP, Toker A, Pandolfi PP, Asara JM, Kirschner MW, Sicinski P, Cantley L & Wei W Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus Nature 508, 541–545, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan F, Ding Y, Zhang Y, Zhou Y, Li M & Wang C Curcumin Suppresses Proliferation and Migration of MDA-MB-231 Breast Cancer Cells through Autophagy-Dependent Akt Degradation PLoS One 11, e0146553, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Bisson JA, Mills B, Paul Helt JC, Zwaka TP & Cohen ED Wnt5a and Wnt11 inhibit the canonical Wnt pathway and promote cardiac progenitor development via the Caspase-dependent degradation of AKT Dev Biol 398, 80–96, (2015). [DOI] [PubMed] [Google Scholar]
- 41.Bachelder RE, Wendt MA, Fujita N, Tsuruo T & Mercurio AM The cleavage of Akt/protein kinase B by death receptor signaling is an important event in detachment-induced apoptosis J Biol Chem 276, 34702–34707, (2001). [DOI] [PubMed] [Google Scholar]
- 42.Chu J, Lauretti E & Pratico D Caspase-3-dependent cleavage of Akt modulates tau phosphorylation via GSK3beta kinase: implications for Alzheimer’s disease Mol Psychiatry 22, 1002–1008, (2017). [DOI] [PubMed] [Google Scholar]
- 43.Li X, Liu J & Gao T beta-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt Mol Cell Biol 29, 6192–6205, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu C, Ji SY, Sha QQ, Sun QY & Fan HY CRL4-DCAF1 ubiquitin E3 ligase directs protein phosphatase 2A degradation to control oocyte meiotic maturation Nat Commun 6, 8017, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oberg EA, Nifoussi SK, Gingras AC & Strack S Selective proteasomal degradation of the B’beta subunit of protein phosphatase 2A by the E3 ubiquitin ligase adaptor Kelch-like 15 J Biol Chem 287, 43378–43389, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmeister M, Prelle C, Kuchler P, Kovacevic I, Moser M, Muller-Esterl W & Oess S The ubiquitin E3 ligase NOSIP modulates protein phosphatase 2A activity in craniofacial development PLoS One 9, e116150, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, Ropers HH, Schneider R & Schweiger S MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation Nat Genet 29, 287–294, (2001). [DOI] [PubMed] [Google Scholar]
- 48.Schoenebeck B, Bader V, Zhu XR, Schmitz B, Lubbert H & Stichel CC Sgk1, a cell survival response in neurodegenerative diseases Mol Cell Neurosci 30, 249–264, (2005). [DOI] [PubMed] [Google Scholar]
- 49.Zhou R & Snyder PM Nedd4–2 phosphorylation induces serum and glucocorticoid-regulated kinase (SGK) ubiquitination and degradation J Biol Chem 280, 4518–4523, (2005). [DOI] [PubMed] [Google Scholar]
- 50.Belova L, Sharma S, Brickley DR, Nicolarsen JR, Patterson C & Conzen SD Ubiquitin-proteasome degradation of serum- and glucocorticoid-regulated kinase-1 (SGK-1) is mediated by the chaperone-dependent E3 ligase CHIP Biochem J 400, 235–244, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao D, Wan L, Inuzuka H, Berg AH, Tseng A, Zhai B, Shaik S, Bennett E, Tron AE, Gasser JA, Lau A, Gygi SP, Harper JW, DeCaprio JA, Toker A & Wei W Rictor forms a complex with Cullin-1 to promote SGK1 ubiquitination and destruction Mol Cell 39, 797–808, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brand C, Horovitz-Fried M, Inbar A, Tamar Brutman B, Brodie C & Sampson SR Insulin stimulation of PKCdelta triggers its rapid degradation via the ubiquitin-proteasome pathway Biochim Biophys Acta 1803, 1265–1275, (2010). [DOI] [PubMed] [Google Scholar]
- 53.Iturrioz X & Parker PJ PKCzetaII is a target for degradation through the tumour suppressor protein pVHL FEBS Lett 581, 1397–1402, (2007). [DOI] [PubMed] [Google Scholar]
