Abstract
Prodrug activation, by exogenously administered enzymes, for cancer therapy is an approach to achieve better selectivity and less systemic toxicity than conventional chemotherapy. However, the short half-lives of the activating enzymes in the bloodstream has limited its success. Demonstrated here is that a tyrosinase-MOF nanoreactor activates the prodrug paracetamol in cancer cells in a long-lasting manner. By generating reactive oxygen species (ROS) and depleting glutathione (GSH), the product of the enzymatic conversion of paracetamol is toxic to drug-resistant cancer cells. Tyrosinase-MOF nanoreactors cause significant cell death in the presence of paracetamol for up to three days after being internalized by cells, while free enzymes totally lose activity in a few hours. Thus, enzyme-MOF nanocomposites are envisioned to be novel persistent platforms for various biomedical applications.
Chemotherapeutics typically interfere with mitosis and demonstrate maximum toxicity towards rapidly dividing cells.1 However, chemotherapy drugs are poorly selective and often result in severe adverse effects that include kidney damage, bone marrow suppression, nerve damage, hair loss, low blood count, and inflammation.2-4 In addition, resistance to chemotherapy occurs in certain cancer cell lines, thereby declining the efficiency of the treatment.5-7 One strategy to approach these problems is to employ prodrugs, compounds that are innocuous until metabolized to give cytotoxic products, in a tumor microenvironment.8-11 However, this approach involves a new challenge, the identification of activation mechanisms specific to cancer cells. To date, enzymes overexpressed in tumor cells have been used to activate prodrugs.10, 12 Nevertheless, because these targeting enzymes may also be present in normal cells, albeit at lower levels, many of these prodrug therapies retained poor selectivity and demonstrated minimal success.13, 14 To circumvent this problem, an alternative strategy involves delivering exogenous “activating” enzymes into cancer cells. Tumor-specific accumulation of the enzyme activator is achieved by conjugation to tumor-directing antibodies or immunoliposomes.15-17 While potentially useful, the biggest drawback is that externally administered enzymes are highly vulnerable to degradation in the bloodstream.18, 19 Thus the feasibility of this strategy remains unclear and strategies that can improve the biocompatibility of externally administered enzymes are needed.
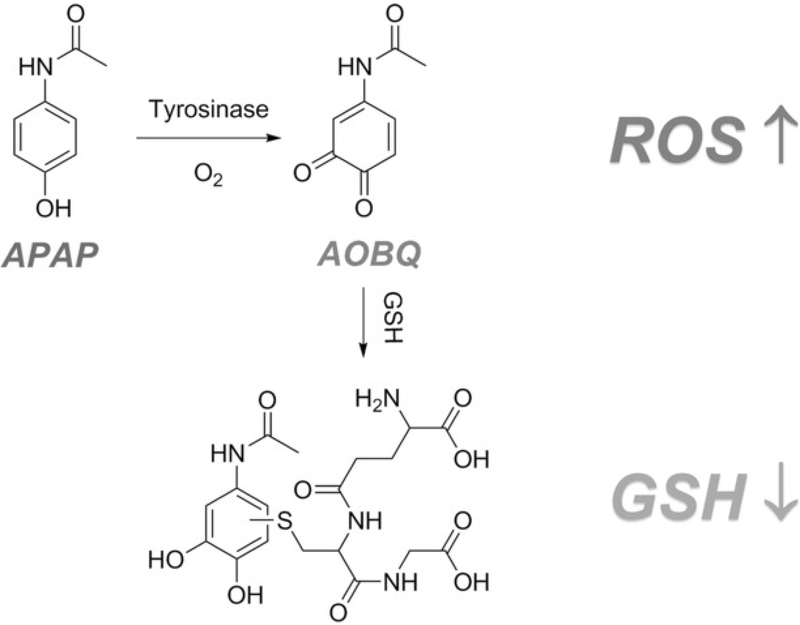
Herein we demonstrate that enzymatic nanoreactors based on metal-organic frameworks (MOFs) can be potent prodrug activators. MOFs are an emerging platform that demonstrates potential for a number of biotechnological applications, including sensing, imaging, drug delivery, and enzyme encapsulation.20-42 Encapsulated enzymes demonstrate well-preserved catalytic activities.21, 24, 25 Encapsulated enzymes also show enhanced stability under protein denaturation conditions, such as organic solvents, extreme pH environments, or high temperatures.20, 26, 43 Moreover, MOFs efficiently protect encapsulated enzymes from proteolytic degradation, presumably by preventing access of proteases to the protein substrate.27, 44 Notably, the porous nature of MOFs allows facile substrate diffusion and product release, a feature typically superior to other well-established carriers.45 In this work, we selected PCN-333(Al) as the enzyme carrier owing to its high enzyme encapsulation capacity, facile modification with fluorophores, and chemical robustness in a cellular environment.25 To achieve tumor-specific prodrug activation, tyrosinase (TYR), which is uniquely localized in melanocytes and melanoma cells, is chosen as the activating enzyme.46 Paracetamol (APAP), the active ingredient of Tylenol, is used as the nontoxic prodrug.47 Both in cellulo and in vivo experiments establish that APAP, along with an enzyme-MOF nanoreactor, induced significant cytotoxicity in drug-resistant cancer cells and led to tumor regression. Mechanistically, cytotoxicity arises from 4-acetamido-o-benzoquinone (AOBQ), the enzymatic conversion product of APAP, and from subsequent generation of reactive oxygen species (ROS) and glutathione (GSH) depletion (Scheme 1).
