Abstract
Genetic factors strongly contribute to the pathogenesis of sporadic Alzheimer's disease (AD). Nevertheless, genome-wide association studies only yielded single nucleotide polymorphism loci of moderate importance. In contrast, microsatellite repeats are functionally less characterized structures within our genomes. Previous work has shown that endothelin-converting enzyme-1 (ECE-1) is able to reduce amyloid β content. Here we demonstrate that a CpG-CA repeat within the human ECE-1c promoter is highly polymorphic, harbors transcriptional start sites, is able to recruit the transcription factors poly(ADP-ribose) polymerase-1 and splicing factor proline and glutamine-rich, and is functional regarding haplotype-specific promoter activity. Furthermore, genotyping of 403 AD patients and 444 controls for CpG-CA repeat length indicated shifted allelic frequency distributions. Sequencing of 245 haplotype clones demonstrated that the overall CpG-CA repeat composition of AD patients and controls is distinct. Finally, we show that human and chimpanzee [CpG]m–[CA]n ECE-1c promoter repeats are genetically and functionally distinct. Our data indicate that a short genomic repeat structure constitutes a novel core promoter element, coincides with human evolution, and contributes to the pathogenesis of AD.
Introduction
Alzheimer's disease (AD) is the most common cause of dementia affecting >10 million people worldwide (Blennow et al., 2006). AD can be grouped into seldom (<1%), familial (monogenetic), and frequent sporadic forms (Blennow et al., 2006; Goedert and Spillantini, 2006). The pathophysiology of AD extracellular deposition of amyloid β (Aβ) plaques, composed of Aβ40/42, as well as intraneuronal neurofibrillary tangles, consisting of hyperphosphorylated tau protein, seem crucial (Blennow et al., 2006; Roberson and Mucke, 2006). In sporadic forms the clearance, but not the production, of Aβ is impaired (Blennow et al., 2006; Zlokovic, 2008), which highlights the importance of Aβ-degrading enzymes such as insulin-degrading enzyme, neutral endopeptidase, and endothelin-converting enzyme-1(ECE-1; Tanzi et al., 2004; Blennow et al., 2006; Eckman et al., 2006).
ECE-1 is the key enzyme in endothelin biosynthesis (Yanagisawa et al., 1998) and is involved in a broad spectrum of (patho)physiologies ranging from heart and gut development (Yanagisawa et al., 1998) to hypertension (Funke-Kaiser et al., 2003c), atherosclerosis (Ihling et al., 2001) and cardiomyopathy (Serneri et al., 2000). Recently, it was demonstrated that ECE-1 also exerts essential roles beyond the endothelin system based on its role in receptor recycling (Roosterman et al., 2007).
Several publications have demonstrated the pathogenetic role of ECE-1 in AD. ECE-1 can degrade Aβ40 and Aβ42 in vitro (Eckman et al., 2001). Furthermore, cerebral Aβ40/42 content is significantly increased in ECE-1 knock-out mice compared with wild-type animals (Eckman et al., 2003). Consistently, induction of ECE-1 activity by transgenic overexpression of protein kinase C strongly reduced plaque and Aβ deposit area (Choi et al., 2006). Furthermore, intracranial viral delivery of ECE-1 caused an ∼50% reduction in Aβ content in mice (Carty et al., 2008).
ECE-1 is expressed in at least four isoforms, termed ECE-1a, ECE-1b, ECE-1c, and ECE-1d, which are generated by alternative promoters (Valdenaire et al., 1999a; Funke-Kaiser et al., 2000). In humans, single nucleotide polymorphisms (SNPs) in the ECE-1b isoform-specific promoter, initially described by our group in the context of arterial hypertension (Funke-Kaiser et al., 2003c), affect prefrontal ECE-1 mRNA expression and are associated with the likelihood of developing AD to a similar degree as the apolipoprotein E2 (APOE2) allele (Funalot et al., 2004). Despite this clinical association, these SNPs in the ECE-1b isoform-specific promoter alter promoter activity only by 30–40% (Funke-Kaiser et al., 2003c). Furthermore, the absolute activity of the ECE-1b promoter is relatively low compared with the promoter of the ECE-1c isoform (Funke-Kaiser et al., 2000; 2003a). ECE-1c is the major ECE-1 isoform, since it shows a strong, ubiquitous expression pattern (Schweizer et al., 1997 Funke-Kaiser et al., 2003b), a broader subcellular distribution (Valdenaire et al., 1999b), and the highest mRNA level (Lindenau et al., 2006) compared with the other ECE-1 isoforms. We previously identified a polymorphic [CpG]m–[CA]n microsatellite repeat in the promoter region of human ECE-1c and demonstrated its functionality in vitro (Funke-Kaiser et al., 2003b), which by far quantitatively exceeds the functionality of the Alzheimer-associated ECE-1b promoter SNP as mentioned above. These data imply a pathophysiological role of the polymorphic microsatellite of the ECE-1c promoter in AD in vivo. Furthermore, in contrast to the epigenetically important CpG dinucleotide-binding proteins such as MBD1, MBD3, or MeCP2 (Ballestar and Wolffe, 2001), CpG microsatellite-binding proteins, i.e., transcription factors with the ability to bind to [CpG]m repeats, have not been characterized yet.
Therefore, our aim was to identify putative, novel CpG-CA microsatellite-binding proteins, and to analyze the association of the corresponding polymorphic cis-element with AD in the context of the ECE-1c promoter.
Materials and Methods
Cell culture.
EA.hy926 (human endothelial; a generous gift from C.-J.S. Edgell, University of North Carolina), SH-SY5Y (human neuronal), and KELLY (human neuronal) cells were cultured as previously described (Schefe et al., 2006; Reinemund et al., 2009). Mouse embryonic fibroblasts (MEFs) derived from wild-type and homozygous poly(ADP-ribose) polymerase-1 (PARP-1) knock-out mice were obtained from Z.-Q. Wang (Leibniz Institute for Age Research, Jena, Germany) and were grown in high glucose DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin, 2 mm l-glutamine, 1 mm sodium pyruvate, and 0.1 mm β-mercaptoethanol (Invitrogen).
Great apes study population.
DNA from unrelated common chimpanzees (Pan troglodytes) was provided by C. Roos (Gene Bank of Primates, German Primate Center, Göttingen, Germany).
Cardiovascular study population (valsartan study).
Sixty patients with treated or untreated hypertension (grade 1; European Society of Hypertension/European Society of Cardiology Guidelines 2003) and type 2 diabetes were randomized to placebo or the angiotensin AT1 receptor (AT1R) blocker valsartan to evaluate the putative anti-inflammatory and antidiabetic potential of valsartan. The participants were aged 44–78 years (mean = 64 a); 25% were females. Blood and adipose tissue biopsies were taken at several points of time during the study. The study protocol including several genetic analyses was approved by the ethical committee of the Charité-Universitätsmedizin Berlin [EK Nr. 2140 (15.5.2004) and EA1/076/05 (18.4.2005)]. Written informed consent was obtained from all participants. Study results regarding inflammatory, lipid, and glucose parameters were published recently (Kintscher et al., 2010).
AD study population.
Human genomic DNA of AD patients (n = 403) and respective nondemented controls (n = 444) was obtained from the German Dementia Competence Network (DCN) (Kornhuber et al., 2009; www.kompetenznetz-demenzen.de) and genotyped as described below. The patients/controls were aged 49–92 years [mean = 73 a; mean = 72.1 a for AD (SD = 8.1 a); mean = 74.8 a for controls (SD = 7.5 a)]; 60% were females (61.7% in the AD group; 58.2% in the control group; no statistical difference between both groups according to χ2 testing) and all were of Caucasian ethnicity. Diagnosis of AD was based, for example, on clinical (e.g., neuropsychological testing), paraclinical (e.g., levels of Aβ42 and tau in CSF) and radiological (e.g., NMR) parameters. Genetic analyses regarding ECE-1 were approved by the ethical committee of the Charité-Universitätsmedizin Berlin (#215–10; 6./7.6.2007).
DNA and RNA isolations.
Regarding the cardiovascular study population genomic DNA was isolated from human blood using the Qiaamp DNA Blood Mini Kit (Qiagen) or the Invisorb Spin Blood Mini Kit (Invitek). RNA from blood of the valsartan study participants was extracted using the Paxgene Blood RNA Kit (Preanalytix; Hombrechtikon); RNA from adipose tissue was isolated using Trisure (Bioline) or the Rneasy Lipid Tissue Mini Kit (Qiagen). RNA isolations from blood and adipose tissue comprised a DNase digest. Quality of RNA was controlled using an Agilent 2100 Bioanalyzer (Agilent Technologies). Only RNAs with an RNA integrity number ≥ 6 (blood cells) or ≥ 4 (adipose tissue) were used for further analyses.
Fractionated protein extraction and gel filtration.
Nuclear and cytosolic proteins were isolated as described previously (Funke-Kaiser et al., 2003c). Cytosolic and nuclear fractions were controlled by Western blotting using antibodies against calpain (Cat. no. 208730; Calbiochem) and TFIID (TATA box-binding protein; TBP) (sc-273, Santa Cruz Biotechnology), respectively.
Size exclusion chromatography (gel filtration) of the nuclear protein extracts was performed at 8°C using a Superdex 200 10/300 column (GE Healthcare) with a bed volume of 24 ml equilibrated in 50 mm Tris, 200 mm NaCl, and 1 mm dithiothreitol (DTT). pH 7.9, on an ÄKTA Purifier UPC 10 System (GE Healthcare). Ten micrograms of nuclear protein extract was loaded and the run was executed with a flow rate of 0.5 ml/min. After 7 ml, fractions of 300 μl each were collected.
Genotyping by fluorescent-labeled genomic PCR.
Genomic DNA isolated from blood was amplified using the following primers spanning the microsatellite of the human ECE-1c promoter: 5′-atatagtcaggactttccttcaca-3′ (sense; with a 5′-FAM modification) and 5′-gcggggaacctggagggcaggagcag-3′ (antisense). Amplification reactions were controlled by agarose gel electrophoresis, denatured (95°C, 2 min; DCN study) or 3 min (valsartan study) and resolved on an ABI 3730xl Analyzer (Applied Biosystems) with a Performance Optimized Polymer (POP)-7 system (DCN study) or an ABI 3130xl Analyzer with a POP-7 system (valsartan study) to determine product lengths. Data were analyzed using the Genemarker 1.6 (Soft Genetics; for the DCN study) or Genemapper 4.0 (ABI; for the valsartan study) software. Determined product lengths were rounded to larger even numbers using Microsoft Excel to reflect that the ABI analyzer resolves nucleotides and considering the dinucleotide structure of the ECE-1c repeat confirmed by sequencing as well as the fact that stutter bands, which are discussed below, are usually one or more repeat units shorter as the true allele (Mulero et al., 2006; Olejniczak and Krzyzosiak, 2006).
Quantitative real-time PCR.
Reverse transcription (RT) was performed using M-MLV RT (RNase H minus; Promega) and 250 ng of RNA (blood) or 100 ng of RNA (adipose tissue). PCR was performed applying a SYBR Green I reaction mix and the following primer pairs: human ECE-1c: 5′-agcacgcgagctatgatgt-3′ (sense), 5′-agagcgagtccaccaggtc-3′ (antisense) and human 18S rRNA: 5′-ccgcagctaggaataatggaata-3′ (sense), 5′-tctagcggcgcaatacgaat-3′ (antisense). A reaction without addition of reverse transcriptase (RT−) served as negative control. The PCRs were run on a Stratagene Mx3000P. Inspection of melting curves served to control for product specificity. Furthermore, the PCR products regarding 18S and ECE-1c, both controlled by an RT− reaction, were subcloned and confirmed by sequencing.
