Abstract
Computational and empirical neuroimaging studies have suggested that the anatomical connections between brain regions primarily constrain their functional interactions. Given that the large-scale organization of functional networks is determined by the temporal relationships between brain regions, the structural limitations may extend to the global characteristics of functional networks. Here, we explored the extent to which the functional network community structure is determined by the underlying anatomical architecture. We directly compared macaque (Macaca fascicularis) functional connectivity (FC) assessed using spontaneous blood oxygen level-dependent functional magnetic resonance imaging (BOLD-fMRI) to directed anatomical connectivity derived from macaque axonal tract tracing studies. Consistent with previous reports, FC increased with increasing strength of anatomical connection, and FC was also present between regions that had no direct anatomical connection. We observed moderate similarity between the FC of each region and its anatomical connectivity. Notably, anatomical connectivity patterns, as described by structural motifs, were different within and across functional modules: partitioning of the functional network was supported by dense bidirectional anatomical connections within clusters and unidirectional connections between clusters. Together, our data directly demonstrate that the FC patterns observed in resting-state BOLD-fMRI are dictated by the underlying neuroanatomical architecture. Importantly, we show how this architecture contributes to the global organizational principles of both functional specialization and integration.
Introduction
Spontaneous brain activity in the absence of an explicit task and the temporal relationships of such activity between regions [or resting-state functional connectivity (FC)] has become a valuable tool for investigating the intrinsic network dynamics of the brain (Snyder and Raichle, 2012). These coherent fluctuations are thought to confer different functional network configurations in support of flexible behavior (Deco et al., 2011). Importantly, resting-state FC may provide a valuable measure of information processing capacity and network topology in both healthy and diseased states (Rowe, 2010).
When assessing resting-state blood oxygen level-dependent functional magnetic resonance imaging (BOLD–fMRI) data, graph theoretical tools have provided a mathematical framework for describing the organizational principles of the brain of functional segregation (Zeki, 1978; Passingham et al., 2002; Preuss, 2007) and functional integration (Bressler, 1995; Tononi et al., 1998; Varela et al., 2001). Resting-state fMRI networks have a modular community structure, whereby a network can be segregated into smaller subnetworks (or modules) (Meunier et al., 2010). Brain areas (or nodes) within each module are highly functionally interconnected but have sparse FC with nodes in other modules. Partitioning therefore allows for highly specialized functions within modules and more comprehensive functions across modules. Functional networks have consistently been decomposed into visual, sensorimotor, attention, default-mode, and subcortical subnetworks (He et al., 2009; Meunier et al., 2009; Rubinov and Sporns, 2011). Functional integration across modules is facilitated by “hub” regions having high intermodular FC (Meunier et al., 2010). A growing literature suggests that the functional integrative capacity of a node may be supported by its anatomical connectedness (Sporns et al., 2007). Structural connectivity, as determined using diffusion-weighted techniques, is correlated with FC (Damoiseaux and Greicius, 2009; Bressler and Menon, 2010; Honey et al., 2010), and FC tends to increase with an increasing number of detected axonal fibers between regions (Hagmann et al., 2008; Honey et al., 2009; see also Honey et al., 2007).
FC, when measured using correlation, lacks information about the directionality of information flow. Because local FC is not a perfect reflection of local structural connectivity (Vincent et al., 2007), it is unclear whether the modular organization of functional networks is a product of the underlying anatomical architecture or whether the influence of structure is limited only to determining local functional connections. Indeed, it has been argued that functional flexibility arises from specific patterns of small sets of anatomical connections (motifs; Sporns and Kötter, 2004), and a recent theoretical study has suggested that FC strengths vary with motif patterns (Adachi et al., 2012). Here, we empirically test the hypothesis that the separate-but-connected organization of functional networks is attributable to distinct and directed patterns of anatomical connections. We derived clusters from resting-state FC patterns in macaque monkeys and then examined the anatomical basis of these functionally defined clusters using data from axonal tract tracing studies in macaques. Using histologically derived anatomical data, as opposed to diffusion-weighted techniques, allowed us to explore motif patterns and assess whether the directionality of specific anatomical connections contributed to the observed resting-state network properties. As expected, we found local functional relationships in the macaque cortex to be related to the strength of their underlying anatomical connectivity. Importantly, we report how the global organization of the functional network is determined by specific structural constraints: dense and bidirectional anatomical connections support information processing within highly clustered functional divisions, whereas sparse unidirectional anatomical connections support the integration of information across divisions.
Materials and Methods
Macaque fMRI.
The fMRI data presented here were the focus of a previous report, and the animal preparation, data acquisition, and image preprocessing have been described previously in detail (Hutchison et al., 2011). They are briefly described below. Data were obtained from six (four female) macaque monkeys (Macaca fascicularis, 3.6–5.3 kg). All surgical and experimental protocols were approved by the Animal Use Subcommittee of the University of Western Ontario Council on Animal Care and were in accordance with the Canadian Council on Animal Care guidelines.
Animal preparation.
Before each scanning session, anesthesia was induced using atropine (0.4 mg/kg, i.m.), ipratropium (0.025 mg/kg, i.m.), and ketamine hydrochloride (7.5 mg/kg, i.m.), followed by an intravenous bolus of propofol (3 ml; 10 mg/ml). After intubation with an endotracheal tube, anesthesia was maintained using 1.5% isoflurane in oxygen. Animals were then placed in a custom-built primate chair, and their heads were immobilized before being inserted into the magnet bore. The isoflurane level was lowered to 1% for image acquisition. Animals were spontaneously breathing throughout the duration of the experiment. Physiological parameters were continuously monitored during image acquisition and were within the normal range (rectal temperature, 36.5°C; respiration, 25–30 breaths/min; end-tidal CO2, 24–28 mmHg). Body temperature was maintained using a heating disk and thermal insulation.
Data acquisition.
Data were obtained using an actively shielded 7 T, 68 cm horizontal bore scanner with a DirectDrive console (Varian) and a Siemens AC84 gradient subsystem operating at a slew rate of 350 mT · m−1 · s−1. A five-channel transceive primate head radio frequency coil, designed and manufactured in-house, was used for all experiments. Optimization of the magnetic field (B0 shimming) was done using an automated three-dimensional mapping procedure over the specific volume of interest. Each monkey's scanning session consisted of two runs of 300 continuous echo-planar imaging (EPI) functional volumes [10 min per run; repetition time (TR), 2000 ms; echo time (TE), 16 ms; flip angle, 70°; 30 slices; 72 × 72 matrix; field of view (FOV), 96 × 96 mm; acquisition voxel size, 1.3 × 1.3 × 1.5 mm]. EPI images were acquired with Generalized Autocalibrating Partially Parallel Acquisition at an acceleration factor of 2. Each image was corrected for physiological fluctuations with navigator echo correction. High-resolution T2-weighted anatomical images were also acquired during each session using a fast spin echo acquisition scheme (TR, 5000 ms; TE, 38.6 ms; echo train length, 5; effective echo, 3; 30 slices; 256 × 250 matrix; FOV, 96 × 96; acquisition voxel size, 375 μm × 384 μm × 1.5 mm).
Image preprocessing.
Functional image preprocessing was performed using the FMRIB (Functional MRI of the Brain) Software Library toolbox (http://www.fmrib.ox.ac.uk). Preprocessing included motion correction, brain extraction, spatial smoothing (full-width at half-maximum, 3 mm), high- and low-pass temporal filtering, and normalization to the F99 atlas template (Van Essen, 2004) (http://sumsdb.wustl.edu/sums/macaquemore.do). Nuisance variables were removed by regression using the AFNI (Analysis of Functional NeuroImages) software package (afni.nimh.nih.gov/afni). These included six motion parameters as well as the global white matter and CSF signals. Regression of the global mean signal was not performed.
Anatomical dataset.
Anatomical data were initially derived from the CoCoMac database (Stephan et al., 2001) (http://cocomac.org, redeveloped at http://cocomac.g-node.org) of axonal tract tracing studies using the Regional Map parcellation of Kötter and Wanke (2005) and specified as connectivity between 82 cortical regions of interest (ROIs; 41 per hemisphere; Table 1). This anatomical connectivity matrix included interhemispheric connections (563 of a possible 1640, or 34.3%), of which 270 were mirrored from one hemisphere to the other. If information about the connectivity between two regions was not available in CoCoMac, the connection was deemed to be absent. The anatomical connectivity matrix also classified the strength of the anatomical connections as weak, moderate or strong. Generally, the classification of label density/strength was made relative to other brain sites labeled by the same injection and based on the descriptions provided in the original literature reports (Bezgin et al., 2012). If a description was not provided, the connection strength was set to “moderate.”
