Abstract
The C-type lectin chondrolectin (chodl) represents one of the major gene products dysregulated in spinal muscular atrophy models in mice. However, to date, no function has been determined for the gene. We have identified chodl and other novel genes potentially involved in motor axon differentiation, by expression profiling of transgenically labeled motor neurons in embryonic zebrafish. To enrich the profile for genes involved in differentiation of peripheral motor axons, we inhibited the function of LIM-HDs (LIM homeodomain factors) by overexpression of a dominant-negative cofactor, thereby rendering labeled axons unable to grow out of the spinal cord. Importantly, labeled cells still exhibited axon growth and most cells retained markers of motor neuron identity. Functional tests of chodl, by overexpression and knockdown, confirm crucial functions of this gene for motor axon growth in vivo. Indeed, knockdown of chodl induces arrest or stalling of motor axon growth at the horizontal myoseptum, an intermediate target and navigational choice point, and reduced muscle innervation at later developmental stages. This phenotype is rescued by chodl overexpression, suggesting that correct expression levels of chodl are important for interactions of growth cones of motor axons with the horizontal myoseptum. Combined, these results identify upstream regulators and downstream functions of chodl during motor axon growth.
Introduction
Motor axons make complex pathway decisions during their growth to their eventual muscle targets (Landmesser, 2001; Guthrie, 2007). At the same time, neurodegenerative diseases of the spinal cord often affect motor neurons. Recognition molecules, necessary for axonal navigation, have been implicated in the disease process (Schmidt et al., 2009). Thus, to identify potential therapeutic targets, there is a need to better understand axonal differentiation of motor neurons and the molecules involved (Bonanomi and Pfaff, 2010).
The zebrafish is ideally suited for such studies because of the unique development of primary motor neurons in the trunk that allows functional analyses of development at the single-cell level (Lewis and Eisen, 2003). In each hemisegment of the trunk, three to four primary motor neurons send out pioneer axons on a joint trajectory to the horizontal myoseptum, an intermediate target. From here, the axon of the caudal primary motor neuron (CaP) continues to grow ventrally, the middle primary motor neuron (MiP) forms a dorsal collateral from a more dorsal position along the axon, and the rostral primary motor neuron (RoP) grows its axon laterally. The CaP axon is the first to grow out, followed by MiP and RoP (Eisen et al., 1986; Myers et al., 1986).
A number of factors have been found that specifically affect interactions of growing motor axons with the horizontal myoseptum choice point (Bernhardt and Schachner, 2000; Rodino-Klapac and Beattie, 2004; Zhang et al., 2004; Schweitzer et al., 2005; Schneider and Granato, 2006; Hilario et al., 2010). Many of these factors are either extracellular matrix (ECM) components at the horizontal myoseptum or genes that modify the ECM. However, relatively little is known about the potential receptors on the axons.
Motor neuron identity, as well as axonal pathfinding, depend on LIM-domain containing transcription factors [LIM homeodomain factors (LIM-HDs)] in vertebrates (Tsuchida et al., 1994; Pfaff et al., 1996; Bhati et al., 2008), including zebrafish (Inoue et al., 1994; Appel et al., 1995; Tokumoto et al., 1995; Hutchinson and Eisen, 2006; Hutchinson et al., 2007). LIM-HDs control the expression of axon pathfinding genes in motor neurons (Kania and Jessell, 2003) and sensory neurons (Miyashita et al., 2004; Yeo et al., 2004) and function in complexes with cofactors, such as CLIM (also known as NLI and Ldb) (Lee and Pfaff, 2003; Güngör et al., 2007). Overexpression of a dominant-negative form of CLIM in zebrafish (DN-CLIM) prevents motor neurons from forming correct projections (Segawa et al., 2001; Becker et al., 2002). Indeed, motor neurons in the ventral spinal cord that overexpress DN-CLIM can still grow axons, but only inside the spinal cord (Segawa et al., 2001; Zhong et al., 2011).
Here, we use these neurons for expression profiling to find genes involved in motor axon navigation. We identify the motor neuron-specific C-type lectin gene chondrolectin (chodl), previously found to be dysregulated in mouse spinal muscular atrophy (SMA) models (Zhang et al., 2008; Bäumer et al., 2009), as an important regulator of motor axon/choice point interactions during development.
Materials and Methods
Animals.
All fish are kept and bred in our laboratory fish facility according to standard methods (Westerfield, 2000), and all experimental procedures have been approved by the British Home Office. We used wild-type (wik) and Tg(mnx1:GFP)ml2 (Flanagan-Steet et al., 2005) and Tg(vsx1:GFP)nn5 (Kimura et al., 2008) embryos of either sex. For simplicity, Tg(mnx1:GFP)ml2 is designated HB9:GFP throughout this manuscript.
Gene knockdown.
We purchased splice site-directed morpholinos to chodl (MO1: exon 2–intron 2 splice site, TATGAAACTCCTCATCTCACCTGAA; MO2: intron 1–exon 2 splice site, CTGACCTGGAGAGACAAAATTCACA), a start codon directed morpholino (MO3: CGCATCCTCGTGTTTCCCTTGAGTC), and standard control morpholino from Gene Tools. For injections, rhodamine dextran (0.8%; Mr = 10 × 103; Invitrogen) was added to morpholino solutions. A glass micropipette was filled with morpholino solution. A volume of 0.5 nl per egg (one- to four-cell stage) was injected as described previously (Feldner et al., 2007). Generally, the morpholino concentration was 1 mm, if not stated otherwise. Control morpholino injections were performed with every experiment on embryos taken from the same clutch or pool of clutches as the embryos receiving specific morpholinos, to avoid any influence of developmental differences. For rescue and synergy experiments, the total morpholino load per egg was kept constant between groups by supplementing with control morpholino. For long-term labeling of single CaP axons, specific or control morpholinos (0.4 mm) were coinjected with the pG1-HB9:mmGFP plasmid (0.4 μg/μl) (Flanagan-Steet et al., 2005) at the single-cell stage. Successful gene knockdown after injection of splice site-directed morpholinos was determined by PCR for chodl (primer 1: forward, 5′-GTGTAAGCCCAGCTCGTTG-3′; reverse primer, 5′-CTATCTTTGGCGTCTTGGAG-3′). To detect an aberrant splice product containing intronic sequences, we used primer 2 (forward, 5′-TACGTATTTGGCTGCCAGTG-3′) located in intron 2 together with a reverse primer in exon 7 (reverse, 5′-GCTAGCAGGAAGGTGCAGAC-3′).
Gene overexpression.
DN-CLIM was overexpressed as described previously (Becker et al., 2002; Zhong et al., 2011) using the myc epitope-tagged DN-CLIM in the CS2-MT plasmid to generate the mRNA. The CS2-MT-NLS plasmid that contains a nuclear localization sequence served to generate the control mRNA.
To overexpress chodl, we amplified full-length chodl from embryonic cDNA (primers: forward, 5′-CTCTCTCCGCATTTCAGAGG-3′; reverse, 5′-GCTAGCAGGAAGGTGCAGAC-3′) and inserted the product into the vector pcDNA3-IRES-GFP (a gift from Dr. Lee Tan, Key Laboratory of Molecular Medicine, Ministry of Education, Fudan University, Shanghai, China) that also drives GFP expression from its second ribosomal entry site. The same vector without the chodl insert was used to generate control mRNA.
We synthesized mRNA with the mMessage mMachine Kit (Ambion) followed by extension of the poly(A) tail with the poly(A) tailing kit (Ambion), each according to the manufacturer's instructions. The mRNA (typically 1–2 μg/μl) was injected in the same way as morpholinos. Successful overexpression was indicated by ubiquitous GFP fluorescence for chodl mRNA or myc immunoreactivity for DN-CLIM overexpression at 24 h postfertilization (hpf).
Immunohistochemistry.
We used the following antibodies: Islet-1/-2 (40.2D6; 1:1000), which labels motor neuron nuclei and Rohon–Beard cells (Feldner et al., 2005), the 4D9 antibody (1:100) to engrailed, which labels muscle pioneer cells at the horizontal myoseptum (Patel et al., 1989), anti-HB9 (MNR2; 81.5C10-a; 1:400), labeling motor neurons (Reimer et al., 2008), and anti-znp1 (1:400), labeling motor axons (Trevarrow et al., 1990), were all obtained as concentrates from the Developmental Studies Hybridoma Bank maintained by Department of Biological Sciences, University of Iowa (Iowa City, IA). We also used anti-GABA (A 2052; 1:1000; Sigma-Aldrich), anti-Pax2 (PRB-276P; 1:300; Covance), anti-choline acetyltransferase (ChAT) (AB144P; 1:500; Millipore), anti-GFP (A 11122; 1:200; Invitrogen), and anti-Chx10 (Kimura et al., 2008) antibodies. Secondary Cy2- and Cy3-conjugated antibodies were purchased from Jackson ImmunoResearch Laboratories. Immunohistochemistry on embryos was performed as described previously (Feldner et al., 2007). For each experiment, control embryos were labeled in parallel for direct comparison.
In situ hybridization.