Data represent the mean expression level (± SD) standardized to 18S rRNA expression of at least three independent measurements per cDNA (technical triplicates) calculated according to the ddCT method. Regarding the quantification of ECE-1c mRNA in blood cells and adipose tissue of the valsartan study population only runs with a CT difference ≥ 5 between RT+ and RT− (i.e., without addition of reverse transcriptase) were used for analyses.
5′-RACE.
An RNA ligase-mediated (RLM) 5′-RACE, which only amplifies capped mRNA due to a procedure involving calf intestinal phosphatase and tobacco acid pyrophosphatase, was performed using the Generacer kit (Invitrogen). The following primers specific for human ECE-1 were used: 5′-ctgcaggccgttggggtatgc-3′ (first round) and 5′-catagctcgcgtgctccgccccggct-3′ or 5′-gcggggaacctggagggcaggagcag-3′ (nested PCR; specific for ECE-1c). The reaction products were subcloned and sequenced.
RNase protection assay.
An RNase protection assay (RPA) to map transcriptional start sites was performed as described previously (Funke-Kaiser et al., 2000, 2001). Genomic RPA probes were subcloned using the following primer pairs: “218 nt”-probe: 5′-ggcaacaaaccccaaatataccg-3′ (sense), 5′-aggagaataggagacagaaacgg-3′ (antisense) and “356 nt”-probe: 5′-atatagtcaggactttccttcaca-3′ (sense), 5′-gcggggaacctggagggcaggagcag-3′ (antisense).
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assay (EMSA) experiments were performed as described previously (Funke-Kaiser et al., 2003c) using 32P-radiolabeled oligonucleotides; respective nucleotide sequences are given in the figure legends. For supershift analyses the following antibodies were used: XRCC6 (= Ku-70; sc-1486; Santa Cruz Biotechnology), XRCC5 (= Ku-86; sc-1484; Santa Cruz Biotechnology), KHDRBS1 (= Sam68; sc-333; Santa Cruz Biotechnology), RecQ1 (sc-25547; Santa Cruz Biotechnology), TATA box-binding protein (sc-204, Santa Cruz Biotechnology), PARP-1 (sc-74470 X and sc-1562; Santa Cruz Biotechnology), TFII-I (sc-46670 X; Santa Cruz Biotechnology), and splicing factor proline and glutamine-rich (SFPQ) (sc-28730; Santa Cruz Biotechnology).
DNA affinity chromatography and mass spectrometry.
Affinity chromatography was performed using the DNA-binding Protein Purification Kit (Roche), oligonucleotides derived from the human ECE-1c promoter CpG-CA repeat as given in the figure legends, and protein fractions after size exclusion chromatography showing the highest oligonucleotide-binding affinity in EMSA experiments. Eluted proteins were prepared for mass spectrometry (MS) by trypsin in-gel digestion. NanoLC-MS/MS analysis was performed with an integrated system that includes an UltiMate 3000 nanoflow liquid chromatography system (Dionex) and a micrOTOF-Q mass spectrometer equipped with a nanoelectrospray ionization source (Bruker Daltonics). Tryptic peptides were separated through a C18 PepMap100, 75 μm i.d. column (Dionex). Peptides were eluted from the column with a gradient of 2.0–80% buffer B (99.9% acetonitrile/0.1% formic acid) in buffer A (1% acetonitrile/98.9% water/0.1% acetic acid) for 40 min. Eluting peptides were continuously analyzed by selecting the five most intense ion signals of a survey scan (m/z 50–2500) for sequential MS/MS fragmentation. Database search was conducted with Mascot 2.1 and the NCBInr database. Protein identification was based on at least two distinct peptides.
Subcloning, reporter gene assays, and protein expression.
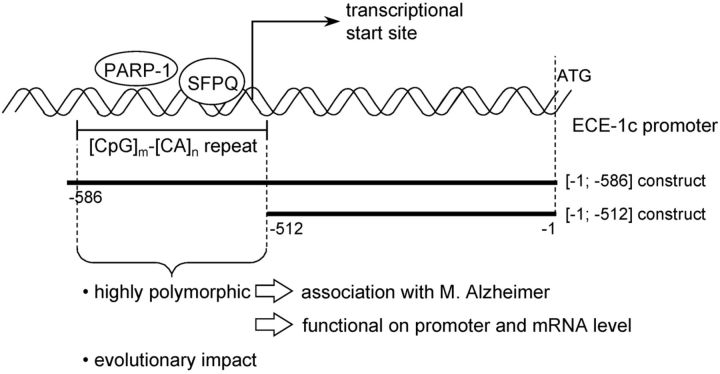
Serial deletion mutants of the ECE-1c promoter were subcloned into the luciferase reporter vector pGL3-basic (Promega) using the following primers: 5′-ttctcctccctcttacacaca-3′ (sense; position −586 relative to the translational start site of the ECE-1 promoter sequence GenBank accession GI:4185238; human and chimpanzee), 5′-acacaagcagctgcagcgc-3′ (sense; position −512 relative to the ATG; human), 5′-acacacgcagctgcagcgc-3′ (sense; −512; chimpanzee), and 5′-agctcgcgtgctccgcc-3′ (antisense; directly upstream of the ATG; human and chimpanzee).
Promoter reporter assays were performed using the Dual-Luciferase Reporter Assay System (Promega). Cotransfection of the phRL-null plasmid (encoding humanized renilla luciferase; Promega) served for standardization. Relative luciferase activity (RLA) is defined as the mean value of the firefly luciferase/renilla luciferase ratios of each construct related to the insertless reporter plasmid pGL3-basic (Funke-Kaiser et al., 2003a).
The subcloning of a pcDNA3.1-based expression vector encoding human PARP-1 was described previously by our group (Reinemund et al., 2009). Recombinant PARP-1 was synthesized using the TNT T7 Coupled Reticulocyte Lysate System (Promega). The complete coding sequence of the human SFPQ, based on GenBank accession no. GI:197209833, was subcloned into the mammalian expression vector pcDNA3.1 myc-His(−)C (Invitrogen) using the following primers: 5′-gaggagctagcgccaccatgtctcgggatcggttccg-3′ (sense, translation start codon underlined) and 5′-tcctcggtaccggtggcctaaaatcggggttttttgtttg-3′ (antisense).
Genotyping by subcloning.
PCR products obtained by the fluorescent-labeled genomic PCR described above were subcloned into the pGEM-T Easy vector (Promega) and sequenced. In addition, human ECE-1c promoter regions subcloned into the pGL3-basic reporter gene vector were sequenced. If not stated otherwise, the short 5′-CA repeat directly upstream of the [CpG]m–[CA]n repeat is 6 bp in length throughout the manuscript (e.g., in Figs. 1B, 2, 3, 4C, 6).
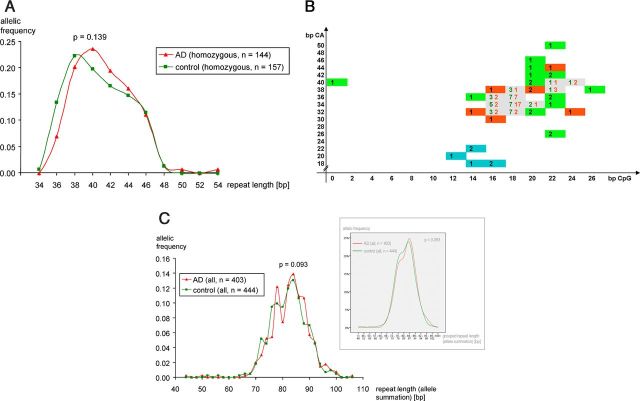
Figure 1.
Association of the ECE-1c promoter microsatellite with AD. A, Homozygous (regarding repeat length and not repeat composition) samples of 144 AD patients (red triangles) and 157 control individuals (green rectangles) were genotyped regarding the total length of the ECE-1c promoter microsatellite polymorphism using a fluorescent-labeled genomic PCR spanning the microsatellite repeat. Reaction products were separated on an ABI analyzer. Repeat length denotes the length of the [CG]m–[CA]n repeat itself of one allele, i.e., PCR product length minus length of the flanking region. B, The ECE-1c promoter region of 47 AD patients (red rectangles or red numbers), 58 nondemented controls (comprising 38 controls of the dementia study and 20 individuals of the valsartan study; green rectangles or green numbers) and of 6 chimpanzees (blue rectangles) was subcloned and sequenced to determine the [CpG]m–[CA]n repeat compositions. Abscissa and ordinate indicate the lengths of the CG repeat and CA repeat in base pairs, respectively. The respective genotypings are based on multiple independent clones per individual and multiple sequencing reactions per clone as depicted in Table 3. C, All (i.e., homozygous and heterozygous) samples of 403 AD patients (red triangles) and 404 control individuals (green rectangles) were genotyped regarding the length of the [CG]m–[CA]n repeat within the ECE-1c promoter. Repeat length denotes the summation of the repeat lengths of both homologous chromosomes per individual. The inset represents the identical distributions but with grouped alleles plotted on the abscissa.
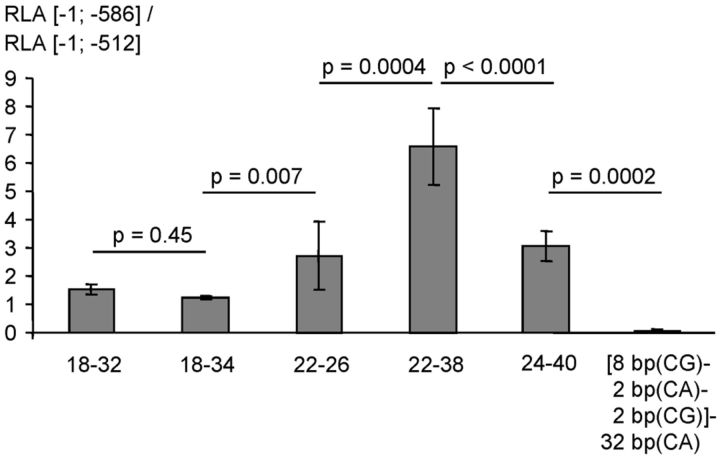
Figure 2.
Functional effects of the human ECE-1c promoter repeat polymorphism. Two serial deletion ([−1; −512] without the repeat, and [−1; −586] comprising the repeat; compare Fig. 7) of multiple human haplotypes of the ECE-1c promoter were transiently transfected into KELLY cells. The ordinate represents the ratio ([−1; −586] construct/[−1; −512] construct) of the renilla-standardized and pGL3-basic-normalized firefly luciferase promoter activities (i.e., RLAs; ±SD), which indicates the contribution of the microsatellite to the total promoter activity. The haplotype matrix (CpG repeat [bp]-CA repeat [bp]) of the transfected plasmid preparation is shown below the columns. The sixth column refers to a haplotype in which a CA dinucleotide is interspersed into the CG repeat. The columns represent the mean of several independent experiments as given in Table 4.
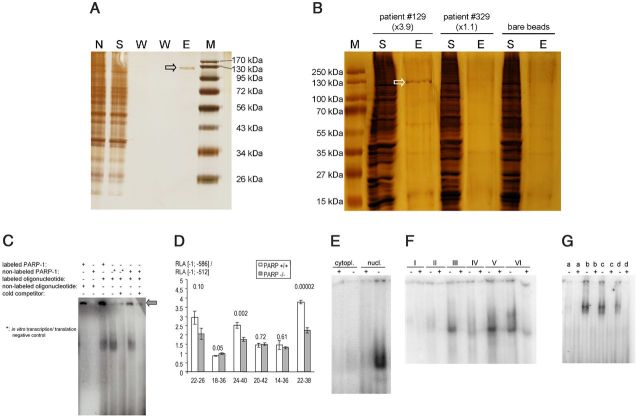
Figure 3.