Table 1.
Cortical regions of interest abbreviations
| Abbreviation | Region |
|---|---|
| PFCpol | Prefrontal polar cortex |
| PFCm | Medial prefrontal cortex |
| PFCol | Orbitolateral prefrontal cortex |
| PFCom | Orbitomedial prefrontal cortex |
| PFCoi | Orbitoinferior prefrontal cortex |
| PFCdl | Dorsolateral prefrontal cortex |
| PFCdm | Dorsomedial prefrontal cortex |
| PFCcl | Centrolateral prefrontal cortex |
| PFCvl | Ventrolateral prefrontal cortex |
| CCa | Anterior cingulate cortex |
| CCs | Subgenual cingulate cortex |
| CCp | Posterior cingulate cortex |
| CCr | Retrosplenial cingulate cortex |
| FEF | Frontal eye field |
| PMCvl | Ventrolateral premotor cortex |
| PMCdl | Dorsolateral premotor cortex |
| PMCm | Medial premotor cortex |
| M1 | Primary motor cortex |
| S1 | Primary somatosensory cortex |
| S2 | Secondary somatosensory cortex |
| G | Gustatory cortex |
| Ia | Anterior insula |
| Ip | Posterior insula |
| A1 | Primary auditory cortex |
| A2 | Secondary auditory cortex |
| HC | Hippocampus |
| PHC | Parahippocampal cortex |
| Amyg | Amygdala |
| TCc | Central temporal cortex |
| TCi | Inferior temporal cortex |
| TCs | Superior temporal cortex |
| TCpol | Temporal polar cortex |
| TCv | Ventral temporal cortex |
| PCi | Inferior parietal cortex |
| PCip | Intraparietal cortex |
| PCm | Medial parietal cortex |
| PCs | Superior parietal cortex |
| VACv | Anterior visual area (ventral) |
| VACd | Anterior visual area (dorsal) |
| V2 | Visual area 2 |
| V1 | Visual area 1 |
The Regional Map parcellation was drawn on the F99 macaque standard cortical surface template (Van Essen et al., 2001; Bezgin et al., 2008) and transformed to voxel space with a 2 mm extrusion using the Caret software package (http://www.nitrc.org/projects/caret/).
Statistical analysis.
The Regional Map parcellation in F99 voxel space was used to define ROIs in the fMRI data. The anatomical and functional parcellations were therefore the same. A weighted average time series was calculated for each ROI using a probabilistic weighting scheme. Each voxel was weighted according to the probability that it was within the specified ROI, favoring voxels that were deeper within the structure. In other words, voxels nearest the center of each ROI were weighted more heavily than those nearest the ROI boundary. The sum of weights of all voxels within each ROI was therefore equal to 1. To obtain an average FC matrix, regionwise correlations were first performed on these weighted average time series for each monkey's scans. The results were averaged across scans after a Fisher z-transform and then across monkeys before conversion back to correlation coefficients. We used a bootstrapping procedure to eliminate outliers from the average Pearson's correlation coefficient matrix (Efron and Tibshirani, 1993). For each correlation coefficient, we sampled with replacement from the six individual coefficients (one from each animal) and calculated a resampled mean. This was repeated 1000 times to approximate a sampling distribution of mean values for each connection. We then determined the mean and SE of this sampling distribution and used them to exclude unreliable coefficients: if the observed grand average correlation coefficient (averaged across six subjects) for that connection fell outside of the mean ± 2 SE of the bootstrapped distribution, its value was set to 0. We repeated this technique for all connections in the FC matrix. All analyses of the average FC matrix were performed after this bootstrapping procedure.
We computed the cosine similarity of each region to describe the correspondence between the anatomical and FC of a region as follows:
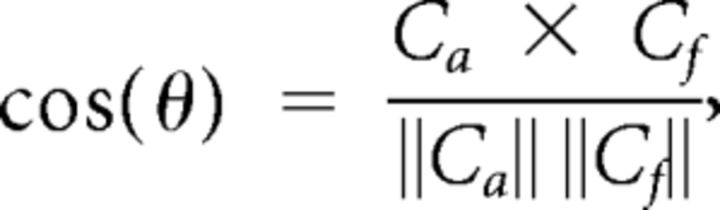 |
where Ca and Cf are the region's anatomical and FC patterns as vectors, respectively. θ is the angle between the anatomical and FC vectors. Because the anatomical connectivity matrix is asymmetric, we treated the pattern of connectivity of each region as a source vector and target vector separately. In other words, the FC vector of each region is compared with a vector of axonal projections that originate from that region as well as a vector of axonal projections that terminate in that region. Similarity can range from −1 to 1, where values approaching −1 indicate that the vectors are nearly opposite and values approaching 1 as highly alike or correlated. A cosine similarity of 0 would indicate that the two vectors are perpendicular to each other, i.e., completely independent. We also obtained a global measure of similarity by reshaping the entire anatomical connectivity and FC matrices into vectors and then computing their cosine similarity.
We used graph theoretical measures derived using the Brain Connectivity Toolbox (BCT; http://www.brain-connectivity-toolbox.net) to evaluate the relationship between structure and function. We describe the community structure of the functional network by applying a method for handling densely connected and weighted graphs to our average FC matrix (BCT function “modularity_louvain_und_sign”). This avoided the need to convert the functional network into a sparse binary graph and allowed us to keep all reliable connections and their weights. This decomposition method essentially maximizes positive connections within modules and negative connections across modules. We chose to make the influence of negative weights less than that of positive weights in determining modularity (Q* option in BCT) because it provides a more neurobiologically plausible decomposition (Rubinov and Sporns, 2011). In this weighting scheme, positive weights have twice the influence as negative ones in a graph with an equal number of positive and negative weights. The resultant Q* modularity measure from this BCT function describes the extent to which the smaller network communities are connected within themselves compared with their connections to each other. A Q* value of 1 means that the modules are fully segregated with no connections between them, and a value of 0 would mean the intermodular and intramodular connections are equivalent. The decomposition was performed using the Louvain optimization algorithm to approximate the highest Q* value possible (Blondel et al., 2008). Using the detected community structure, we derived the functional participation coefficient for positive and negative weights separately (BCT function “participation_coef_sign”). The participation coefficient describes the distribution of each node's connections to nodes in other modules, with values close to 1 indicating that the connections are uniformly distributed among other modules, whereas values close to 0 indicate that the connections are mostly within its own module (Guimer à and Nunes Amaral, 2005). Finally, we generated 1000 fully connected and weighted functional null models for comparison (BCT function “randmio_und_signed”).
Using the community structure of the functional network, we partitioned the anatomical network into matching modules. The network participation indices (NPIs; Kötter and Stephan, 2003) of each node were calculated. These included density (proportion of existing connections relative to all possible connections), symmetry (proportion of bidirectional connections relative to unidirectional ones), and transmission [proportion of efferent connections relative to all (efferent + afferent) connections]. Next, we determined the modularity indices (MIs) of each node for the NPI measures. The MI for each of these measures was taken as the difference between the NPI of a node within its respective module and its corresponding NPI across the entire structural network.
For comparison, we generated 1000 anatomical null models using the BCT by randomly rewiring the anatomical network while preserving the degree distributions (BCT function “randmio_dir”). We additionally generated 1000 anatomical null models with preserved connection length distributions. We estimated connection lengths by calculating the direct Euclidean distance between the centers of each ROI. We then divided the observed connection length distribution into quartiles (delimited by 24.0, 34.6, and 43.2 mm) and rewired the network by randomly swapping connections within each quartile. All of the fixed-degree and fixed-connection length null models were partitioned into four modules as defined by the functional community structure, and their MIs for each of density, symmetry, and transmission were determined.
Finally, motif frequencies were calculated using the BCT (functions “motif3struct_bin” and “motif4struct_bin”) for both the modules and across the whole network in the observed anatomical network as well as for the two sets of anatomical null models. To account for the differences in size between modules as well as their size differences from the whole network, motif frequencies for each node were normalized by dividing by the sum of all motif occurrences for that node (i.e., motif probability).
Results
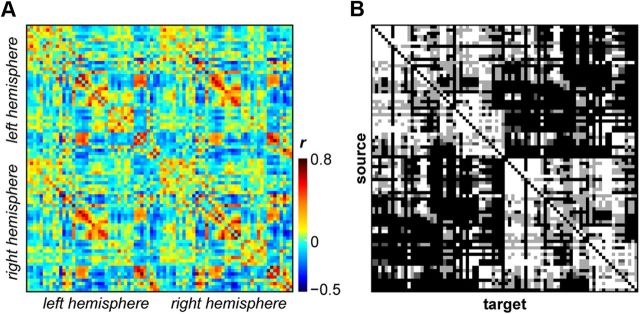
We derived resting-state cortical FC measures using regionwise correlations of BOLD time series between 82 anatomically defined ROIs (41 per hemisphere; Table 1) for each animal. Figure 1A shows the average FC across all animals, with correlation coefficients ranging between −0.436 and 0.773. FC was strong both within (range, −0.437 to 0.704) and across (range, −0.377 to 0.773) hemispheres. Figure 1B depicts the anatomical connectivity of the same 82 regions as derived from the CoCoMac database.
