Abstract
Millions of diverse molecules constituting the lipidome act as important signals within cells. Of these, cardiolipin (CL) and phosphatidylethanolamine (PE) participate in apoptosis and ferroptosis, respectively. Their subcellular distribution is largely unknown. Imaging mass spectrometry is capable of deciphering the spatial distribution of multiple lipids at subcellular levels. Here we report the development of a unique 70 keV gas-cluster ion beam that consists of (CO2)n+(n > 10000) projectiles. Coupled with direct current beam buncher-time-of-flight secondary-ion mass spectrometry, it is optimized for sensitivity towards high-mass species (up to m/z 3000) at high spatial resolution (1 mm). In combination with immunohistochemistry, phospholipids, including PE and CL, have been assessed in subcellular compartments of mouse hippocampal neuronal cells and rat brain tissue.
Keywords: cardiolipin, ferroptosis, gas-clusterion beam secondary-ion mass spectrometry, subcellular imaging
Lipids are abundant small molecules of cells and tissues in all three domains of life: Bacteria, Archaea, and Eukaryotes. Their primary role, as building blocks of the lipid bilayer in biological membranes with embedded protein components, has been appreciated for decades. Contemporary mass spectrometry (MS) has demonstrated that the lipidome includes millions of species of lipids. This extraordinary diversity has led to new concepts on lipid signaling in health and disease.
Selective and specific oxygenation of two classes of lipids is associated with two programs of regulated cell death—apoptosis and ferroptosis.[1] Cardiolipin (CL), a mitochondrial-specific phospholipid, is indispensable for the integrity of the inner mitochondrial membrane (IMM) and possesses vital roles in signaling.[2] CL oxidation is a required stage of intrinsic apoptosis.[3] Hydroperoxy-arachidonoyl-phosphatidyl-ethanolamines (HOO-AA-PE) generated in the endoplasmic reticulum (ER) are strong predictive biomarkers of ferroptosis.[4] However, specific cell types that contribute to CL and phosphatidylethanolamine (PE) oxidation in apoptotic/ferroptotic death remain unknown and information on their intracellular localization/distribution is lacking. We developed a novel matrix-assisted laser desorption/ionization (MALDI) protocol for mass spectrometry imaging (MSI) of CL molecular species[5] and demonstrated selective decreases in CL species that contain oxidizable polyunsaturated fatty acids (PUFA) in an animal model of traumatic brain injury (TBI).[5b] However, the cellular and subcellular distribution of individual CL and PE species in programed cell death remains undetermined.[6] Although recent advances for MALDI imaging have allowed spatial resolution to approach 1.4 mm under special circumstances,[7] sensitivity and matrix application issues associated with targeted lipids in our samples currently do not allow such resolutions to be routinely achieved.
Secondary-ion mass spectrometry (SIMS) offers unprecedented spatial resolution. However, this poses great challenges since the number of molecules available for detection decreases as the sampling size decreases. For example, only 5 × 106 lipid molecules are present in an area of 1 × 1 μm2 of the cellular membrane.[8] Given a typical ionization efficiency of 10−4 for biomolecules, approximately 5 × 102 molecules remain to be detected.[9] Moreover, in-source fragmentation has made the detection of intact biomolecules, including lipids, very difficult. Consequently, SIMS imaging is known for high spatial resolution but a low mass range (< m/z 1000), especially with the widely used liquid-metal ion-gun (LMIG) sources.[10]
The evolution of primary-ion beams has shaped SIMS development due to their vital role in determining spatial resolution and chemical sensitivity. Particularly, the introduction of gas-cluster ion beams (GCIB) has shifted the SIMS field from fragment detection to molecular profiling. The mass range been greatly expanded, and the intact molecularion yield shows a 100-fold increase compared with C60+ beams, especially after a certain dose of sputter (approximately 5 × 1013 ionscm−2).[11] However, technical difficulties have limited the focusing of GCIB for high-resolution imaging. One obstacle lies in the instrument design, which usually pulses the primary-ion beam on a nanosecond scale. This deteriorates the beam focus by swiping the beam across the aperture, and prolonged acquisition is required due to low signal. Several groups have started to explore the imaging capability of the GCIB. The Winograd and Kagan/Bayır labs have demonstrated that a 20 keV Ar 3500+ cluster beam yields large intact lipid species, such as CL and gangliosides up to approximately m/z 3000 on rat-brain tissue at a spatial resolution of approximately 9 μm.[12] Angerer et al. have shown that the beam focus can be improved to approximately 3 μm by increasing the beam energy to 40 keV[13] and have successfully imaged gangliosides, CL, and other intact lipids on various biological tissues/cells.[13,14] Matsuo et al. have used a GCIB to image grid samples at 4 mm resolution.[15] Passarelli et al. have imaged rat-brain tissue at approximately 2 mm resolution using 10 keV Ar3000+ High mass-resolving power has also been also demonstrated, but no ions above m/z 1000 were imaged.[16]
The Winograd lab has been developing high spatialresolution GCIBs to take full advantage of low chemical damage and larger molecule detection. Here, the high-voltage GCIB is introduced that operates at energies up to 70 keV with gas-cluster sizes up to approximtely 30000 molecules (Supporting Information, Figure S1). In conjunction with the J105 3D Chemical Imager SIMS system (Ionoptika, UK),[17] these advances allow a spatial resolution of 1.0–1.6 μm to be achieved for lipid imaging at the cellular and subcellular levels. Immunohistochemical (IHC) confocal fluorescence microscopy has also been exploited after SIMS measurement to assign specific lipid species to a particular cellular/subcellular location. Combined with depth profiling, this allows the exploration of lipid asymmetry in 3D. These advances dramatically improve the ability to investigate the distribution of lipid molecules and their functional activity in single cells.