PARP-1 binds to and regulates the human ECE-1c promoter repeat. A, DNA affinity chromatography using the oligonucleotide 5′-ttacacacacgcgcgcgcgcgcgcgcacacttacacacacgcgcgcgcgcgcgcgcacac-3′ and 2 μg of nuclear proteins (not size fractionated) of human endothelial EA.hy926 cells (N). Binding reaction was performed in a buffer containing 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm DTT, 5% glycerol, 1 mm MgCl2, and 300 mm KCl. The identical buffer but with 2 m KCl was used to obtain the eluate (E). Indicated fractions (without further dialysis) were resolved by SDS-PAGE followed by silver staining. S, binding supernatant; W, wash fractions; M, molecular weight marker; open arrow, band identified as PARP-1 by MS. B, DNA affinity chromatography using nuclear proteins (not size fractionated) of human neuronal KELLY cells and oligonucleotides derived from human ECE-1c promoters in which the repeat per se confers high-activating (patient #129: 5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′) or low-activating (patient #329: 5′-tcttacacacacgcgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacaagcag-3′) effects in luciferase reporter gene assays. Bare beads served as further negative control. Indicated fractions (abbreviations as in A) were resolved by SDS-PAGE followed by silver staining. Open arrow, band identified as PARP-1 by MS. “×3.9” and “×1.1” are the ratios ([−1; −586] construct/[−1; −512] construct) of the firefly luciferase promoter activities standardized to the renilla luciferase activities in the corresponding reporter gene assays, which reflect the promoter-regulating property of the microsatellite per se. C, EMSA using nuclear proteins of KELLY cells and the human ECE-1c promoter-derived oligonucleotide 5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacac acacacacacaagcag-3′. The recombinant PARP-1 protein and/or the oligonucleotide were radiolabeled with 35S-methionine or [γ-32P]ATP, respectively, as indicated. The in vitro transcription/translation negative control was performed without radiolabeling. Arrow, DNA-PARP-1 complex. D, Haplotype-specific ECE-1c promoter activities in PARP-1-deficient cells. Two serial deletion mutants ([−1; −512] and [−1; −586]) of multiple haplotypes of the human ECE-1c promoter were transiently transfected into homozygous PARP-1-deficient (PARP −/−; gray columns) or wild-type control MEFs (PARP +/+; white columns) (up to 11 independent experiments; each n = 3–4). The ordinate indicates the ratio ([−1; −586] construct/[−1; −512] construct) of the renilla-standardized and pGL3-basic-normalized firefly luciferase promoter activities (i.e., RLAs) ± SEM. The p value of the comparison PARP +/+ versus PARP −/− is given by the number above the column. The matrix of the haplotype (CpG repeat [bp]-CA repeat [bp]) is shown below the columns. E, EMSA using nuclear (nucl.) and cytoplasmic (cytopl.) proteins of human endothelial EA.hy926 cells and the oligonucleotide 5′-ttacacacacgcgcgcgcgcgcgcgcacac-3′. +, Binding reaction with unlabeled competitor oligonucleotide; −, binding reaction without competitor. F, EMSA using nuclear proteins of human endothelial EA.hy926 cells and the following distinct repeat oligonucleotides: I, 5′-cacgcgcgcgcgcgcgcgca-3′; II, 5′-cacgcgcgcgcgcgcgcgcgcgca-3′; III, 5′-cacgcgcgcgcgcgcgcgcgcgcgcgca-3′; IV, 5′-cacgcgcgcgcgcgcgcgcgcgcgcgcgcgca-3′; V, 5′-ttacacacacgcgcgcgcgcgcgcgcacac-3′; and VI, 5′-ttacacacacgcgcgcgcgcgcgcgcgcgcgcgcacac-3′. + and − as in A. G, EMSA using size-fractionated nuclear proteins of human neuronal KELLY cells and the oligonucleotide 5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′. a–d indicate the gel filtration fractions ranging from 54 to 424 kDa. + and − as in A.
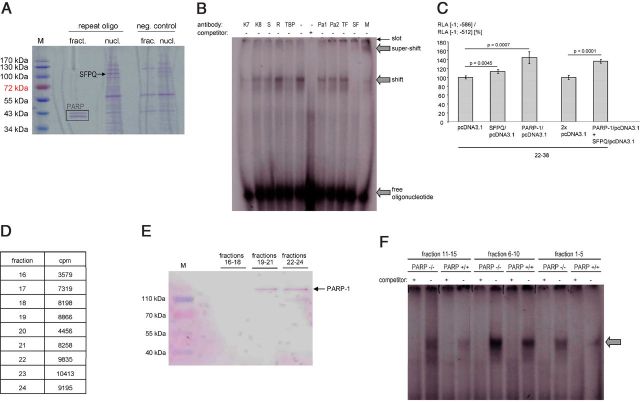
Figure 4.
Analyses of interactions between SFPQ and PARP-1 with the ECE-1c promoter. A, SDS-PAGE (Coomassie staining) after DNA affinity chromatography using the ECE-1c promoter-derived oligonucleotide 5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′ (repeat oligo) and proteins from KELLY cells (nucl., total nuclear proteins; fract., gel-filtration fraction no. 28 of nuclear proteins). Beads without oligonucleotides served as negative (neg.) control. M, molecular weight marker; PARP, PARP-1 (degradation) fragments as identified by MS. B, EMSA using nuclear proteins from KELLY cells after gel filtration and the oligonucleotide as in A. For supershift analyses antibodies against Ku-70 (K7), Ku-86 (K8), Sam68 (S), RecQ1 (R), TBP, PARP-1 (Pa1, Pa2), TFII-I (TF), or SFPQ (SF) were used. These proteins represent candidates for ECE-1c promoter binding based on, for example, MS data. The specificity of the SFPQ supershift is further illustrated by the disappearance of the shifted band. M, mixture of anti-PARP-1, anti-TFII-I, and anti-SFPQ antibodies. C, The luciferase promoter constructs [−1; −512] (i.e., without the repeat) or [−1; −586] (comprising a human repeat consisting of 6 bp(CA)-22 bp(CpG)-38 bp(CA)) and two expression vectors encoding PARP-1 and SFPQ, respectively, were transiently (co)transfected into KELLY cells (n = 4 wells). Transient transfections of an insertless expression vector (pcDNA3.1) served as negative controls. The ratio (RLA [−1; −586] construct/RLA [−1; −512] construct) indicates the contribution of the microsatellite to the total promoter activity. D, Confirmation of the PARP-1-DNA interaction by gel filtration. A double-stranded, 32P-labeled deoxyoligonucleotide derived from the human ECE-1c promoter repeat (5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′) was incubated with nuclear proteins of KELLY cells followed by gel filtration. Radioactivity was measured in all 40 gel filtration fractions and proteins were precipitated by trichloroacetic acid in fractions 16—24, which exhibited the highest counts per minute (cpm) values. E, Western blotting after gel filtration. Fractions from D were pooled as indicated followed by Western blotting using an anti-PARP-1 antibody. M, molecular weight marker. F, EMSA using PARP-1-deficient cells. Nuclear proteins were extracted from MEFs of homozygous PARP-1 knock-out mice (PARP−/−) or respective wild-type controls (PARP+/+) followed by gel filtration. Subsequent EMSA was performed with the radiolabeled oligonucleotide 5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′ derived from the human ECE-1c promoter. The arrow indicates a specific, i.e., competeable, [CpG]m[CA]n-transcription factor complex.
Figure 6.
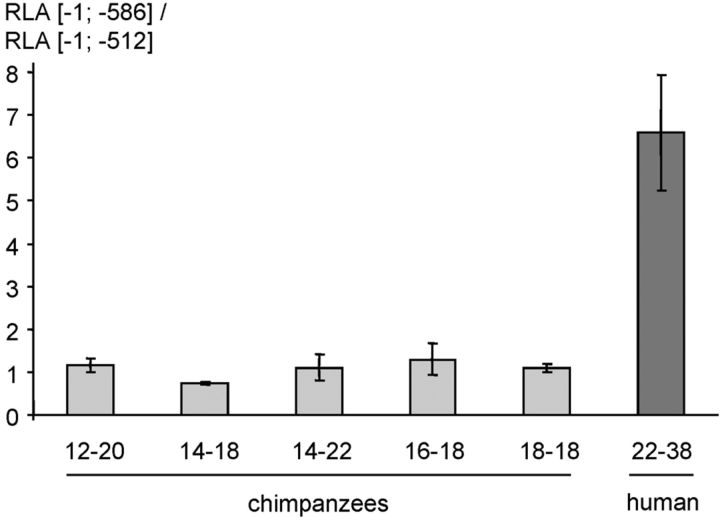
Species-specific functional effects of the ECE-1c promoter repeat. Two serial deletion mutants ([−1; −512] without the repeat and [−1; −586] comprising the repeat; compare Fig. 7) of multiple chimpanzee and one human haplotypes of the ECE-1c promoter were transiently transfected into KELLY cells (3 independent experiments regarding the chimpanzee and one regarding the human species; n = 4 for each experiment). Ordinate and haplotype matrix of the transfected plasmid preparation as in Figure 2. For all chimpanzee–human comparisons the p value is <0.001.
Western blotting.
Immunoblotting was performed as described recently (Seidel et al., 2011) using the anti-PARP-1 antibody sc-74470 X (Santa Cruz Biotechnology).
Statistical analysis.
Regarding reporter gene assays, a two-tailed, unpaired t test was applied and statistical significance was assumed at p < 0.05. Concerning the association study of AD patients and controls, a two-tailed Mann–Whitney test was applied regarding repeat length (Fig. 1A,C) and a Fisher′s exact test regarding repeat composition (Fig. 1B). Values are given as ± SD unless otherwise stated.
Results
The ECE-1c promoter microsatellite is polymorphic and associated with different ECE-1c mRNA levels
Initially, a pilot study was performed to confirm our previous observation (Funke-Kaiser et al., 2003b) that the CpG-CA repeat of the human ECE-1c promoter is highly polymorphic, and to analyze whether this polymorphism is functional on the mRNA level. Fifty-eight patients of a cardiovascular study performed at our institute (Kintscher et al., 2010) were genotyped with respect to the absolute length of the [CpG]m–[CA]n microsatellite by fluorescent-labeled genomic PCR. Results demonstrated interindividual differences of up to 24 bp per chromosome concerning the repeat structure (Table 1). In addition, real-time PCR analyses of blood (samples of 55 patients measured in 276 triplicate runs) and adipose tissues (samples of 19 patients measured in 22 triplicate runs) indicated that the ECE-1c mRNA level is characterized by markedly large interindividual differences (39- and 3.2-fold regarding blood cells and adipose tissue, respectively; Table 2).
Table 1.