Scheme 1.
Nontoxic prodrug APAP oxidation is catalyzed by tyrosinase, thus yielding cytotoxic AOBQ, which induces ROS generation and GSH depletion.
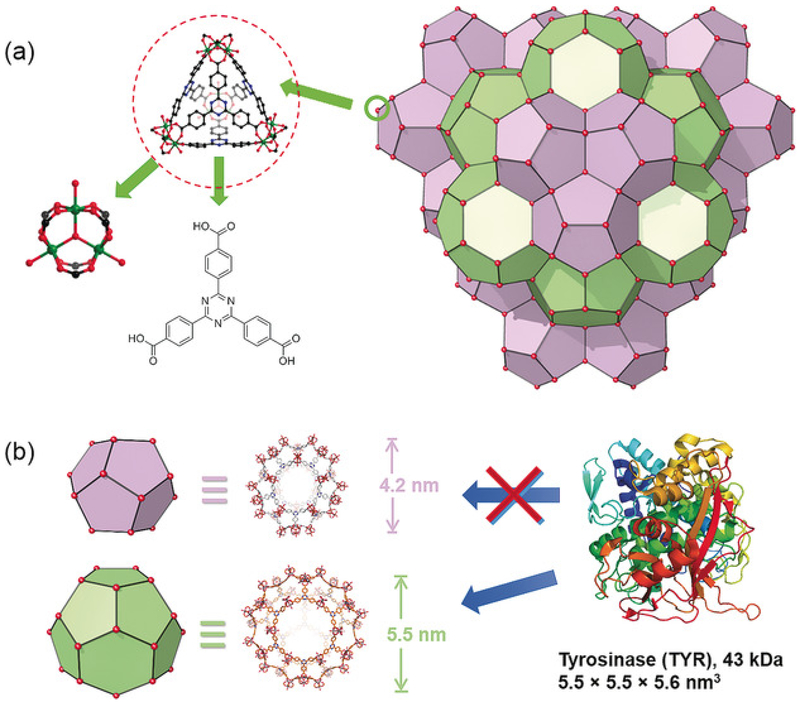
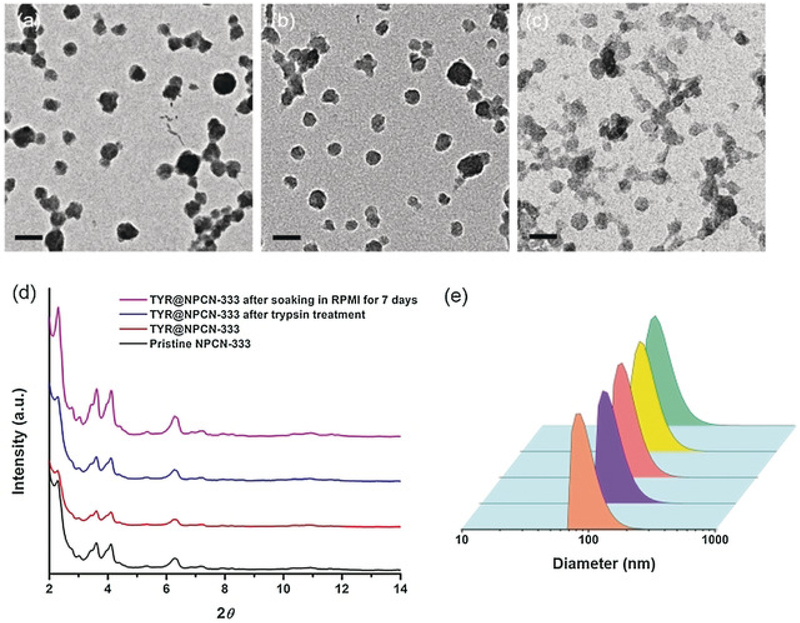
PCN-333 is composed of a trimeric-oxo cluster and a planar triangular ligand, 4,4′,4″-s-triazine-2,4,6-triyl-tribenzoic acid (TATB), which self-assembles into a supertetrahedron (STH) with metal clusters on the corner and TATB on the face (Figure 1a). A 1.5 nm cavity is present in the STH. Two types of mesoporous cavities with diameters of 4.2 and 5.5 nm are composed of STHs through vertex sharing (Figure 1b). PCN-333 nanoparticles (NPCN-333) were prepared by solvothermal reaction of AlCl3 and TATB in DMF at 95 °C. The particle size determined by transmission electron microscopy (TEM) was around 100 nm (Figure 1a). A fluorescent version of NPCN-333 was prepared by a ligand metathesis reaction with a BTB ligand functionalized with a green fluorescent BODIPY fluorophore (see Figure S1 in the Supporting Information). The well-maintained crystallinity of NPCN-333 after enzyme loading and after soaking in the cell culture medium Roswell Park Memorial Institute 1640 (RPMI 1640), for up to 7 days, was confirmed by the TEM images (Figure 1b,c) and by the nearly unchanged PXRD pattern (Figure 1d). The monodispersity of NPCN-333 immersed in RPMI 1640 over an extended period of time was verified by the particle-size distribution diagram derived from dynamic light scattering (DLS) data collected on day 0 and day 7 (Figure 1e). One molecule of TYR contains a single 43 kDa polypeptide with dimensions of 5.5×5.5×5.6 nm3.48 Therefore TYR may be accommodated in the 5.5 nm cavity, but presumably not in the 1.5 and 4.2 nm cavity. After incubating TYR with a NPCN-333 slurry for 20 minutes, the enzyme loading capacity was estimated to be 0.80 gg-1 according to the BCA method, which is in consistent with the theoretical value (1.08 gg-1). The distribution of TYR in the material was studied by an N2 isotherm at 77 K. The pore-size distribution diagram of TYR encapsulated within NPCN-333 (TYR@NPCN-333) clearly showed the presence of a microporous cavity around 1.5 nm and a mesoporous cavity at around 4 nm. Notably, the 5 nm cavity disappeared, and is consistent with the cavity being occupied by TYR (see Figure S5). Thermal gravimetric analysis (TGA) also indicated the high loading of TYR in NPCN-333 (see Figure S6).