- 54.Han S, Witt RM, Santos TM, Polizzano C, Sabatini BL & Ramesh V Pam (Protein associated with Myc) functions as an E3 ubiquitin ligase and regulates TSC/mTOR signaling Cell Signal 20, 1084–1091, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chano T, Saji M, Inoue H, Minami K, Kobayashi T, Hino O & Okabe H Neuromuscular abundance of RB1CC1 contributes to the non-proliferating enlarged cell phenotype through both RB1 maintenance and TSC1 degradation Int J Mol Med 18, 425–432, (2006). [PubMed] [Google Scholar]
- 56.Guo P, Ma X, Zhao W, Huai W, Li T, Qiu Y, Zhang Y & Han L TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex Oncogene 37, 478–488, (2018). [DOI] [PubMed] [Google Scholar]
- 57.Seo G, Kim SK, Byun YJ, Oh E, Jeong SW, Chae GT & Lee SB Hydrogen peroxide induces Beclin 1-independent autophagic cell death by suppressing the mTOR pathway via promoting the ubiquitination and degradation of Rheb in GSH-depleted RAW 264.7 cells Free Radic Res 45, 389–399, (2011). [DOI] [PubMed] [Google Scholar]
- 58.Harraz MM, Tyagi R, Cortes P & Snyder SH Antidepressant action of ketamine via mTOR is mediated by inhibition of nitrergic Rheb degradation Mol Psychiatry 21, 313–319, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Araujo ME, Stasyk T, Taub N, Ebner HL, Furst B, Filipek P, Weys SR, Hess MW, Lindner H, Kremser L & Huber LA Stability of the endosomal scaffold protein LAMTOR3 depends on heterodimer assembly and proteasomal degradation J Biol Chem 288, 18228–18242, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E & Sabatini DM The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1 Mol Cell 52, 495–505, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagashima K, Fukushima H, Shimizu K, Yamada A, Hidaka M, Hasumi H, Ikebe T, Fukumoto S, Okabe K & Inuzuka H Nutrient-induced FNIP degradation by SCFbeta-TRCP regulates FLCN complex localization and promotes renal cancer progression Oncotarget 8, 9947–9960, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M & Sabatini DM A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1 Science 340, 1100–1106, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J, Ou Y, Yang Y, Li W, Xu Y, Xie Y & Liu Y KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing Nature 557, 585–589, (2018). [DOI] [PubMed] [Google Scholar]
- 64.Shimizu K, Fukushima H, Ogura K, Lien EC, Nihira NT, Zhang J, North BJ, Guo A, Nagashima K, Nakagawa T, Hoshikawa S, Watahiki A, Okabe K, Yamada A, Toker A, Asara JM, Fukumoto S, Nakayama KI, Nakayama K, Inuzuka H & Wei W The SCFbeta-TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis Sci Signal 10, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamidi A, Song J, Thakur N, Itoh S, Marcusson A, Bergh A, Heldin CH & Landstrom M TGF-beta promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85alpha Sci Signal 10, (2017). [DOI] [PubMed] [Google Scholar]
- 66.Wang B, Jie Z, Joo D, Ordureau A, Liu P, Gan W, Guo J, Zhang J, North BJ, Dai X, Cheng X, Bian X, Zhang L, Harper JW, Sun SC & Wei W TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling Nature 545, 365–369, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J & Diaz-Meco MT K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells Mol Cell 51, 283–296, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghosh P, Wu M, Zhang H & Sun H mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase Cell Cycle 7, 373–381, (2008). [DOI] [PubMed] [Google Scholar]
- 69.Hussain S, Feldman AL, Das C, Ziesmer SC, Ansell SM & Galardy PJ Ubiquitin hydrolase UCH-L1 destabilizes mTOR complex 1 by antagonizing DDB1-CUL4-mediated ubiquitination of raptor Mol Cell Biol 33, 1188–1197, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park D, Lee MN, Jeong H, Koh A, Yang YR, Suh PG & Ryu SH Parkin ubiquitinates mTOR to regulate mTORC1 activity under mitochondrial stress Cell Signal 26, 2122–2130, (2014). [DOI] [PubMed] [Google Scholar]