Genotyping of the absolute ECE-1c promoter repeat length by fluorescent-labeled genomic PCR within a cardiovascular study population
| Patient identifier n = 58 | Length of CpG-CA-repeat [bp] (1. chromosome) | Length of CpG-CA-repeat [bp] (sister chromosome) | Summation homologous chromosomes | Zygosity |
|---|---|---|---|---|
| 130 | 22 | 44 | 66 | Heterozygous |
| 114 | 28 | 40 | 68 | Heterozygous |
| 116 | 30 | 38 | 68 | Heterozygous |
| 101 | 36 | 36 | 72 | Homozygous |
| 126 | 40 | 46 | 86 | Heterozygous |
| 121 | 42 | 46 | 88 | Heterozygous |
| 103 | 44 | 44 | 88 | Homozygous |
| 306 | 46 | 52 | 98 | Heterozygous |
| Minimum (total population) | 22 | 36 | 66 | |
| Maximum (total population) | 46 | 52 | 98 |
This table only displays an extract from all analyses focusing on minimal and maximal repeat lengths.
Table 2.
Interindividual human ECE-1c mRNA expression in blood cells and adipose tissue as measured by real-time PCR within a cardiovascular study population
| Patient identifier | Blood cells |
Adipose tissue |
||
|---|---|---|---|---|
| Number of real-time PCR measurements (each in technical triplicates) | ECE-1c mRNA expression (mean value standardized to EA.hy926 in %) | Number of real-time PCR measurements | ECE-1c mRNA expression (mean value standardized to EA.hy926 in %) | |
| 126 | 1 | 1.0 | 3 | 72.9 |
| 307 | 1 | 1.5 | 1 | 31.1 |
| 317 | 1 | 1.5 | 1 | 56.6 |
| 101 | 4 | 1.6 | 3 | 100.2 |
| 111 | 5 | 8.7 | n.d. | n.d. |
| 329 | 3 | 9.0 | n.d. | n.d. |
| 304 | 4 | 9.3 | n.d. | n.d. |
| 323 | 5 | 9.4 | n.d. | n.d. |
| 325 | 4 | 19.3 | n.d. | n.d. |
| 128 | 7 | 20.0 | 2 | 65.8 |
| 130 | 3 | 20.1 | n.d. | n.d. |
| 119 | 1 | 39.3 | n.d. | n.d. |
| Total number of triplicate PCR runs | 276 | 22 | ||
To control for quality, only RNAs with a RNA integrity number ≥6 (blood cells) and ≥4 (adipose tissue) and PCRs with a CT difference ≥5 between RT+ and RT− (i.e., control without addition of reverse transcriptase) were used as described in the materials and methods section. n.d., not determined. This table only displays an extracts from all analyses focusing on low, medium, and high mRNA levels.
In addition, a subanalysis of 21 patients from the cardiovascular population indicated that a high ECE-1c mRNA expression in blood cells is associated with low and high, but not intermediate, total CpG-CA repeat lengths (i.e., U-shaped relationship; data not shown) suggesting that this promoter polymorphism is also functional on the mRNA level.
The polymorphic ECE-1c promoter microsatellite is associated with AD
To analyze the functional relevance of the ECE-1c repeat polymorphism in AD, 403 patients and 444 respective controls of the German DCN, the largest German multicenter study regarding AD, were genotyped. Determination of the absolute [CG]m–[CA]n repeat length of homozygous samples by genomic PCR followed by separation on a capillary analyzer demonstrated that the distributions of the allelic frequencies—which are based on product lengths rounded to larger even numbers considering, e.g., the resolution of the capillary analyzer and the dinucleotide structure of the repeat—of AD patients and controls are horizontally shifted with maxima at 40 and 38 bp, respectively (Fig. 1A). Furthermore, the 36 bp allele is associated with a 1.9-fold (= 0.070/0.134) reduced risk of AD. Statistical analysis indicated a p value of 0.139 regarding the difference of the homozygous AD and control distributions. In this context it is important to note that homozygosity refers to the absolute repeat length and not to the [CG]m–[CA]n composition.
Interestingly, when all (i.e., homozygous plus heterozygous) 847 samples were analyzed a p value of 0.093 was derived (Fig. 1C). Moreover, graphical illustration of allelic frequencies of grouped alleles again showed the horizontally shifted distributions regarding AD and controls (Fig. 1C, inset).
Since the total length of the [CG]m-–[CA]n repeat only mirrors a minor fraction of the information content present in this polymorphism, the ECE-1c promoter region of 47 AD patients and 38 controls of the German DCN, in addition to 20 patients of our cardiovascular population without overt dementia, were subcloned and sequenced to determine the variables m and n of the [CG]m–[CA]n structure (Fig. 1 B; Table 3). As discussed below, the respective ECE-1c promoter region of six chimpanzees was also included. The results, which are based on 245 haplotype clones and 462 sequencing reactions, demonstrate that both the CpG and the CA repeat are highly polymorphic. Furthermore, some repeat compositions could only be observed in AD or nondemented controls (Fig. 1B).
Table 3.
Genotyping of the ECE-1c promoter repeat region by subcloning and sequencing
| Internal designation of human patient/control or nonhuman sample | CA [bp] | CpG [bp] | CA [bp] | Total length [bp] | Number of independent clones | Number sequencing reactions of all clones | Study population/diagnosis | Zygosity |
|---|---|---|---|---|---|---|---|---|
| MND/E1 945 Pl. 3 D11 | 6 | 16 | 30 | 52 | 1 | 2 | AD | Homoz. |
| E1/H120 Pl. 5 H10 | 6 | 14 | 32 | 52 | 1 | 2 | AD | Homoz. |
| MND/E1 946 Pl. 3 D12 | 6 | 16 | 32 | 54 | 1 | 4 | AD | Homoz. |
| MND/E1 109 Pl. 1 B10 | 6 | 16 | 32 | 54 | 3 | 5 | AD | Homoz. |
| MND/E1 928 Pl. 3 D1 | 6 | 16 | 34 | 56 | 3 | 4 | AD | Homoz. |
| E1 M642 Pl. 7 H3 | 6 | 18 | 34 | 56 | 3 | 5 | AD | Homoz. |
| E1/H1 Pl. 5 F9 | 6 | 18 | 34 | 56 | 3 | 7 | AD | Homoz. |
| E1 M518 Pl. 7 C10 | 6 | 18 | 32 | 56 | 1 | 2 | AD | Homoz. |
| MND/E1 308 Pl. 1 B6 | 6 | 18 | 32 | 56 | 1 | 2 | AD | Homoz. |
| MND/E1 59 Pl. 1 B1 | 6 | 18 | 34 | 58 | 1 | 2 | AD | Homoz. |
| MND/E1 86 Pl. 1 B5 | 6 | 18 | 34 | 58 | 1 | 2 | AD | Homoz. |
| MND/E1 920 Pl. 3 C8 | 6 | 18 | 34 | 58 | 5 | 3 | AD | Homoz. |
| MND/E1 129 Pl. 1 C4 | 6 | 18 | 34 | 58 | 1 | 2 | AD | Homoz. |
| E1 M219 Pl. 7 A12 | 6 | 18 | 34 | 58 | 3 | 5 | AD | Homoz. |
| MND/E1 923 Pl. 3 C11 | 6 | 18 | 34 | 58 | 3 | 5 | AD | Homoz. |
| E1 M224 Pl. 7 B2 | 6 | 18 | 34 | 58 | 3 | 3 | AD | Homoz. |
| E1/H5 Pl. 5 F10 | 6 | 18 | 34 | 58 | 1 | 2 | AD | Homoz. |
| E1/H114 Pl. 5 H7 | 6 | 18 | 34 | 58 | 5 | 10 | AD | Homoz. |
| E1 M641 Pl. 2 F4 | 6 | 18 | 34 | 58 | 1 | 2 | AD | Homoz. |
| E1/H94 Pl. 5 G12 | 6 | 18 | 34 | 58 | 5 | 10 | AD | Homoz. |
| E1/H16, Pl. 5 F12 | 6 | 18 | 34 | 58 | 4 | 7 | AD | Homoz. |
| MND/E1 598, Pl. 2 B6 | 6 | 18 | 34 | 58 | 1 | 2 | AD | Homoz. |
| E1 M230 Pl. 7 B3 | 8 | 16 | 34 | 58 | 1 | 2 | AD | Homoz. |
| E1 M506, Pl. 2 D11 | 6 | 18 | 34 | 58 | 3 | 4 | AD | Homoz. |
| MND/E1 63, Pl. 1 B3 | 6 | 16 | 36 | 58 | 1 | 2 | AD | Homoz. |
| E1 M569 Pl. 2 E8 | 6 | 16 | 36 | 58 | 1 | 2 | AD | Homoz. |
| MND/E1 28 Pl. 1 A2 | 6 | 20 | 34 | 60 | 2 | 4 | AD | Homoz. |
| MND/E1 321 Pl. 1 F2 | 6 | 18 | 36 | 60 | 1 | 2 | AD | Homoz. |
| MND/E1114, Pl. 1 C1 | 6 | 18 | 36 | 60 | 1 | 2 | AD | Homoz. |
| E1/H123 Pl. 5 H12 | 6 | 18 | 36 | 60 | 2 | 3 | AD | Homoz. |
| E1 M 545 Pl. 7 D3 | 6 | 18 | 36 | 60 | 3 | 2 | AD | Homoz. |
| MND/E1 952 Pl. 3 E4 | 6 | 16 | 38 | 60 | 1 | 2 | AD | Homoz. |
| MND 527 Pl. 5 A3 | 6 | 18 | 38 | 60 | 3 | 3 | AD | Homoz. |
| E1 M672 Pl. 7 H12 | 6 | 18 | 36 | 60 | 4 | 6 | AD | Homoz. |
| E1 M544, Pl. 2 E4 | 6 | 18 | 36 | 60 | 1 | 2 | AD | Homoz. |
| MND/E1 538, Pl. 2 A7 | 6 | 18 | 38 | 62 | 1 | 2 | AD | Homoz. |
| MND/E1 521, Pl. 2 A5 | 6 | 18 | 38 | 62 | 1 | 2 | AD | Homoz. |
| E1 M456 Pl. 2 D9 | 6 | 24 | 32 | 62 | 1 | 2 | AD | Homoz. |
| MND/E1 887 Pl. 3 A10 | 6 | 20 | 38 | 64 | 4 | 4 | AD | Homoz. |
| MND/E1 979 Pl. 3 F10 | 6 | 20 | 38 | 64 | 2 | 2 | AD | Homoz. |
| MND/E1 917 Pl. 3 C5 | 6 | 22 | 38 | 66 | 3 | 5 | AD | Homoz. |
| MND/E1 350 Pl. 1 B8 | 6 | 22 | 38 | 66 | 3 | 6 | AD | Homoz. |
| MND/E1 715, Pl. 2 D12 | 6 | 22 | 38 | 66 | 2 | 4 | AD | Homoz. |