Figure 1.
Structure of PCN-333. a) NPCN-333 is composed of aluminum trimeric cluster and TATB ligand, which self-assembles into supertetrahedrons as the secondary building block of NPCN-333. b) Two types of mesoporous cavities in NPCN-333: 4.2 nm dodecahedral cage (light purple) and 5.5 nm hexacaidecahedral cage (light green). Based on the size of tyrosinase, it can only be accommodated in the 5.5 nm cage.
TYR catalyzes the oxidation of APAP to an o-quinone compound, 4-acetamido-o-benzoquinone (AOBQ), which absorbs at 450 nm. The enzymatic activities of free TYR and the TYR@NPCN-333 nanoreactor in 50 mm Tris-HCl buffer (pH 6.8) at ambient temperature was monitored by UV-vis spectrometry. The TYR@NPCN-333 nanoreactor followed Michaelis-Menten kinetics and showed a kcat value of 0.377 s-1 and a Km value of 1.119 mm(see Figure S7). In comparison, the free enzyme displayed a kcat of 0.405 s-1 and a Km of 1.338 mm (see Figure S8). The catalytic efficiencies, kcat/Km, were 0.302 s-1 mm-1 for free TYR versus 0.337 s-1 mm-1 for TYR@NPCN-333.
Resistance of the enzyme-MOF nanoreactor towards degradation is critical for practical applications in living biosystems. To elucidate whether NPCN-333 could protect encapsulated TYR, free TYR and TYR@NPCN-333 were incubated with trypsin solution for 2 hours at 37 °C. Then, the enzyme was released by dissolving TYR@NPCN-333 with HCl and the supernatant was subjected to SDS-PAGE analysis. As shown in Figure S9, free TYR was readily digested by trypsin as illustrated by the presence of small-molecular-weight bands. In contrast, bands corresponding to intact TYR were predominantly present for TYR@NPCN-333. Additionally, the enzymatic activity of TYR@NPCN-333 was almost unchanged after trypsin treatment (see Figure S10). Moreover, to study whether MOFs protect encapsulated enzymes in an acidic medium, free TYR and TYR@NPCN-333 were incubated in pH 5 buffer, and enzymatic activities were measured at 2 hours and 12 hours. While a 2 hour incubation only resulted in a 15% activity loss on free enzyme, 75% of the original activity was lost after a 12 hour incubation. In contrast, TYR@NPCN-333 maintained over 75 % activity after the 12 hour incubation at pH 5 (see Figure S11).