Figure 1.
Connectivity matrices of 82 macaque cortical regions. A, Resting-state BOLD–fMRI FC (Pearson's correlation coefficients) averaged across six monkeys. B, Structural connectivity derived from the CoCoMac database of macaque axonal tract tracing studies. Strengths of anatomical connections are denoted by shading (strong, white; moderate, light gray; weak, dark gray; absent connection, black). Matrices are organized generally from anterior to posterior regions, for left then right hemispheres. Labels and ordering of regions within each hemisphere are specified in Table 1.
Local FC varies with anatomical connectivity
We found the strength of FC to vary significantly with the strength of anatomical connectivity when anatomical connections were present (Kruskal–Wallis test, p < 0.001; Fig. 2A). These results suggest that the density of the underlying axonal tracts dictate, to some degree, the strength of the functional connections. Consistent with previous studies (Koch et al., 2002; Honey et al., 2009; Adachi et al., 2012), we also found strong FC in the absence of direct axonal connections (Fig. 2A). When two regions were not anatomically connected, median FC was −0.126, which was significantly different from the median correlation coefficients of pairs with direct anatomical connections (see above; Kruskal–Wallis test, p < 0.001), and this was driven by a large proportion (0.69) of negative correlations. Inferring FC from anatomical connectivity therefore resulted in a number of false-positive and false-negative detections. True-positive detections made up for 52.6% (3492 of 6642), false-negatives for 22.3% (1479 of 6642), and false-positives for 25.2% (1671 of 6642) of all connections. Notably, the vast majority of false-positive detections (72.3%) were found in intrahemispheric connections. Together, these results suggest that FC is additionally mediated by indirect anatomical connections.
Figure 2.
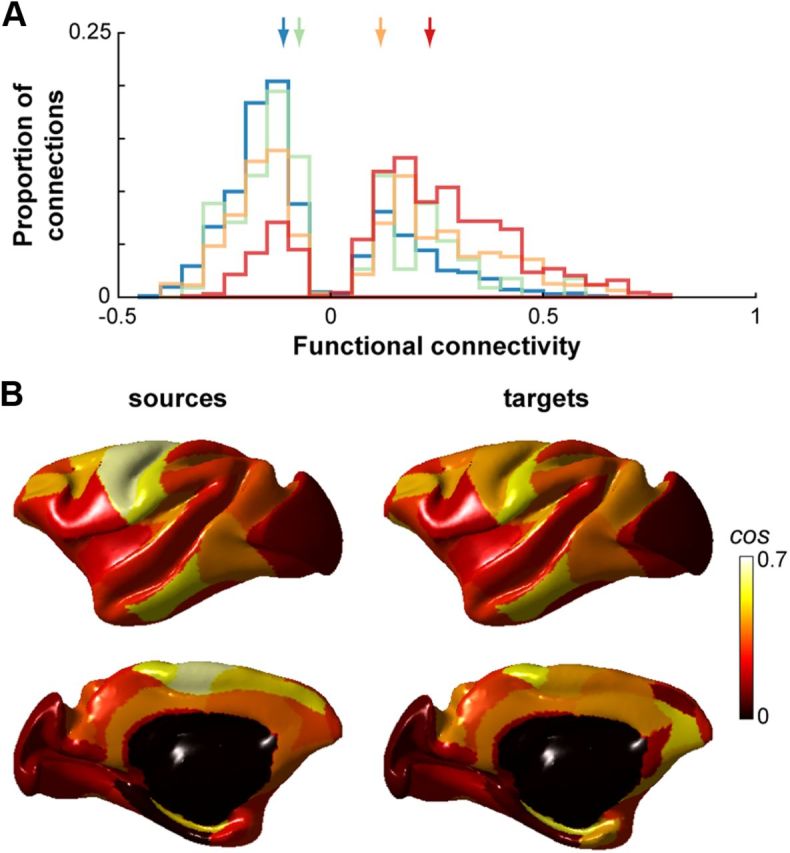
FC varies with anatomical connectivity. A, Distributions of FC strengths for region pairs with no direct anatomical connections (blue) and weak (green), moderate (orange), and strong (red) anatomical connections. Colored arrows indicate the medians of the corresponding distributions (no direct anatomical connection, −0.126; weak −0.090; moderate, 0.092; strong, 0.201). Only reliable coefficients as determined by a bootstrapping procedure are shown. B, Cosine similarity between functional and structural connectivity for each cortical region of the left hemisphere projected onto the F99 surface. Values are displayed for regions as sources and targets separately. Right hemisphere cosine similarities were similar (data not shown).
To further examine the relationship between local FC and the underlying anatomical structure, we determined the cosine similarity between the FC and anatomical connectivity matrices. Akin to a correlation, the cosine similarity describes the correspondence between two vectors. When considering the whole cortical network, the cosine similarity was 0.32. When considering regions individually, however, cosine similarity ranged extensively from −0.19 to 0.63 when regions were treated as sources and from −0.13 to 0.54 when they were treated as targets (Fig. 2B). Cosine similarity was greater for intrahemispheric comparisons (0.34 and 0.33 for left and right hemispheres, respectively) than for interhemispheric comparisons (0.25 and 0.23 for left-to-right and right-to-left, respectively), owing mostly to sparser anatomical connections between hemispheres.
Anatomical connections support global functional community structure
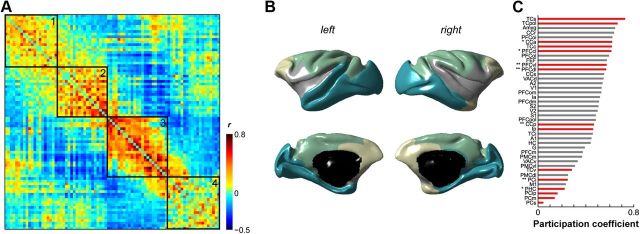
To determine the extent to which the neuroanatomical architecture constrains functional organization, we examined the connections within and across anatomical subnetworks that support various functional subnetworks. We first partitioned the functional network into modules using previously described graph theoretical tools (Rubinov and Sporns, 2011). The macaque functional network was highly modular compared with null models (Q* = 0.503 vs 0.1995 ± 0.0008) and partitioned into four subnetworks (Fig. 3A,B; Table 2): (1) an anterior module predominantly comprising prefrontal and cingulate regions; (2) a ventrolateral module that included the secondary somatosensory and auditory cortices, the ventrolateral premotor cortex, and the insulae; (3) a dorsomedial module including both primary somatosensory and motor cortices as well as extrastriate and parietal areas; and (4) a posterior module that included visual areas V1 and V2 as well as limbic and paralimbic regions (amygdala, hippocampus, parahippocampus). When considering the functional networks across individual animals, the partitions varied slightly, but the networks were highly modular and most regions were consistently clustered into similar subnetworks (Table 3).
Figure 3.
Community structure of macaque resting-state fMRI networks. A, The functional network partitioned into four modules. Node ordering as specified in Table 2. B, Functional community structure projected onto the F99 surface. Modules 1 (yellow), 2 (gray), 3 (green), and 4 (blue) as outlined in Table 2. C, Rank-ordered functional participation coefficients for positive weights in left hemisphere. Right hemisphere data were similar (data not shown). Nodes identified as hubs in the structural network using apex ratio are denoted by red bars (Fig. 7). * denote structural hubs identified using degree measures. ** denote structural hubs identified using both degree and betweenness centrality measures. For abbreviations, see Table 1.
Table 2.