We used the indicated primers to generate probes for chodl (forward, 5′-AGTCGTGTTGCGTTCTGGGA-3′; reverse, 5′-CTGTCTATCTTTGGCGTCTTGG-3′), tachykinin 1 (tac1) (forward, 5′-AAGGGAAAGTTACTGGGAGC-3′; reverse, 5′-GGGAGCGAATGTGAAGATGA-3′), calcitonin/calcitonin-related polypeptide, α (calca) (forward, 5′-CCTACGCTCTGATTATTTGCC-3′; reverse, 5′-TTCCTCCCTCCTTCGGTTC-3′), and NK1 transcription factor-related 2-like, b (nkx1.2lb) (forward, 5′-ATCACACGATCGAGCACAAG-3′; reverse, 5′-TTAGCACGTATTGCCGAATG-3′). Nonradioactive in situ hybridization was performed as described previously (Feldner et al., 2007). In some cases, in situ hybridization was followed by GFP immunohistochemistry for transgene visualization, as above.
FACS and expression profiling.
All fish embryos (26 hpf; n > 200 per group) were dechorionated and washed with calcium-free Ringer's buffer for 15 min. Embryos or tails only were then mechanically dissociated in 0.25% trypsin in 1 mm EDTA in PBS and incubated for 30–60 min at 28.5°C. Embryo digestion was stopped by adding an equal volume of 2 mm CaCl2 with 20% fetal calf serum and centrifugation at 300 × g for 5 min at room temperature. The tissue was resuspended in 500 μl of L-15 medium, and sorted by a BD FACSAria II Flow Cytometer (BD Biosciences). Viability was tested by propidium iodide staining and found to be >90%. Sorted cells were immediately frozen in lysis buffer at a concentration of 1000 cells/μl and volumes of at least 10 μl. A typical yield for 100 embryo tails was 2.5 × 105 cells, 19% of which were GFP positive. Samples were then sent to Miltenyi Biotech for RNA isolation and gene expression profiling on one-color chip hybridization on zebrafish gene expression microarrays (V2) provided by Agilent Technology. These arrays contain 43,803 probes per chip. From the pool of isolated RNA, cDNA was amplified twice, fluorescently labeled, and separately hybridized on a chip (details available from Agilent Technology). Significance was calculated using the ratio error model (Weng et al., 2006). Regulation was assumed with p < 0.01 and a cutoff of at least twofold upregulation or downregulation.
Time-lapse video microscopy.
Dechorionated 18 hpf embryos were mounted on coverslips with 2% low-melting agarose prepared in E3 embryo medium with 0.02% w/v 3-aminobenzoic acid ethyl ester (MS222) (Sigma-Aldrich). Image stacks were taken by confocal microscopy (LSM710; Zeiss) with a 20× lens at 28°C every 10 min between 18 and 27 hpf. CaP axon growth speed (axon length quantification) was analyzed based on time-lapse still picture series prepared with the ZEN2009 software (Zeiss).
Quantifications and statistical analyses.
To quantify double-labeled neurons in control and DN-CLIM-injected embryos, specimens were fixed at 24 hpf, and their trunk region was scanned with a 20× objective by confocal microscopy. All fluorescent neurons in trunk segments 8 and 9 were counted in z-projections of confocal image stacks.
The number of trunk segments in which ChAT immunoreactivity was detectable in the ventral spinal cord was determined for segments 7–14 on one side of the embryo.
To correlate axonal differentiation with the onset of gene expression in the tail segments 16–26, the last 10 segments were assessed for in situ hybridization signal and presence of a motor axon.
The length of ventral motor axons in segments 7–14 after morpholino or overexpression manipulations was assessed in znp-1-labeled embryos or HB9:GFP transgenic embryos by determining whether an axon was shorter than in wild-type embryos, in which axons had reached the ventral edge of the ventral myotome. Additional categories were as follows: shorter than the position of the horizontal myoseptum (HM−), at the horizontal myoseptum (HM), or grown beyond the horizontal myoseptum but still shorter than wild-type length (HM+). The horizontal myoseptum was either identified by 4D9 immunoreactivity (znp-1-labeled preparations) or by the characteristic thickening of the CaP axon at the horizontal myoseptum (HB9:GFP transgenic embryos). The observer was always blinded to the treatment of the embryos.
Data are presented as mean, and error bars indicate the SEM. Groups were compared by Mann–Whitney U test (for two groups), one-way ANOVA, or two-way ANOVA followed by the Holm–Sidac post hoc test (SigmaStat 3.5) for multiple comparisons.
Results
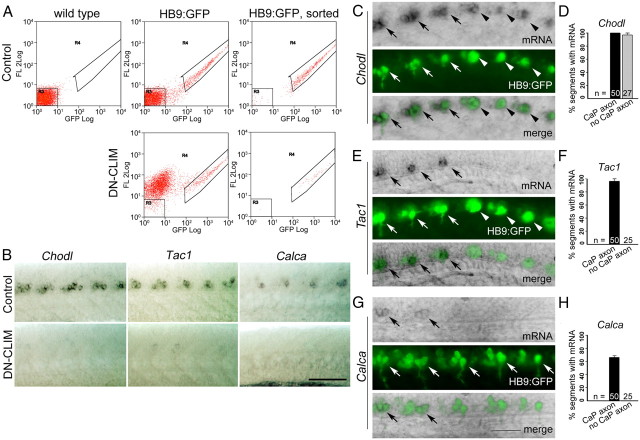
To identify new genes that are involved in motor axon growth, we decided to use expression profiling of transgenically labeled primary trunk motor neurons (Fig. 1A) in which LIM-HD signaling is compromised. LIM-HD transcription factors are pivotal regulators of motor neuron development during embryogenesis of many species (Tsuchida et al., 1994; Pfaff et al., 1996; Bhati et al., 2008). To exert these functions, LIM-HDs require the association of CLIM cofactors (Lee and Pfaff, 2003). Inhibition of LIM-HDs was achieved via overexpression of a DN-CLIM protein. While lacking motifs that mediate interactions with other classes of proteins, DN-CLIM only contains the LIM-interaction domain (LID) and a nuclear localization signal, thus specifically targeting LIM-HD transcription factors (Segawa et al., 2001; Güngör et al., 2007) (Fig. 1B). Overexpression of DN-CLIM in HB9:GFP transgenic zebrafish resulted in axons emerging from GFP-positive neurons, but these axons grew along the ventral edge of the spinal cord instead of exiting it (Fig. 1C) (Segawa et al., 2001; Zhong et al., 2011). The presence of GFP-expressing neurons in these embryos indicated that the HB9 promoter fragment used to generate this fish was still active and suggested that GFP+ cells might have retained motor neuron characteristics. Hence, expression profiles from motor neurons versus GFP+ cells in DN-CLIM-treated embryos could be enriched for genes that are involved specifically in motor axon pathfinding and not in cell fate decisions or axon growth per se. A similar approach has led to the identification of genes involved in formation of the midbrain–hindbrain boundary downstream of LIM-HDs (Hirate et al., 2001). However, transfating of motor neurons into interneuron cell types has been described after compromising LIM-HD signaling (Segawa et al., 2001), warranting further investigations into the identity of HB9:GFP+ cells in control and DN-CLIM-treated animals.
Figure 1.
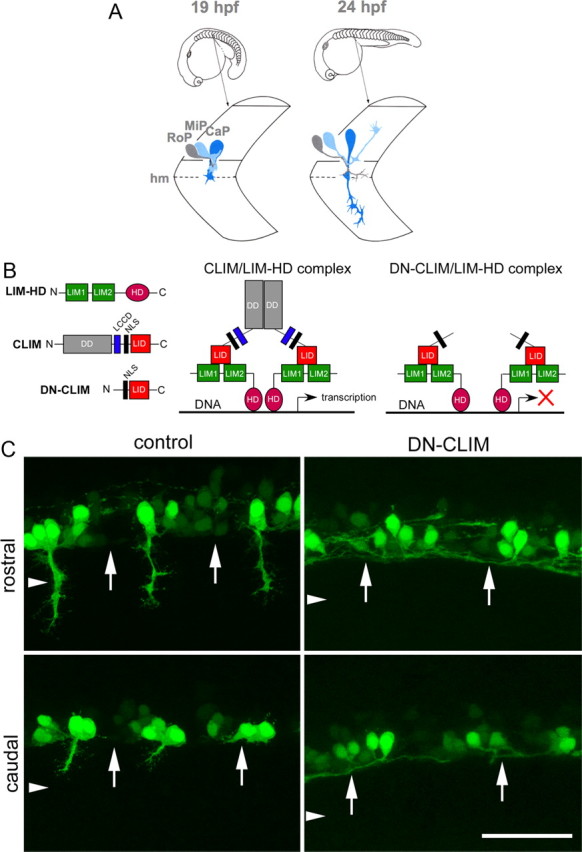
DN-CLIM overexpression inhibits LIM-HD-dependent gene transcription and prevents HB9:GFP+ motor neurons from growing axons out of the spinal cord. A, Schematic presentation of axon growth by the primary motor neurons CaP, MiP, and RoP. Axons grow ventrally out of the spinal cord on a joint pathway and diverge at the horizontal myoseptum (hm) choice point [modified after the study by Westerfield and Eisen (1988) with permission from Elsevier]. B, Structures of typical LIM-HDs, containing LIM domains and homeodomain (HD), and CLIM, containing dimerization domain (DD), Ldb1/Chip conserved domain (LCCD), nuclear localization sequence (NLS), and LIM interaction domain (LID) are shown. DN-CLIM lacks the DD and LCCD domains and inhibits the formation of transcriptionally active LIM-HD complexes. C, Lateral trunk views of control mRNA or DN-CLIM mRNA-injected HB9:GFP transgenic embryos are shown (orientation as in A). In rostral and caudal segments, HB9:GFP+ cells with axons are visible, but axons do not manage to grow out of the spinal cord after DN-CLIM injection. The ventral border of the spinal cord is indicated by arrows; the approximate position of the horizontal myoseptum is indicated by arrowheads. Scale bar, 50 μm.