| E1 H111 Pl. 2 C5 | 6 | 22 | 38 | 66 | 1 | 2 | AD | Homoz. |
| MND/E1 623 Pl. 1 D8 | 6 | 22 | 40 | 68 | 1 | 2 | AD | Homoz. |
| MND/E1 519, Pl. 2 A4 | 6 | 24 | 40 | 70 | 1 | 2 | AD | Homoz. |
| MND/E1 972 Pl. 3 F7 | 6 | 22 | 44 | 72 | 1 | 2 | AD | Homoz. |
| MND 31 Pl. 3 G9 | 6 | 16 | 32 | 54 | 4 | 4 | Control | Homoz. |
| MND1328 Pl. 2 H5 | 6 | 18 | 32 | 56 | 1 | 2 | Control | Homoz. |
| MND326 Pl. 2 G8 | 6 | 16 | 34 | 56 | 1 | 2 | Control | Homoz. |
| MND/E1 912 Pl. 3 C1 | 6 | 18 | 32 | 56 | 3 | 5 | Control | Homoz. |
| MND 755 Pl. 5 C5 | 6 | 18 | 32 | 56 | 3 | 2 | Control | Homoz. |
| MND 840 Pl. 5 D2 | 8 | 16 | 32 | 56 | 1 | 2 | Control | Homoz. |
| MND 404 Pl. 4 G1 | 6 | 18 | 32 | 56 | 4 | 7 | Control | Homoz. |
| MND 616 Pl. 5 B6 | 6 | 18 | 32 | 56 | 3 | 5 | Control | Homoz. |
| MND 9 Pl. 3 G4 | 6 | 16 | 34 | 56 | 3 | 3 | Control | Homoz. |
| MND 487 Pl. 4 H4 | 6 | 16 | 34 | 56 | 1 | 2 | Control | Homoz. |
| MND/E1 971 Pl. 3 F6 | 6 | 16 | 34 | 56 | 3 | 6 | Control | Homoz. |
| MND 1400 Pl. 5 F4 | 6 | 14 | 36 | 56 | 1 | 4 | Control | Homoz. |
| MND/E1 638 Pl. 1 D11 | 8 | 16 | 32 | 56 | 1 | 2 | Control | Homoz. |
| MND 851 Pl. 5 D3 | 6 | 18 | 34 | 58 | 2 | 3 | Control | Homoz. |
| MND 566 Pl. 5 A10 | 6 | 18 | 34 | 58 | 1 | 2 | Control | Homoz. |
| MND 443 Pl. 4 D8 | 6 | 16 | 36 | 58 | 1 | 2 | Control | Homoz. |
| H 166 Pl. 6 B8 | 6 | 18 | 34 | 58 | 4 | 6 | Control | Homoz. |
| MND/E1 903 Pl. 3 B7 | 6 | 18 | 34 | 58 | 4 | 7 | Control | Homoz. |
| MND/E1 881 Pl. 3 A5 | 6 | 16 | 36 | 58 | 2 | 8 | Control | Homoz. |
| MND 14 Pl. 3 G5 | 6 | 18 | 34 | 58 | 4 | 8 | Control | Homoz. |
| MND 534 Pl. 5 A4 | 6 | 20 | 34 | 60 | 1 | 2 | Control | Homoz. |
| MND/E1 922 Pl. 3 C10 | 6 | 20 | 34 | 60 | 5 | 10 | Control | Homoz. |
| MND 1120, Pl. 5 E1 | 6 | 16 | 36 | 60 | 1 | 2 | Control | Homoz. |
| M55 0208_8 B4 | 6 | 18 | 36 | 60 | 1 | 2 | Control | Homoz. |
| MND 830 Pl. 5 C12 | 6 | 18 | 36 | 60 | 2 | 4 | Control | Homoz. |
| MND 839 Pl. 5 D1 | 6 | 18 | 36 | 60 | 3 | 5 | Control | Homoz. |
| MND 114 Pl. 4 A6 | 6 | 18 | 36 | 60 | 3 | 5 | Control | Homoz. |
| MND/E1 907 Pl. 3 B10 | 6 | 18 | 38 | 60 | 5 | 10 | Control | Homoz. |
| MND/E1 948 Pl. 3 E1 | 6 | 18 | 36 | 60 | 2 | 2 | Control | Homoz. |
| MND 70 Pl. 3 H6 | 6 | 18 | 36 | 60 | 3 | 6 | Control | Homoz. |
| MND/E1 734 Pl. 2 E7 | 6 | 18 | 36 | 60 | 2 | 4 | Control | Homoz. |
| MND 1022 Pl. 5 D7 | 6 | 18 | 38 | 62 | 4 | 7 | Control | Homoz. |
| MND 46 Pl. 3 H1 | 6 | 22 | 36 | 64 | 6 | 11 | Control | Homoz. |
| MND/E1 910 Pl. 3 B11 | 6 | 20 | 40 | 66 | 1 | 2 | Control | Homoz. |
| MND 6 Pl. 3 G3 | 6 | 20 | 40 | 66 | 3 | 5 | Control | Homoz. |
| MND 1582 Pl. 2 H10 | 6 | 20 | 42 | 68 | 1 | 2 | Control | Homoz. |
| MND/E1 810 Pl. 2 A8 | 6 | 24 | 40 | 70 | 1 | 2 | Control | Homoz. |
| MND287 Pl. 2 G5 | 6 | 26 | 38 | 70 | 1 | 2 | Control | Homoz. |
| Pat. 114 | 0 | 0 | 40 | 40 | 4 | 9 | Valsartan study | Heteroz. |
| Pat. 116 | 6 | 22 | 26 | 54 | 1 | 4 | Valsartan study | Heteroz. |
| Pat. 120 | 6 | 22 | 26 | 54 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 305 | 6 | 18 | 32 | 56 | 1 | 5 | Valsartan study | Heteroz. |
| Pat. 129 | 6 | 16 | 34 | 56 | 1 | 3 | Valsartan study | Homoz. |
| Pat. 127 | 6 | 20 | 32 | 58 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 101 | 6 | 18 | 32 | 58 | 1 | 3 | Valsartan study | Homoz. |
| Pat. 114 | 6 | 18 | 34 | 58 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 330 | 6 | 20 | 32 | 58 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 329 | 6 | 18 | 34 | 59 | 1 | 3 | Valsartan study | Homoz. |
| Pat. 322 | 6 | 22 | 34 | 62 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 324 | 6 | 18 | 38 | 62 | 2 | 5 | Valsartan study | Heteroz. |
| Pat. 117 | 6 | 22 | 36 | 64 | 2 | 15 | Valsartan study | Homoz. |
| Pat. 120 | 6 | 22 | 38 | 66 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 306 | 6 | 22 | 40 | 68 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 305 | 6 | 22 | 42 | 70 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 322 | 6 | 22 | 42 | 70 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 103 | 6 | 20 | 44 | 70 | 1 | 3 | Valsartan study | Homoz. |
| Pat. 127 | 6 | 20 | 46 | 72 | 1 | 3 | Valsartan study | Heteroz. |
| Pat. 306 | 6 | 22 | 50 | 78 | 1 | 3 | Valsartan study | Heteroz. |
| 1123 | 6 | 12 | 20 | 38 | 5 | 8 | Chimpanzee | — |
| 1124 | 6 | 14 | 18 | 38 | 4 | 7 | Chimpanzee | — |
| 1126 | 6 | 16 | 18 | 40 | 7 | 14 | Chimpanzee | — |
| 1125 | 6 | 16 | 18 | 40 | 4 | 8 | Chimpanzee | — |
| 1119 | 6 | 14 | 22 | 42 | 5 | 7 | Chimpanzee | — |
| 1118 | 6 | 14 | 22 | 42 | 7 | 11 | Chimpanzee | — |
The ECE-1c promoter region of 47 AD patients, 58 nondemented controls (comprising 38 controls of the dementia study and 20 individuals of the valsartan study) and of 6 chimpanzees was subcloned and sequenced to determine the [CpG]m–[CA]n repeat compositions and the lengths of an additional, short 5′-CA repeat. “Total length” is the summation of the CA, CpG, and CA lengths. The “number of independent clones” refers to haplotype-specific ECE-1c promoter regions subcloned into plasmids. The given base pair values are the means of the independent clones, which in turn are calculated as the mean of the respective sequencing reactions. AD, patient with Alzheimer's disease; control, control sample within the dementia study; homoz., homozygous; heteroz., heterozygous.
To statistically test differences in the frequencies of [CpG]m–[CA]n combinations between human healthy controls and patients with AD (Fig. 1B) (i.e., to statistically assess at once all human repeat compositions observed in contrast to simply limit analyses to a certain repeat composition thereby neglecting multiple testing) we used Fisher's exact test and analyzed first all possible repeat combinations (58 controls, 47 AD; p = 0.123) and second only those combinations with at least two observed individuals (47 controls, 39 AD; p = 0.006) to reduce the noise by repeat compositions observed only once. The Bonferroni-corrected significance level for those two tests is 0.05/2 = 0.025 indicating that the [CpG]m–[CA]n repeat compositions of the AD patients and the control population are significantly different.
Because so-called stutter bands (or shadow bands) are a known phenomenon concerning bacterial plasmid replication and DNA synthesis (i.e., PCR or sequencing) of repeat structures (Murray et al., 1993; Walsh et al., 1996; Olejniczak and Krzyzosiak, 2006), we addressed the issue of whether the difference of two base pairs regarding the maxima of the allele frequency distributions (Fig. 1A) can be confirmed via sequencing. Within the data presented above, the ECE-1c promoter regions of 21 plus 16 AD patients with a total repeat length of 38 bp or 40 bp, respectively, as well as of 17 plus 15 corresponding controls were subcloned (multiple clones per individual) and sequenced (multiple sequencing reactions per clone) yielding a difference of 2.2 bp between 38 bp and 40 bp alleles, thereby confirming the results of the genotyping by fluorescent-labeled PCR.
The polymorphic ECE-1c promoter repeat is functional on the promoter level
To characterize the functionalities of the different repeat haplotypes we performed promoter reporter gene assays. Thirty-five different haplotype-specific luciferase-encoding plasmids were transfected into human neuronal cells. As shown in Figure 2 and Table 4 the promoter repeat consisting of 6 bp(CA)-22 bp(CpG)-38 bp(CA) increased promoter activity 6.6-fold. In contrast, several other microsatellite haplotypes (e.g., 6-18-34, 6-16-38) did not function as positive or negative regulatory elements. Moreover, a haplotype with a CA insertion within the CpG repeat strongly decreased promoter activity 20-fold (i.e., to a RLA [−1; −586]/RLA [−1; −512] ratio of 0.05; Fig. 2).
Table 4.
