Abstract
Background
N6-Methyladenosine (m6A), the most prevalent modification in mammalian mRNA, plays important roles in numerous biological processes. Several m6A associated proteins such as methyltransferase like 3 (METTL3), methyltransferase like 14 (METTL14), α-ketoglutarate-dependent dioxygenase AlkB homolog 5 (ALKBH5) and YTH domain containing 2 (YTHDC2) are involved in the regulation of spermatogenesis and oogenesis. However, the role of the first detected m6A demethylase, fat mass and obesity associate protein (FTO), in germ cells remains elusive. Elucidation of FTO roles in the regulation of germ cell fate will provide novel insights into the mammalian reproduction.
Methods
Mouse GC-1 spg cells were treated with the ester form of meclofenamic acid (MA2) to inhibit the demethylase activity of FTO. The cellular m6A and m6Am level were analyzed through high performance liquid chromatography combined with tandem mass spectrometry (HPLC/MS-MS). The cell apoptosis was detected via TUNEL and flow cytometry. The cell proliferation was detected through EdU and western blot. The mRNA level of core cyclin dependent kinases (CDKs) was quantified via q-PCR. RNA decay assay were performed to detect RNA stability. Dual fluorescence assay was conducted to study whether MA2 affects the expression of CDK2 dependent on the m6A modification at 3’UTR.
Results
MA2 significantly increased the cellular m6A level and down-regulated the expression of CDK1, CDK2, CDK6 and CdC25a, resulting in arrest of G1/S transition and decrease of cell proliferation. MA2 downregulated CDK2 mRNA stability. Additionally, mutation of the predicted m6A sites in the Cdk2–3’UTR could mitigated the degradation of CDK2 mRNA after MA2 treatment.
Conclusion
MA2 affected CDKs expression through the m6A-dependent mRNA degradation pathway, and thus repressed spermatogonial proliferation.
Electronic supplementary material
The online version of this article (10.1186/s40104-019-0361-6) contains supplementary material, which is available to authorized users.
Keywords: Cell cycle, FTO, Meclofenamic acid, N6-methyladenosine, Spermatogonial proliferation
Background
Over 140 chemical modifications have been discovered in RNA, of which N6-methyladenosine (m6A) is the most abundant one that distributes extensively in about 60% of mammalian mRNA. m6A mainly occurs at a common RR (m6A) CH motif, in the 3’UTR and near the stop codon [1, 2]. m6A modification is catalyzed by the methyltransferase complex composed by METTL3, METTL14, Wilms’ tumor 1-associating protein (WTAP), vir like m6A methyltransferase associated protein (KIAA1429), zinc finger CCCH domain-containing protein 13 (ZC3H13), the CCHC zinc finger-containing protein (ZCCHC4) and other undefined proteins [3–5], which are called “m6A writers”. Among these writers, METTL3 is the sole catalytic subunit and the others act as adaptors to keep the stable structure and to enhance RNA-substrate recognition. m6A can be erased by “m6A erasers” FTO and ALKBH5 [6], and be recognized by “m6A readers” such as YTH-domain containing proteins YTHDF1, YTHDF2, YTHDF3 and YTHDC2 [7, 8]. As an epitranscriptomic biomarker, m6A participates in the regulation of several post-transcriptional events, including translation, RNA stability, alternative splicing and pre-miRNA processing [9–11]. Increasing evidences have demonstrated that m6A plays critical roles in many biological processes, such as differentiation of embryonic stem cells [12], cancer development [13, 14], virus infection [15, 16], neurogenesis, oocyte maturation [17, 18] spermatogenesis [19–21], etc.
Spermatogenesis is a complex process that contains proliferation and differentiation of spermatogonia, meiosis of spermatocytes and post-meiotic spermiogenesis of round spermatids, resulting in generating millions of spermatozoa daily in mammalian testes [22]. This process requires accurately, spatially and temporally regulated patterns of gene expression. Investigating the underlying mechanisms of spermatogenesis will provide theoretical basis for development of novel approaches for male infertility therapy and male contraception. Epigenetic factors, such as histone modifications and DNA modifications, have been reported to play important roles in spermatogenesis [23–26]. However, as an epitranscriptomic biomarker, the roles of m6A in the regulation of spermatogenesis remain largely unknown [6]. Importantly, depletion of ALKBH5 leads to retarded spermatogenesis, serious apoptosis in pachytene spermatocytes in mice. Moreover, adult male Mettl3 flox/− Vasa-Cre mice were infertile, with no spermatids in the seminiferous tubules [21]. Similarly, depletion of YTHDC2 leads to infertility as well [19]. Meiosis was arrested at zygotene stage in METTL3 or YTHDC2 knockout mice. Furthermore, simultaneously deletion of METTL14 and METTL3 in advanced mouse germ cells resulted in decrease of sperm motility and increase of abnormal sperm ratio, indicating the significance of m6A in spermiogenesis [20].
Fto is identified as a candidate gene contributing to the obesity and type II diabetes [27, 28]. FTO belongs to the AlkB related non-haem iron and 2-oxoglutarate-dependent oxygenase superfamily [29], which is able to oxidatively demethylate RNA and DNA. The m6A demethylase activity of FTO has been demonstrated to be associated with the splicing of RUNX1 translocation partner 1 (RUNX1T1) [28], an adipogenic regulatory factor. Deficiency of FTO results in reduced adipocyte size and body weight [30–32]. Researches on the phenotypes of FTO knockout mice were mainly focused on the fat mass accumulation, body composition, energy expenditure and other obesity relative phenotypes in these studies. On one hand, the demethylation activity of FTO have not been mentioned in these knockout studies. On the other hand, though FTO showed high expression level in testis, however, effects of FTO on spermatogenesis have been overlooked. Given that another m6A demethylase ALKBH5 is involved in the regulation of spermatogenesis [6], we speculated that FTO may also regulate the fate of male germ cells.
Recent studies have reported that meclofenamic acid can selectively compete with FTO for binding to nucleic acid in vitro. The ethyl ester form of meclofenamic acid (MA2) selectively inhibited FTO activity and elevated cellular m6A level in Hela cells [33]. The objective of this study was to elucidate the effects of MA2 on FTO demethylase activity and the fate of male germ cells. In the present study, we confirmed that MA2 specifically inhibited FTO demethylase activity in spermatogonia. We found that MA2 repressed cell proliferation through prolonging G1 stage. We further demonstrated that the 3’UTR m6A sites regulated CDK2 expression through directly affecting its mRNA stability. In summary, our findings suggested that MA2 regulates proliferation and cell cycle progression in spermatogonia through the m6A demethylase activity.
Methods
Cell culture and drug treatment
The mouse spermatogonia cell line GC-1 spg and HEK293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (GE, SH30022) with 10% fetal bovine serum (Gibco, 12484028). Cells were kept in humidified atmosphere, at 37 °C in the presence of 5% CO2. MA2 was kindly provided by the Chinese Academy of Sciences Shanghai Institute of Materia Medica. MA2 powder was dissolved with DMSO to 50 mmol/L and storage at − 20 °C. Spermatogonia were grown to 40% confluence. Subsequently, the culture medium was replaced by medium containing different concentration of MA2. After 24 h and 48 h of culture, cells were washed with Phosphate Buffer Saline (PBS, GE, SH30028) and collected for analysis.
Antibodies
The primary and secondary antibodies were purchased from commercial sources as follows: Mouse anti-FTO (Santa Cruz Biotechnology, sc-271713), Rabbit anti-Bax (CST, 2772), Rabbit anti-Bcl-2 (CST, 2876), Mouse anti-PCNA (Santa Cruz Biotechnology, sc-56), Mouse anti-Ki67 (Santa Cruz Biotechnology, sc-23900), Rabbit anti m6A (Synaptic Systems, 202003), Rabbit anti-Actin (Sigma Aldrich, A2547), Mouse anti Tubulin (Santa Cruz Biotechnology, sc-365791). HRP conjugated goat anti-rabbit IgG (CWbio, CW0156) and HRP conjugated goat anti-mouse IgG (CWbio, CW0102S).