Identity of HB9:GFP+ cells in DN-CLIM-injected embryos
In control embryos, the HB9:GFP transgene was clearly expressed in motor neurons in the ventral spinal cord with axons exiting the spinal cord. Furthermore, HB9 immunoreactivity was found in 61% of HB9:GFP+ cells, and 60% of the cells showed immunoreactivity for Islet-1/-2 (Fig. 2A–D). Some of the GFP+ cells in each hemisegment were positive for ChAT, indicating a mature motor neuron phenotype for some of the HB9:GFP+ motor neurons (Fig. 2E,F). The cells that were not labeled for HB9 or Islet-1/-2 antibodies displayed generally weaker GFP expression. To determine whether these cells could be interneurons, we used double labeling with markers of interneurons. However, nkx1.2lb mRNA (Bae et al., 2004), Chx10 protein (Kimura et al., 2008), Pax2 protein (Batista and Lewis, 2008), and GABA (Segawa et al., 2001) revealed no or little overlap with GFP labeling (Fig. 3). Thus, the majority of HB9:GFP+ cells were likely motor neurons on the basis of axonal projections out of the spinal cord and expression of motor neuron markers (HB9; Islet-1/-2). Some cells that expressed lower levels of GFP might have been interneurons of undetermined identity or developing motor neurons in which incipient transgene expression was detectable, but immunohistochemistry was not sufficiently sensitive to detect low levels of endogenous protein.
Figure 2.
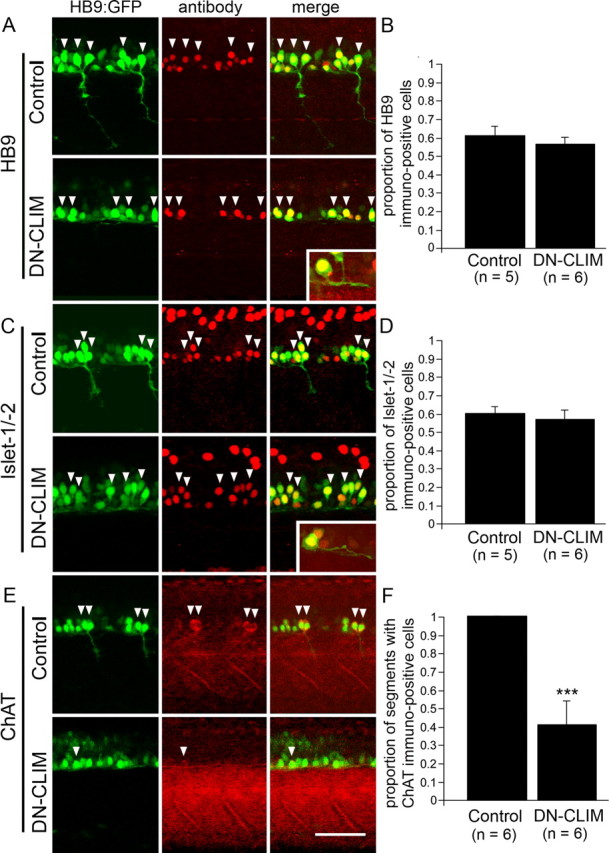
DN-CLIM treatment differentially affects motor neuron marker expression in spinal HB9:GFP+ cells at 24 hpf. Trunk views are shown; for orientation see Figure 1A. Double-labeled cells are indicated by arrowheads. HB9:GFP+ cells do not show a significant reduction in the proportion of HB9 (A, B)- or Islet-1/-2 (C, D)-immunolabeled cells. The insets depict double-labeled cells with intraspinal axons at higher magnification. The proportion of segments with ChAT-expressing HB9:GFP+ cells is significantly reduced (E, F). ***p < 0.001. Scale bar: 50 μm; insets, 25 μm. Error bars indicate SEM.
Figure 3.
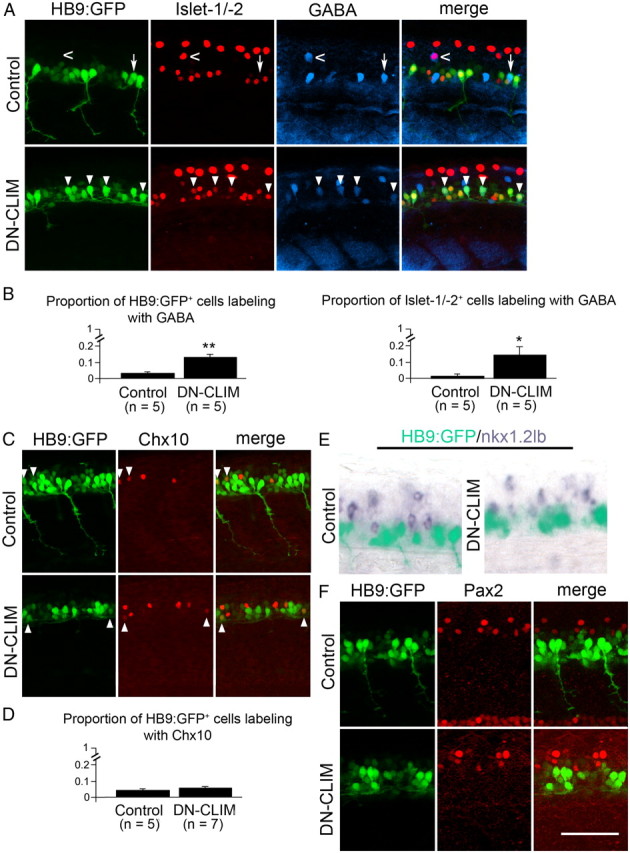
DN-CLIM treatment differentially affects interneuron marker expression in HB9:GFP+ cells. Trunk views are shown; for orientation, see Figure 1A. Double/triple-labeled cells are indicated by arrowheads. A, B, Double labeling of Islet-1/-2 and GABA in HB9:GFP+ cells indicates a small, but significant increase in HB9:GFP/GABA and Islet-1/-2/GABA double-labeled cells after DN-CLIM treatment. The arrow indicates a cell that is only labeled by the transgene and GABA antibody; the open arrow indicates a cell that is only labeled by Islet-1/-2 and GABA antibodies. The dorsal location of this cell suggests interneuron identity. C, D, The proportion of HB9:GFP+ cells that are colabeled with Chox10 immunolabeling remains very small after DN-CIM treatment. E, F, There is no apparent overlap of nkx1.2lb mRNA, nor Pax2 immunoreactivity, and HB9:GFP transgene in control and DN-CLIM-treated animals. *p < 0.05 (one-sided); **p < 0.01. Scale bar, 50 μm. Error bars indicate SEM.
In DN-CLIM-injected embryos, 29% fewer HB9:GFP+ cells were observed (control: 25.5 ± 1.59 cells, n = 6 embryos; DN-CLIM: 18.2 ± 0.65 cells, n = 6 embryos; p < 0.01). This could indicate that some cells adopted a different fate. However, these cells would not skew a further analysis of gene expression, as they would not be sorted into the pool of fluorescent cells. The cells that still expressed GFP also retained immunoreactivity for the motor neuron markers HB9 (56%) and Islet-1/-2 (56%) both to an extent that was not significantly different from controls (Fig. 2A–D). This is consistent with the hypothesis that fate changes of GFP+ cells were rare. Single HB9:GFP+ cells were observable in the ventral spinal cord that could be colabeled with HB9 or Islet-1/-2 antibodies and grew an axon within the spinal cord of DN-CLIM-treated embryos (Fig. 2A,C), indicating that motor neuron-like cells still grew (intraspinal) axon. However, the proportion of segments that contained motor neurons expressing the mature marker ChAT was reduced by 65% (Fig. 2E,F), suggesting that motor neuron maturation was attenuated by DN-CLIM.
After DN-CLIM injection, the proportion of HB9:GFP+ cells that were colabeled by Chx10 antibodies remained unchanged (4% in controls and 5% in DN-CLIM-treated embryos), suggesting that HB9:GFP+ cells did not change their fate into interneurons that express Chx10 (Fig. 3C,D). Moreover, HB9:GFP+ cell did not acquire expression of nkx1.2lb mRNA (Fig. 3E) or Pax2 immunoreactivity after DN-CLIM injection (Fig. 3F). However, it has previously been shown that Islet-2+ motor neurons acquire the interneuron marker GABA when cofactor interactions of LIM-HDs are disrupted (Segawa et al., 2001). Therefore, we quantified coexpression in triple-labeling studies of HB9:GFP, Islet-1/-2, and GABA. Indeed, the proportion of Islet-1/-2+ cells in the ventral spinal cord that were also GABA+ was significantly increased from 1% in controls to 14% in DN-CLIM-treated embryos. Likewise, the proportion of HB9:GFP+ cells that were also GABA+ was significantly increased from 2 to 12% (Fig. 3A,B). In summary, the majority of HB9:GFP+ cells retained motor neuron identity to some extent, as indicated by HB9 and Islet-1/-2 immunoreactivity, whereas full maturation into ChAT+ motor neurons was attenuated and some HB9:GFP+ cells acquired interneuron characteristics (i.e., GABA immunoreactivity).