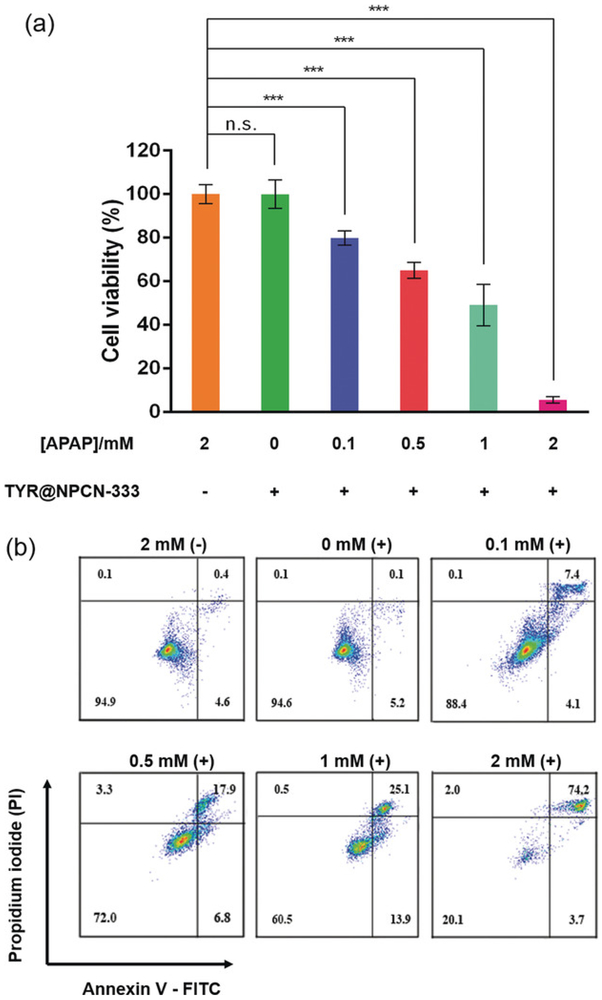
In vitro results indicated that TYR@NPCN-333 is enzymatically active and relatively resistant to perturbations. Cell-based experiments were therefore conducted to examine whether TYR@NPCN-333 could activate APAP in cancer cells. To test the performance of TYR@NPCN-333, we chose SKOV3-TR, an ovarian adenocarcinoma cell that is resistant to cisplatin, tumor necrosis factor, diphtheria toxin, adriamycin, and taxane.49-51 First, the cellular internalization of the enzyme-MOF nanoreactor was established using confocal microscopy. A fluorescent particle, TYR@FNPCN-333, was used to monitor cellular trafficking. After incubation with cells, TYR@FNPCN-333 displayed an intracellular punctate distribution around the nucleic area that colocalizes with LysoTracker, a marker for late endosomes and lysosomes (see Figure S12).52 This data indicates that the nanoreactor accumulated within endocytic organelles. Next, the cytotoxicity induced by TYR@NPCN-333 was quantified by the MTT assay. The prodrug, or the enzyme-MOF nanoreactor, did not display any noticeable cytotoxicity (Figure 3a). In contrast, cells that were pretreated with TYR@NPCN-333 for 6 hours, and then subsequently exposed to increasing concentrations of APAP (0.1 to 2 mm) displayed a correlated decrease in viability. Besides, similar cytotoxicity was observed in other cancer cell lines, including non-small lung carcinoma cell H1299 and cervical carcinoma cell HeLa (see Figure S13 and S14). In complementary assays, the cytotoxicity of the TYR@NPCN-333-APAP combination was assessed by flow cytometry. After different treatments, cells were stained with annexin V-FITC, a fluorescent marker of surface-exposed phosphatidyl serine, and with propidium iodide (PI), a DNA stain that detects the permeabilization of the plasma membrane. As observed previously, neither APAP nor TYR@NPCN-333 caused an increase in annexin V or PI staining, thus indicating that these conditions are not toxic to cells (Figure 3b). However, cells pretreated with TYR@NPCN-333 and exposed to APAP displayed dual labeling, which is indicative of cell death.
Figure 3.
Cytotoxicity of TYR@NPCN-333-APAP. a) Cell proliferation of SKOV3-TR cells upon different treatments measured by MTT assay. The experiment was performed in quintuplicate in 96 well plates (n=5), mean ± s.d., *** represents p≤0.001, multiple t test. b) Cell viability of SKOV3-TR cells stained with annexin V-FITC/propidium iodide (PI) and measured by flow cytometry after different treatments. Cells with (+) or without (−) 6 h TYR@NPCN-333 pretreatment was incubated with 0–2 mmAPAP for 2 h. The experiment was performed three times and representative results from one test are displayed.
Because activating enzymes must be administered before treatment with a prodrug, it is important to ensure that their activity can be maintained over a prolonged period of time.53To study how the enzyme-MOF nanoreactor performs in this respect, SKOV3-TR cells pretreated with TYR@NPCN-333 for 6 hours were cultured in fresh media for several days. An MTT assay revealed that a 2 mm APAP treatment on day 3 could lead to 50 % cell proliferation inhibition (see Figure S15). On the contrary, free TYR showed slight inhibition effect (<20 %) on cells after APAP treatment on day 0. However, the activity of free TYR totally disappeared from day 1 (see Figure S15).
Next, we sought to characterize how TYR@NPCN-333-APAP induces toxicity. When used as the sole therapeutic, neither TYR@NPCN-333 nor APAP display cytotoxicity. We hypothesized that the cytotoxicity observed was a result of the in situ generation of AOBQ as an enzymatic product. Based on the properties of p-quinone compounds, we suspected that AOBQ would undergo single-electron redox reactions and induce the generation of reactive oxygen species (ROS) in cells.54-57 Real-time monitoring of superoxide (O2−) concentration using a superoxide detection reagent in SKOV3-TR cells upon different treatments was performed to test this hypothesis. The intensity of red fluorescence is proportional to the concentration of intracellular O2−. Cells treated with TYR@NPCN-333-APAP displayed elevated levels of O2−whereas the O2− level in cells treated with either TYR@NPCN-333 or APAP remained unchanged (see Figure S16).