Detection of cell viability
Cell viability was detected using a CCK-8 cell counting kit (Vazyme, A311–01) following the manufacturer’s instructions. In brief, cells were cultured in 96-well plates and treated with 40, 80 and 120 μmol/L of MA2 and the DMSO control. After incubation for 24 or 48 h, the culture medium containing MA2 was replaced by fresh medium. Ten microliters of CCK-8 were added to each well and incubated for 2 h at 37 °C. The OD values were quantified by Microplate Spectrophotometer.
Apoptosis assay
TUNEL assay was employed to detect apoptosis. Ten thousands of cells were seeded to each well of the 96-well plate and cultured overnight. After treated with 0, 40, 80 and 120 μmol/L of MA2, cells were fixed with 4% paraformaldehyde for 30 min followed by incubating with 0.1% Triton X-100. The fixed cells were labeled with TUNEL BrightGreen (Vazyme, A112–01). The nuclei were counterstained by [Hoechst 33342 (Thermo, 62249)]. TUNEL positive cells were observed and counted under an inverted fluorescence microscope (Olympus, IX71).
Cell proliferation assay
Cell proliferation was detected using an EdU Apollo 567 In Vitro Imaging Kit (RiboBio, C10310–1). Briefly, the cells were seeded into 96-well plates (1 × 104 cells per well) and cultured overnight. After incubating with 0, 40, 80 and 120 μmol/L MA2 for 48 h, cells were labeled with 10 μmol/L EdU at 37 °C, 5% CO2 for 2 h, fixed in 4% paraformaldehyde for 30 min, followed by staining with fluorescent dye. Cell nuclei were counterstained by Hoechest 33342 (Thermo, 62249). EdU positive cells were observed and counted under an inverted fluorescence microscope (Olympus, IX71).
Flow cytometric analysis
Spermatogonia were treated with 0, 40, 80 and 120 μmol/L of MA2 for 48 h. The Cells were detached with 0.25% (w/v) trypsin and washed with PBS. Subsequently, cells were kept in 70% cold ethanol at − 20 °C overnight. The cells were incubated with 50 μg/mL of propidium iodide (Sigma Aldrich, 81845) at 37 °C in dark for 30 min, and analyzed with a flow Cytometer (Cyflow Cube). The cell cycle data were analyzed using the software Modfit.
RNA isolation, cDNA synthesis and quantitative RT-PCR
Total RNA was extracted using Trizol reagent (TAKARA, 9108), followed by Chloroform centrifugation and isopropanol precipitation. CDNA was synthesized using Primescript RT reagent Kit (Roche, 04896866001). Q-PCR was performed using the SYBR Green II PCR Mix (TAKARA) and the fluorescence ration PCR instrument IQ5 (Bio-Rad). The qRT-PCR primers used were listed in Additional file 2: Table S1.
Western blot assay
Cells were lysed with RIPA lysis buffer (Solarbio Life Sciences, R0020) containing 1% PMSF followed by ultrasonication. Cell lysates were incubated on ice for 30 min, followed by centrifuging at 4 °C for 10 min. The protein concentration of the supernatant was quantified using a BCA Protein Assay Kit (TAKARA, T9300A). The proteins were separated through SDS-PAGE using the electrophoresis apparatus (Bio-Rad). Subsequently, the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, IBFP0785C) using a semi dry transfer instrument (Bio-Rad). The membranes were blocked with 5% non-fat milk at room temperature for 1 h, and incubated with primary antibodies at 4 °C overnight. After overnight incubation, the membranes were washed with PBST and incubated with HRP-conjugated secondary antibodies at room temperature for 1 h, followed by incubation with the Immobilon Western Chemiluminescent HRP Substrate (Millipore) and photographed using the ECL imaging system (Bio-Rad).
m6A/m6Am dot-blot
mRNA was purified using PolyATtract® mRNA Isolation Systems (Promega, Z5310) following the manufacturer’s instructions. mRNA was denatured at 95 °C for 5 min and loaded onto a Hybond-N+ membrane (GE HealthCare, RPN303B). After closslinking under ultraviolet radiation, the membrane was blocked with 5% non-fat milk (Bio-Rad) for 1 h, incubated with rabbit anti-m6A polyclone antibody (Synaptic Systems, 202003) at 4 °C overnight. Then the membrane was incubated with HRP-Conjugated goat anti-rabbit IgG (CWbio, CW0156) at room temperature for 2 h. After being incubated with a Immobilon Western Chemiluminescent HRP Substrate (Millipore, WBKLS0500), the immunocomplex was photographed using the ECL imaging system. Finally, the membrane was stained with 0.02% methylene blue to eliminate the difference in mRNA amount.
Plasmids construction and transfection
cDNA of the cells was synthesized as mentioned above. The DNA fragments of CREBBP-3’UTR and CDK2-3’UTR were amplified via PCR. Mutation of CREBBP-3’UTR and CDK2-3’UTR were generated through the overlapping PCR followed by homologous recombination using a NovoRec recombinase (Novoprotein, NR001). The purified 3’UTR fragments were constructed to pCMV-mCherry-EF1α-GFP vector. For dual-fluorescence protein report assay, plasmids were transfected to HEK-293 T Cells using the TurboFect™ Transfection Reagent (Thermo Fisher, R0534) following the manufacturer’s instructions. After 48 h, the fluorescence intensity was detected by a Synergy Muti-Mode Microplate Reader (BioTek). The relative fluorescence intensity (RFI) was calculated as follows: RFI = FI (red) / FI (green).
Quantitative analysis of RNA modification levels
The quantitative analysis of m6A and m6Am level was performed according to previously described [34]. Briefly, total RNA was isolated from cells treated with 0 μmol/L, 40 μmol/L, 80 μmol/L and 120 μmol/L MA2, respectively. mRNA was purified using the PolyATtract® mRNA Isolation Systems (Promega, Z5310) following the manufacturer’s instructions. Three hundred micrograms of mRNA was further treated with 3 μL RppH (NEB) at 37 °C for 2 h. After the removel of cap, mRNA was treated with nuclease P1 (Sigma, N8630) at 42 °C for 2 h. Afterwards, the ribonucleotides were treated with alkaline phosphatase (Sigma, P5931) at 37 °C to remove the phosphate group. The amount of adenosine (A), m6A and m6Am was measured according to HPLC-tandem mass spectrometry. The ratio of m6A to A and m6Am to A were calculated based on the calibrated concentrations.
RNA-decay assay
Cells were treated with 0, 40 and 80 μmol/L MA2 for 24 h. Then the culture medium were added 5 μg/mL actinomycin D and incubated for 0 h, 3 h and 6 h, respectively. Cells were harvested and subjected to RNA extraction. Real-time quantitative PCR were used to analyze the mRNA level of the target genes in each group.
FTO-RIP-PCR
For FTO-RIP, the full CDS of Fto gene was amplified by high fidelity PCR. The Flag sequence was inserted to the 5′ end of Fto-CDS. The Flag-Fto sequence was inserted into the CD513B-CMV plasmid. Flag-GFP was used as negative control. Anti-FLAG-based RIP was performed with modifications according to [35]. Breifly, CD513B-Flag-FTO and CD513B-Flag-GFP were transfected to the GC-1 cells, respectively. After 24 h culture, cells were collected and lysed by the lysis buffer, while 1/10 volume of cell lysates were preserved as input. Protein A beads were coated with 10 μg rabbit anti-FLAG antibody. Cell lysates and beads were mixed and rotated at 4 °C overnight. After washed for five times, beads and input lysates were incubated with proteinase K at 55 °C for 30 min followed by centrifugation. IP RNA was extracted from the supernatant using TRIZOL followed by chloroform/ isopropanol centrifugation. RT-PCR was performed to normalize the mRNA level of target genes.