Effect of the ECE-1c repeat polymorphisms on the promoter activity in neuronal cells
| Diagnosis | RLA [−1; −586]/RLA[−1; −512] mean | RLA [−1; −586]/RLA [−1; −512] SD | Matrix (CA[bp]-CpG[bp]-CA[bp]) of the transfected plasmid |
|---|---|---|---|
| Valsartan study | 1.12 | 0.11 | 6-0-34 |
| Control | 0.95 | 0.38 | 6-14-36 |
| Valsartan study | 1.36 | 0.72 | 6-14-40 |
| AD | 1.13 | 0.18 | 6-16-30 |
| Control | 0.89 | 0.23 | 6-16-32 |
| AD | 1.34 | 0.17 | 6-16-32 |
| Control | 1.48 | 0.20 | 6 bp(CA)-16 bp(CpG)-[14 bp(CA)-2 bp(CG)-18 bp(CA)] |
| AD | 1.00 | 0.02 | 6-16-38 |
| Control | 1.51 | 0.16 | 6-18-32 |
| Control | 1.36 | 0.52 | 6-18-32 |
| AD | 1.04 | 0.30 | 6-18-32 |
| Valsartan study | 1.09 | 0.30 | 6-18-34 |
| Valsartan study | 1.23 | 0.04 | 6-18-34 |
| AD | 1.05 | 0.07 | 6-18-34 |
| Valsartan study | 1.25 | 0.18 | 6-18-36 |
| Control | 1.08 | 0.36 | 6-18-36 |
| Valsartan study | 1.26 | 0.03 | 6-18-38 |
| Control | 1.68 | 0.76 | 6-18-38 |
| AD | 1.41 | 1.25 | 6-18-38 |
| Control | 1.13 | 0.15 | 6-18-38 |
| Control | 0.90 | 0.24 | 10-18-42 |
| Valsartan study | 1.39 | 0.19 | 6-20-32 |
| Control | 0.95 | 0.22 | 6-20-40 |
| Control | 1.44 | 0.31 | 6-20-42 |
| Valsartan study | 2.73 | 1.19 | 6-22-26 |
| AD | 6.60 | 1.35 | 6-22-38 |
| Valsartan study | 0.84 | 0.15 | 6-22-42 |
| AD | 1.13 | 0.06 | 6-22-44 |
| Valsartan study | 1.66 | 0.91 | 6-22-50 |
| Control | 3.08 | 0.54 | 6-24-40 |
| Control | 0.05 | 0.04 | 6 bp(CA)-[8 bp(CG)-2 bp(CA)-2 bp(CG)]-32 bp(CA) |
| AD | 1.08 | 0.19 | 8-16-34 |
Two serial deletion mutants ([−1; −512] without the repeat and [−1; −586] comprising the repeat (compare Fig. 7) of multiple human haplotypes of the ECE-1c promoter were transiently transfected into KELLY cells (2–5 independent transfections, each n = 4). RLAs are defined as the renilla-standardized and pGL3-basic-normalised firefly luciferase promoter activities, e.g., RLA [−1; −586] = ([−1; −586]-pGL3-basic(firefly)/phRL-null(renilla))/(insertless pGL3-basic(firefly)/phRL-null(renilla)). The ratio RLA [−1; −586] construct/RLA [−1; −512] construct indicates the contribution of the microsatellite to the total promoter activity. “Control” refers to the German Dementia Competence Network study. “Matrix” denotes the individual haplotype (CA repeat [length in bp]-CpG repeat [length in bp]-CA repeat [length in bp]) of the transfected plasmid preparation. Please note that different individuals can share an identical matrix. Bold characters represent unusual repeat compositions or extreme RLA ratios, respectively. The identity of the region [−1; −512] in both constructs ([−1; −512]/pGL3-basic and [−1; −586]/ pGL3-basic) was confirmed by sequencing to exclude effects of SNPs located downstream of the microsatellite.
PARP-1 is able to functionally bind the ECE-1c promoter microsatellite
As discussed above, transcription factors with the ability to bind to [CpG]m repeats have not been characterized in the literature so far to our knowledge. In contrast, EMSA experiments using endothelial and neuronal cells indicated that oligonucleotides derived from the ECE-1c promoter CpG-CA repeat are able to bind (a) nuclear protein(s) (Fig. 3,E–G). However, the molecular identity of this DNA-binding activity remained to be established since the bioinformatic database query using Transfac (Heinemeyer et al., 1999) did not return any known transcription factor with the ability to bind a [CG]m–[CA]n matrix. Therefore, we performed DNA affinity chromatographies using nuclear proteins and oligonucleotides derived from the ECE-1c promoter microsatellite (Fig. 3A,B). Identification of the respective eluated bands by MS revealed PARP-1 as a CpG-CA-binding protein in endothelial (Fig. 3A) as well as in neuronal (Fig. 3B) human cells. The binding of PARP-1 to the promoter repeat was confirmed by an EMSA in which recombinant PARP-1 protein itself was radiolabeled in addition to the oligonucleotide and which indicated a high molecular weight of the PARP-1-DNA complex (Fig. 3C). In addition, the interaction between PARP-1 and the ECE-1c promoter repeat was validated by gel filtration (Fig. 4D,E). The functionality of PARP-1 regarding ECE-1c regulation was analyzed by haplotype-specific luciferase reporter gene assays (Fig. 3D). These experiments indicated that the cis-acting activity of the CA-CpG repeat per se, which is given by the promoter activity of a construct comprising the repeat (i.e., [−1; −586]) divided by the promoter activity of a construct without the repeat (i.e., [−1; −512]) (compare Fig. 7), can be reduced by PARP-1 ablation. Furthermore, the contribution of PARP-1 to promoter activation depends on the [CG]m–[CA]n haplotype since only promoters with a higher basal activity of the repeat are affected by the cellular PARP-1 knock-out (Fig. 3D). Regarding these constructs it is important to note that PARP-1 ablation does not completely abolish the cis-acting activity of the CA-CpG repeat. Consistently, EMSA demonstrated a transcription factor [CG]m–[CA]n interaction even in PARP-1-deficient cells (Fig. 4F).
Figure 7.
Identification of SFPQ and PARP-1 as promoter microsatellite-binding transcription factors and association of the respective polymorphic and functional cis-element with AD and evolution. The transcriptional start site within the microsatellite repeat is indicative of a novel core promoter element. The locations of the promoter reporter gene constructs [−1; −512] and [−1; −586] are shown.
SFRQ is able to bind the ECE-1c promoter microsatellite
Since the experiments described before imply (an) additional transcription factor(s) regarding the regulation of and the binding to the promoter repeat, we performed further affinity chromatographies using nuclear proteins fractionated by gel filtration. As shown in Figure 4A, SFPQ was identified as a [CG]m–[CA]n-binding protein by MS. Moreover, the binding of SFPQ to the ECE-1c promoter repeat was confirmed by supershift EMSA (Fig. 4B).
Finally, overexpression of SFPQ and PARP-1 alone or in combination further significantly increased the positive regulatory function mediated by the microsatellite element (Fig. 4C).
The ECE-1c promoter microsatellite is able to initiate transcription
Since PARP is, as discussed below, involved in the basal transcriptional machinery, we analyzed whether the ECE-1c promoter repeat is able to initiate transcription. To that purpose, an RNA ligase-mediated-5′-RACE, which only detects start sites of capped RNAs but not of degraded transcripts, was performed (Fig. 5A,B). Sequencing of the respective RACE amplification products was able to demonstrate transcriptional start sites within the [CG]m–[CA]n repeat of the ECE-1c promoter in human epithelial cells, human neuronal cells, and human blood cells (Fig. 5B). To further confirm this novel finding that a microsatellite harbors transcriptional start sites, we performed a genomic RNase protection assay (Fig. 5C). Hybridization with probe overspanning the [CG]m–[CA]n microsatellite yielded a protected fragment with a length indicative of a start site within the repeat, whereas no protected signal was observable using a negative control probe derived from the genomic region directly upstream of the repeat.
Figure 5.
The polymorphic microsatellite of the human ECE-1c promoter harbors transcriptional start sites. A, RNA ligase-mediated-5′-RACE. A 5′-RACE was performed on human neuronal SH-SY5Y cells and reaction products were separated in an agarose gel. Lane 1, DNA ladder; lane 2, 5′-RACE reaction; lane 3, negative (no template) control. B, RLM-5′-RACE reaction products from different cell types were subcloned and sequenced. The [CG]m–[CA]n microsatellite is underlined. Transcriptional start sites in SH-SY5Y cells (green), in EA.hy926 cells (yellow), and in human blood (red) are indicated. Blue, translational start site of exon 1c. C, Genomic RNase protection assay to analyze transcriptional start sites in EA.hy926 cells. Total RNA was hybridized with different antisense RNA probes derived from the human ECE-1c promoter region. The “218 nt”-probe was complementary to a genomic region directly upstream of the [CG]m–[CA]n microsatellite, whereas the “356 nt”-probe overspanned the [CG]m–[CA]n microsatellite. No protected signal was observable using the “218 nt”-probe whereas hybridization with the “356 nt”-probe yielded a protected fragment between 200 and 300 nt (arrow). A transcriptional start site within the [CG]m–[CA]n microsatellite corresponds to a protected fragment between 202 and 258 nt. M, molecular size marker; -, control reaction without RNase; 1, yeast RNA; 2, RNA from EA.hy926 cells.
Evolutionary significance of the ECE-1c promoter microsatellite
As discussed below, promoter sequences are important drivers of evolution. Furthermore, diseases with involvement of ECE-1 such as AD and myocardial infarction are human-specific to a certain degree with respect to prevalence and pathological features (see Discussion). Therefore, we subcloned and sequenced the ECE-1c promoter region of six chimpanzees (Table 3). Interestingly, the [CG]m–[CA]n repeat compositions were completely distinct from the human ones with lower repeat numbers regarding both dinucleotides (Fig. 1B). To analyze the functional effects of this genetic difference, reporter gene assays were performed, which demonstrated that the ape [CG]m–[CA]n promoter repeat does not contribute to the ECE-1c promoter activity in contrast to a human one (Fig. 6).
Discussion
In this study we have demonstrated that human and chimpanzee ECE-1c promoter regions contain a highly polymorphic microsatellite repeat, which is functional regarding promoter activity and is associated with extreme (i.e., 39-fold) interindividual differences in ECE-1c mRNA levels in the human species. The repeat structure per se, despite being relatively short with its 40–78 bp (Table 3), strongly contributes to promoter activity since the high RLA of the [−1; −512] construct of ∼40 (i.e., 40-fold higher compared with an insertless control plasmid) in neuronal cells (Funke-Kaiser et al., 2003b) can be increased severalfold by certain haplotypes of this microsatellite (Figs. 3D, 2; Table 4).
In addition to these effects on gene regulation, our genomic association study comprising >800 individuals revealed that the allelic distribution regarding repeat length is distinct in AD patients and respective controls (Fig. 1A,C). Moreover, the overall [CG]m–[CA]n repeat compositions of AD patients and controls are significantly distinct based on a Fisher's exact test (Fig. 1B). In this context, it is of interest to note that the single, most frequently observed repeat composition CpG(18 bp)-CA(34 bp), which is significantly associated with AD (p = 0.003 using a χ2 test; Fig. 1B), is not able to increase ECE-1c promoter activity (Fig. 2) implying a relatively decreased ECE-1 enzyme activity and, therefore, a reduced Aβ clearance.
Sporadic AD has a strong genetic component of ∼80% (Blennow et al., 2006; Gatz et al., 2006). Nevertheless, genome-wide association studies (GWAS), which focus on SNPs, have only revealed loci with modest effects on AD risk (Bertram and Tanzi, 2008). Furthermore, the two most powerful GWAS on SNPs performed so far yielded only odds ratios in the range of 0.7 to 1.3 unlinked to APOE (Harold et al., 2009; Hollingworth et al., 2011). On the other hand, polymorphic microsatellites confer a much higher information content compared with typically biallelic SNPs since they comprise multiple alleles (Lindholm et al., 2004; Fig. 1B). Therefore, polymorphic repeats within a single gene can mimic a complex/polygenic pattern of inheritance (Gatchel and Zoghbi, 2005). This implies that, beyond the relevance of the ECE-1c promoter repeat presented here, promoter microsatellites in general are of greater importance in conferring genetic risk in complex human diseases than currently appreciated. Nevertheless, larger association studies concerning promoter repeat polymorphisms in AD are rare (Li et al., 2008).