Cortical regions comprising each partition of the functional network (see Fig. 3A)
| Module | Cortical regions |
|---|---|
| 1 | Dorsolateral prefrontal cortex |
| Prefrontal polar cortex | |
| Centrolateral prefrontal cortex | |
| Medial prefrontal cortex | |
| Dorsomedial prefrontal cortex | |
| Orbitomedial prefrontal cortex | |
| Anterior cingulate cortex | |
| Orbitolateral prefrontal cortex | |
| Subgenual cingulate cortex | |
| Amygdala, R | |
| Temporal polar cortex, R | |
| 2 | Orbitoinferior prefrontal cortex |
| Anterior insula | |
| Superior temporal cortex, R | |
| Ventrolateral prefrontal cortex | |
| Posterior insula | |
| Gustatory cortex | |
| Primary auditory cortex | |
| Ventrolateral premotor cortex | |
| Secondary somatosensory cortex | |
| Secondary auditory cortex | |
| 3 | Primary motor cortex |
| Primary somatosensory cortex | |
| Medial premotor cortex | |
| Dorsolateral premotor cortex | |
| Superior parietal cortex | |
| Inferior parietal cortex | |
| Intraparietal cortex | |
| Medial parietal cortex | |
| Posterior cingulate cortex | |
| Frontal eye field | |
| Anterior visual area (dorsal) | |
| Retrosplenial cingulate cortex, L | |
| 4 | Visual area 2 |
| Retrosplenial cingulate cortex, R | |
| Central temporal cortex | |
| Anterior visual area (ventral) | |
| Superior temporal cortex, L | |
| Visual area 1 | |
| Ventral temporal cortex | |
| Inferior temporal cortex | |
| Hippocampus | |
| Amygdala, L | |
| Temporal polar cortex, L | |
| Parahippocampal cortex |
Regions of both left (L) and right (R) hemispheres appear together in the same module unless otherwise stated.
Table 3.
Functional network partitions for individual monkeys
| Cortical region | Module |
|||||
|---|---|---|---|---|---|---|
| M1 Q = 0.5435 | M2 Q = 0.5216 | M3 Q = 0.4522 | M4 Q = 0.5213 | M5 Q = 0.4908 | M6 Q = 0.3329 | |
| PFCpol | 1 | 1 | 1 | 1 (L), 2 (R) | 1 | 1 |
| PFCm | 1 | 1 | 5 | 1 | 1 | 1 |
| PFCol | 1 | 1 | 4 | 1 | 1 | 1 |
| PFCom | 1 | 1 | 5 | 1 | 1 | 1 |
| PFCoi | 1 | 3 | 4 | 1 | 1 | 1 |
| PFCdl | 1 | 1 | 1 | 2 | 2 (L), 1 (R) | 2 |
| PFCdm | 1 | 1 | 5 | 2 | 1 | 1 |
| PFCcl | 1 | 1 | 1 | 2 | 4 (L), 2 (R) | 1 |
| PFCvl | 4 | 1 | 4 | 1 | 1 | 3 |
| CCa | 2 | 1 | 5 | 1 | 1 | 1 |
| CCs | 1 (L), 4 (R) | 1 | 5 (L), 4 (R) | 1 | 1 | 1 |
| CCp | 2 | 1 (L), | 1 | 2 | 2 | 2 |
| CCr | 3 (L), 2 (R) | 1 | 1 | 1 (L), 2 (R) | 1 (L), 2 (R) | 2 (L), 1 (R) |
| FEF | 1 | 1 (L), | 1 | 2 | 2 | 1 (L), 2 (R) |
| PMCvl | 4 | 3 | 4 | 1 | 1 | 3 |
| PMCdl | 2 (L), 1 (R) | 2 | 2 (L), 1 (R) | 2 | 2 | 2 |
| PMCm | 2 | 2 | 2 | 2 | 2 | 2 (L), 3 (R) |
| M1 | 2 | 2 (L), | 2 | 2 | 2 | 2 |
| S1 | 4 | 3 | 4 | 2 | 2 | 5 (L), 2 (R) |
| S2 | 4 | 3 | 4 | 1 | 1 | 3 |
| G | 4 | 3 | 4 | 1 | 1 | 3 |
| Ia | 4 | 3 | 4 | 1 | 1 | 1 |
| Ip | 4 | 3 | 4 | 1 | 1 | 1 (L), 3 (R) |
| A1 | 4 | 3 | 4 | 1 | 1 | 3 |
| A2 | 4 | 3 | 4 | 1 | 1 | 3 (L), 1 (R) |
| HC | 3 | 3 | 3 (L), 1 (R) | 1 | 1 | 3 (L), 1 (R) |
| PHC | 3 | 3 | 3 (L), 1 (R) | 1 | 1 | 3 |
| Amyg | 1 | 3 | 3 (L), 4 (R) | 1 | 1 | 1 |
| TCc | 1 | 1 | 1 | 1 (L), 2 (R) | 5 | 1 (L), 3 (R) |
| TCi | 3 | 3 | 3 | 1 (L), 2 (R) | 1 (L), 3 (R) | 3 |
| TCs | 1 | 3 | 1 (L), 4 (R) | 1 (L), 2 (R) | 5 (L), 1 (R) | 1 (L), 3 (R) |
| TCpol | 1 | 3 | 4 | 1 | 1 | 1 |
| TCv | 3 | 3 | 3 (L), 1 (R) | 1 | 1 (L), 3 (R) | 3 |
| PCi | 2 | 2 | 2 | 2 | 2 | 2 |
| PCip | 2 | 2 | 2 (L), 6 (R) | 2 | 2 | 2 |
| PCm | 2 | 2 | 2 (L), 1 (R) | 2 | 2 | 2 |
| PCs | 2 | 2 | 2 | 2 | 2 | 2 |
| VACv | 3 | 3 (L), | 3 (L), | 1 | 2 | 3 |
| VACd | 2 | 1 | 1 | 2 | 2 | 2 (L), 4 (R) |
| V2 | 3 (L), 2 (R) | 1 | 3 (L), | 1 | 2 | 2 |
| V1 | 3 | 1 | 3 | 1 | 1 (L), 2 (R) | 2 |
Modularity (Q) is indicated for each monkey. Regions of both left (L) and right (R) hemispheres appear together in the same module unless otherwise stated. For cortical region abbreviations, see Table 1.
We can further describe the community structure of the functional network by computing a participation coefficient for each region. This measure characterizes the extent to which each region participates across the entire network, with larger coefficients indicative of a more diverse set of intermodular connections. Figure 3C shows the functional participation coefficient for positive weights. Some regions—such as those in parietal cortex, parahippocampal cortex, and primary motor cortex—have low participation coefficients, suggesting that they are more functionally embedded within their own module. Other regions—such as superior and polar temporal cortices, the amygdala, retrosplenial and anterior cingulate cortices, as well as orbito-prefrontal cortex—have high participation coefficients, suggesting that they have a more diverse set of functional connections to regions outside of their own modules.
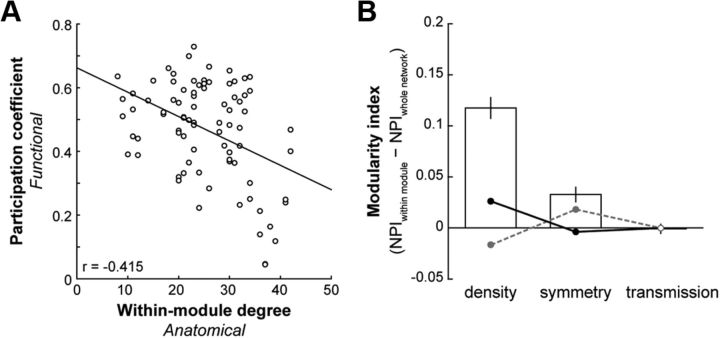
To determine whether there is a relationship between structure and function at the network level, we correlated the functional participation coefficient to the within-module degree in the anatomical network. We found that the functional participation coefficient for positive weights varied with the anatomical within-module degree (Pearson's correlation, r = −0.415, p < 0.0001; Fig. 4A): the fewer intramodular anatomical connections that a region had, the more likely it was to have more intermodular functional connections. We found no such relationship for the functional participation coefficient calculated with negative weights (r = 0.127, p = 0.25).
Figure 4.
Functional modules are supported by specific anatomical connectivity. A, Participation coefficient for positive weights in the functional network as a function of the anatomical network within-module degree (p < 0.0001). B, Average ± SEM MI of NPIs. NPIs (density, symmetry, transmission) for each node in the anatomical connectivity matrix were calculated in the context of its functionally defined module as well as the entire network. The MI was taken as the difference in NPI between these two contexts. Circular data points represent the average ± SEM MI of NPIs for 1000 null models with fixed-degree (gray dashed line) and fixed-connection length (black solid line) distributions. Filled circles represent significant differences from the observed in at least 75% of null models (t tests, p < 0.05).