- 71.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG & Lin HK The E3 ligase TRAF6 regulates Akt ubiquitination and activation Science 325, 1134–1138, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fan CD, Lum MA, Xu C, Black JD & Wang X Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4–1, in the insulin-like growth factor-1 response J Biol Chem 288, 1674–1684, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI & Lin HK The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis Cell 149, 1098–1111, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li W, Peng C, Lee MH, Lim D, Zhu F, Fu Y, Yang G, Sheng Y, Xiao L, Dong X, Ma W, Bode AM, Cao Y & Dong Z TRAF4 is a critical molecule for Akt activation in lung cancer Cancer Res 73, 6938–6950, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Yang Z, Ou J, Xia X, Zhi F & Cui J The F-box protein FBXL18 promotes glioma progression by promoting K63-linked ubiquitination of Akt FEBS Lett 591, 145–154, (2017). [DOI] [PubMed] [Google Scholar]
- 76.Deng L, Jiang C, Chen L, Jin J, Wei J, Zhao L, Chen M, Pan W, Xu Y, Chu H, Wang X, Ge X, Li D, Liao L, Liu M, Li L & Wang P The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation Mol Cell 58, 804–818, (2015). [DOI] [PubMed] [Google Scholar]
- 77.Jin G, Lee SW, Zhang X, Cai Z, Gao Y, Chou PC, Rezaeian AH, Han F, Wang CY, Yao JC, Gong Z, Chan CH, Huang CY, Tsai FJ, Tsai CH, Tu SH, Wu CH, Sarbassov dos D, Ho YS & Lin HK Skp2-Mediated RagA Ubiquitination Elicits a Negative Feedback to Prevent Amino-Acid-Dependent mTORC1 Hyperactivation by Recruiting GATOR1 Mol Cell 58, 989–1000, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li T, Qin JJ, Yang X, Ji YX, Guo F, Cheng WL, Wu X, Gong FH, Hong Y, Zhu XY, Gong J, Wang Z, Huang Z, She ZG & Li H The Ubiquitin E3 Ligase TRAF6 Exacerbates Ischemic Stroke by Ubiquitinating and Activating Rac1 J Neurosci 37, 12123–12140, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshihara H, Fukushima T, Hakuno F, Saeki Y, Tanaka K, Ito A, Yoshida M, Iemura S, Natsume T, Asano T, Chida K, Girnita L & Takahashi S Insulin/insulin-like growth factor (IGF) stimulation abrogates an association between a deubiquitinating enzyme USP7 and insulin receptor substrates (IRSs) followed by proteasomal degradation of IRSs Biochem Biophys Res Commun 423, 122–127, (2012). [DOI] [PubMed] [Google Scholar]
- 80.Wu Y, Zhou H, Wu K, Lee S, Li R & Liu X PTEN phosphorylation and nuclear export mediate free fatty acid-induced oxidative stress Antioxid Redox Signal 20, 1382–1395, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao L, Wang X, Yu Y, Deng L, Chen L, Peng X, Jiao C, Gao G, Tan X, Pan W, Ge X & Wang P OTUB1 protein suppresses mTOR complex 1 (mTORC1) activity by deubiquitinating the mTORC1 inhibitor DEPTOR J Biol Chem 293, 4883–4892, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bridges CR, Tan MC, Premarathne S, Nanayakkara D, Bellette B, Zencak D, Domingo D, Gecz J, Murtaza M, Jolly LA & Wood SA USP9X deubiquitylating enzyme maintains RAPTOR protein levels, mTORC1 signalling and proliferation in neural progenitors Sci Rep 7, 391, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim JH, Seo D, Kim SJ, Choi DW, Park JS, Ha J, Choi J, Lee JH, Jung SM, Seo KW, Lee EW, Lee YS, Cheong H, Choi CY & Park SH The deubiquitinating enzyme USP20 stabilizes ULK1 and promotes autophagy initiation EMBO Rep 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang WL, Jin G, Li CF, Jeong YS, Moten A, Xu D, Feng Z, Chen W, Cai Z, Darnay B, Gu W & Lin HK Cycles of ubiquitination and deubiquitination critically regulate growth factor-mediated activation of Akt signaling Sci Signal 6, ra3, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Agrawal