Statistical analysis
All data were collected from at least three independent experiments. Data were analyzed using two-tailed student’s t-test or One-way ANOVA followed by a Duncan’s multiple range test (SPSS 22 for windows), with P < 0.05 Considered statistically significant.
Results
MA2 elevates cellular m6A and m6Am level and decreases cell viability
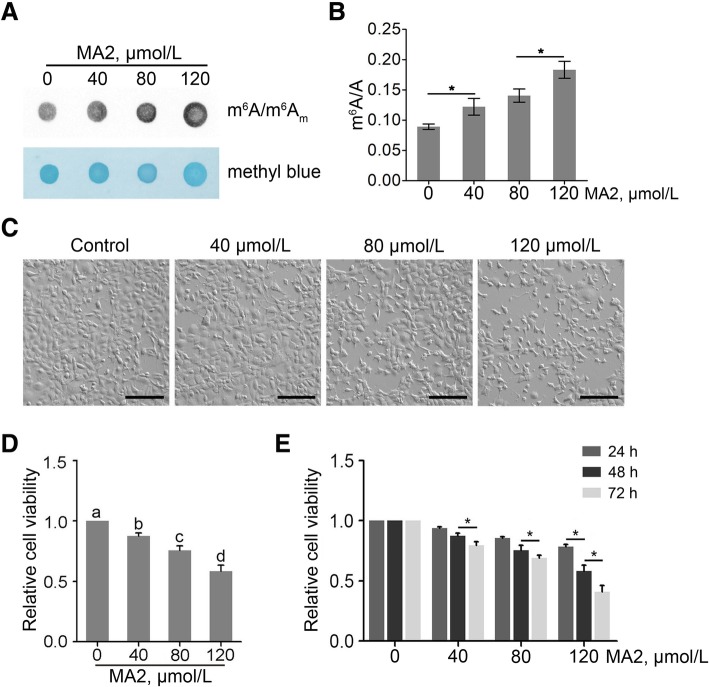
To investigate whether MA2 affect the m6A level in spermatogonia, we treated the cells with 0, 40, 80 and 120 μmol/L of MA2 for 48 h, respectively. The m6A/m6Am level was detected by dot blot and HPLC/MS-MS. As shown in Fig. 1a and b, MA2 enhanced cellular m6A level in a dose-dependent manner. Additionally, MA2 did not affect the m6Am level of polyA RNA. However, MA2 increased the m6Am level of small nuclear RNA in a dose- dependent manner (Additional file 1: Figure S1). We then detected the cell morphology under the microscopy. As shown in Fig. 1c, MA2 did not affect cell morphology, but led to a decrease in cell density. We next detected the cell viability via CCK-8 assay, and found that the cell viability significantly decreased in both dose and time dependent manner (Fig. 1c and d). Thus, MA2 efficiently elevated the m6A and m6Am level and significantly reduced cell viability of spermatogonia.
Fig. 1.
Phenotypes of spermatogonia treated with MA2. a Effect of MA2 on the cellular m6A level. GC-1 cells were treated with 0, 40, 80 and 120 μmol/L of MA2 for 48 h. Cellular m6A level was analyzed via m6A dot blot. b Quantification of cellular m6A level through HPLC/MS-MS. Data were presented as the mean ± SEM, n = 3. *P < 0.05. c Images of the GC-1 spg cells. Cells were treated with 40, 80 and 120 μmol/L of MA2 or the carrier DMSO for 48 h in triplicates. The morphology of cells was photographed. Bar = 50 μm. d Dose-dependent effect of MA2 on cell viability. GC-1 cells were treated with MA2 for 48 h. Different lower-case letters denoted significant differences (P < 0.05). e Time-dependent effect of MA2 on cell viability. GC-1 cells were treated with 0, 40, 80 and 120 μmol/L MA2 in triplicates. Cell viability was detected via CCK-8 assay. Data were presented as the mean ± SEM, n = 3. *P < 0.05
Effects of MA2 on spermatogonial apoptosis and proliferation
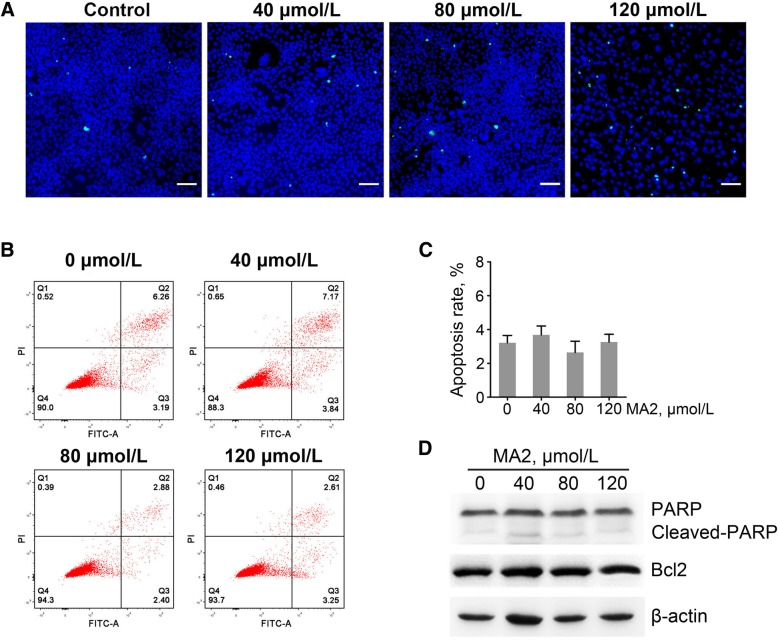
To investigate whether the treatment of MA2 led to cell apoptosis, we treated the GC-1 cells with 0, 40, 80 and 120 μmol/L of MA2 for 48 h. TUNEL assay showed that positive rate was not different between the MA2 treatments and the control (Fig. 2a and c), indicating that MA2 did not cause apoptosis. To further verify the phenotype, we performed the PI/Annexin V staining followed by flow cytometry. As shown in Fig. 2b, apoptosis rates (quantitative value in quadrant 3) in all treated groups were not different, which was accordant to the TUNEL assay result.
Fig. 2.
Effect of MA2 on cell apoptosis. a GC-1 cells were treated with 0, 40, 80 and 120 μmol/L MA2 for 48 h. TUNEL assay was used to analyze apoptosis. TUNEL positive cells were indicated by green fluorescence. Cell nucleic were stained by DAPI (blue). Bar = 50 μm. b GC-1 cells were double-stained with PI/Annexin V and analyzed by flow cytometry. Q4 represents non-apoptotic cells, Q3 represents apoptotic cells, and Q2 represents dead cells. c Apoptotic rates of all groups were showed in column diagram. Data were represented by mean ± SEM, (n = 3 independent replicates).d Effects of MA2 on the abundance of PARP, cleaved PARP and Bcl2. β-Actin was used as the loading control
We further detected the expression of apoptosis relative protein PARP and anti-apoptosis protein Bcl2. As shown in Fig. 2d, the level of PARP in treated groups was comparable to the control, and expression of the cleaved-PARP in each group was hard to observe. Additionally, the level of anti-apoptosis protein Bcl2 was similar in all the groups. Taken together, these results suggest that MA2 did not induce cell apoptosis.