Expression profiling reveals motor neuron expressed genes that are downstream of LIM-HDs
Since most HB9:GFP+ cells in DN-CLIM-injected embryos retained at least some characteristics of motor neurons, we next used DN-CLIM-injected versus uninjected HB9:GFP transgenic embryos for successful FACS of HB9:GFP+ cells (Fig. 4A) followed by expression profiling. Thus, we obtained a list of >160 sequences that were downregulated at least twofold in GFP+ cells in DN-CLIM-injected embryos compared with GFP+ motor neurons in uninjected embryos. Comparing expression of GFP+ motor neurons with GFP− cells in wild-type embryos from embryonic trunks and tails yielded >2100 at least twofold overrepresented sequences in HB9:GFP+ motor neurons (data not shown). The intersection of these two lists contained 28 sequences that were both downregulated after DN-CLIM treatment and overrepresented in GFP+ cells of the trunk and tail of uninjected embryos (Table 1). Among the top 10 regulated genes, we found chodl, tac1, and calca to be expressed in clusters in the ventral spinal cord in the position of motor neurons, using the available online expression patterns (ZFIN.org). In situ hybridization for these genes in HB9:GFP transgenic embryos at 24 hpf confirmed expression in motor neurons with axons growing out of the spinal cord and absence of a signal in DN-CLIM-injected embryos (Fig. 4B–H). These results showed that expression of chodl, tac1, and calca is downstream of LIM-HDs in motor neurons and validated these screen results.
Figure 4.
Expression profiling reveals LIM-HD-dependent genes expressed in motor neurons. Orientations of photomicrographs are as in Figure 1A. A, FACS shows a population of cells (boxed in the far right of each panel) that are only present in control or DN-CLIM-injected HB9:GFP embryos, but not in wild-type, and can be successfully sorted (right column). B, chodl, tac1, and calca are expressed specifically in motor neuron clusters and expression is abolished in DN-CLIM-injected embryos. C–H, Lateral views of the tail region of in situ hybridizations in HB9:GFP transgenic embryos are shown. HB9:GFP+ motor neurons in caudal, younger, segments without axons (no CaP axons) are indicated by white arrowheads, and motor neurons with axons by white arrows in fluorescence illumination. Expression of mRNA is indicated by black arrows in light microscopic and merged images. All motor neurons express chodl (C, D). tac1 expression coincides with axon growth (E, F). calca is only expressed by motor neurons with longer axons (G, H). Scale bars: B, 100 μm; C, E, G, 50 μm. Error bars indicate SEM.
Table 1.
Sequences that are downregulated after DN-CLIM treatment and enriched in HB9:GFP+ neurons of wild-type embryos (sorted by fold downregulation after DN-CLIM treatment)
| Fold downregulation after DN-CLIM treatment | Fold higher in HB9:GFP+ versus HB9:GFP− | Sequence designation on gene array | Gene name |
|---|---|---|---|
| −4.0901 | 3.0531 | vsx2 | vsx2 |
| −3.3734 | 18.8099 | nkx1.2lb | nkx1.2lb |
| −3.2371 | 8.68663 | TC285423 | chodl |
| −3.127 | 2.79665 | flj13639 | 36k |
| −2.9492 | 7.22032 | tal2 | tal2 |
| −2.8507 | 8.51966 | zgc:92886 | calca |
| −2.8159 | 3.47611 | wu:fk57g06 | — |
| −2.7056 | 7.35989 | hlxb9 | mnx1 |
| −2.6079 | 34.3453 | BC083533 | tac1 |
| −2.5928 | 12.2849 | slc6a5 | glyt2 |
| −2.5894 | 7.01896 | BI864161 | syt6 |
| −2.5822 | 2.78151 | TC282185 | lmo1 |
| −2.5659 | 2.06127 | try | try |
| −2.4778 | 2.22156 | TC299721 | zgc:165474 |
| −2.4615 | 7.0457 | BI476673 | — |
| −2.3383 | 2.90834 | ENSDART00000013302 | slc6a17 |
| −2.3125 | 2.26726 | CK397088 | — |
| −2.2928 | 10.3374 | CK705301 | st18 |
| −2.2926 | 2.30332 | tpbgl | tpbgl |
| −2.2236 | 7.93957 | ENSDART00000022233 | — |
| −2.1145 | 13.2798 | zgc:101108 | nxph1 |
| −2.1115 | 2.11138 | zgc:92208 | mllt10 |
| −2.0953 | 2.05984 | BC055562 | tada2 |
| −2.0502 | 26.233 | gad1 | gad1 |
| −2.0441 | 3.32192 | tlx1 | tlx1 |
| −2.0315 | 3.02975 | BI840491 | — |
| −2.0157 | 2.3796 | AL725137 | eif3ha |
| −2.0045 | 2.13384 | TC273590 | cbx1b |
chodl, tac1, and calca regulation could be relatively proximal to the LIM-HD regulatory network or their lack of expression after DN-CLIM overexpression could be a consequence of axons not exiting the spinal cord and therefore not receiving inductive signals. To determine whether such inductive signals could play a role, we determined the onset of chodl, tac1, and calca expression relative to axon outgrowth. To do this, we analyzed the caudal trunk segments of 22 hpf embryos, because trunk segments and their motor neurons are progressively younger toward the tail (Sato-Maeda et al., 2008). chodl expression was detectable in almost all segments with HB9:GFP+ motor neurons, including the most caudal ones that did not yet show axon outgrowth (Fig. 4C,D). tac1 expression was detectable in all segments with visible HB9:GFP+ motor axon, and in none of the segments with HB9:GFP+ neurons without axons in more caudal segments (Fig. 4E,F). calca expression was only detectable in more rostral segments, in which motor axons were of some length. In more caudal HB9:GFP+ motor neurons with shorter or no axons, calca mRNA was not detectable (Fig. 4G,H). Thus, chodl is expressed very early during motor neuron differentiation, whereas tac1 expression correlates with axon outgrowth and calca is only detectably expressed after motor axons have grown for some distance into the muscle periphery. These data indicated that chodl regulation is most likely to be proximal to the LIM-HD network, whereas extrinsic inductive signals could play a more prominent role for expression of tac1 and calca.
chodl is a C-type lectin gene expressed in developing motor neurons
Because chodl has been implicated in motor neuron disease, but its function has not been elucidated (Zhang et al., 2008; Bäumer et al., 2009), we focused our further analysis on this gene. Cloning of a zebrafish cDNA encoding chodl in full length revealed a sequence coding for a predicted protein of 296 aa with a signal peptide (amino acids 1–20), a C-type lectin domain (amino acids 30–187), and a transmembrane domain (amino acids 238–260), suggesting cell surface localization (Fig. 5A). Comparisons with other vertebrate groups revealed a high degree of overall conservation (e.g., 94% identity with Xenopus and chicken and 92% with mouse and human; Fig. 5B).
Figure 5.
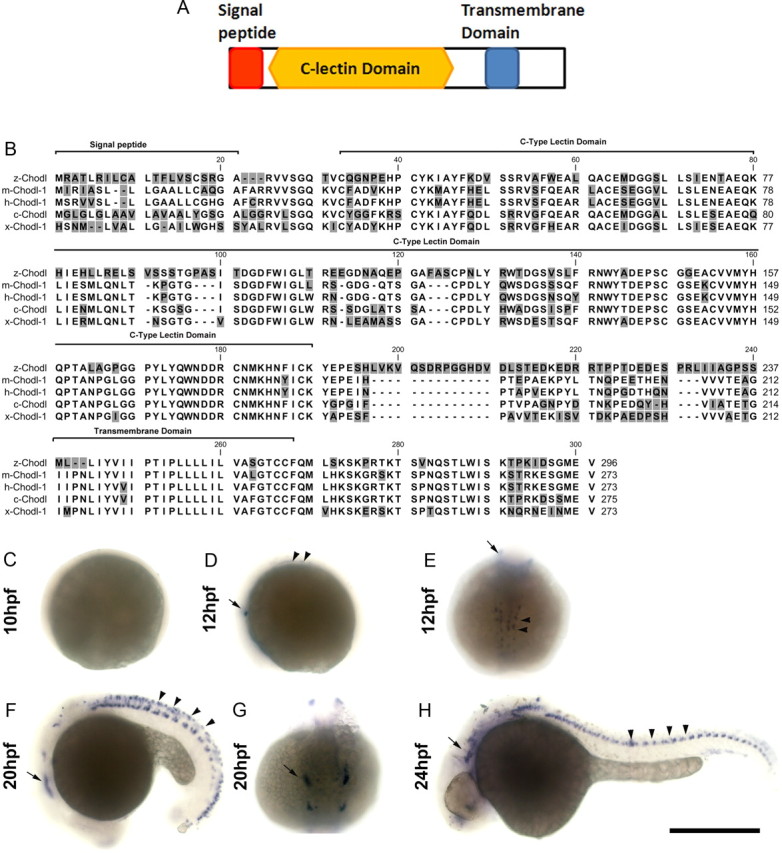
Cloning and expression of chodl. A, Domain analysis of the chodl sequence predicts the presence of a signal peptide, a C-type lectin domain and a transmembrane domain. B, The predicted amino acid sequence of chodl shows conservation with chodl of other vertebrates (m, mouse; h, human; c, chicken; x, Xenopus laevis). C–H, Whole-mount in situ hybridization indicates a signal from 12 hpf. The arrows depict the trigeminal ganglion in D–H. The arrowheads in D indicate cells in the trunk. The arrowheads in F indicate a row of cells in the trunk dorsal to the motor neurons. In H, the arrowheads point to motor neurons. Scale bar, 500 μm.