As mentioned in the Introduction, transcription factors with the ability to bind to [CpG]m repeats have not been characterized yet. Here we were able to demonstrate that PARP-1 and SFPQ bind to and regulate the CA-CpG microsatellite of the human ECE-1c promoter. PARP-1 is a ubiquitous nuclear protein catalyzing the attachment of poly(ADP-ribose) (PAR) polymers to several different acceptor proteins such as histones, RNA polymerase II, p53, and PARP itself (Kraus and Lis, 2003). PARP-1 can act as a transcriptional coactivator, consistent with the decreased ECE-1c promoter activities in PARP-1-deficient cells observed here, and also as a repressor (Meisterernst et al., 1997; Kraus and Lis, 2003; Jagtap and Szabó, 2005). PARP-1 effects transcription in the absence of DNA damage due to several different mechanisms ranging from direct sequence-specific DNA binding, via recruitment by transcription factors, to poly(ADP-ribosyl)ation of other transcription factors and histones (D'Amours et al., 1999; Kraus and Lis, 2003). The important role of PARP concerning the basal transcription is known since 1983 when the identity of TFIIC with PARP was shown (Slattery et al., 1983). In addition, RNA polymerase II can serve as a substrate of the poly(ADP-ribosyl)ation reaction (D'Amours et al., 1999) and PARP can enhance transcription by acting during pre-initiation complex formation (Meisterernst et al., 1997). These observations suggest that our novel finding of transcription initiation within the ECE-1c promoter repeat discussed below is mediated by PARP-1. PARP-1 is involved in a broad spectrum of (patho)physiologies such as necrosis, apoptosis, inflammation, DNA repair, hypertension, diabetes, stroke, and AD (Love et al., 1999; Jagtap and Szabó, 2005). Consistent with the latter and the results of our association study, PARP-1 seems to play a role in learning and memory (Kim et al., 2005). Moreover, ECE-1c (Funke-Kaiser et al., 2003b) and PARP-1 (Ziegler and Oei, 2001) are ubiquitously expressed and both enzymes are implicated in similar diseases such as hypertension (Funke-Kaiser et al., 2003c; Jagtap and Szabó, 2005) and AD (Love et al., 1999; Eckman et al., 2003). Therefore, the first molecular link between the endothelin system and PARP-1 illustrated here might help to further elucidate the pathophysiology of these diseases and to consider the examination of combination therapies using the already available endothelin receptor blockers (Rich and McLaughlin, 2003) and PARP inhibitors (Jagtap and Szabó, 2005).
The second transcription factor identified in this study by MS based on its ability to bind the ECE-1c promoter repeat is SFPQ, also called polypyrimidine tract-binding protein-associated splicing factor (PSF). SFPQ is a multifunctional nuclear protein that is involved in neuronal splicing but also can act as a transcription factor. Consistently, SFPQ contains RNA-binding and also DNA-binding domains (Shav-Tal and Zipori, 2002; Tapia-Páez et al., 2008). Interestingly, PARP-1 and SFPQ have been described as parts of a complex regulating the dyslexia susceptibility gene DYX1C1 (Tapia-Páez et al., 2008) further confirming our findings. To the best of our knowledge, the link between SFPQ and AD demonstrated here has not been described so far.
Transcriptional initiation at core promoter regions is the key process in transforming genomic information into structure. Several core promoter elements, such as the TATA box, the initiator (Inr), the TFIIB recognition element (BRE), and the downstream core promoter element (DPE), have been described that can recruit the basal transcriptional machinery (Smale, 2001; Butler and Kadonaga, 2002). Approximately one-fourth of all promoters do not have any of these four core promoter elements, suggesting the existence of other yet undiscovered elements (Gershenzon and Ioshikhes, 2005). Our CA-CG repeat region is distinct from the known core promoter elements mentioned above. Furthermore, we were able to demonstrate transcriptional start sites directly within the promoter microsatellite using two independent methods and different cell lines as well as human material (Fig. 5). Therefore, the ECE-1 repeat structure seems to function as a novel core promoter element, in which PARP-1 might recruit the RNA polymerase II transcriptional machinery.
A further major finding of this study is its evolutionary aspect. Ubiquitously expressed genes, i.e., housekeeping genes, in general exhibit a higher evolutionary conservation compared with tissue-specific ones (Khaitovich et al., 2005). This also applies to the housekeeping gene ECE-1 (Funke-Kaiser et al., 2003b), which is evolutionarily conserved (Oetjen et al., 2001; Zhang et al., 2001). Consistently, the KA/KS value, which indicates the standardized number of coding base substitutions, of ECE-1 is relatively low compared with genes such as angiotensin-converting enzyme, AT1R, and prion protein indicating selective constraints in evolution (Chimpanzee Sequencing and Analysis Consortium, 2005). Regarding transcriptome diversity (i.e., differential mRNA expression) it was previously shown that ubiquitously expressed genes differ less among individuals within a species than tissue-specific genes (Khaitovich et al., 2005). ECE-1c is an exception to this observation considering its ubiquitous expression (Schweizer et al., 1997; Funke-Kaiser et al., 2003b) but the strong interindividual differences in the human species described here. This unusual “behavior” of the ECE-1 gene (i.e., high intrahuman mRNA variability of an evolutionarily conserved housekeeping gene) might be explained by the CpG-CA microsatellite repeat itself. This satellite is not present in the rodent lineage and strongly shortened in the chimpanzee lineage based on our own results (Fig. 1B) and according to bioinformatics (Basic Local Alignment Search Tool) at the National Center for Biotechnology Information (NCBI; data not shown), and seems, therefore, quasi-human specific. In addition, it does not contribute to promoter activity in the chimpanzee species in contrast to the human one (Fig. 6). Furthermore, due to its polymorphic nature the CpG-CA promoter repeat confers variability to ECE-1c mRNA levels in the human species. Consistently, it was shown that CpG island-associated promoters, in contrast to the coding sequence discussed above, are in general less constrained and might contribute to evolutionary plasticity (Carninci et al., 2006). Additionally, a recent publication suggests that CpG islands in promoter regions can contribute to warm-blooded vertebrate evolution (Sharif et al., 2010).
In the evolutionary context it is important to note that, as discussed above, ECE-1 is involved in human cardiovascular diseases and AD. Interestingly, myocardial infarction and AD pathology with tangles are rare in great apes (Gearing et al., 1994; Olson and Varki, 2003; Chimpanzee Sequencing and Analysis Consortium, 2005). Therefore, we hypothesize that humans whose ECE-1c CpG-CA microsatellite resembles the one in the chimpanzee genome might be less prone to AD and coronary artery disease, due to altered binding of the transcription factor complex comprising SFPQ and PARP-1.
In addition to evolutionary pathomechanisms, ECE-1 is also involved in pathophysiological mechanisms of AD. Our in vitro analyses using human neuronal cells, the expression of ECE-1 in murine and human brain neurons (Davenport et al., 1998; Sluck et al., 1999), and the ECE-1b haplotype-dependent neocortical mRNA expression (Funalot et al., 2004) as well as the increased Aβ burden in ECE-1 knock-out mice (Eckman et al., 2003) imply that ECE-1 is involved in the neuronal clearance of amyloid. Furthermore, due to its additional cerebrovascular expression (Palmer and Love, 2011) and its cardiovascular importance ECE-1 might be a molecular link between vascular and neuronal contributors in AD, considering the importance of the cerebral vasculature and cardiovascular risk factors in this disease (Zlokovic et al., 2008).
We are aware that the p values of our AD association study are beyond the border of significance with regard to absolute repeat lengths (Fig. 1A,C) and that an extreme number of alleles exists (Fig. 1B). Nevertheless, the shifted allele frequency distributions (Fig. 1A,C) and the statistical significance observed regarding repeat compositions of AD patients versus controls (Fig. 1B) clearly suggest a biological significance. The power of this study is relatively high concerning absolute numbers (e.g., >800 individuals in the AD study and 245 haplotype subclonings) but relatively small considering all the allelic possibilities of the repeat structure. Therefore, we consider this publication as a nonfinalizing step in the research of complex and fascinating promoter microsatellites with respect to basal gene regulation— considering novel transcriptional start site within the repeat—to DNA-protein interactions, to evolution, and to disease.
To conclude, our results demonstrate that a highly polymorphic promoter microsatellite is able to bind the transcription factors PARP-1 and SFQP, constitutes a novel core promoter element, strongly affects haplotype-specific promoter activity, exhibits evolutionary plasticity, and is associated with AD (Fig. 7). Since these phenomena are centered around a DNA region shorter than 100 bp, they illustrate the immense complexity of our genomes in addition to the medical relevance of repeat structures.
Footnotes
This work was supported by grants from the Deutsche Forschungsgemeinschaft to H.F.-K. (FU 463/2-1), the Dr. Werner Jackstädt Foundation (H.F.-K. and T.U.), and the German Federal Ministry of Education and Research to the German Dementia Competence Network (01GI0420). We thank Achim Kramer (Institute of Medical Immunology, Charité) for contributing personnel and equipment for the MS analyses and Dr. Christian Roos (Gene Bank of Primates, German Primate Center, Göttingen, Germany) for providing the chimpanzee DNA.
The authors declare no competing financial interests.