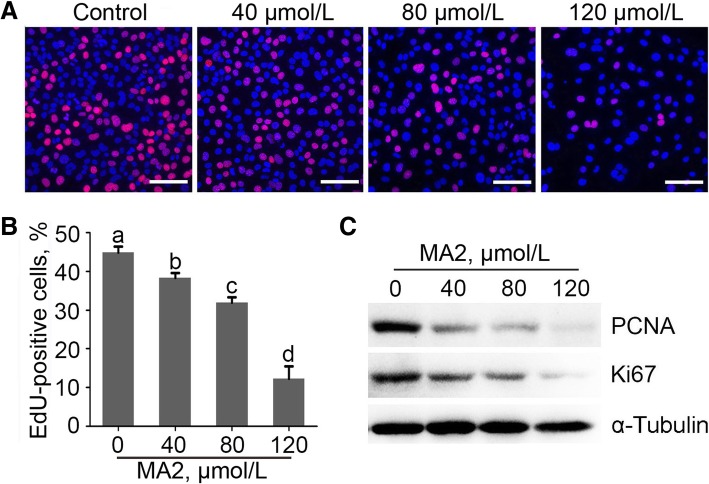
To detect cell proliferation, we performed EdU assay, which measures the rate of DNA replication and labels S-phase cells [36]. Cells were treated with 0, 40, 80 and 120 μmol/L of MA2 for 48 h. EdU assay showed that MA2 caused an intensive decrease of EdU positive cells (Fig. 3a and b). Meanwhile, the expression of proliferation markers PCNA and Ki67, that are expressed in active phases of cell cycle [37, 38], drastically decreased in the MA2 treatments (Fig. 3c), indicating that cell proliferation was dramatically repressed by MA2. These observations suggested that MA2 repressed cell proliferation.
Fig. 3.
Effect of MA2 on cell proliferation. a GC-1 cells were treated with 0, 40, 80 and 120 μmol/L of MA2 for 48 h. EdU assay was performed to analyze cell proliferation. EdU positive cells were indicated by red fluorescence. Nuclei were stained by DAPI (blue), bar = 50 μm. b EdU positive rates were showed in column diagram. Data were represented by the mean ± SEM, n = 3. Different lower-case letters denoted significant differences (P < 0.05). c Effects of MA2 on expression of the proliferation markers. GC-1 cells were treated with different concentration of MA2 for 48 h. Expression of PCNA and Ki67 were detected by western blot. α-Tubulin was used as the loading control
MA2 arrests cell cycle via down-regulating mRNA level of CDKs
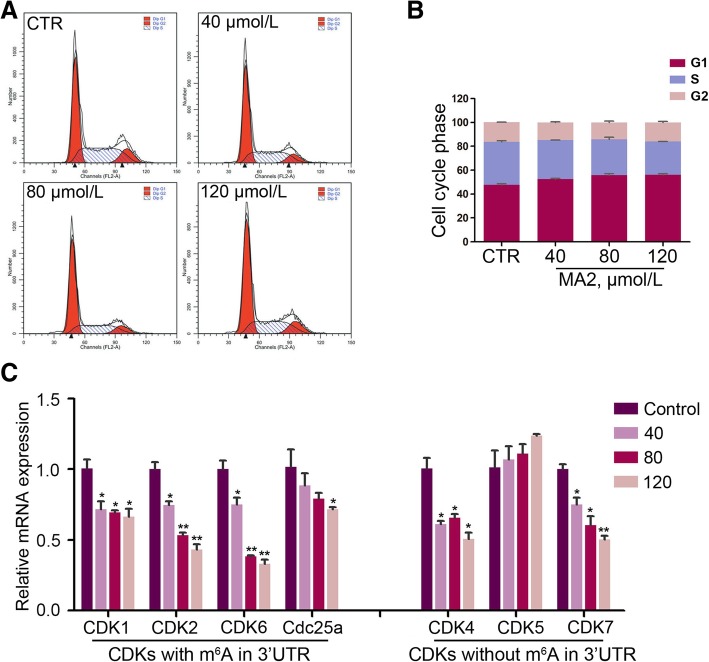
Previous studies showed that m6A was involved in the regulation of cell cycle during oocyte maturation in Xenopus laevis oocytes [17]. To determine whether MA2 affects the cell cycle transition, we analyzed the cell cycle through flow cytometry. Interestingly, MA2 treatment led to a significant increase of cell numbers in G0/G1 phase and a dramatic decrease in S and G2/M phase (Fig. 4a and b), indicating that MA2 arrested the G1/S transition.
Fig. 4.
Effects of MA2 on progression of cell cycle. a Cells were treated with 40, 80 and 120 μmol/L of MA2 or the carrier DMSO for 48 h. Subsequently, the cells were harvested and stained with propidium iodide and analyzed using a flow cytometry. (A) Cell cycle phase was analyzed by ModFit. b Quantification of percentage of cells in each cycle phase. Data were represented by the mean ± SEM, n = 3. c Relative mRNA expression of CDKs involved in G1/S transition. *P < 0.05, **P < 0.01. Data were represented by mean ± SEM, n = 3
As the cell cycle progression is elaborately regulated by the cyclin-dependent kinases (CDKs), to investigate whether MA2 affect CDK expression, we searched the online database of RNA methylation (http://rna.sysu.edu.cn/rmbase/) and refered the RNA methylation information of the core CDKs that are involved in G1/S transition. As shown in Additional file 2: Table S2, CDK1, CDK2, CDK6 and CdC25a contained m6A modified sites in the 3’UTR, while CDK4, CDK5 and CDK7 did not. Subsequently, the mRNA level of the seven CDKs was measured through q-PCR. Interestingly, the mRNA level of CDK1, 2, 6 and CdC25a was significantly down-regulated by MA2. However, the mRNA level of CDK4 and CDK7 was also down-regulated by MA2, while expression of CDK5 showed no difference (Fig. 4c). Together, these results suggested that MA2 affected the expression of CDKs involved in G1/S transition in spermatogonia.
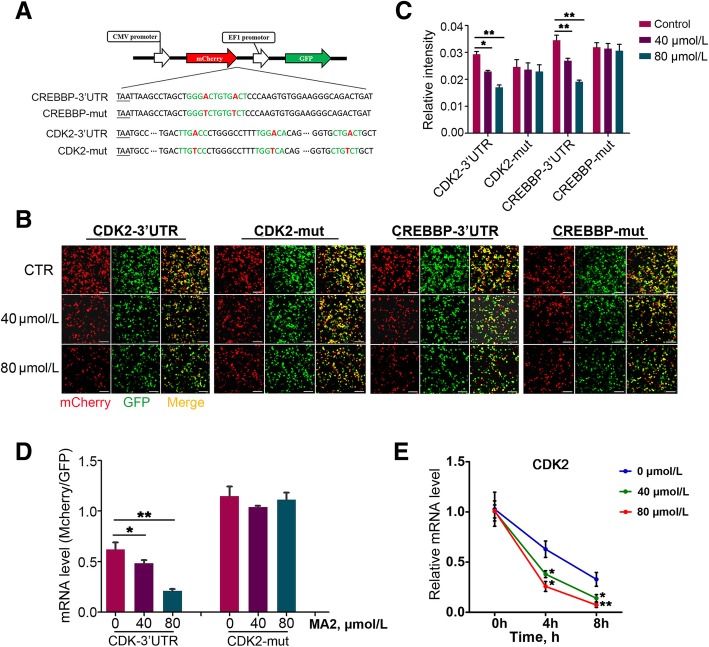
CDK2 mRNA level dependent on 3’UTR m6A modification
Previous studies have demonstrated that the 3’UTR m6A modification can accelerate mRNA degradation [7]. Considering that the CDK2 mRNA harbored three m6A sites in the 3’UTR, we assumed that MA2 down-regulated the CDK2 mRNA level through accelerating mRNA degradation. To verify the hypothesis, we performed a dual fluorescence assay. Three predicted methylated adenines in the 3’UTR of CDK2 were mutated to thymine (Fig. 5a). Considering that the m6A sites within the CREBBP-3’UTR have been reported to downregulated the stability of CREBBP mRNA [7], we employed the CREBBP-3’UTR as positive control. Briefly, CDK2-3’UTR and CREBBP-3’UTR were respectively constructed to the downstream of a coding sequence (CDS) that encoded a red fluorescence protein (RFP). The relative fluorescence intensity (RFI, red to green) indicated the expression of CDK2 and CREBBP. Interestingly, MA2 caused a significant decrease of the RFI in the cells that were transfected with CDK2-3’UTR. Whereas, in CDK2-mut cells, it was similar between the control and MA2 treated groups. In addition, RFI of the CREBBP-3’UTR group and CREBBP-mut group was consistent with that in CDK2 (Fig. 5b and c). We further quantified the mRNA level through RT-PCR. As shown in Fig. 5d, the tendency of mRNA level was similar to that of the RFI, indicating that MA2 reduced the mRNA level of CDK2 through elevating the 3’UTR m6A modifications.