Whole-mount in situ hybridization indicated no expression of chodl at 8 and 10 hpf, but at 12 hpf expression was detected in lateral cells of the head and ventral cells in the midtrunk region, consistent with the positions of the trigeminal ganglia and motor neurons, respectively. At 20 hpf, expression was detected in cranial ganglia, motor neurons, and a row of cells in the dorsal spinal cord. By 24 hpf, this expression was downregulated again, and in the spinal cord expression of chodl appeared to be restricted to HB9:GFP+ motor neurons, many of which had axons that left the spinal cord. Vsx1 is a marker for interneurons that emerge from the ventricular zone adjacent and directly dorsal to motor neurons. In transgenic fish, in which GFP expression is driven by the vsx1 promoter (Kimura et al., 2008), no overlap of GFP labeling with chodl mRNA expression was observed at 24 hpf (data not shown), lending further support to expression of chodl in the trunk mainly in motor neurons. In addition, cranial ganglia and neurons in the forebrain were labeled by the probe (Fig. 5C–H). Thus chodl expression is restricted to specific cells types in the nervous system and is expressed in primary motor neurons from early stages of their differentiation to at least 24 hpf.
chodl is important for motor axon growth
To determine the function of chodl, we designed two splice-blocking morpholinos, both targeting exon 2. MO1 (1 mm), targeting the exon 2/intron 2 boundary, and MO2 (1 mm), targeting the intron 1/exon 2 boundary, modestly reduced the intensity of the wild-type PCR band in injected embryos at 24 hpf. Combining the two morpholinos at one-half the concentration each (0.5 mm) had a more profound reducing effect on the wild-type band and produced an extra band, potentially due to a cryptic splice site being revealed (Draper et al., 2001) (Fig. 6A). Sequencing this ectopic band indicated precise excision of exon 2, leading to premature stop codons, such that it is unlikely that the aberrantly spliced message led to translation of functional protein. Using another primer combination, in which the upstream primer was located in intron 2, 49 bp upstream of exon 3, amplified a transcript when MO2 was used. This transcript was much more abundant, when MO1 and MO2 were injected together. Sequencing of this band revealed that it indeed contained intronic sequences. This aberrant splice product could potentially interfere with chodl gene function.
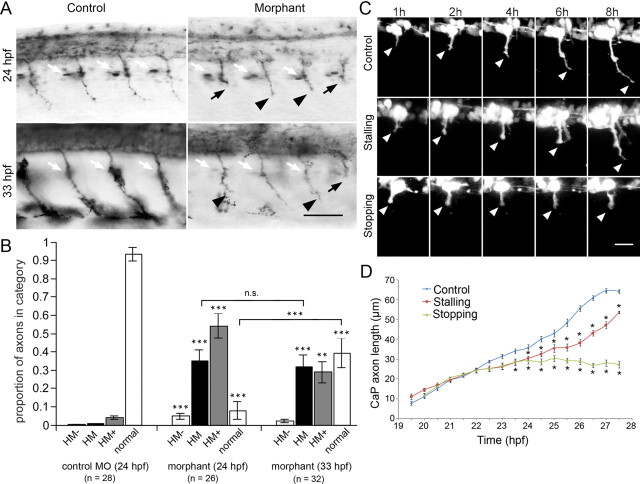
Figure 6.
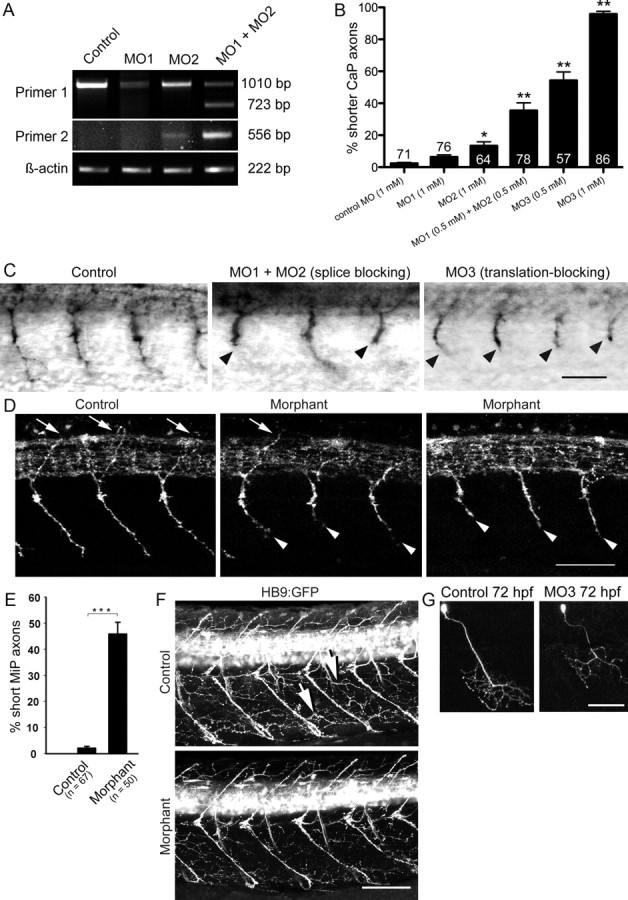
Knockdown of chodl leads to shorter motor axons and reduced innervation of myotomes at later developmental stages. A, A primer pair located in exons 1 (primer 1) and 7 yields a weaker wild-type band for chodl after application of two splice-site directed morpholinos in PCR. Coapplication of the morpholinos additionally leads to an ectopic band (MO1, 1 mm; MO2, 1 mm; MO1 plus MO2, 0.5 mm each). Using a primer pair in intron 2 (primer 2) and exon 7 reveals a transcript that includes intronic sequences after treatment with MO2. In embryos injected with both morpholinos, this abnormal transcript is amplified much more strongly. B, Quantification of axonal aberrations shows different magnitudes of effects on CaP axons that depend on morpholino combinations and concentrations (numbers of embryos are given on top of bars). C, Examples of morpholino actions on znp-1-labeled motor axons at 24 hpf are shown (orientation as in Fig. 1A). Shorter axons are indicated by arrowheads (control, 1 mm control morpholino; MO1 plus MO2, 0.5 mm each; MO3, 1 mm). D, Lateral trunk views at 33 hpf are shown; orientation as in Figure 1A. Many znp-1-labeled MiP axons do not reach the level of the dorsal border of the spinal cord in morphants. MiPs of normal length are pointed out by arrows. Shorter CaP axons in morphants are indicated by arrowheads (control, 1 mm control morpholino; morphant, 1 mm MO3). E, The number of shorter MiP axons is significant in morphants (control, 1 mm control morpholino; morphant, 1 mm MO3). F, Lateral trunk views at 72 hpf are shown; orientation is as in Figure 1A. Motor axons exhibit reduced myotomal branching in morphants. Two examples of myotome branches are depicted by arrows in the control (control, 1 mm control morpholino; morphant, 1 mm MO3). G, Single CaP axon labeling reveals a shorter ventral axon at 72 hpf. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars: C, 50 μm; D, 50 μm; F, 80 μm; G, 50 μm. Error bars indicate SEM.
The differences in the degrees of alterations of the wild-type transcript by these different treatments were reflected in the penetrance of the phenotype. MO1 (1 mm) alone had no significant effect on motor axon growth at 24 hpf, and MO2 (1 mm) led to only a small (13%) but significant proportion of motor axons that were shorter than in control morpholino-injected embryos (2.4%). However, the combination of MO1 and MO2 at one-half the concentration each (0.5 mm) led to a synergistic and significant effect of 34% shorter motor axons, supporting that knockdown of chodl impaired motor axon growth.
Since even the combination of MO1 and MO2 did not fully reduce the transcription of wild-type mRNA, we designed a translation-blocking morpholino, MO3. This morpholino showed a significant and dose-dependent shortening of axons with a very high penetrance of up to 95% of motor axons at a 1 mm concentration (Fig. 6B,C). This morpholino was used in all following experiments.
To determine whether morpholino treatment could nonspecifically retard general development, we analyzed the segmental position of the primordium of the lateral line nerve, because this nerve grows continually from rostral to caudal along the trunk and is, therefore, also used to stage embryonic development (Kimmel et al., 1995). Trunk segment 4.8 ± 0.18 was reached in control morpholino-treated embryos and trunk segment 4.4 ± 0.15 in chodl morphants at 24 hpf (n > 40 for each group; p > 0.05). Thus, the position of the lateral line primordium in morphants did not show a statistically significant difference from controls. Moreover, the shape and size of trunk segments was not altered compared with control morpholino-injected embryos (data not shown). In our hands (Feldner et al., 2005, 2007; Schweitzer et al., 2005), a number of specific and control morpholinos have never produced the present phenotype, which consists of unbranched shorter motor axons. Thus, a number of observations strongly suggest that morpholino effects were specific (Eisen and Smith, 2008): (1) different morpholinos elicited the same very specific phenotype; (2) they acted synergistically; (3) general morphology and developmental progress were undisturbed; (4) importantly, morpholino effects were rescued by mRNA overexpression (see below).
To elucidate whether MiP axons were also affected by chodl knockdown, we determined the trajectories of MiP axons in whole-mounted znp-1-labeled embryos at 33 hpf. Almost all (98%) dorsally growing wild-type axons reached or exceeded the dorsal edge of the medially located spinal cord, whereas significantly fewer axons (54%) reached the dorsal edge of the spinal cord in morphants (Fig. 6D,E). As a control, we measured the thickness of the spinal cord, which could have skewed our quantification. Morpholino treatment did not affect the thickness of the spinal cord at 33 hpf (uninjected control, 49.3 ± 1.15 μm, n = 6; morphant, 49.8 ± 1.66 μm, n = 6; p = 0.8). Thus, MiP axon growth was impaired. This suggests that chodl is important for CaP as well as MiP axon growth.