P, Chen YT, Schilling B, Gibson BW & Hughes RE Ubiquitin-specific peptidase 9, X-linked (USP9X) modulates activity of mammalian target of rapamycin (mTOR) J Biol Chem 287, 21164–21175, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tatebe H & Shiozaki K Evolutionary Conservation of the Components in the TOR Signaling Pathways Biomolecules 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sacco JJ, Yau TY, Darling S, Patel V, Liu H, Urbe S, Clague MJ & Coulson JM The deubiquitylase Ataxin-3 restricts PTEN transcription in lung cancer cells Oncogene 33, 4265–4272, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ & Xiong Y WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase Genes Dev 22, 866–871, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meszaros B, Kumar M, Gibson TJ, Uyar B & Dosztanyi Z Degrons in cancer Sci Signal 10, (2017). [DOI] [PubMed] [Google Scholar]
- 90.Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, Matsuno T, Xu X, Huang Y, Zhang W, Park SH, Kim YI, Choi YD, Shen H, Heo KS, Xu H, Bourne P, Koga T, Xu H, Yan C, Wang B, Chen LF, Feng XH & Li JD CYLD negatively regulates transforming growth factor-beta-signalling via deubiquitinating Akt Nat Commun 3, 771, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, Xing G, Kong X, Wang L, Li Y, Zhou T, Gao D, Xiao ZX, Yin Y, Wei W, He F & Zhang L Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis Nat Cell Biol 17, 1169–1181, (2015). [DOI] [PubMed] [Google Scholar]
- 92.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, Wang M, Chen D, Sun Y, Hung MC, Chen J & Ma L Deubiquitylation and stabilization of PTEN by USP13 Nat Cell Biol 15, 1486–1494, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, Yen HC & Elledge SJ Global identification of modular cullin-RING ligase substrates Cell 147, 459–474, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bruning Ulrike, M.-R. F, Kalucka Joanna, Goveia Jermaine, Taverna Federico, Queiroz Karla C.S., Dubois Charlotte, Cantelmo Anna Rita, Chen Rongyuan, Loroch Stefan, Timmerman Evy, Caixeta Vanessa, Bloch Katarzyna, Conradi Lena-Christin, Treps Lucas, Staes An, Gevaert Kris, Tee Andrew, Dewerchin Mieke, Semenkovich Clay F., Impens Francis, Schilling Birgit, Verdin Eric, Swinnen Johannes V., Meier Jordan L., Kulkarni Rhushikesh A., Sickmann Albert, Ghesquière Bart, Schoonjans Luc, Li Xuri, Mazzone Massimiliano, Carmeliet Peter. Impairment of Angiogenesis by Fatty Acid Synthase Inhibition Involves mTOR Malonylation Cell Metabolism 28, 1–15, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang M, Wang C, Zhu X, Tang S, Shi L, Cao X & Chen T E3 ubiquitin ligase CHIP facilitates Toll-like receptor signaling by recruiting and polyubiquitinating Src and atypical PKC{zeta} J Exp Med 208, 2099–2112, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE & Pagano M S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth Science 314, 467–471, (2006). [DOI] [PubMed] [Google Scholar]
- 97.Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y & Pagano M mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR Mol Cell 44, 317–324, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton J & Pagano M betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis Mol Cell 33, 109–116, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu C, Ning Z, Wang A, Chen D, Liu X, Xia T, Tekcham DS, Wang W, Li T, Liu X, Liu J, Qi H, Luo H, Du J, Ma C, Yan Q, Liu J, Xu G, Piao HL & Tan G USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma Cancer Lett, (2018). [DOI] [PubMed] [Google Scholar]
- 100.Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H & Jiang X USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN Mol Cell Biochem 441, 1–7, (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.