Fig. 5.
MA2 downregulated CDK2 expression and cell viability through the 3’UTR-m6A mediated mRNA degradation. a Schematic diagram of plasmids used in the 3’UTR fluorescence reporter assay. Letter A and T (red) represented the mutation of predicted m6A to T. b Effects of CDK2-3’UTR and CDK2-mut on the expression of mCherry. Cells were treated with 40 and 80 μmol/L of MA2, and simultaneously transfected with CDK2-3’UTR and CDK2-mut plasmids. After 48 h, the cells were photographed under the fluorescence microscope. Bar = 50 μm. c Relative fluorescence intensity was showed in column diagram. Data were represented by the mean ± SEM, n = 3. *P < 0.05, **P < 0.01. d Relative mRNA level of mCherry to GFP in each group. n = 3 independent duplications, *P < 0.05, **P < 0.01. e Cells were treated with 0, 40 and 80 μmol/L MA2 for 24 h. After MA2 treatment, cells were treated with 5 μmol/L actinomycin D for 4 h and 8 h, respectively. Remained RNA level was quantified by RT-PCR. Data were represented by the mean ± SEM, n = 3. *P < 0.05, **P < 0.01
Previous studies have demonstrated the important role of m6A in mRNA degradation [7]. Therefore, we speculated that MA2 down-regulated CDK2 expression through suppressing mRNA stability. To this end, we performed the RNA decay experiment. Cells were treated with 0, 40 and 80 μmol/L of MA2 for 24 h, then subjected to 5 μmol/L of actomycin D to abolish the synthesis of new transcripts. After 4 h and 8 h treatment, the remained mRNA level of Cdk2 was quantified by RT-PCR. As shown in Fig. 5e, the Cdk2 mRNA level in the MA2 treated cells was significantly lower than the control, indicating that MA2 repressed CDK2 stability.
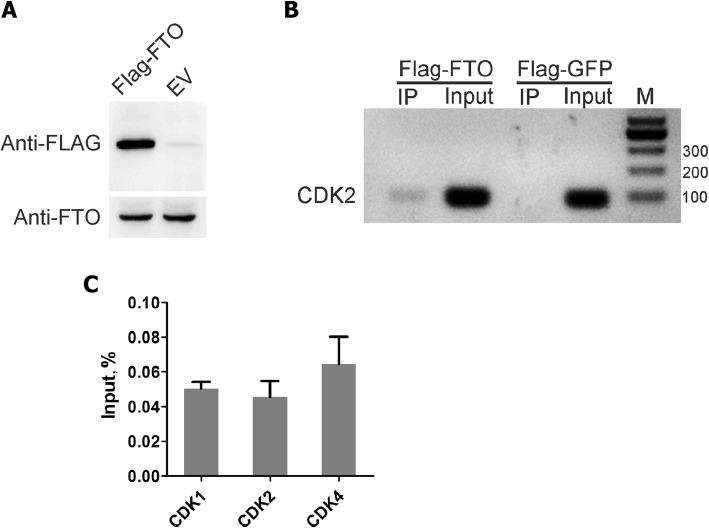
To further validate the interaction between FTO and CDKs mRNA, we performed the Flag based FTO-RIP. CD513B-Flag-FTO and CD513B-Flag-GFP were transfected to GC-1 cells, respectively. Cells were collected and subjected to RIP using anti-FLAG antibody. We found that mRNA of CDK1, CDK2 and CDK4 could be detected in the IP RNA, while CDK5, CDK6 and CDK7 could not (Fig. 6b and c), indicating that FTO directly interacted with CDK1, CDK2 and CDK4.
Fig. 6.
Anti-FLAG-based RNA immunoprecipitation coupled to RT-qPCR. a Validation of Flag tagged FTO expression by Western blot. β-actin was used as the loading control. b Electrophoretogram of the RT-PCR products using immunoprecipitated RNA and Cdk2 primer. c RNA IP confirmation of FTO binding to RNAs identifed by RT-qPCR
Together, these results suggested that MA2 regulated CDK2 expression, at least partially, through the m6A/mRNA degradation pathway.
Discussion
In the present study, we found that MA2, by inhibiting FTO demethylase activity, arrested G1/S transition and suppressed spermatogonial proliferation through downregulating CDK expression. We further demonstrated that MA2 accelerated the degradation of CDK2 mRNA, thus led to down-regulation of CDK2 expression through elevating the 3’UTR m6A level.
Growing evidences have indicated the significance of m6A in regulation of cell proliferation [39–42]. Deletion of METTL3 disrupted homeostatic proliferation of mouse naïve T cells [39]. Similarly, silencing ALKBH5 in glioblastoma stem cells suppressed cell proliferation via enhancing FOXM1 expression [41]. Moreover, FTO promotes proliferation of AML cells through targeting of the ASB2 and RARA in acute myeloid leukemia cells [42]. In addition, FTO regulated proliferation and differentiation of adult neural stem cells through modulating the BDNF/Akt signaling pathway [40]. Here, we revealed the regulatory role of m6A demethylase in the proliferation of spermatogonia.
MA, as a member of the non-steroidal anti-inflammatory drug, is well known for its inhibitory role in prostaglandins synthesis [43]. MA can inhibit the activity of cytochrome c oxidase subunit 2 [44], which promotes proliferation of various cancer cells. As such, MA possesses the therapeutic potential in different types of cancer patients [45, 46]. In the present study, we showed that MA2, the ester form of MA, elevated cellular m6A level in spermatogonia, which was accordant to that in Hela cells [33]. We found that MA2 repressed cell viability through inhibiting the demethylase activity of FTO, providing new insights into the mechanisms of MA suppressing cancer cell proliferation.
The transition from G1 to S stage is essential for cell proliferation. In the present study, we showed that inhibiting the demethylase activity of FTO down-regulated the CDKs that were associated with G1/S transition. Among the detected genes, CDKs 1, 2, 6 and CdC25a possessed m6A sites in the 3’UTR. Previous studies have adequately demonstrated that increased m6A modifications in the 3’UTR will reduce mRNA stability through interacting with the m6A reader YTHDF2, YTHDF3 or YTHDC2 [7, 8, 47]. In the present study, we found that MA2 treatment significantly accelerated the degradation of CDK2 mRNA, resulting in downregulation of CDK2 expression and spermatogonial proliferation. In the dual fluorescence assay, we mutated the predicted methylated adenosine in the CDK2 3’UTR. Actually, the mutation of adenosine could potentially also prevent the binding of RNA binding proteins that depend on adenosine, and the contribution of other binding proteins in the phenotypes should not be overlooked.