To assess later effects of chodl knockdown, we analyzed branching of motor nerves onto the myotomes in HB9:GFP transgenic animals at 48 hpf (data not shown) and 72 hpf (Fig. 5F), when motor nerves consist of many axons from primary and secondary motor neurons. Even though the main motor nerves formed, the density of motor branches on trunk muscles was reduced compared with control morpholino-injected animals at both time points.
To determine whether this phenotype was a consequence of reduced axon growth or missing motor neurons, we followed individual CaP axons over time by coinjection of morpholino and an HB9:GFP plasmid, which randomly labels few individual motor neurons. In controls injected with the plasmid and control morpholino (n = 4 embryos), all 10 observed CaP axons had grown beyond the horizontal myoseptum region at 33 hpf, reached the ventral edge of the trunk and started to branch at 48 hpf, and were still present at 72 hpf (Fig. 6G). In experimental animals (n = 6), we observed eight axons at different time points. Only one of eight CaP axons displayed the growth patterns observed in controls. Some axons were still at the level of the horizontal myoseptum at 33 hpf (three of seven axons) and only two of eight axons had reached the ventral edge of the trunk at 48 hpf. At 72 hpf, axons were still short (two of seven axons; Fig. 6G) or showed signs of degeneration, such as axon retraction, or loss of cells (four of seven axons). Thus hypo-innervation of myotomes in chodl morphants at 72 hpf was due to long-term impairment of axon growth and/or eventual degeneration.
CaP motor axons are specifically affected at the horizontal myoseptum in chodl morphants
Motor axons pause at the horizontal myoseptum during their growth and in the case of the CaP axon later continue to grow into the ventral myotome (Eisen et al., 1986). To determine which of the different phases of axon growth were affected by the knockdown resulting in shorter axons, we categorized axon lengths. Already at 24 hpf almost all axons had grown ventrally beyond the horizontal myoseptum. At 24 hpf (4.3%) and 33 hpf (1.7%), few axons had not reached the horizontal myoseptum in morphants. Strikingly, 34.6% of the axons had extended only to the position of the horizontal myoseptum, significantly more than in control morpholino-injected embryos (0.2%) at 24 hpf. At 33 hpf, 31.3% of the segments still showed axons at the level of the horizontal myoseptum. This was not significantly different from the proportion at 24 hpf, suggesting that almost one-third of the CaP axons were completely arrested at the horizontal myoseptum over the observation period. At 24 hpf, 54% of the axons had grown beyond the horizontal myoseptum but were not of wild-type length and only 8% appeared to be of normal length. At 33 hpf, 39% of the axons—almost five times more than at 24 hpf—were of normal length, suggesting that axons grew relatively quickly once they passed the horizontal myoseptum. Combined, this distribution of axon lengths indicates that axons reach the horizontal myoseptum similar to controls and then spend a disproportionally long time at the horizontal myoseptum to either continue growth or stop at the horizontal myoseptum (Fig. 7A,B).
Figure 7.
Axons stall preferentially at the horizontal myoseptum in chodl morphants. (control, 1 mm control morpholino; morphant, 1 mm MO3, for all panels). A, Lateral trunk views at 24 and 33 hpf are shown (orientation as in Fig. 1A). The position of the horizontal myoseptum is revealed by engrailed immunolabeled muscle pioneer cells and axons are labeled by znp-1. The position of the horizontal myoseptum is indicated by the white arrows. At 33 hpf, muscle pioneer cells are not always visible because they are located in a different focal plane. Axons that are stalled at the horizontal myoseptum are indicated by black arrows. Axons that have grown beyond it, but are still shorter than in controls, are indicated by arrowheads. B, Categorizing axon lengths (HM−, not reaching the horizontal myoseptum in histological preparations; HM, at the horizontal myoseptum; HM+, grown beyond the horizontal myoseptum, but still shorter than wild-type axon; normal, wild-type length) reveals a high proportion of axons that stall at the horizontal myoseptum in morphants at 24 and 33 hpf. Significances are tested against control morpholino, except for bracketed comparisons. C, Time-lapse analysis shows that chodl morphant axons stall for a disproportionally long time at the horizontal myoseptum. Two behaviors are observed, either axons stall longer than controls at the horizontal myoseptum (stalling), or they do not grow beyond the horizontal myoseptum during the observation period (stopping). D, Quantification of CaP axon lengths indicates that morphant axons grow toward the horizontal myoseptum at a velocity that is similar to that of control axons, but stall longer in the horizontal myoseptum. Those axons that manage to grow beyond the choice point grow again at similar velocity as control axons. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars: A, 50 μm; C, 20 μm. Error bars indicate SEM.
To directly test this hypothesis, we visualized growth of ventral motor axons by time-lapse analysis in morpholino-injected HB9:GFP transgenic embryos (Fig. 7C,D). Results revealed that, up to 21 hpf, morphant and wild-type axons grew at comparable velocities (control: 7.1 ± 0.7 μm/h, n = 4 axons; morphant: 8.3 ± 1.2 μm/h, n = 8 axons; p = 0.54), similar to previously reported growth speeds (7.1 ± 1.3 μm/h) (Myers et al., 1986). After that time, wild-type and morphant axons slowed down at the horizontal myoseptum (growth velocity wild-type: 5.4 ± 1.3 μm/h, n = 4; morphant: 4.1 ± 0.8 μm/h, n = 6; p = 0.359). Around 24 hpf, wild-type axons continued to grow into the ventral myotome at a velocity of 9.6 ± 1.2 μm/h (n = 4), whereas morphant axons displayed two types of behaviors. Either axons stalled at the horizontal myoseptum until 27 hpf, the end of the observation period, or they resumed growth and entered the ventral myotome a short time (∼1.5 h) after the wild-type axons. When morphant axons entered the ventral myotome, their growth speed (10.5 ± 2.6 μm/h; p = 0.7) was again comparable with that of wild-type axons. Both types of morphant axon behaviors resulted in significantly shorter axons when compared with controls after the horizontal myoseptum was reached (Fig. 7D). Thus, histology and in vivo time-lapse analysis showed normal growth velocity of morphant axons before and after the horizontal myoseptum was reached. However, the normal stalling period at the horizontal myoseptum was either significantly prolonged or axons were completely arrested at the horizontal myoseptum.
Overexpression of chodl leads to shorter motor axons and rescues knockdown effects
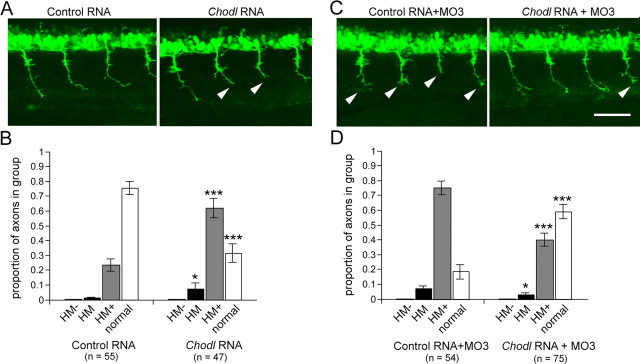
Next, we determined whether increasing the levels of chodl expression would perturb motor axon growth. After injection of mRNA transcribed in vitro from a bicistronic vector coding for chodl and GFP, we observed ubiquitous GFP expression in the embryo. In control RNA-injected embryos, 75% of the axons were of wild-type length, whereas significantly fewer axons (31%) were of wild-type length in mRNA-injected embryos (Fig. 8A,B). This result indicates that overexpression of chodl impairs growth in a proportion of axons and suggests that specific levels of chodl expression are important for optimal axon elongation.
Figure 8.
chodl overexpression affects axon growth and rescues morphants. Lateral trunk views of HB9:GFP transgenic embryos are shown (orientation as in Fig. 1A; short axons are indicated by arrowheads; categories in B and D are the same as in Fig. 7). A, B, Injection of chodl mRNA alone induces shorter CaP axons. C, D, Shorter axons induced by chodl MO3 (0.5 mm; C, arrowheads) are rescued by chodl RNA. *p < 0.05; ***p < 0.001. Scale bar: C, 50 μm. Error bars indicate SEM.
If specific levels of chodl expression were necessary for correct interactions of motor axons with the horizontal myoseptum, mRNA overexpression should rescue effects of gene knockdown. Indeed, when the highly efficient start codon-directed morpholino MO3 was injected at a concentration of 0.5 μm, together with control RNA, only 16.5% of the axons were of wild-type length. In contrast, coinjection with chodl mRNA led to approximately three times more axons of wild-type length (57%), which was statistically highly significant (Fig. 8C,D). It is unlikely that the morpholino inadvertently interacted directly with the injected RNA, since a sequence of only 5 bases of the morpholino matched the sequence of the injected RNA and morpholinos become ineffective when as few as 5 of their 20–21 bases are not matched to the target RNA (Eisen and Smith, 2008). Combined, these results indicate that levels of chodl expression are critical for correct interactions of motor axons with the horizontal myoseptum.