We next examined whether the anatomical connectivity within the functionally defined partitions were different from that across the entire network. We determined the MI of each anatomical node for a set of NPIs (see Materials and Methods). A positive MI value indicates a greater participation of that anatomical node within its functionally defined module than across the whole network, and a negative MI indicates a greater participation of that node across the whole network than within its functionally defined module. The MI was significantly >0 for both density and symmetry (Fig. 4B; t test, p < 0.001). This suggests that nodes were more likely to be connected to other nodes within their own modules than to nodes outside of their modules. Moreover, their connections within their own modules were more likely to be bidirectional compared with their connections to nodes outside their modules. The MI for transmission, however, was not significantly different from 0 (Fig. 4B; p = 0.87), indicating that the proportion of afferent and efferent connections of a node was the same within and outside of its module. Notably, the MI for density was significantly greater than that for all null models with preserved degree distributions (Fig. 4B; t tests, all p < 0.001). The MI for symmetry was significantly greater than that for the majority of these null models (Fig. 4B; 791 of 1000 with p < 0.05). The observed MI for transmission, however, was no different from the fixed degree null models (Fig. 4B; all p > 0.11).
Previous studies have shown how FC increases with decreasing distance between regions (Honey et al., 2009), and we found a similar, but weak, relationship between distance and positive FC weights (Pearson's correlation, r = −0.27, p < 0.001) but not negative FC weights (r = 0.043, p = 0.20). Anatomical connectivity, when measured using diffusion imaging techniques, is also known to increase with decreasing distance (Honey et al., 2009; Lewis et al., 2009), and we observed a similar pattern of anatomical connectivity in our histologically derived dataset, with the distribution of connection lengths skewed toward shorter distances (data not shown) (Kaiser and Hilgetag, 2004). Because our functional divisions were spatially contiguous (Fig. 3B), it is possible that the increased density and symmetry of anatomical connections we observed within modules were simply because of the greater proximity of regions within modules. We tested the hypothesis that asymmetrical connections are more likely to exist between distant regions by examining the connection lengths of both unidirectional and bidirectional connections in our anatomical data. Within hemispheres, we found no differences in the distances between regions that were unidirectionally connected (median, 26.5 mm), those that were bidirectionally connected (median, 28.2 mm), and those that were not connected (median, 27.8 mm) (Kruskall–Wallis test, p = 0.318). The only difference we observed was when we also considered connections between hemispheres (p < 0.001). A post hoc comparison indicated that interhemispheric regions that were not connected were significantly more distant from each other (median, 37.9 mm) than either unidirectional (median, 31.4 mm) or bidirectional (median, 31.4 mm) connections across hemispheres (Tukey–Kramer test, p < 0.05), and no differences existed between interhemispheric unidirectional and bidirectional connection lengths. Anatomical connection symmetry was therefore not a function of distance. We also compared our observed anatomical network measures with those from 1000 null models whose connection length distributions were preserved. The MI for density was significantly >0 for most null models (771 of 1000; t tests, p < 0.05), suggesting that proximity does play a role in determining the density of connections within modules. However, we still found a large and significant difference in the MI for density between the observed network and the null models (Fig. 4B; average MI, 0.12 vs 0.026; t tests, all p < 0.01). The same was true for symmetry for most null models (946 of 1000; t tests, p < 0.05), additionally substantiating our above analyses that showed how symmetry did not increase with decreasing distance.
Distinct structural motifs support functional organization
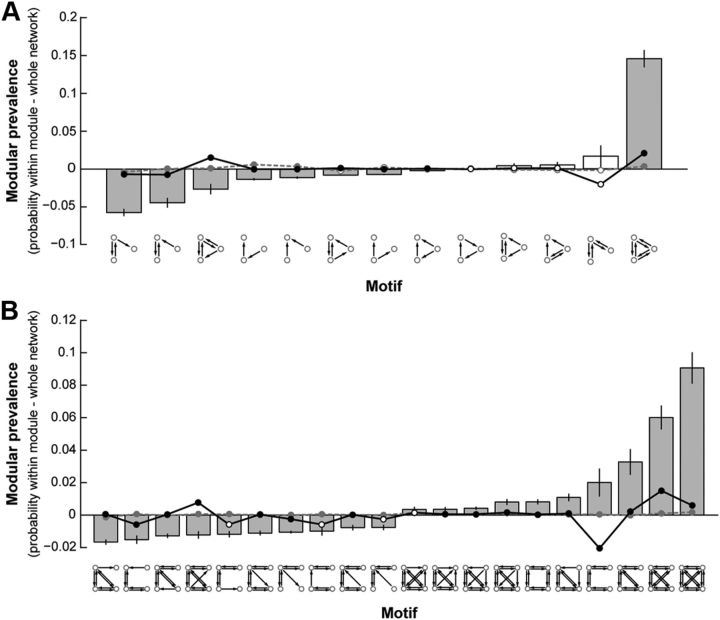
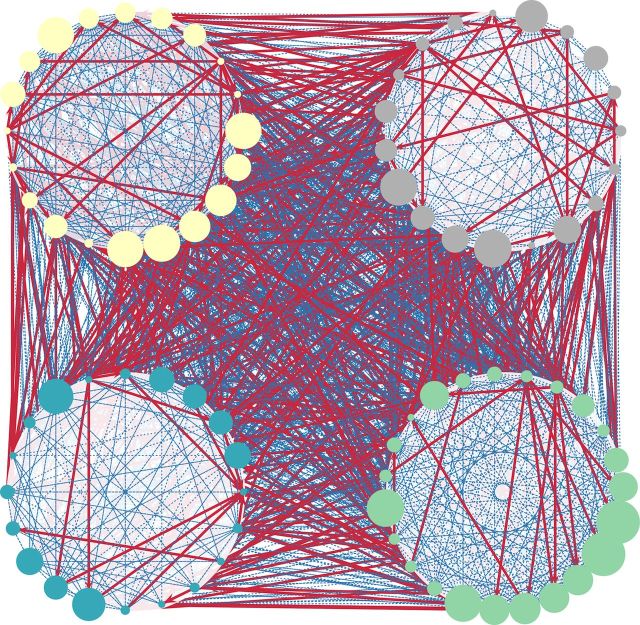
The symmetry of anatomical connections within functionally defined modules suggests that information processing is supported by bidirectional anatomical connections within modules that allow for greater functional integration. To explore this further, we compared structural motifs found within modules with those found across the whole network. Specifically, for each unique motif, we computed the probability of occurrence for each node, considering only its respective module or considering the network as a whole. We describe the prevalence of a motif within modules as the difference between these two probabilities. Figure 5A shows the modular prevalence for motifs with three nodes. The fully connected bidirectional motif had the highest modular prevalence, on average being 14.6% more likely to appear within functionally defined modules than when considering the whole network. Interestingly, the motifs with the highest probability of occurrence outside of the modules (having a negative modular prevalence) were those with a single asymmetrically connected node. Examining the prevalence of motifs having four nodes yielded similar results (Fig. 5B). Together with the NPIs, these data suggest that functional modules are made up of dense, bidirectional structural connections and that functional communication between modules is supported by unidirectional connections via specific nodes (Fig. 6).
Figure 5.
Modular prevalence of motif classes with three (A) and four (B) nodes. Average ± SEM difference between the probability of motif occurrence within module and across the whole network. Filled bars represent significant differences from 0 (t test, p < 0.001). Circular data points represent the average ± SEM modular prevalence of motif classes for 1000 null models with fixed-degree (gray dashed line) and fixed-connection length (black solid line) distributions. Filled circles represent significant differences from the observed in at least 75% of null models (t tests, p < 0.05). In B, only the top 10 most and least prevalent motif classes are shown.
Figure 6.
Functional community structure is supported by differential anatomical wiring within and across modules. Anatomical connections are depicted for functionally defined modules. Unidirectional connections (red) are predominantly between modules, whereas bidirectional connections (blue) are more abundant within modules. Modules 1–4 are shown clockwise starting from the top left (colors as in Fig. 3B). The size of each node is scaled according to its apex ratio (see Fig. 7).