The mRNA level of several CDKs without m6A modifications in the 3’UTR, such as CDK4 and CDK7, was also downregulated by MA2. These results suggested that MA2 regulates the transition of G1/S probably through other pathways. Indeed, previous studies have reported that MA in the inhibited cancer cell proliferation [45, 48]. Sekine et.al reported that combination of simvastatin and MA suppressed the proliferation of prostate cancer cells through inhibition of the expression of aldo-keto reductase family 1 member C3 (AKR1C3). The MA dosage was 50 μmol/L, which was comparable to the dosage of MA2 used in the present study. Considering that MA2 have a stronger ability to pass through the cell membrane, it is obvious that one of the potential off-target proteins in the present study may be AKR1C3. Additionally, partial of MA2 potential targets were listed in Additional file 2: Table S3. The other m6A demethylase ALKBH5 has been reported involves in the regulation of spermatogenesis. Deficiency of ALKBH5 induces serious apoptosis in male germ cells in mouse. On one hand, the function of ALKBH5 in spermatogonial proliferation has not been mentioned. On the other hand, the selective activity of MA towards FTO has been shown in recent study. Huang et.al reported that MA could not inhibit the ALKBH5-mediated dm6A conversion to adenine in RNA even at high concentrations up to 0.5 mmol/L in vivo. Therefore, we suggest that the concentration of MA2 used in our present study have no effect on ALKBH5, and the phenotypes induced by MA2 was not associated with ALKBH5.
Recent studies have reported that m6A was not the unique substrate of FTO. FTO can also demethylate at the N6,2′-O-dimethyladenosine (m6Am). (2) Mauer et al. reported that FTO preferentially demethylated m6Am rather than m6A, and reduced the stability of m6Am mRNAs [49]. Recently, (2) Wei et al. reported that the FTO-mediated demethylation has a greater effect on the transcript levels of mRNAs possessing internal m6A than the ones with cap m6Am [50]. In the present study, we detected both m6A and m6Am level of cells treated with MA2, and found that MA2 treatment significantly increased the m6A level of mRNA and the m6Am level of small nuclear RNA. Since these two types of RNA modifications affect the expression of target genes [51], indicating that MA2 suppressed spermatogonial proliferation probably not only through m6A, but also via other pathways such as m6Am.
Since the CDK2-Cyclin E complex plays important roles in metaphase II (MII) arrest in oocytes [52] and formation of synaptonemal complex in spermatocytes [53], the CDK2 is important for both mitosis and meiosis. Here, we demonstrate that the m6A modification in Cdk2 3’UTR is related with the CDK2 mRNA stability. This finding is accordant to the recent research on Xenopus laevis, which demonstrated that m6A was associated with cell cycle and protein translation during oocyte maturation [17]. Recent studies reported that the meiosis was arrested at zygotene stage, with abnormal dissociation of synaptonemal complex when METTL3 or YTHDC2 was depleted in spermatocytes [54, 55]. This phenotype is similar to that of CDK2 deletion [21, 56]. Collectively, we propose that FTO also may play key regulatory roles in meiosis through regulating CDK2 expression. Since the FTO depletion mice exhibits developmental retardation, FTO conditional KO mouse model is needed to further investigate the functions in detail.
Conclusion
Our findings suggest that MA2 affects male germ cell proliferation through an RNA methylation pathway, thus put forward new insights into the regulatory mechanisms of RNA methylation in spermatogenesis.
Additional files
Figure S1. Detection of m6Am level in polyA RNA and small nuclear RNA. Cells were treated with 0, 40, 80 and 120 μmol/L MA2. Content of m6Am and A was detected by LC/MS-MS. Relative m6Am level was normalized by m6Am/A. (A) Relative m6Am level of PolyA RNA. Data were represented by the mean ± SEM, n = 3. n,s means P > 0.05. (B) Relative m6Am level of snRNA. Data were represented by the mean ± SEM, n = 3. (JPG 648 kb)
Table S1. Oligo information. Table S2. m6A modification information of CDKs. Table S3. Information of potential MA2 targets. (DOCX 19 kb)
Acknowledgments
We thank Dr. Caiguang Yang, at Shanghai Institute of Materia Medica, Chinese Academy of Sciences, for the generous gift of MA2. We thank Dr. Bin Wu (Arizona Center for Reproductive Endocrinology and Infertility, Tucson, AZ 85712, USA) for editing the manuscript.
Funding
This study was supported in part by the National Natural Science Foundation of China (Grant No. 31572401) to WZ.
Availability of data and materials
All data supporting our findings are included in the manuscript.
Abbreviations
- ALKBH5
α-ketoglutarate-dependent dioxygenase AlkB homolog 5
- CdC25a
Cell division cycle 25A
- CDK
Cyclin-dependent kinase
- CREBBP
CREB binding protein
- FTO
Fat mass and obesity protein
- Ki67
Marker of proliferation Ki-67
- m6A
N6-methyladenosine
- MA
Meclofenamic acid
- MA2
Ester form of meclofenamic acid
- METTL14
Methyltransferase like 14
- METTL3
Methyltransferase like 3
- PARP1
Poly (ADP-ribose) polymerase family member 1
- PCNA
Proliferating cell nuclear antigen
- RFI
Relative fluorescent intensity
- RUNX1T1
Runt-related transcription factor 1
- YTHDC2
YTH domain containing 2
Authors’ contributions
Conceived and designed the experiments: TH, JYG and WXZ. Performed the experiments: TH, JYG, YHL, TYF and QG. Analyzed the data: TH and JYG. Wrote the paper: TH, YZ and WXZ. All read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Tao Huang, Email: huangtao_199112@163.com.
Jiayin Guo, Email: guojiayin89kza@163.com.
Yinghua Lv, Email: hongriyinghua@163.com.
Yi Zheng, Email: y.zheng@nwafu.edu.cn.
Tongying Feng, Email: fengtongying1994@163.com.
Qiang Gao, Email: gaoqiang1151866427@126.com.
Wenxian Zeng, Phone: +86 029 87092120, Phone: 15229247918, Email: zengwenxian2015@126.com.