Besides inducing ROS generation, o-quinone compounds are notorious for consuming glutathione (GSH) in living cells.58, 59 Therefore we sought to measure the GSH concentration in cells upon TYR@NPCN-333-APAP treatments. GSH reacts with DNTB [5,5′-dithio-bis-2-(nitrobenzoic acid)] and produces a yellow-colored compound (5-thio-2-nitrobenzoic acid), which can be quantitatively measured by UV-vis spectroscopy at 405 nm.60 Because GSH is the most abundant nonprotein thiol in eukaryotic cells, this measurement yields a good estimation on intracellular GSH concentration.61 GSH levels remained unchanged in cells treated with either TYR@NPCN-333 or APAP. However, when cells were pretreated with TYR@NPCN-333 for 6 hours, treatment with an APAP solution resulted in GSH depletion, which showed a strong correlation with APAP concentration (see Figure S17). Notably, after 2 mm APAP treatment for only 2 hours, intracellular GSH concentration decreased to about 50 % of its original level (cells were not permeabilized at this time point, as shown by the absence of PI staining in Figure S18). Mass spectrometry analysis of the cell lysate revealed the presence of a species with molecular weight of 492, which is in consistent with that of APAP-GSH conjugate (see Figure S19).58 In addition, to further confirm the role of ROS generation and GSH depletion in AOBQ-induced cytotoxicity, we did two control experiments in which ascorbic acid (AA) and GSH were supplemented to cell culture medium along with TYR@NPCN-333-APAP treatment (APAP concentration was 500 μm). Compared with the negative control group, the cell viability showed 20 % and 30 % increase for AA and GSH supplement groups, respectively (see Figure S20). Overall, these results have established that TYR@NPCN-333-APAP exhibits cytotoxicity by inducing oxidative stress, as manifested by a combination of ROS induction and GSH depletion.
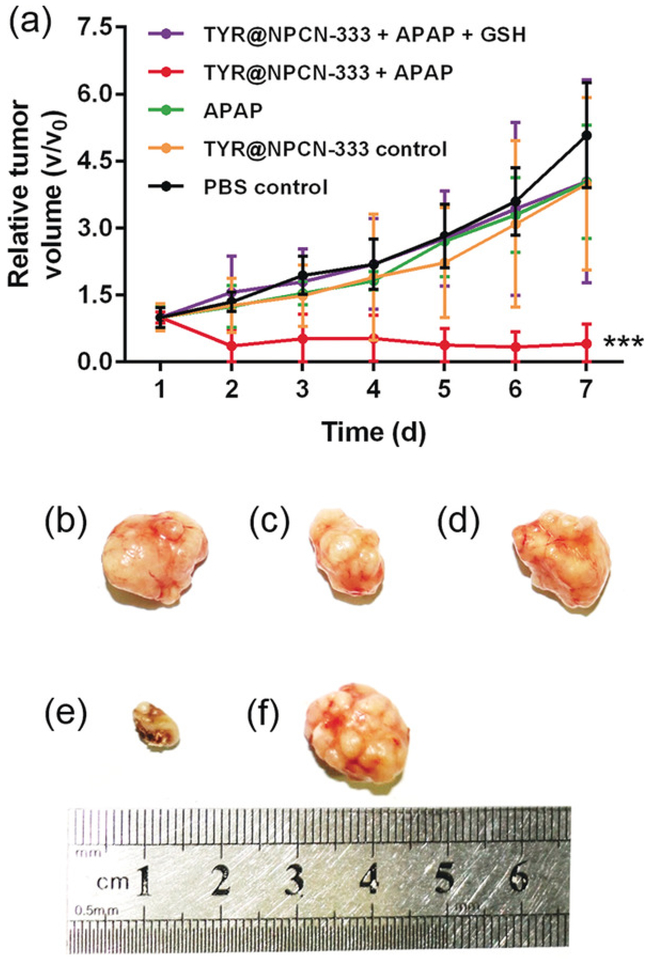
To evaluate the antitumor efficacy of APAP in the presence of an MOF nanoreactor, in vivo experiments were performed on a HeLa subcutaneous xenograft model. Mice were treated with PBS, TYR@NPCN-333 (1.5 mgkg-1), APAP (200 mgkg-1), TYR@NPCN-333 (1.5 mgkg-1), and APAP (200 mgkg-1), respectively. Compared with PBS control, tumor regression only occurred in the group receiving TYR@NPCN-333-APAP treatment (tumor size decreased from ≈50 to ≈20 mm3), while neither the nanoreactor nor the prodrug alone induced any antitumor efficacy (Figure 4). More importantly, in the TYR@NPCN-333-APAP group, two out of five tumors were completely eradicated by the end of the experiment upon a single treatment. Additionally, tumor regression failed in mice treated with TYR@NPCN-333-APAP, but supplemented with GSH (700 mgkg-1), further confirming that tumor regression is attributable to oxidative stress. Hematoxylin and eosin (H&E) staining of tumor tissues showed that only the group receiving TYR@NPCN-333-APAP therapy demonstrated cell apoptosis/necrosis (see Figures S22 and S23).