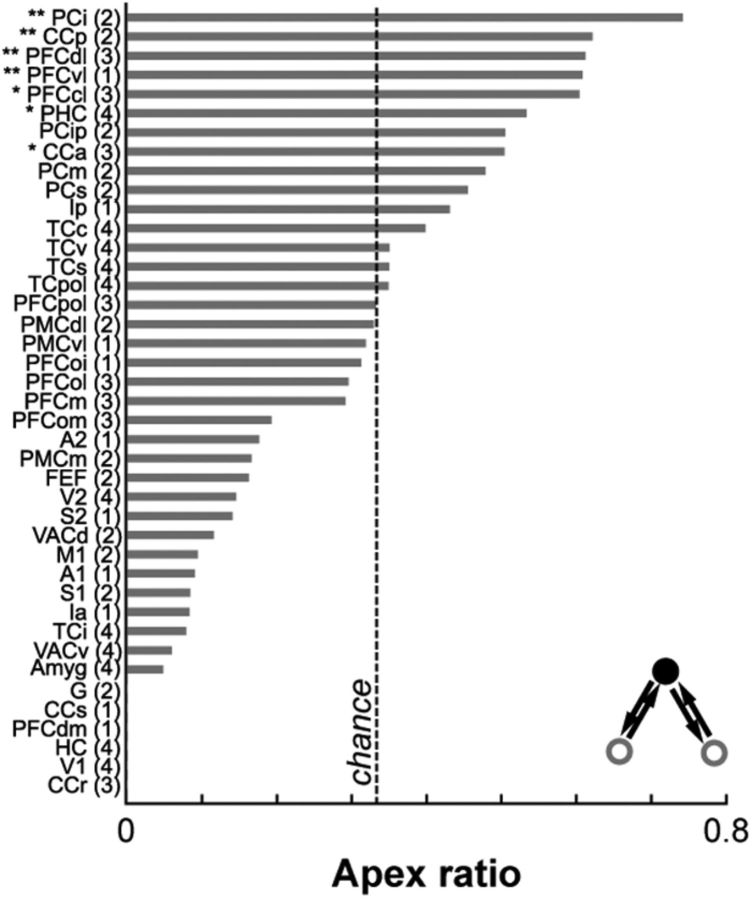
To determine which anatomical nodes act as hubs to support functional integration both within (provincial hubs) and across (connector hubs) modules, we computed the apex ratio for each region in the structural network (Sporns et al., 2007). This measure gives an estimate of the “hubness” of a region by determining how often each region is the central node in a specific hub-like motif in which two unconnected nodes are bidirectionally connected to a third apical node (Fig. 7, inset). When considering the whole anatomical network, the apex ratio was high for regions in the parietal, prefrontal, cingulate, and temporal cortices, as well as in the posterior insula and the parahippocampal cortex (Fig. 7). Using degree or betweenness centrality measures to identify hub regions gave similar results. These structural hubs spanned all functionally defined modules. Notably, regions identified as structural hubs also tended to be those regions with the highest and lowest functional participation coefficients (Fig. 3B). Structural hubs with a greater diversity of FC across modules included regions in the temporal and prefrontal cortices, as well as the anterior cingulate cortex. In contrast, structural hubs whose FC was limited mostly to its own module included regions in the parietal cortex and the parahippocampal cortex. These areas were generally consistent with areas previously identified as functional hubs in humans (Buckner et al., 2009; Cole et al., 2010). Specific anatomical connections therefore determine the degree to which a region is functionally connected to other regions within and across modules.
Figure 7.
Hubs in the structural network span all functional modules. Rank-ordered apex ratio for nodes in the anatomical network for the left hemisphere. Right hemisphere data were similar (data not shown). The functional module assignment of each node is denoted in parentheses. * denote hubs identified using degree measures. ** denote hubs identified using both degree and betweenness centrality measures. For abbreviations, see Table 1.
Discussion
In the present study, we examined the relationship between structure and function in the primate brain by comparing directed anatomical connectivity derived from axonal tract tracing studies in macaques to resting-state BOLD–fMRI FC obtained from anesthetized macaques. We found that the strength of FC varied with the strength of the anatomical connections between regions and that FC was robustly present even between regions that had no direct anatomical connection. We also demonstrated moderate correspondence between the FC and anatomical connectivity of each region. Importantly, we showed how functional modularity was associated with denser and highly bidirectional anatomical connections within modules. An examination of the motif structures revealed that symmetrically connected structural motifs occurred more frequently within functionally defined modules compared with the whole network, whereas asymmetrically connected motifs occurred more frequently between modules. Finally, we identified structural hubs that facilitate communication both within and between functional modules. Together, these results suggest that local neuroanatomical connectivity helps to establish the global community structure of the functional network.
The relationship between brain structure and BOLD–fMRI has drawn much interest in recent years. In humans, studies using diffusion imaging-based white matter tractography in combination with BOLD–fMRI have qualitatively described how functionally connected regions have corresponding white matter connectivity (Greicius et al., 2009; van den Heuvel et al., 2009a). In addition, quantitative comparisons have shown how the strength of FC varies with the density of the detected fiber tract (Koch et al., 2002; Hagmann et al., 2008; Skudlarski et al., 2008; Honey et al., 2009). As in the present study, however, FC is also consistently reported between regions that have no direct anatomical connections (Koch et al., 2002; Vincent et al., 2007; Skudlarski et al., 2008; Habas et al., 2009; Honey et al., 2009; Buckner et al., 2011), suggesting that FC between pairs of regions is also mediated by polysynaptic as well as shared afferent/efferent connections with other regions. The similarity we observed between FC and anatomical connectivity was ∼0.3 overall and in line with previous reports in humans (Hagmann et al., 2008; Skudlarski et al., 2008; Honey et al., 2009). The relationship between structure and function is, therefore, non-trivial and BOLD–fMRI FC analyses serve to complement rather than replace diffusion imaging or anatomical tracer studies in determining anatomical connectivity. Constructing functional networks using coarser timescales yields better matches between structure and function (Honey et al., 2007), whereas finer timescales result in highly variable functional networks (Chang and Glover, 2010; Hutchison et al., 2012a). This variability in functional networks is believed to emerge from the intrinsic dynamics of individual nodes, allowing the system to maintain a broad repertoire of states (Bressler and Kelso, 2001; Deco et al., 2011). Thus, different configurations of coactive regions (i.e., subnetworks) can occur in different functional contexts despite a fixed structural backbone (McIntosh, 2000, 2004; Sporns and Kötter, 2004), leading to a somewhat weak correspondence between structural and functional networks.
In the present study, resting-state BOLD–fMRI was acquired from macaques under anesthesia, and the resulting FC patterns may not reflect the network dynamics of the awake brain. However, homologous resting-state networks have been reported in both awake (Moeller et al., 2009; Mantini et al., 2011) and anesthetized (Vincent et al., 2007; Hutchison et al., 2011; Adachi et al., 2012) monkeys, suggesting a conserved functional architecture between monkeys and humans (Margulies et al., 2009; Kelly et al., 2010; Hutchison et al., 2012b). Our resting-state macaque cortical network formed four functional clusters. The first (anterior) module included regions in the lateral prefrontal cortex thought to be involved in cognitive control (Miyashita and Hayashi, 2000; Miller and Cohen, 2001). This module was also composed of regions important in reward-related behavior, such as the amygdala, medial and orbito-prefrontal cortices, as well as the anterior cingulate cortex (Cardinal et al., 2002; Holland and Gallagher, 2004). The second (ventrolateral) module included ventrolateral prefrontal cortex, ventrolateral premotor cortex, the insulae, gustatory cortex, as well as both primary and secondary auditory cortices. These regions make up parts of the ventral motor system, a network thought to be involved in understanding observed actions and visually guided grasping (Nelissen et al., 2005; Davare et al., 2011). This network is additionally involved in controlling the movements of the mouth and tongue and is considered the precursor to the human language system (Binkofski and Buccino, 2006; Gentilucci and Dalla Volta, 2008). The third (dorsomedial) module was primarily composed of regions involved in sensorimotor integration. For example, the dorsal anterior visual area and regions in the posterior parietal cortex form the visuomotor networks for eye movements (Moore et al., 2003; Johnston and Everling, 2008) and limb movements (Picard and Strick, 2001; Hoshi and Tanji, 2007) via the frontal eye fields and premotor/motor cortices, respectively. A fourth (posterior) module comprised visual and extrastriate regions responsible for visual processing (Felleman and Van Essen, 1991; Tsao et al., 2003), as well as the amygdala and hippocampus whose interactions are indicated in both declarative and emotional memory (Rolls et al., 1993; Chudasama et al., 2009). We also identified structural hubs that serve to aid functional communication either across modules or within modules by showing how these regions tended to have either high or low functional participation coefficients (Fig. 3B). These regions may therefore be considered putative functional connector and provincial hubs, respectively.
Most empirical studies of the structural contribution to functional networks have used diffusion imaging-based tractography to measure anatomical connectivity. These methods, however, suffer from several limitations (Mori and Zhang, 2006). First, they cannot provide information on the directionality of axons. Second, crossing fibers are difficult to detect in diffusion tensor imaging as fiber orientations within a given voxel are averaged. Diffusion spectrum imaging may be able to resolve the fiber orientation complexities within voxels, but it is still constrained by the acquisition resolution which limits the size of detectable fiber tracts, as well as the ability to discern bundles that cross each other at small angles (Hagmann et al., 2007; Wedeen et al., 2008). Using data from invasive axonal tract tracing studies provided us a more direct measure of the anatomical connectivity of the macaque brain. Importantly, it allowed us to investigate whether the directionality of axonal connections plays a role in determining the organization of functional networks. Connectivity data collated in CoCoMac, however, lacks information about some interhemispheric connections because tracing studies predominantly investigate ipsilateral connections. This incompleteness of the anatomical connectivity matrix likely had little effect on our results, because computing the cosine similarity for only region pairs whose anatomical connectivity is explicitly specified in CoCoMac resulted in only a slightly higher correspondence between structure and function than when we consider all region pairs (0.37 vs 0.32).