Discussion
Here, we used a novel expression profiling paradigm to find factors expressed by motor neurons necessary for correct axon growth. We find the C-type lectin chodl as crucial for interactions of motor axons with the horizontal myoseptum, an important navigational choice point. Our study is the first to demonstrate a developmental function of chodl, which has been implicated in SMA (Zhang et al., 2008; Bäumer et al., 2009).
chodl has a specific role in intermediate target recognition
chodl knockdown leads to a very specific phenotype in motor neurons. Motor axons exit the spinal cord and approach the horizontal myoseptum correctly and at normal velocity, as determined in histological preparations and by time-lapse video analysis. Axon growth was even unimpeded for those growth cones that were able to pass the horizontal myoseptum. It was only in the position of this choice point that axons stalled for a disproportionally long time or stopped altogether, supporting that chodl is necessary for axons to correctly interpret guidance cues in the horizontal myoseptum. The fact that overexpression of chodl induced a similar phenotype as knockdown supports that the level of chodl expression needs to be optimal for efficient interactions with the horizontal myoseptum. Similarly, loss of function and gain of function for the ECM component collagen XIXa1 (colXIX) (also known as stumpy) at the horizontal myoseptum both elicit the same phenotype—shorter motor axons (Hilario et al., 2010). Although motor nerves eventually form in chodl morphants, the observation that muscle innervation is still sparse at later time points of development indicates that chodl might be important not only at the horizontal myoseptum choice point, but at choice points or intermediate targets that exist at more distal positions along motor axon pathways (Beattie, 2000). The observation in single CaP axon tracing that some axons that managed to navigate the horizontal myoseptum did not show normal extension at 48 and 72 h support this interpretation. To our knowledge, chodl is the first C-type lectin to be implicated in axon pathfinding in vertebrates. In C. elegans another C-type lectin, CLEC-38, has been shown to be involved in dorsal axon guidance (Kulkarni et al., 2008).
What are the potential ligands of Chodl?
The chodl structure carries the hallmarks of an ECM receptor, with a signal peptide, a transmembrane domain, and a C-type lectin domain, shown to interact with ECM molecules (Lundell et al., 2004). Interestingly, the phenotypes of chodl morphants and mutants for the ECM molecule colXIX at the horizontal myoseptum are quite similar (Beattie et al., 2000; Hilario et al., 2010). Both conditions show normal growth of axons along the common pathway, stalling and stopping of axons at the horizontal myoseptum, impaired growth of the CaP and MiP axon, and reduced innervation of the musculature at later developmental stages. However, there need not be direct physical interactions between Chodl and ColXIX, as the ECM at the horizontal myoseptum is complex and ColXIX could simply anchor other potential interaction partners to the horizontal myoseptum. For example, Tenascin-C (Schweitzer et al., 2005) and chondroitin sulfates (Bernhardt and Schachner, 2000) are accumulated at the horizontal myoseptum and play a role in motor axon growth. In the mutant for the muscle-specific tyrosine kinase unplugged, in which deposition of Tenascin-C and chondroitin sulfates is defective, motor axons make inappropriate pathway decisions at the horizontal myoseptum (Zhang et al., 2004; Schweitzer et al., 2005). Interestingly, C-type lectin domains, such as that in Chodl, have been shown to interact with Tenascin-C (Lundell et al., 2004). Moreover, enzymatic modification of glycosylation of the myotome-derived type XVIII collagen is also important for motor axon extension (Schneider and Granato, 2006). Finally, the topped mutation represents an unknown muscle-derived activity (Rodino-Klapac and Beattie, 2004), which also leads to stalling of CaP axons at the horizontal myoseptum. While it is obvious that motor axons encounter a complex ECM at the horizontal myoseptum, the identity of specific interaction partners of chodl are presently unclear.
Possible connection of chodl to SMA
We show here that chodl is evolutionarily highly conserved, with an amino acid identity of >92% across vertebrate classes. chodl is specifically expressed in fast motor neurons in mice (Enjin et al., 2010). SMA is caused by reduced levels of the “survival of motor neurons” (SMN) genes, which are involved in RNA splicing among other functions (for recent review, see Sleigh et al., 2011). Using exon profiling in mouse models of SMA, specific isoform changes for chodl have been found already at early symptomatic stages, whereas most genes were dysregulated only at late symptomatic stages (Zhang et al., 2008; Bäumer et al., 2009). Reduced expression of SMN leads to shorter mouse motor axons in vitro (Rossoll et al., 2003), and one of the phenotypes in developing zebrafish (McWhorter et al., 2003) and mice (Liu et al., 2010) is also shorter axons. Thus, part of the phenotypes could be mediated through altered chodl expression, which when missing also leads to arrested or delayed motor axon growth, as we show here. Almost one-third of the CaP motor axons appear to be completely arrested in the horizontal myoseptum in chodl morphants. Such a defect could eventually lead to degeneration of these motor neurons, possibly due to a lack of trophic support from the muscle targets (Gould and Oppenheim, 2011). Indeed, retracted axons and missing CaP neurons observed at later developmental stages suggest that some motor neurons that are depleted in chodl expression eventually degenerate. Notably, in the colXIX mutant, showing a similar phenotype as our chodl morphants, CaP axons that are permanently stalled at the horizontal myoseptum also show signs of degeneration at 72 hpf (Beattie et al., 2000). However, studies in other mouse SMA models do not find abnormal development of motor axons, but degeneration of axons once axonal connections are established (McGovern et al., 2008; Murray et al., 2010). Hence, chodl could have roles in axon maintenance as well as pathfinding.
Expression profiling of HB9:GFP+ cells from DN-CLIM-injected embryos reveals genes potentially involved in motor axon differentiation
Our gene array expression profiling has revealed three genes that are specifically expressed in motor neurons, and we demonstrate a role in axon growth for one of these. This supports that this screening approach identifies axonal factors that may be involved in motor axon growth.
In our approach, we reasoned that HB9:GFP+ neurons may retain expression of genes needed to maintain motor neuron identity to a certain degree and general axon growth after DN-CLIM treatment. Thus, genes involved in motor axon navigation should be enriched in the list of downregulated genes after DN-CLIM treatment. The fact that many HB9:GFP+ cells retained HB9 and Islet-1/-2 immunoreactivity and only few acquire interneuron marker expression supported this notion. However, we confirm here that some motor neurons acquire the interneuron marker GABA when LIM-HD signaling is blocked (Segawa et al., 2001), and we show that many motor neurons lose expression of the mature motor neuron marker ChAT and fewer cells expressed GFP in DN-CLIM-injected embryos. Given that combinatorial expression of LIM-HD factors controls motor neuron fate in vertebrates (Tsuchida et al., 1994; Pfaff et al., 1996), including zebrafish (Inoue et al., 1994; Appel et al., 1995; Tokumoto et al., 1995; Hutchinson and Eisen, 2006; Hutchinson et al., 2007), it is likely that HB9:GFP+ neurons lose some aspects of their motor neuron identity under DN-CLIM treatment.
Nevertheless, using our approach, we identified chodl, tac1, and calca with motor neuron-specific expression, which are downstream of the LIM-HD regulatory network. The fact that chodl is expressed very early during motor neuron differentiation supports relatively proximal regulation of the gene by the LIM-HD network. Although the promoter regions of all three genes contain putative recognition motifs for LIM-HDs (data not shown), it is presently unclear whether direct regulation occurs. Future research will have to show whether tac1, coding for the neuropeptides substance P/neurokinin A (Nawa et al., 1984), and calca, coding for calcitonin and the calcitonin-related neuropeptide (Amara et al., 1982), have specific roles in motor axon navigation or synapse formation.
We conclude that our newly designed expression profiling approach to motor axon pathfinding has revealed the LIM-HD regulated C-type lectin chodl as pivotal in choice point interactions for growing motor axons. Discovering the genes involved in axonal differentiation and analyzing their function in zebrafish may ultimately lead to a better understanding of motor neuron diseases such as SMA.
Footnotes
This work was supported by an Overseas Research Students Award and a PhD studentship from the College of Medicine and Veterinary Medicine at the University of Edinburgh (Z.Z.) and a PhD studentship from MND Scotland (J.O.). The microarray study was funded by the Moray Endowment Fund. We thank Dr. Christine Beattie for critically reading this manuscript, Xibei Jia for help with the gene chip analysis, Lee Tan for the pCDNA3-IRES-GFP vector, Drs. Dirk Meyer and Shin-ichi Higashijima for transgenic fish and reagents, Dr. Angela Scott for help with statistics, Dr. Simon Monard (Institute for Stem Cell Research, King's Buildings, University of Edinburgh, Edinburgh, UK) for help with FACS, Dr. Jan Soetaert for help with time-lapse confocal microscopy, and Maria Rubio for expert fish care.