Figure 4.
In vivo antitumor efficiency of TYR@NPCN-333-APAP. a) Time-dependent relative tumor growth upon different treatments. The experiment was performed on five bilateral tumor-bearing mice (n=5), mean ± s.d., *** represents p<0.001, multiple t test. b-f) Tumor images at day 7 upon different treatments: b) PBS, c) APAP, d) TYR@NPCN-333, e) APAP + TYR@NPCN-333, f) APAP + TYR@NPCN-333 + GSH. The experiment was performed on five bilateral tumor-bearing mice (n=5) and representative results from one test are displayed.
In summary, we demonstrate that the enzyme-MOF nanoreactor can be a highly efficient activator for cancer therapy coupled with a prodrug. Encapsulated enzymes are well protected by MOFs in proteolytic conditions and in acidic environments, wherein free enzymes would quickly lose activity. The prodrug can be efficiently activated by the enzyme-MOF nanoreactor, thus generating a cytotoxic compound which inhibits cell proliferation and induces apoptosis/necrosis by promoting oxidative stress. In vivo experiment shows a 2.5-time regression of tumor volume after a single treatment. Overall, we envision that the enzyme-MOF nanoreactor system as a new platform for vast biomedical and biotechnological applications due to the ability to cater for intrinsic diversities in structure and functionality.
Supplementary Material
Figure 2.
Structural characterization of NPCN-333. TEM images of NPCN-333 (a), TYR@NPCN-333 (b), and TYR@NPCN-333 after soaking in RPMI-1640 for 7 days (c). Scale bar: 200 nm. d) Powder X-ray diffraction (PXRD) patterns of pristine NPCN-333 (black), TYR@NPCN-333 (red), TYR@NPCN-333 after trypsin treatment (blue), and TYR@NPCN-333 after soaking in RPMI 1640 media for 7 days (magenta). e) Particle-size distribution measured by dynamic light scattering (DLS): (from front to back) pristine NPCN-333, TYR@NPCN-333, TYR@NPCN-333 after trypsin treatment, and TYR@NPCN-333 after soaking in RPMI 1640 media for 0 and 7 days.
Acknowledgements
This work was supported by the Welch Foundation through a Robert A. Welch Chair in Chemistry to H.C.Z. (A-0030), by a grant from the National Institutes of Health (to J.P.P., award number R01GM110137), by a grant from Strategic Transformative Research Program, College of Science, Texas A&M University to X.L., by China Scholarship Council and by Youth Innovation Promotion Association CAS.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Xizhen Lian, Department of Chemistry, Texas A&M University, College Station, TX 77843-3255 (USA).
Yanyan Huang, Beijing National Laboratory for MolecularSciences; CAS Key, Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy Of Sciences, Beijing, 100190(China).
Yuanyuan Zhu, Beijing National Laboratory for MolecularSciences; CAS Key, Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy Of Sciences, Beijing, 100190(China).
Yu Fang, Department of Chemistry, Texas A&M University, College Station, TX 77843-3255 (USA).
Rui Zhao, Beijing National Laboratory for MolecularSciences; CAS Key, Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy Of Sciences, Beijing, 100190(China).
Elizabeth Joseph, Department of Chemistry, Texas A&M University, College Station, TX 77843-3255 (USA).
Jialuo Li, Department of Chemistry, Texas A&M University, College Station, TX 77843-3255 (USA).
Jean-Philippe Pellois, Department of Biochemistry and Biophysics, Texas A&M University College Station, TX 77843-2128 (USA); Department of Chemistry, Texas A&M University, College Station, TX 77843-3255 (USA).
Hong-Cai Zhou, Department of Chemistry, Texas A&M University, College Station, TX 77843-3255 (USA).