References
- 1.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini Dan, Moshitch-Moshkovitz Sharon, Schwartz Schraga, Salmon-Divon Mali, Ungar Lior, Osenberg Sivan, Cesarkas Karen, Jacob-Hirsch Jasmine, Amariglio Ninette, Kupiec Martin, Sorek Rotem, Rechavi Gideon. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 3.Liu Jianzhao, Yue Yanan, Han Dali, Wang Xiao, Fu Ye, Zhang Liang, Jia Guifang, Yu Miao, Lu Zhike, Deng Xin, Dai Qing, Chen Weizhong, He Chuan. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chemical Biology. 2013;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ping Xiao-Li, Sun Bao-Fa, Wang Lu, Xiao Wen, Yang Xin, Wang Wen-Jia, Adhikari Samir, Shi Yue, Lv Ying, Chen Yu-Sheng, Zhao Xu, Li Ang, Yang Ying, Dahal Ujwal, Lou Xiao-Min, Liu Xi, Huang Jun, Yuan Wei-Ping, Zhu Xiao-Fan, Cheng Tao, Zhao Yong-Liang, Wang Xinquan, Danielsen Jannie M Rendtlew, Liu Feng, Yang Yun-Gui. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Research. 2014;24(2):177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen Jing, Lv Ruitu, Ma Honghui, Shen Hongjie, He Chenxi, Wang Jiahua, Jiao Fangfang, Liu Hang, Yang Pengyuan, Tan Li, Lan Fei, Shi Yujiang Geno, He Chuan, Shi Yang, Diao Jianbo. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Molecular Cell. 2018;69(6):1028-1038.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Guanqun, Dahl John Arne, Niu Yamei, Fedorcsak Peter, Huang Chun-Min, Li Charles J., Vågbø Cathrine B., Shi Yue, Wang Wen-Ling, Song Shu-Hui, Lu Zhike, Bosmans Ralph P.G., Dai Qing, Hao Ya-Juan, Yang Xin, Zhao Wen-Ming, Tong Wei-Min, Wang Xiu-Jie, Bogdan Florian, Furu Kari, Fu Ye, Jia Guifang, Zhao Xu, Liu Jun, Krokan Hans E., Klungland Arne, Yang Yun-Gui, He Chuan. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Molecular Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Xiao, Lu Zhike, Gomez Adrian, Hon Gary C., Yue Yanan, Han Dali, Fu Ye, Parisien Marc, Dai Qing, Jia Guifang, Ren Bing, Pan Tao, He Chuan. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2013;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Chao, Wang Xiao, Liu Ke, Roundtree Ian A, Tempel Wolfram, Li Yanjun, Lu Zhike, He Chuan, Min Jinrong. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nature Chemical Biology. 2014;10(11):927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. [DOI] [PMC free article] [PubMed]
- 10.Alarcón Claudio R., Goodarzi Hani, Lee Hyeseung, Liu Xuhang, Tavazoie Saeed, Tavazoie Sohail F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Zhe, Li Jiong, Feng Guoxing, Gao Shan, Wang Yuan, Zhang Shuqin, Liu Yunxia, Ye Lihong, Li Yueguo, Zhang Xiaodong. MicroRNA-145 ModulatesN6-Methyladenosine Levels by Targeting the 3′-Untranslated mRNA Region of theN6-Methyladenosine Binding YTH Domain Family 2 Protein. Journal of Biological Chemistry. 2017;292(9):3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista Pedro J., Molinie Benoit, Wang Jinkai, Qu Kun, Zhang Jiajing, Li Lingjie, Bouley Donna M., Lujan Ernesto, Haddad Bahareh, Daneshvar Kaveh, Carter Ava C., Flynn Ryan A., Zhou Chan, Lim Kok-Seong, Dedon Peter, Wernig Marius, Mullen Alan C., Xing Yi, Giallourakis Cosmas C., Chang Howard Y. m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Shuibin, Choe Junho, Du Peng, Triboulet Robinson, Gregory Richard I. The m 6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Molecular Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su R, Li ZJ, Weng HY, Weng XC, Zuo ZX, Li CY, et al. Fto plays an oncogenic role in acute myeloid leukemia as a N-6-Methyladenosine RNA demethylase. Blood. 2016(22):128. [DOI] [PMC free article] [PubMed]
- 15.Gokhale Nandan S., McIntyre Alexa B.R., McFadden Michael J., Roder Allison E., Kennedy Edward M., Gandara Jorge A., Hopcraft Sharon E., Quicke Kendra M., Vazquez Christine, Willer Jason, Ilkayeva Olga R., Law Brittany A., Holley Christopher L., Garcia-Blanco Mariano A., Evans Matthew J., Suthar Mehul S., Bradrick Shelton S., Mason Christopher E., Horner Stacy M. N6 -Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host & Microbe. 2016;20(5):654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy Edward M., Bogerd Hal P., Kornepati Anand V.R., Kang Dong, Ghoshal Delta, Marshall Joy B., Poling Brigid C., Tsai Kevin, Gokhale Nandan S., Horner Stacy M., Cullen Bryan R. Posttranscriptional m 6 A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host & Microbe. 2016;19(5):675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi ST, Ma JY, Wang ZB, Guo L, Hou Y, Sun QY. N6-Methyladenosine sequencing highlights the involvement of mRNA methylation in oocyte meiotic maturation and embryo development by regulating translation in Xenopus laevis. J Biol Chem. 2016;291(44):23020–23026. doi: 10.1074/jbc.M116.748889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanova Ivayla, Much Christian, Di Giacomo Monica, Azzi Chiara, Morgan Marcos, Moreira Pedro N., Monahan Jack, Carrieri Claudia, Enright Anton J., O’Carroll Dónal. The RNA m 6 A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Molecular Cell. 2017;67(6):1059-1067.e4. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017. [DOI] [PMC free article] [PubMed]
- 20.Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, et al. Mettl3−/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017. [DOI] [PMC free article] [PubMed]
- 21.Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017. [DOI] [PMC free article] [PubMed]
- 22.Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972;52(1):198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- 23.Trasler JM. Epigenetics in spermatogenesis. Mol Cell Endocrinol. 2009;306(1–2):33–36. doi: 10.1016/j.mce.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Tseng YT, Liao HF, Yu CY, Mo CF, Lin SP. Epigenetic factors in the regulation of prospermatogonia and spermatogonial stem cells. Reproduction. 2015;150(3):R77–R91. doi: 10.1530/REP-14-0679. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Li X, Guo J, Zhang P, Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis. J Anim Sci Biotechnol. 2017;8:35. doi: 10.1186/s40104-017-0166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge SQ, Lin SL, Zhao ZH, Sun QY. Epigenetic dynamics and interplay during spermatogenesis and embryogenesis: implications for male fertility and offspring health. Oncotarget. 2017;8(32):53804–53818. doi: 10.18632/oncotarget.17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–5. [DOI] [PMC free article] [PubMed]
- 28.Zhao Xu, Yang Ying, Sun Bao-Fa, Shi Yue, Yang Xin, Xiao Wen, Hao Ya-Juan, Ping Xiao-Li, Chen Yu-Sheng, Wang Wen-Jia, Jin Kang-Xuan, Wang Xing, Huang Chun-Min, Fu Yu, Ge Xiao-Meng, Song Shu-Hui, Jeong Hyun Seok, Yanagisawa Hiroyuki, Niu Yamei, Jia Gui-Fang, Wu Wei, Tong Wei-Min, Okamoto Akimitsu, He Chuan, Danielsen Jannie M Rendtlew, Wang Xiu-Jie, Yang Yun-Gui. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Research. 2014;24(12):1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerken T., Girard C. A., Tung Y.-C. L., Webby C. J., Saudek V., Hewitson K. S., Yeo G. S. H., McDonough M. A., Cunliffe S., McNeill L. A., Galvanovskis J., Rorsman P., Robins P., Prieur X., Coll A. P., Ma M., Jovanovic Z., Farooqi I. S., Sedgwick B., Barroso I., Lindahl T., Ponting C. P., Ashcroft F. M., O'Rahilly S., Schofield C. J. The Obesity-Associated FTO Gene Encodes a 2-Oxoglutarate-Dependent Nucleic Acid Demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer Julia, Koch Linda, Emmerling Christian, Vierkotten Jeanette, Peters Thomas, Brüning Jens C., Rüther Ulrich. Inactivation of the Fto gene protects from obesity. Nature. 2009;458(7240):894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 31.Gao Xue, Shin Yong-Hyun, Li Min, Wang Fei, Tong Qiang, Zhang Pumin. The Fat Mass and Obesity Associated Gene FTO Functions in the Brain to Regulate Postnatal Growth in Mice. PLoS ONE. 2010;5(11):e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMurray Fiona, Church Chris D., Larder Rachel, Nicholson George, Wells Sara, Teboul Lydia, Tung Y. C. Loraine, Rimmington Debra, Bosch Fatima, Jimenez Veronica, Yeo Giles S. H., O'Rahilly Stephen, Ashcroft Frances M., Coll Anthony P., Cox Roger D. Adult Onset Global Loss of the Fto Gene Alters Body Composition and Metabolism in the Mouse. PLoS Genetics. 