References
- Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–4125. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- Bae YK, Shimizu T, Muraoka O, Yabe T, Hirata T, Nojima H, Hirano T, Hibi M. Expression of sax1/nkx1.2 and sax2/nkx1.1 in zebrafish. Gene Expr Patterns. 2004;4:481–486. doi: 10.1016/j.modgep.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Batista MF, Lewis KE. Pax2/8 act redundantly to specify glycinergic and GABAergic fates of multiple spinal interneurons. Dev Biol. 2008;323:88–97. doi: 10.1016/j.ydbio.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie CE. Control of motor axon guidance in the zebrafish embryo. Brain Res Bull. 2000;53:489–500. doi: 10.1016/s0361-9230(00)00382-8. [DOI] [PubMed] [Google Scholar]
- Beattie CE, Melancon E, Eisen JS. Mutations in the stumpy gene reveal intermediate targets for zebrafish motor axons. Development. 2000;127:2653–2662. doi: 10.1242/dev.127.12.2653. [DOI] [PubMed] [Google Scholar]
- Becker T, Ostendorff HP, Bossenz M, Schlüter A, Becker CG, Peirano RI, Bach I. Multiple functions of LIM domain-binding CLIM/NLI/Ldb cofactors during zebrafish development. Mech Dev. 2002;117:75–85. doi: 10.1016/s0925-4773(02)00178-8. [DOI] [PubMed] [Google Scholar]
- Bernhardt RR, Schachner M. Chondroitin sulfates affect the formation of the segmental motor nerves in zebrafish embryos. Dev Biol. 2000;221:206–219. doi: 10.1006/dbio.2000.9673. [DOI] [PubMed] [Google Scholar]
- Bhati M, Lee C, Nancarrow AL, Lee M, Craig VJ, Bach I, Guss JM, Mackay JP, Matthews JM. Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J. 2008;27:2018–2029. doi: 10.1038/emboj.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanomi D, Pfaff SL. Motor axon pathfinding. Cold Spring Harb Perspect Biol. 2010;2:a001735. doi: 10.1101/cshperspect.a001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Myers PZ, Westerfield M. Pathway selection by growth cones of identified motoneurones in live zebra fish embryos. Nature. 1986;320:269–271. doi: 10.1038/320269a0. [DOI] [PubMed] [Google Scholar]
- Enjin A, Rabe N, Nakanishi ST, Vallstedt A, Gezelius H, Memic F, Lind M, Hjalt T, Tourtellotte WG, Bruder C, Eichele G, Whelan PJ, Kullander K. Identification of novel spinal cholinergic genetic subtypes disclose Chodl and Pitx2 as markers for fast motor neurons and partition cells. J Comp Neurol. 2010;518:2284–2304. doi: 10.1002/cne.22332. [DOI] [PubMed] [Google Scholar]
- Feldner J, Becker T, Goishi K, Schweitzer J, Lee P, Schachner M, Klagsbrun M, Becker CG. Neuropilin-1a is involved in trunk motor axon outgrowth in embryonic zebrafish. Dev Dyn. 2005;234:535–549. doi: 10.1002/dvdy.20520. [DOI] [PubMed] [Google Scholar]
- Feldner J, Reimer MM, Schweitzer J, Wendik B, Meyer D, Becker T, Becker CG. PlexinA3 restricts spinal exit points and branching of trunk motor nerves in embryonic zebrafish. J Neurosci. 2007;27:4978–4983. doi: 10.1523/JNEUROSCI.1132-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan-Steet H, Fox MA, Meyer D, Sanes JR. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- Gould TW, Oppenheim RW. Motor neuron trophic factors: therapeutic use in ALS? Brain Res Rev. 2011;67:1–39. doi: 10.1016/j.brainresrev.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güngör C, Taniguchi-Ishigaki N, Ma H, Drung A, Tursun B, Ostendorff HP, Bossenz M, Becker CG, Becker T, Bach I. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc Natl Acad Sci U S A. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie S. Patterning and axon guidance of cranial motor neurons. Nat Rev Neurosci. 2007;8:859–871. doi: 10.1038/nrn2254. [DOI] [PubMed] [Google Scholar]
- Hilario JD, Wang C, Beattie CE. Collagen XIXa1 is crucial for motor axon navigation at intermediate targets. Development. 2010;137:4261–4269. doi: 10.1242/dev.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Mieda M, Harada T, Yamasu K, Okamoto H. Identification of ephrin-A3 and novel genes specific to the midbrain-MHB in embryonic zebrafish by ordered differential display. Mech Dev. 2001;107:83–96. doi: 10.1016/s0925-4773(01)00467-1. [DOI] [PubMed] [Google Scholar]
- Hutchinson SA, Eisen JS. Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development. 2006;133:2137–2147. doi: 10.1242/dev.02355. [DOI] [PubMed] [Google Scholar]
- Hutchinson SA, Cheesman SE, Hale LA, Boone JQ, Eisen JS. Nkx6 proteins specify one zebrafish primary motoneuron subtype by regulating late islet1 expression. Development. 2007;134:1671–1677. doi: 10.1242/dev.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38:581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Satou C, Higashijima S. V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord. Development. 2008;135:3001–3005. doi: 10.1242/dev.024802. [DOI] [PubMed] [Google Scholar]
- Kulkarni G, Li H, Wadsworth WG. CLEC-38, a transmembrane protein with C-type lectin-like domains, negatively regulates UNC-40-mediated axon outgrowth and promotes presynaptic development in Caenorhabditis elegans. J Neurosci. 2008;28:4541–4550. doi: 10.1523/JNEUROSCI.5542-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser LT. The acquisition of motoneuron subtype identity and motor circuit formation. Int J Dev Neurosci. 2001;19:175–182. doi: 10.1016/s0736-5748(00)00090-3. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. From cells to circuits: development of the zebrafish spinal cord. Prog Neurobiol. 2003;69:419–449. doi: 10.1016/s0301-0082(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Liu H, Shafey D, Moores JN, Kothary R. Neurodevelopmental consequences of Smn depletion in a mouse model of spinal muscular atrophy. J Neurosci Res. 2010;88:111–122. doi: 10.1002/jnr.22189. [DOI] [PubMed] [Google Scholar]
- Lundell A, Olin AI, Mörgelin M, al-Karadaghi S, Aspberg A, Logan DT. Structural basis for interactions between tenascins and lectican C-type lectin domains: evidence for a crosslinking role for tenascins. Structure. 2004;12:1495–1506. doi: 10.1016/j.str.2004.05.021. [DOI] [PubMed] [Google Scholar]
- McGovern VL, Gavrilina TO, Beattie CE, Burghes AH. Embryonic motor axon development in the severe SMA mouse. Hum Mol Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Yeo SY, Hirate Y, Segawa H, Wada H, Little MH, Yamada T, Takahashi N, Okamoto H. PlexinA4 is necessary as a downstream target of Islet2 to mediate Slit signaling for promotion of sensory axon branching. Development. 2004;131:3705–3715. doi: 10.1242/dev.01228. [DOI] [PubMed] [Google Scholar]
- Murray LM, Lee S, Bäumer D, Parson SH, Talbot K, Gillingwater TH. Pre-symptomatic development of lower motor neuron connectivity in a mouse model of severe spinal muscular atrophy. Hum Mol Genet. 2010;19:420–433. doi: 10.1093/hmg/ddp506. [DOI] [PubMed] [Google Scholar]
- Myers PZ, Eisen JS, Westerfield M. Development and axonal outgrowth of identified motoneurons in the zebrafish. J Neurosci. 1986;6:2278–2289. doi: 10.1523/JNEUROSCI.06-08-02278.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawa H, Kotani H, Nakanishi S. Tissue-specific generation of two preprotachykinin mRNAs from one gene by alternative RNA splicing. Nature. 1984;312:729–734. doi: 10.1038/312729a0. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Reimer MM, Sörensen I, Kuscha V, Frank RE, Liu C, Becker CG, Becker T. Motor neuron regeneration in adult zebrafish. J Neurosci. 2008;28:8510–8516. doi: 10.1523/JNEUROSCI.1189-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino-Klapac LR, Beattie CE. Zebrafish topped is required for ventral motor axon guidance. Dev Biol. 2004;273:308–320. doi: 10.1016/j.ydbio.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kröning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Maeda M, Obinata M, Shoji W. Position fine-tuning of caudal primary motoneurons in the zebrafish spinal cord. Development. 2008;135:323–332. doi: 10.1242/dev.007559. [DOI] [PubMed] [Google Scholar]
- Schmidt ER, Pasterkamp RJ, van den Berg LH. Axon guidance proteins: novel therapeutic targets for ALS? Prog Neurobiol. 2009;88:286–301. doi: 10.1016/j.pneurobio.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Schneider VA, Granato M. The myotomal diwanka (lh3) glycosyltransferase and type XVIII collagen are critical for motor growth cone migration. Neuron. 2006;50:683–695. doi: 10.1016/j.neuron.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Schweitzer J, Becker T, Lefebvre J, Granato M, Schachner M, Becker CG. Tenascin-C is involved in motor axon outgrowth in the trunk of developing zebrafish. Dev Dyn. 2005;234:550–566. doi: 10.1002/dvdy.20525. [DOI] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–436. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Sleigh JN, Gillingwater TH, Talbot K. The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy. Dis Model Mech. 2011;4:457–467. doi: 10.1242/dmm.007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto M, Gong Z, Tsubokawa T, Hew CL, Uyemura K, Hotta Y, Okamoto H. Molecular heterogeneity among primary motoneurons and within myotomes revealed by the differential mRNA expression of novel islet-1 homologs in embryonic zebrafish. Dev Biol. 1995;171:578–589. doi: 10.1006/dbio.1995.1306. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- Westerfield M. Ed 4. Eugene, OR: University of Oregon; 2000. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio) [Google Scholar]
- Westerfield M, Eisen JS. Neuromuscular specificity: pathfinding by identified motor growth cones in a vertebrate embryo. Trends Neurosci. 1988;11:18–22. doi: 10.1016/0166-2236(88)90044-6. [DOI] [PubMed] [Google Scholar]
- Yeo SY, Miyashita T, Fricke C, Little MH, Yamada T, Kuwada JY, Huh TL, Chien CB, Okamoto H. Involvement of Islet-2 in the Slit signaling for axonal branching and defasciculation of the sensory neurons in embryonic zebrafish. Mech Dev. 2004;121:315–324. doi: 10.1016/j.mod.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lefebvre JL, Zhao S, Granato M. Zebrafish unplugged reveals a role for muscle-specific kinase homologs in axonal pathway choice. Nat Neurosci. 2004;7:1303–1309. doi: 10.1038/nn1350. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Ma H, Taniguchi-Ishigaki N, Nagarajan L, Becker CG, Bach I, Becker T. Regulation of LIM-HDs complexes by SSDP1 during early zebrafish nervous system development. Dev Biol. 2011;349:213–224. doi: 10.1016/j.ydbio.2010.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]