Invasive axonal tract tracing studies, in conjunction with recent advances in macaque resting-state BOLD–fMRI, can therefore provide a more thorough understanding of the contribution of anatomical connectivity to resting-state FC in the primate brain. A qualitative comparison of resting-state FC in monkeys to known axonal tracts has demonstrated some correspondence between the two (Mantini et al., 2011). Additionally, Adachi et al. (2012) reported how resting-state FC in monkeys increased with increasing symmetry of the anatomical connections between regions. Using computational modeling, they showed how the pattern of structural connectivity had a significant impact on the FC between two anatomically unconnected regions. Randomly rewired anatomical networks that preserved motif frequency had the greatest correspondence to the empirical data than when preserving the degree distributions, modularity, or clustering coefficient of nodes within the macaque anatomical network. Here, we add to this existing literature by demonstrating empirically that network-level functional segregation is supported by the underlying anatomical connectivity. Notably, we observed no differences between the unidirectional and bidirectional connection length distributions despite a general decreasing probability of anatomical connection with distance. This result suggests that the extra metabolic cost of maintaining a long-range bidirectional connection as opposed to a unidirectional one is outweighed by the functional advantage of increased information flow, further supporting our findings that the directionality of connections drive functional segregation and specialization.
Most previous human studies of functional modularity have reported the decomposition of average whole-brain networks into five modules (Fair et al., 2009; He et al., 2009; Meunier et al., 2009), whereas we detected only four. The differences in decomposition could be attributable to inherent differences between species or differences in parcellation schemes, because we did not include subcortical structures that are commonly included in human networks. Decomposition differences could also be attributable to differences in network construction, because previous studies used sparse and binary adjacency matrices. Interestingly, Rubinov and Sporns (2011) showed how increasing the influence of negative weights led to a decreasing number of modules, whereas increasing the threshold of a binarized matrix resulted in an increasing number of modules, as is expected as the graph becomes more and more disconnected. A recent study examining individual human functional networks reported the detection of between 4 and 5 (mean, 4.6) using the same algorithm as in the present study (Lord et al., 2012), suggesting that some variability exists across individuals (Table 3). Of note, the decomposition of average human functional networks using the techniques we used resulted in only four consistent modules (Rubinov and Sporns, 2011), which closely match the functional divisions we observed.
By characterizing the community structure of a macaque resting-state functional network, we showed how this functional clustering was a consequence of the underlying structural organization. This is in line with previous work examining the modular structure of anatomical networks. Partitioning of both monkey and human structural networks result in anatomical subnetworks that may underlie different sensory, motor, and cognitive functions (Hilgetag et al., 2000; Chen et al., 2008; Hagmann et al., 2008). Our data support the hypothesis that both functional specialization (Passingham et al., 2002) and functional integration (Sporns et al., 2007) are attributable to patterns of local afferent and efferent connections. The notable presence of fully connected symmetric motifs within modules suggests that these connections support greater functional integration within modules. The higher prevalence of asymmetric motifs outside of the modules suggests that functional information flow between modules is supported by unidirectional anatomical connections.
Resting-state FC is increasingly considered as a measure of functional capacity. For example, graph measures that characterize the efficiency of the functional network are correlated with intellectual ability (van den Heuvel et al., 2009b). Moreover, altered FC is consistently reported in a growing number of neurodegenerative and neuropsychiatric disorders (Buckner et al., 2008; Greicius, 2008; Bassett and Bullmore, 2009). Little is known, however, about the underlying neuroanatomy in those states. Given the highly conserved nature of neuroanatomical connections across primate species (Katz and Harris-Warrick, 1999; Preuss, 2000), it is reasonable to infer that homologous resting-state networks arise from the shared constraints of homologous anatomical architectures. As such, the macaque brain provides a powerful model for studying the relationship between structure and function in disease states.
Footnotes
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) (S.E.) and the J. S. McDonnell Foundation (A.R.M.). K.S. holds a Fellowship Award from the CIHR.
The authors declare no competing financial interests.
References
- Adachi Y, Osada T, Sporns O, Watanabe T, Matsui T, Miyamoto K, Miyashita Y. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb Cortex. 2012;22:1586–1592. doi: 10.1093/cercor/bhr234. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET. Human brain networks in health and disease. Curr Opin Neurol. 2009;22:340–347. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezgin G, Wanke E, Krumnack A, Kötter R. Deducing logical relationships between spatially registered cortical parcellations under conditions of uncertainty. Neural Netw. 2008;21:1132–1145. doi: 10.1016/j.neunet.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Bezgin G, Vakorin VA, van Opstal AJ, McIntosh AR, Bakker R. Hundreds of brain maps in one atlas: Registering coordinate-independent primate neuro-anatomical data to a standard brain. Neuroimage. 2012;62:67–76. doi: 10.1016/j.neuroimage.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Binkofski F, Buccino G. The role of ventral premotor cortex in action execution and action understanding. J Physiol Paris. 2006;99:396–405. doi: 10.1016/j.jphysparis.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008:P10008. [Google Scholar]
- Bressler SL. Large-scale cortical networks and cognition. Brain Res Brain Res Rev. 1995;20:288–304. doi: 10.1016/0165-0173(94)00016-i. [DOI] [PubMed] [Google Scholar]
- Bressler SL, Kelso JA. Cortical coordination dynamics and cognition. Trends Cogn Sci. 2001;5:26–36. doi: 10.1016/s1364-6613(00)01564-3. [DOI] [PubMed] [Google Scholar]
- Bressler SL, Menon V. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn Sci. 2010;14:277–290. doi: 10.1016/j.tics.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:2322–2345. doi: 10.1152/jn.00339.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinal RN, Parkinson JA, Hall J, Everitt BJ. Emotion and motivation: the role of the amygdala, ventral striatum, and prefrontal cortex. Neurosci Biobehav Rev. 2002;26:321–352. doi: 10.1016/s0149-7634(02)00007-6. [DOI] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Germann J, Evans AC. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cereb Cortex. 2008;18:2374–2381. doi: 10.1093/cercor/bhn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudasama Y, Izquierdo A, Murray EA. Distinct contributions of the amygdala and hippocampus to fear expression. Eur J Neurosci. 2009;30:2327–2337. doi: 10.1111/j.1460-9568.2009.07012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Pathak S, Schneider W. Identifying the brain's most globally connected regions. Neuroimage. 2010;49:3132–3148. doi: 10.1016/j.neuroimage.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Greicius MD. Greater than the sum of its parts: a review of studies combining structural connectivity and resting-state functional connectivity. Brain Struct Funct. 2009;213:525–533. doi: 10.1007/s00429-009-0208-6. [DOI] [PubMed] [Google Scholar]
- Davare M, Kraskov A, Rothwell JC, Lemon RN. Interactions between areas of the cortical grasping network. Curr Opin Neurobiol. 2011;21:565–570. doi: 10.1016/j.conb.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci. 2011;12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. New York: Chapman and Hall; 1993. [Google Scholar]
- Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, Schlaggar BL, Petersen SE. Functional brain networks develop from a “local to distributed” organization. PLoS Comput Biol. 2009;5:e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Gentilucci M, Dalla Volta R. Spoken language and arm gestures are controlled by the same motor control system. Q J Exp Psychol (Hove) 2008;61:944–957. doi: 10.1080/17470210701625683. [DOI] [PubMed] [Google Scholar]