References
- 1.Corrie PG, Medicine 2008, 36, 24–28. [Google Scholar]
- 2.Loehrer PJ, Einhorn LH, Ann. Intern. Med. 1984, 100, 704–713. [DOI] [PubMed] [Google Scholar]
- 3.Marupudi NI, Han JE, Li KW, Renard VM, Tyler BM, Brem H, Expert Opin. Drug Saf. 2007, 6, 609–621. [DOI] [PubMed] [Google Scholar]
- 4.Milosavljevic N, Duranton C, Djerbi N, Puech PH, Gounon P, Lagadic-Gossmann D, Dimanche-Boitrel MT, Rauch C, Tauc M, Counillon L, Poët M, Cancer Res. 2010, 70, 7514–7522. [DOI] [PubMed] [Google Scholar]
- 5.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG, Nat. Rev. Cancer 2013, 13,714–726. [DOI] [PubMed] [Google Scholar]
- 6.Liu F-S, Taiwan J Obstet. Gynecol. 2009, 48, 239–244.CrossrefPubMedWeb of Science®Google ScholarTexas A&M [DOI] [PubMed] [Google Scholar]
- 7.Alfarouk KO, Stock C-M, Taylor S, Walsh M, Muddathir AK, Verduzco D, Bashir AHH, Mohammed OY, Elhassan GO, Harguindey S, Reshkin SJ, Ibrahim ME, Rauch C, Cancer Cell Int. 2015, 15, 71. CrossrefPubMedWeb of Science®Google ScholarTexas A&M [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC, Proc. Natl. Acad. Sci. USA 2011, 108, 1850–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vingerhoeds MH, Haisma HJ, van Muijen M, van de Rijt RBJ, Crommelin DJA, Storm G, FEBS Lett. 1993, 336, 485–490. [DOI] [PubMed] [Google Scholar]
- 10.Giang I, Boland EL, Poon GMK, AAPS J 2014, 16, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny WA, Eur. J. Med. Chem. 2001, 36, 577–595. [DOI] [PubMed] [Google Scholar]
- 12.Rooseboom M, Commandeur JN, Vermeulen NP, Pharmacol. Rev. 2004, 56, 53–102. [DOI] [PubMed] [Google Scholar]
- 13.Choi KY, Swierczewska M, Lee S, Chen X, Theranostics 2012, 2, 156–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turk B, Nat. Rev. Drug Discovery 2006, 5, 785–799. [DOI] [PubMed] [Google Scholar]
- 15.Knox RJ in Enzyme-Prodrug Strategies for Cancer Therapy (Eds.: ), Springer US, Boston, 1999, pp. 97–131. [Google Scholar]
- 16.Bagshawe KD, Sharma SK, Begent RHJ, Expert Opin. Biol. Ther. 2004, 4, 1777–1789. [DOI] [PubMed] [Google Scholar]
- 17.Bagshawe KD, Sharma SK, Springer CJ, Rogers GT, Ann. Oncol. 1994, 5, 879–891. [DOI] [PubMed] [Google Scholar]
- 18.Harris TO, Shelver DW, Bohnsack JF, Rubens CE, J. Clin. Invest. 2003, 111, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nolan DP, Rolin S, Rodriguez JR, Van Den Abbeele J, Pays E, Eur. J. Biochem. 2000, 267,18–27. [DOI] [PubMed] [Google Scholar]
- 20.He H, Han H, Shi H, Tian Y, Sun F, Song Y, Li Q, Zhu G, ACS Appl. Mater. Interfaces 2016, 8, 24517–24524. [DOI] [PubMed] [Google Scholar]
- 21.Lian X, Fang Y, Joseph E, Wang Q, Li J, Banerjee S, Lollar C, Wang X, Zhou H-C, Chem. Soc. Rev. 2017, 46, 3386–3401. [DOI] [PubMed] [Google Scholar]
- 22.Liu WL, Wu CY, Chen CY, Singco B, Lin CH, Huang HY, Chem. Eur. J. 2014, 20, 8923–8928. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Lykourinou V, Hoang T, Ming LJ, Ma S, Inorg. Chem. 2012, 51, 9156–9158. [DOI] [PubMed] [Google Scholar]
- 24.Lykourinou V, Chen Y, Wang XS, Meng L, Hoang T, Ming LJ, Musselman RL, Ma S, J. Am. Chem. Soc. 2011, 133, 10382–10385. [DOI] [PubMed] [Google Scholar]
- 25.Feng D, Liu TF, Su J, Bosch M, Wei Z, Wan W, Yuan D, Chen YP, Wang X, Wang K, Lian X, Gu ZY, Park J, Zou X, Zhou HC, Nat. Commun. 2015, 6, 5979. [DOI] [PubMed] [Google Scholar]
- 26.Liang K, Ricco R, Doherty CM, Styles MJ, Bell S, Kirby N, Mudie S, Haylock D, Hill AJ, Doonan CJ, Falcaro P, Nat. Commun. 2015, 6, 7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lian X, Chen Y-P, Liu T-F, Zhou H-C, Chem. Sci. 2016, 7, 6969–6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huxford RC, Della Rocca J, Lin W, Curr. Opin. Chem. Biol. 2010, 14, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D, Lu K, Poon C, Lin W, Inorg. Chem. 2014, 53, 1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Della Rocca J, Liu D, Lin W, Acc. Chem. Res. 2011, 44, 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi F-Y, Chen D, Wu M-K, Han L, Jiang H-L, ChemPlusChem 2016, 81, 675–690. [DOI] [PubMed] [Google Scholar]