2013;9(1):e1003166. doi: 10.1371/journal.pgen.1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Yue, Yan Jingli, Li Qi, Li Jiafei, Gong Shouzhe, Zhou Hu, Gan Jianhua, Jiang Hualiang, Jia Gui-Fang, Luo Cheng, Yang Cai-Guang. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Research. 2014;43(1):373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Qin, Sun Baofa, Liu Qing, Cai Min, Wu Ruifan, Wang Fengqin, Yao Yongxi, Wang Yizhen, Wang Xinxia. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m6A-YTHDF1–dependent mechanism. The FASEB Journal. 2019;33(2):2971–2981. doi: 10.1096/fj.201801393RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017;45(19):11356–11370. doi: 10.1093/nar/gkx778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu YM, Arora A, Min WX, Roifman CM, Grunebaum E. EdU incorporation is an alternative non-radioactive assay to [H-3]thymidine uptake for in vitro measurement of mice T-cell proliferations. J Immunol Methods. 2009;350(1–2):29–35. doi: 10.1016/j.jim.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Thaler J, Fechner F, Herold M, Huber H. Interleukin-6 in multiple myeloma: correlation with disease activity and Ki-67 proliferation index. Leuk Lymphoma. 1994;12(3–4):265–271. doi: 10.3109/10428199409059598. [DOI] [PubMed] [Google Scholar]
- 38.Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272(39):24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 39.Li Hua-Bing, Tong Jiyu, Zhu Shu, Batista Pedro J., Duffy Erin E., Zhao Jun, Bailis Will, Cao Guangchao, Kroehling Lina, Chen Yuanyuan, Wang Geng, Broughton James P., Chen Y. Grace, Kluger Yuval, Simon Matthew D., Chang Howard Y., Yin Zhinan, Flavell Richard A. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Liping, Zang Liqun, Zhang Feiran, Chen Junchen, Shen Hui, Shu Liqi, Liang Feng, Feng Chunyue, Chen Deng, Tao Huikang, Xu Tianlei, Li Ziyi, Kang Yunhee, Wu Hao, Tang Lichun, Zhang Pumin, Jin Peng, Shu Qiang, Li Xuekun. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Human Molecular Genetics. 2017;26(13):2398–2411. doi: 10.1093/hmg/ddx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Sicong, Zhao Boxuan Simen, Zhou Aidong, Lin Kangyu, Zheng Shaoping, Lu Zhike, Chen Yaohui, Sulman Erik P., Xie Keping, Bögler Oliver, Majumder Sadhan, He Chuan, Huang Suyun. m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31(4):591-606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Zejuan, Weng Hengyou, Su Rui, Weng Xiaocheng, Zuo Zhixiang, Li Chenying, Huang Huilin, Nachtergaele Sigrid, Dong Lei, Hu Chao, Qin Xi, Tang Lichun, Wang Yungui, Hong Gia-Ming, Huang Hao, Wang Xiao, Chen Ping, Gurbuxani Sandeep, Arnovitz Stephen, Li Yuanyuan, Li Shenglai, Strong Jennifer, Neilly Mary Beth, Larson Richard A., Jiang Xi, Zhang Pumin, Jin Jie, He Chuan, Chen Jianjun. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell. 2017;31(1):127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishiguro H, Kawahara T. Nonsteroidal anti-inflammatory drugs and prostatic diseases. Biomed Res Int. 2014;2014:436123. doi: 10.1155/2014/436123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalgutkar AS, Rowlinson SW, Crews BC, Marnett LJ. Amide derivatives of meclofenamic acid as selective cyclooxygenase-2 inhibitors. Bioorg Med Chem Lett. 2002;12(4):521–524. doi: 10.1016/S0960-894X(01)00792-2. [DOI] [PubMed] [Google Scholar]
- 45.Sekine Y, Nakayama H, Miyazawa Y, Kato H, Furuya Y, Arai S, et al. Simvastatin in combination with meclofenamic acid inhibits the proliferation and migration of human prostate cancer PC-3 cells via an AKR1C3 mechanism. Oncol Lett. 2018;15(3):3167–72. [DOI] [PMC free article] [PubMed]
- 46.Duffy C.P, Elliott C.J, O’Connor R.A, Heenan M.M, Coyle S, Cleary I.M, Kavanagh K, Verhaegen S, O’Loughlin C.M, NicAmhlaoibh R, Clynes M. Enhancement of chemotherapeutic drug toxicity to human tumour cells in vitro by a subset of non-steroidal anti-inflammatory drugs (NSAIDs) European Journal of Cancer. 1998;34(8):1250–1259. doi: 10.1016/S0959-8049(98)00045-8. [DOI] [PubMed] [Google Scholar]
- 47.Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. Structural basis for the discriminative recognition of N6-Methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. 2015;290(41):24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soriano-Hernández Alejandro D., Galvan-Salazar Hector R., Montes-Galindo Daniel A., Rodriguez-Hernandez Alejandrina, Martinez-Martinez Rafael, Guzman-Esquivel Jose, Valdez-Velazquez Laura L., Baltazar-Rodriguez Luz M., Espinoza-Gómez Francisco, Rojas-Martinez Augusto, Ortiz-Lopez Rocio, Gonzalez-Alvarez Rafael, Delgado-Enciso Ivan. Antitumor effect of meclofenamic acid on human androgen-independent prostate cancer: a preclinical evaluation. International Urology and Nephrology. 2011;44(2):471–477. doi: 10.1007/s11255-011-0012-0. [DOI] [PubMed] [Google Scholar]
- 49.Mauer Jan, Luo Xiaobing, Blanjoie Alexandre, Jiao Xinfu, Grozhik Anya V., Patil Deepak P., Linder Bastian, Pickering Brian F., Vasseur Jean-Jacques, Chen Qiuying, Gross Steven S., Elemento Olivier, Debart Françoise, Kiledjian Megerditch, Jaffrey Samie R. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2016;541(7637):371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71(6):973–985 e975. [DOI] [PMC free article] [PubMed]
- 51.Mauer Jan, Sindelar Miriam, Despic Vladimir, Guez Théo, Hawley Ben R., Vasseur Jean-Jacques, Rentmeister Andrea, Gross Steven S., Pellizzoni Livio, Debart Françoise, Goodarzi Hani, Jaffrey Samie R. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nature Chemical Biology. 2019;15(4):340–347. doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabrielli BG, Roy LM, Maller JL. Requirement for Cdk2 in cytostatic factor-mediated metaphase II arrest. Science. 1993;259(5102):1766–1769. doi: 10.1126/science.8456304. [DOI] [PubMed] [Google Scholar]
- 53.Ortega Sagrario, Prieto Ignacio, Odajima Junko, Martín Alberto, Dubus Pierre, Sotillo Rocio, Barbero Jose Luis, Malumbres Marcos, Barbacid Mariano. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nature Genetics. 2003;35(1):25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 54.Xu Kai, Yang Ying, Feng Gui-Hai, Sun Bao-Fa, Chen Jun-Qing, Li Yu-Fei, Chen Yu-Sheng, Zhang Xin-Xin, Wang Chen-Xin, Jiang Li-Yuan, Liu Chao, Zhang Ze-Yu, Wang Xiu-Jie, Zhou Qi, Yang Yun-Gui, Li Wei. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Research. 2017;27(9):1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu Phillip J, Zhu Yunfei, Ma Honghui, Guo Yueshuai, Shi Xiaodan, Liu Yuanyuan, Qi Meijie, Lu Zhike, Shi Hailing, Wang Jianying, Cheng Yiwei, Luo Guanzheng, Dai Qing, Liu Mingxi, Guo Xuejiang, Sha Jiahao, Shen Bin, He Chuan. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Research. 2017;27(9):1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu P, Zhu YF, Ma HH, Cui YQ, Shi XD, Luo GZ, et al. YTHDC2 regulates spermatogenesis through promoting the translation of N-6-methyladenosine-modified RNA. FASEB J. 2017:31.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Detection of m6Am level in polyA RNA and small nuclear RNA. Cells were treated with 0, 40, 80 and 120 μmol/L MA2. Content of m6Am and A was detected by LC/MS-MS. Relative m6Am level was normalized by m6Am/A. (A) Relative m6Am level of PolyA RNA. Data were represented by the mean ± SEM, n = 3. n,s means P > 0.05. (B) Relative m6Am level of snRNA. Data were represented by the mean ± SEM, n = 3. (JPG 648 kb)
Table S1. Oligo information. Table S2. m6A modification information of CDKs. Table S3. Information of potential MA2 targets. (DOCX 19 kb)
Data Availability Statement
All data supporting our findings are included in the manuscript.