- Greicius M. Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol. 2008;21:424–430. doi: 10.1097/WCO.0b013e328306f2c5. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimer à R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas C, Kamdar N, Nguyen D, Prater K, Beckmann CF, Menon V, Greicius MD. Distinct cerebellar contributions to intrinsic connectivity networks. J Neurosci. 2009;29:8586–8594. doi: 10.1523/JNEUROSCI.1868-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen VJ, Meuli R, Thiran JP. Mapping human whole-brain structural networks with diffusion MRI. PLoS One. 2007;2:e597. doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, Zhu C, Gong Q, Zang Y, Evans AC. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PLoS One. 2009;4:e5226. doi: 10.1371/journal.pone.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag CC, Burns GA, O'Neill MA, Scannell JW, Young MP. Anatomical connectivity defines the organization of clusters of cortical areas in the macaque monkey and the cat. Philos Trans R Soc Lond B Biol Sci. 2000;355:91–110. doi: 10.1098/rstb.2000.0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PC, Gallagher M. Amygdala-frontal interactions and reward expectancy. Curr Opin Neurobiol. 2004;14:148–155. doi: 10.1016/j.conb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Honey CJ, Kötter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc Natl Acad Sci U S A. 2007;104:10240–10245. doi: 10.1073/pnas.0701519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Thivierge JP, Sporns O. Can structure predict function in the human brain? Neuroimage. 2010;52:766–776. doi: 10.1016/j.neuroimage.2010.01.071. [DOI] [PubMed] [Google Scholar]
- Hoshi E, Tanji J. Distinctions between dorsal and ventral premotor areas: anatomical connectivity and functional properties. Curr Opin Neurobiol. 2007;17:234–242. doi: 10.1016/j.conb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Leung LS, Mirsattari SM, Gati JS, Menon RS, Everling S. Resting-state networks in the macaque at 7 T. Neuroimage. 2011;56:1546–1555. doi: 10.1016/j.neuroimage.2011.02.063. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp. 2012a doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Gallivan JP, Culham JC, Gati JS, Menon RS, Everling S. Functional connectivity of the frontal eye fields in humans and macaque monkeys investigated with resting-state fMRI. J Neurophysiol. 2012b;107:2463–2474. doi: 10.1152/jn.00891.2011. [DOI] [PubMed] [Google Scholar]
- Johnston K, Everling S. Neurophysiology and neuroanatomy of reflexive and voluntary saccades in non-human primates. Brain Cogn. 2008;68:271–283. doi: 10.1016/j.bandc.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC. Modelling the development of cortical systems networks. Neurocomputing. 2004;58–60:297–302. [Google Scholar]
- Katz PS, Harris-Warrick RM. The evolution of neuronal circuits underlying species-specific behavior. Curr Opin Neurobiol. 1999;9:628–633. doi: 10.1016/S0959-4388(99)00012-4. [DOI] [PubMed] [Google Scholar]
- Kelly C, Uddin LQ, Shehzad Z, Margulies DS, Castellanos FX, Milham MP, Petrides M. Broca's region: linking human brain functional connectivity data and non-human primate tracing anatomy studies. Eur J Neurosci. 2010;32:383–398. doi: 10.1111/j.1460-9568.2010.07279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Norris DG, Hund-Georgiadis M. An investigation of functional and anatomical connectivity using magnetic resonance imaging. Neuroimage. 2002;16:241–250. doi: 10.1006/nimg.2001.1052. [DOI] [PubMed] [Google Scholar]
- Kötter R, Stephan KE. Network participation indices: characterizing component roles for information processing in neural networks. Neural Netw. 2003;16:1261–1275. doi: 10.1016/j.neunet.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Kötter R, Wanke E. Mapping brains without coordinates. Philos Trans R Soc Lond B Biol Sci. 2005;360:751–766. doi: 10.1098/rstb.2005.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Theilmann RJ, Sereno MI, Townsend J. The relation between connection length and degree of connectivity in young adults: a DTI analysis. Cereb Cortex. 2009;19:554–562. doi: 10.1093/cercor/bhn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord A, Horn D, Breakspear M, Walter M. Changes in community structure of resting state functional connectivity in unipolar depression. PLoS One. 2012;7:e41282. doi: 10.1371/journal.pone.0041282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Gerits A, Nelissen K, Durand JB, Joly O, Simone L, Sawamura H, Wardak C, Orban GA, Buckner RL, Vanduffel W. Default mode of brain function in monkeys. J Neurosci. 2011;31:12954–12962. doi: 10.1523/JNEUROSCI.2318-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci U S A. 2009;106:20069–20074. doi: 10.1073/pnas.0905314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AR. Towards a network theory of cognition. Neural Netw. 2000;13:861–870. doi: 10.1016/s0893-6080(00)00059-9. [DOI] [PubMed] [Google Scholar]
- McIntosh AR. Contexts and catalysts: a resolution of the localization and integration of function in the brain. Neuroinformatics. 2004;2:175–182. doi: 10.1385/NI:2:2:175. [DOI] [PubMed] [Google Scholar]
- Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. Neuroimage. 2009;44:715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Front Neurosci. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Hayashi T. Neural representation of visual objects: encoding and top-down activation. Curr Opin Neurobiol. 2000;10:187–194. doi: 10.1016/s0959-4388(00)00071-4. [DOI] [PubMed] [Google Scholar]
- Moeller S, Nallasamy N, Tsao DY, Freiwald WA. Functional connectivity of the macaque brain across stimulus and arousal states. J Neurosci. 2009;29:5897–5909. doi: 10.1523/JNEUROSCI.0220-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Armstrong KM, Fallah M. Visuomotor origins of covert spatial attention. Neuron. 2003;40:671–683. doi: 10.1016/s0896-6273(03)00716-5. [DOI] [PubMed] [Google Scholar]
- Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51:527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Nelissen K, Luppino G, Vanduffel W, Rizzolatti G, Orban GA. Observing others: multiple action representation in the frontal lobe. Science. 2005;310:332–336. doi: 10.1126/science.1115593. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kötter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3:606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Picard N, Strick PL. Imaging the premotor areas. Curr Opin Neurobiol. 2001;11:663–672. doi: 10.1016/s0959-4388(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Preuss TM. Taking the measure of diversity: comparative alternatives to the model-animal paradigm in cortical neuroscience. Brain Behav Evol. 2000;55:287–299. doi: 10.1159/000006664. [DOI] [PubMed] [Google Scholar]
- Preuss TM. Evolutionary specializations of primate brain systems. In: Ravosa MJ, Dagosto M, editors. Primate origins: adaptations and evolution. Chicago: Springer; 2007. pp. 625–675. [Google Scholar]
- Rolls ET, Cahusac PM, Feigenbaum JD, Miyashita Y. Responses of single neurons in the hippocampus of the macaque related to recognition memory. Exp Brain Res. 1993;93:299–306. doi: 10.1007/BF00228398. [DOI] [PubMed] [Google Scholar]
- Rowe JB. Connectivity analysis is essential to understand neurological disorders. Front Syst Neurosci. 2010;4.pii:144. doi: 10.3389/fnsys.2010.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011;56:2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- Skudlarski P, Jagannathan K, Calhoun VD, Hampson M, Skudlarska BA, Pearlson G. Measuring brain connectivity: diffusion tensor imaging validates resting state temporal correlations. Neuroimage. 2008;43:554–561. doi: 10.1016/j.neuroimage.2008.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AZ, Raichle ME. A brief history of the resting state: the Washington University perspective. Neuroimage. 2012;62:902–910. doi: 10.1016/j.neuroimage.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Kötter R. Motifs in brain networks. PLoS Biol. 2004;2:e369. doi: 10.1371/journal.pbio.0020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kötter R. Identification and classification of hubs in brain networks. PLoS One. 2007;2:e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Kamper L, Bozkurt A, Burns GA, Young MP, Kötter R. Advanced database methodology for the Collation of Connectivity data on the Macaque brain (CoCoMac) Philos Trans R Soc Lond B Biol Sci. 2001;356:1159–1186. doi: 10.1098/rstb.2001.0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, McIntosh AR, Russell DP, Edelman GM. Functional clustering: identifying strongly interactive brain regions in neuroimaging data. Neuroimage. 1998;7:133–149. doi: 10.1006/nimg.1997.0313. [DOI] [PubMed] [Google Scholar]
- Tsao DY, Freiwald WA, Knutsen TA, Mandeville JB, Tootell RB. Faces and objects in macaque cerebral cortex. Nat Neurosci. 2003;6:989–995. doi: 10.1038/nn1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RC, Kahn RS, Hulshoff Pol HE. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp. 2009a;30:3127–3141. doi: 10.1002/hbm.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE. Efficiency of functional brain networks and intellectual performance. J Neurosci. 2009b;29:7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. Surface-based approaches to spatial localization and registration in primate cerebral cortex. Neuroimage. 2004;23(Suppl 1):S97–S107. doi: 10.1016/j.neuroimage.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analyses of cerebral cortex. J Am Med Inform Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela F, Lachaux JP, Rodriguez E, Martinerie J. The brainweb: phase synchronization and large-scale integration. Nat Rev Neurosci. 2001;2:229–239. doi: 10.1038/35067550. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Hagmann P, D'Arceuil H, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41:1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Zeki SM. Functional specialisation in the visual cortex of the rhesus monkey. Nature. 1978;274:423–428. doi: 10.1038/274423a0. [DOI] [PubMed] [Google Scholar]