- 32.Kreno LE, Leong K, Farha OK, Allendorf M, Van Duyne RP, Hupp JT, Chem. Rev. 2012, 112, 1105–1125. [DOI] [PubMed] [Google Scholar]
- 33.Tan L-L, Li H, Zhou Y, Zhang Y, Feng X, Wang B, Yang Y-W, Small 2015, 11, 3807–3813. [DOI] [PubMed] [Google Scholar]
- 34.Meng X, Gui B, Yuan D, Zeller M, Wang C, Sci. Adv. 2016, 2, e 1600480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan L-L, Li H, Qiu Y-C, Chen D-X, Wang X, Pan R-Y, Wang Y, Zhang SX-A, Wang B, Yang Y-W, Chem. Sci. 2015, 6, 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu M-X, Yang Y-W, Adv. Mater. 2017, 29, 1606134. [DOI] [PubMed] [Google Scholar]
- 37.Vilela SMF, Salcedo-Atraira P, Colinet I, Salles F, de Koning MC, Joosen MJA, Serre C, Horcajada P, Nanomaterials 2017, 7: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon-Yarza T, Giménez-Marqués M, Mrimi R, Mielcarek A, Gref R, Horcajada P, Serre C, Couvreur P, Angew. Chem. Int. Ed. 2017, 49, 15565–15569. [DOI] [PubMed] [Google Scholar]
- 39.Marcos-Almaraz MT, Gref R, Agostoni V, Kreuz C, Clayette P, Serre C, Couvreur P, Horcajada P, J. Mater. Chem. B 2017, 5, 8563–8569. [DOI] [PubMed] [Google Scholar]
- 40.Rojas S, Wheatley PS, Quartapelle-Procopio E, Gil B, Marszalek B, Morris RE, Barea E, CrystEngComm 2013, 15, 9364–9367. [Google Scholar]
- 41.Giménez-Marqués M, Hidalgo T, Serre C, Horcajada P, Coord. Chem. Rev. 2016, 307, 342–360. [Google Scholar]
- 42.Hidalgo T, Giménez-Marqués M, Bellido E, Avila J, Asensio MC, Salles F, Lozano MV, Guillevic M, Simón-Vázquez R, González-Fernández A, Serre C, Alonso MJ, Horcajada P, Sci. Rep. 2017, 7: 43099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao S-L, Yue D-M, Li X-H, Smith TJ, Li N, Zong M-H, Wu H, Ma Y-Z, Lou W-Y, ACS Sustainable Chem. Eng. 2016, 4, 3586–3595. [Google Scholar]
- 44.Shieh FK, Wang SC, Yen CI, Wu CC, Dutta S, Chou LY, Morabito JV, Hu P, Hsu MH, Wu KC, Tsung CK, J. Am. Chem. Soc. 2015, 137, 4276–4279. [DOI] [PubMed] [Google Scholar]
- 45.Lian X, Erazo-Oliveras A, Pellois J-P, Zhou H-C, Nat. Commun. 2017, 8, 2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawelek J, Korner A, Bergstrom A, Bologna J, Nature 1980, 286, 617–619. [DOI] [PubMed] [Google Scholar]
- 47.Hodgman MJ, Garrard AR, Crit. Care Clin. 2012, 28, 499–516. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, van Leeuwen J, Wichers HJ, Flurkey WH, Agric J. Food Chem. 1999, 47, 374–378. [DOI] [PubMed] [Google Scholar]
- 49.Hua W, Christianson T, Rougeot C, Rochefort H, Clinton GM, Steroid Biochem J. Mol. Biol. 1995, 55, 279–289. [DOI] [PubMed] [Google Scholar]
- 50.Yan X-D, Li M, Yuan Y, Mao N, Pan L-Y, Oncol. Rep. 2007, 17, 1163–1169. [PubMed] [Google Scholar]
- 51.Lee HH, Bellat V, Law B, PLoS One 2017, 12, e 0171044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pol A, Luetterforst R, Lindsay M, Heino S, Ikonen E, Parton RG, J. Cell Biol 2001, 152,1057–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerr DE, Vrudhula VM, Svensson HP, Siemers NO, Senter PD, Bioconjugate Chem. 1999, 10, 1084–1089. [DOI] [PubMed] [Google Scholar]
- 54.Raymond KS, Grafton AK, Wheeler RA, J. Phys. Chem. B 1997, 101, 623–631. [Google Scholar]
- 55.Guo C, Wang W, Feng W, Li P, RSC Adv. 2017, 7, 12775–12782. [Google Scholar]
- 56.Shahidzadeh M, Ghandi M, J. Organomet. Chem. 2001, 625, 108–111. [Google Scholar]
- 57.Guin PS, Das S, Mandal PC, Int. J. Electrochem. Sci. 2011, 2011, 816202. [Google Scholar]
- 58.Moridani MY, Moore M, Bartsch RA, Yang Y, Heibati-Sadati S, J. Pharm. Pharm. Sci. 2005, 8, 348–360. [PubMed] [Google Scholar]
- 59.Moridani MY, Moore M, Bartsch R, Yang Y, Heibati-Sadati S, Cancer Res. 2005, 65, 1387–1387. [Google Scholar]
- 60.Baker MA, Cerniglia GJ, Zaman A, Anal. Biochem. 1990, 190, 360–365. [DOI] [PubMed] [Google Scholar]
- 61.Jiang X, Chen J, Bajić A, Zhang C, Song X, Carroll SL, Cai Z-L, Tang M, Xue M, Cheng N, Schaaf CP, Li F, MacKenzie KR, Ferreon ACM, Xia F, Wang MC, Maletić-Savatić M, Wang J, Nat. Commun. 2017, 8, 16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.