References
- Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Carty NC, Nash K, Lee D, Mercer M, Gottschall PE, Meyers C, Muzyczka N, Gordon MN, Morgan D. Adeno-associated viral (AAV) serotype 5 vector mediated gene delivery of endothelin-converting enzyme reduces Abeta deposits in APP + PS1 transgenic mice. Mol Ther. 2008;16:1580–1586. doi: 10.1038/mt.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Choi DS, Wang D, Yu GQ, Zhu G, Kharazia VN, Paredes JP, Chang WS, Deitchman JK, Mucke L, Messing RO. PKCepsilon increases endothelin converting enzyme activity and reduces amyloid plaque pathology in transgenic mice. Proc Natl Acad Sci U S A. 2006;103:8215–8220. doi: 10.1073/pnas.0509725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- Davenport AP, Kuc RE, Plumpton C, Mockridge JW, Barker PJ, Huskisson NS. Endothelin-converting enzyme in human tissues. Histochem J. 1998;30:359–374. [Google Scholar]
- Eckman EA, Reed DK, Eckman CB. Degradation of the Alzheimer's amyloid beta peptide by endothelin-converting enzyme. J Biol Chem. 2001;276:24540–24548. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- Eckman EA, Watson M, Marlow L, Sambamurti K, Eckman CB. Alzheimer's disease beta-amyloid peptide is increased in mice deficient in endothelin-converting enzyme. J Biol Chem. 2003;278:2081–2084. doi: 10.1074/jbc.C200642200. [DOI] [PubMed] [Google Scholar]
- Eckman EA, Adams SK, Troendle FJ, Stodola BA, Kahn MA, Fauq AH, Xiao HD, Bernstein KE, Eckman CB. Regulation of steady-state beta-amyloid levels in the brain by neprilysin and endothelin-converting enzyme but not angiotensin-converting enzyme. J Biol Chem. 2006;281:30471–30478. doi: 10.1074/jbc.M605827200. [DOI] [PubMed] [Google Scholar]
- Funalot B, Ouimet T, Claperon A, Fallet C, Delacourte A, Epelbaum J, Subkowski T, Léonard N, Codron V, David JP, Amouyel P, Schwartz JC, Helbecque N. Endothelin-converting enzyme-1 is expressed in human cerebral cortex and protects against Alzheimer's disease. Mol Psychiatry. 2004;9:1122–1128. 1059. doi: 10.1038/sj.mp.4001584. [DOI] [PubMed] [Google Scholar]
- Funke-Kaiser H, Bolbrinker J, Theis S, Lemmer J, Richter CM, Paul M, Orzechowski HD. Characterization of the c-specific promoter of the gene encoding human endothelin-converting enzyme-1 (ECE-1) FEBS Lett. 2000;466:310–316. doi: 10.1016/s0014-5793(00)01086-3. [DOI] [PubMed] [Google Scholar]
- Funke-Kaiser H, Theis S, Behrouzi T, Thomas A, Scheuch K, Zollmann FS, Paterka M, Paul M, Orzechowski HD. Functional characterization of the human prion protein promoter in neuronal and endothelial cells. J Mol Med. 2001;79:529–535. doi: 10.1007/s001090100270. [DOI] [PubMed] [Google Scholar]
- Funke-Kaiser H, Lemmer J, Langsdorff CV, Thomas A, Kovacevic SD, Strasdat M, Behrouzi T, Zollmann FS, Paul M, Orzechowski HD. Endothelin-converting enzyme-1 (ECE-1) is a downstream target of the homeobox transcription factor Nkx2-5. FASEB J. 2003a;17:1487–1489. doi: 10.1096/fj.02-0700fje. [DOI] [PubMed] [Google Scholar]
- Funke-Kaiser H, Thomas A, Bremer J, Kovacevic SD, Scheuch K, Bolbrinker J, Theis S, Lemmer J, Zimmermann A, Zollmann FS, Herrmann SM, Paul M, Orzechowski HD. Regulation of the major isoform of human endothelin-converting enzyme-1 by a strong housekeeping promoter modulated by polymorphic microsatellites. J Hypertens. 2003b;21:2111–2124. doi: 10.1097/00004872-200311000-00021. [DOI] [PubMed] [Google Scholar]
- Funke-Kaiser H, Reichenberger F, Köpke K, Herrmann SM, Pfeifer J, Orzechowski HD, Zidek W, Paul M, Brand E. Differential binding of transcription factor E2F-2 to the endothelin-converting enzyme-1b promoter affects blood pressure regulation. Hum Mol Genet. 2003c;12:423–433. doi: 10.1093/hmg/ddg040. [DOI] [PubMed] [Google Scholar]
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Gearing M, Rebeck GW, Hyman BT, Tigges J, Mirra SS. Neuropathology and apolipoprotein E profile of aged chimpanzees: implications for Alzheimer disease. Proc Natl Acad Sci U S A. 1994;91:9382–9386. doi: 10.1073/pnas.91.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon NI, Ioshikhes IP. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 2005;21:1295–1300. doi: 10.1093/bioinformatics/bti172. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG. A century of Alzheimer's disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihling C, Szombathy T, Bohrmann B, Brockhaus M, Schaefer HE, Loeffler BM. Coexpression of endothelin-converting enzyme-1 and endothelin-1 in different stages of human atherosclerosis. Circulation. 2001;104:864–869. doi: 10.1161/hc3301.094742. [DOI] [PubMed] [Google Scholar]
- Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kintscher U, Marx N, Martus P, Stoppelhaar M, Schimkus J, Schneider A, Walcher D, Kümmel A, Winkler R, Kappert K, Dörffel Y, Scholze J, Unger T. Effect of high-dose valsartan on inflammatory and lipid parameters in patients with Type 2 diabetes and hypertension. Diabetes Res Clin Pract. 2010;89:209–215. doi: 10.1016/j.diabres.2010.04.018. [DOI] [PubMed] [Google Scholar]
- Kornhuber J, Schmidtke K, Frolich L, Perneczky R, Wolf S, Hampel H, Jessen F, Heuser I, Peters O, Weih M, Jahn H, Luckhaus C, Hüll M, Gertz HJ, Schröder J, Pantel J, Rienhoff O, Seuchter SA, Rüther E, Henn F, et al. Early and differential diagnosis of dementia and mild cognitive impairment: design and cohort baseline characteristics of the German Dementia Competence Network. Dement Geriatr Cogn Disord. 2009;27:404–417. doi: 10.1159/000210388. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Li K, Dai D, Yao L, Gu X, Luan K, Tian W, Zhao Y, Wang B. Association between the macrophage inflammatory protein-l alpha gene polymorphism and Alzheimer's disease in the Chinese population. Neurosci Lett. 2008;433:125–128. doi: 10.1016/j.neulet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Lindenau S, von Langsdorff C, Saxena A, Paul M, Orzechowski HD. Genomic organisation of the mouse gene encoding endothelin-converting enzyme-1 (ECE-1) and mRNA expression of ECE-1 isoforms in murine tissues. Gene. 2006;373:109–115. doi: 10.1016/j.gene.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Lindholm E, Hodge SE, Greenberg DA. Comparative informativeness for linkage of multiple SNPs and single microsatellites. Hum Hered. 2004;58:164–170. doi: 10.1159/000083543. [DOI] [PubMed] [Google Scholar]
- Love S, Barber R, Wilcock GK. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer's disease. Brain. 1999;122:247–253. doi: 10.1093/brain/122.2.247. [DOI] [PubMed] [Google Scholar]
- Meisterernst M, Stelzer G, Roeder RG. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc Natl Acad Sci U S A. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero JJ, Chang CW, Hennessy LK. Characterization of the N+3 stutter product in the trinucleotide repeat locus DYS392. J Forensic Sci. 2006;51:1069–1073. doi: 10.1111/j.1556-4029.2006.00227.x. [DOI] [PubMed] [Google Scholar]
- Murray V, Monchawin C, England PR. The determination of the sequences present in the shadow bands of a dinucleotide repeat PCR. Nucleic Acids Res. 1993;21:2395–2398. doi: 10.1093/nar/21.10.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen J, Fives-Taylor P, Froeliger E. Characterization of a streptococcal endopeptidase with homology to human endothelin-converting enzyme. Infect Immun. 2001;69:58–64. doi: 10.1128/IAI.69.1.58-64.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak M, Krzyzosiak WJ. Genotyping of simple sequence repeats–factors implicated in shadow band generation revisited. Electrophoresis. 2006;27:3724–3734. doi: 10.1002/elps.200600136. [DOI] [PubMed] [Google Scholar]
- Olson MV, Varki A. Sequencing the chimpanzee genome: insights into human evolution and disease. Nat Rev Genet. 2003;4:20–28. doi: 10.1038/nrg981. [DOI] [PubMed] [Google Scholar]
- Palmer J, Love S. Endothelin receptor antagonists: potential in Alzheimer's disease. Pharmacol Res. 2011;63:525–531. doi: 10.1016/j.phrs.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Reinemund J, Seidel K, Steckelings UM, Zaade D, Klare S, Rompe F, Katerbaum M, Schacherl J, Li Y, Menk M, Schefe JH, Goldin-Lang P, Szabo C, Olah G, Unger T, Funke-Kaiser H. Poly(ADP-ribose) polymerase-1 (PARP-1) transcriptionally regulates angiotensin AT2 receptor (AT2R) and AT2R binding protein (ATBP) genes. Biochem Pharmacol. 2009;77:1795–1805. doi: 10.1016/j.bcp.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Rich S, McLaughlin VV. Endothelin receptor blockers in cardiovascular disease. Circulation. 2003;108:2184–2190. doi: 10.1161/01.CIR.0000094397.19932.78. [DOI] [PubMed] [Google Scholar]
- Roberson ED, Mucke L. 100 years and counting: prospects for defeating Alzheimer's disease. Science. 2006;314:781–784. doi: 10.1126/science.1132813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosterman D, Cottrell GS, Padilla BE, Muller L, Eckman CB, Bunnett NW, Steinhoff M. Endothelin-converting enzyme 1 degrades neuropeptides in endosomes to control receptor recycling. Proc Natl Acad Sci U S A. 2007;104:11838–11843. doi: 10.1073/pnas.0701910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schefe JH, Menk M, Reinemund J, Effertz K, Hobbs RM, Pandolfi PP, Ruiz P, Unger T, Funke-Kaiser H. A novel signal transduction cascade involving direct physical interaction of the renin/prorenin receptor with the transcription factor promyelocytic zinc finger protein. Circ Res. 2006;99:1355–1366. doi: 10.1161/01.RES.0000251700.00994.0d. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Valdenaire O, Nelböck P, Deuschle U, Dumas Milne Edwards JB, Stumpf JG, Löffler BM. Human endothelin-converting enzyme (ECE-1): three isoforms with distinct subcellular localizations. Biochem J. 1997;328:871–877. doi: 10.1042/bj3280871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K, Kirsch S, Lucht K, Zaade D, Reinemund J, Schmitz J, Klare S, Li Y, Schefe JH, Schmerbach K, Goldin-Lang P, Zollmann FS, Thöne-Reineke C, Unger T, Funke-Kaiser H. The promyelocytic leukemia zinc finger (PLZF) protein exerts neuroprotective effects in neuronal cells and is dysregulated in experimental stroke. Brain Pathol. 2011;21:31–43. doi: 10.1111/j.1750-3639.2010.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serneri GG, Cecioni I, Vanni S, Paniccia R, Bandinelli B, Vetere A, Janming X, Bertolozzi I, Boddi M, Lisi GF, Sani G, Modesti PA. Selective upregulation of cardiac endothelin system in patients with ischemic but not idiopathic dilated cardiomyopathy: endothelin-1 system in the human failing heart. Circ Res. 2000;86:377–385. doi: 10.1161/01.res.86.4.377. [DOI] [PubMed] [Google Scholar]
- Sharif J, Endo TA, Toyoda T, Koseki H. Divergence of CpG island promoters: a consequence or cause of evolution? Dev Growth Differ. 2010;52:545–554. doi: 10.1111/j.1440-169X.2010.01193.x. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Slattery E, Dignam JD, Matsui T, Roeder RG. Purification and analysis of a factor which suppresses nick-induced transcription by RNA polymerase II and its identity with poly(ADP-ribose) polymerase. J Biol Chem. 1983;258:5955–5959. [PubMed] [Google Scholar]
- Sluck JM, Lin RC, Katolik LI, Jeng AY, Lehmann JC. Endothelin converting enzyme-1-, endothelin-1-, and endothelin-3-like immunoreactivity in the rat brain. Neuroscience. 1999;91:1483–1497. doi: 10.1016/s0306-4522(98)00692-7. [DOI] [PubMed] [Google Scholar]
- Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Moir RD, Wagner SL. Clearance of Alzheimer's Abeta peptide: the many roads to perdition. Neuron. 2004;43:605–608. doi: 10.1016/j.neuron.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Tapia-Páez I, Tammimies K, Massinen S, Roy AL, Kere J. The complex of TFII-I, PARP1, and SFPQ proteins regulates the DYX1C1 gene implicated in neuronal migration and dyslexia. FASEB J. 2008;22:3001–3009. doi: 10.1096/fj.07-104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdenaire O, Lepailleur-Enouf D, Egidy G, Thouard A, Barret A, Vranckx R, Tougard C, Michel JB. A fourth isoform of endothelin-converting enzyme (ECE-1) is generated from an additional promoter molecular cloning and characterization. Eur J Biochem. 1999a;264:341–349. doi: 10.1046/j.1432-1327.1999.00602.x. [DOI] [PubMed] [Google Scholar]
- Valdenaire O, Barret A, Schweizer A, Rohrbacher E, Mongiat F, Pinet F, Corvol P, Tougard C. Two di-leucine-based motifs account for the different subcellular localizations of the human endothelin-converting enzyme (ECE-1) isoforms. J Cell Sci. 1999b;18:3115–3125. doi: 10.1242/jcs.112.18.3115. [DOI] [PubMed] [Google Scholar]
- Walsh PS, Fildes NJ, Reynolds R. Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res. 1996;24:2807–2812. doi: 10.1093/nar/24.14.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, de Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- Zhang J, Leontovich A, Sarras MP., Jr Molecular and functional evidence for early divergence of an endothelin-like system during metazoan evolution: analysis of the Cnidarian, hydra. Development. 2001;128:1607–1615. doi: 10.1242/dev.128.9.1607. [DOI] [PubMed] [Google Scholar]
- Ziegler M, Oei SL. A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays. 2001;23:543–548. doi: 10.1002/bies.1074. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV. New therapeutic targets in the neurovascular pathway in Alzheimer's disease. Neurotherapeutics. 2008;5:409–414. doi: 10.1016/j.nurt.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]