Initially, the (CO2)n+ cluster-ion beam characteristics were optimized for ionization and resolution. Depth profiling on Irganox 1010 thin films was performed to investigate the effects of kinetic energy and beam energy per molecule (E/n) on ion yield, sputter rate, and fragmentation. Specifically, 20, 40, 60, and 70 keV (CO2)n+ beams were explored with varying E/n of 2.5, 5, 10, and 20 eV/n. The largest cluster used was (CO2)28000+ at 70 keV (2.5 eV/n).
The spectrum of Irganox 1010 using 70 keV (CO)2 28000 + is shown in Figure S2 in the Supporting Information, in which the deprotonated ion [M-H]− at m/z 1175.8 is dominant over the other fragment ions. The intensities of [M H]− at m/z 1175.8 with various beam settings were plotted against cumulative ion dose during depth profiling, as shown in Figure S3 in the Supporting Information. A rather constant [M-H]− signal, the so-called steady state, was observed during depth profiling until the film/Si interface was reached. In contrast, depth profilings utilizing atomic beams or C60+ show a huge initial signal drop before reaching the steady state due to the severe chemical damage.[11]
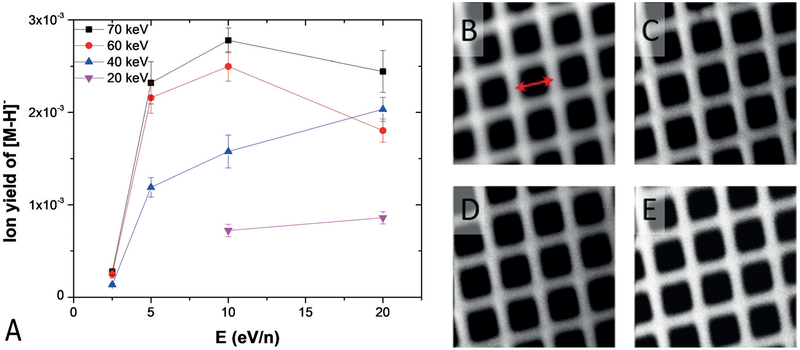
The ionization, sputter rate, and fragmentation with various beam conditions were extracted from the depth- profiling data (Figure 1A and Supporting Information, Figure S4). The ionization is enhanced as the beam kinetic energy increases (Figure 1A). For example, giving a certain E/n at 10 eV/n, the ionization is enhanced by 2.2 to 3.9 fold when the kinetic energy increases from 20 to 70 keV. At the same kinetic energy, there are two different trends: the ionization increases along with the E/n, such as for the 40 keV beams, or the ionization increases along with the E/n then drops when E/n is above 10 eV/n for beams above 60 keV. The trend is unclear for 20 keV beams, owing to a very low sputter rate of below 5 eV/n, resulting in unsuccessful depth profiling. The optimum ionization zones are clearly in the range of 5–10 eV/n for 60–70 keV beams.
Figure 1.
Irganox 1010 molecular-ion yields and beam focus in response to various kinetic energies and E/n of the gas-cluster ion beam (CO2)n+ (n = 1000–28000). A) The molecular-ion yields of [M-H]− m/z 1175.8 at the steady-state region of the depth-profiling curve in Figure S4 in the Supporting Information; error bars are the standard deviations. B–E) Secondary electron scanning electron microscopy images of 1000 mesh Au grid (pitch =25 μm, as indicated by the arrow in (B) using 70 keV (CO2)n (n = 2000, 4000, 8000, and 10000) with the best focus at 1.8, 1.3, 1.0, and 1.0 mm in B, C, D, and E respectively.
The contribution of sputter rate to the ionization enhancement was investigated (Supporting Information, Figure S4A), and it increases as the E/n and kinetic energy of the beams increase. For the 10 eV/n beams, the sputter rate is increased by 2.9 to 5.4 fold as the kinetic energy increases from 20 to 70 keV. This indicates that the enhanced ionization can be attributed to the increasing sputter rate at higher kinetic energies, which matches the observations using 10≈40 keV Ar +n clusters (n = 1000, 2000, 4000) by Angerer et al.[13]
Two different fragmentation trends were observed (Supporting Information, Figure S4B,C). First, at the same kinetic energy, greater E/n produces greater fragment/[M H]− ratios. Second, some fragment ions increase during depth profiling at a constant E/n and kinetic energy, such as for m/z 231.2 in Figure S4B and S4C in the Supporting Information. In contrast, the intensity of the m/z 957.6 is constant during depth profiling (Supporting Information, Figure S4B and S4D). This systematic study has shown that the higher kinetic energy beams yield more intense [M H]− rather than fragments. Therefore, it is advantageous to employ the beam above 60 keV for higher ionization.
A high-energy GCIB is beneficial for beam focusing. The 70 keV beams were further investigated by imaging a 1000 mesh grid. Figure 1B–E displays the images acquired with 70 keV (CO2)n+ beams at varying cluster sizes. The current of each beam was trimmed through a 20 μm aperture along the beam optics to 5–7 pA at the sample stage. The line scan across the metal bar of the grid indicates that a spot of 1 mm can be achieved with 70 keV (CO2)10000+ through fine adjustments of the beam parameters. It is speculated that cluster breakdown through the beam column is lower at higher kinetic energies. Consequently, the beam-energy spread is reduced. Considering that the optimum ionization zone of E/n is between 5–10 eV/n, 70 keV (CO2)10000+ was employed to study the biological samples.
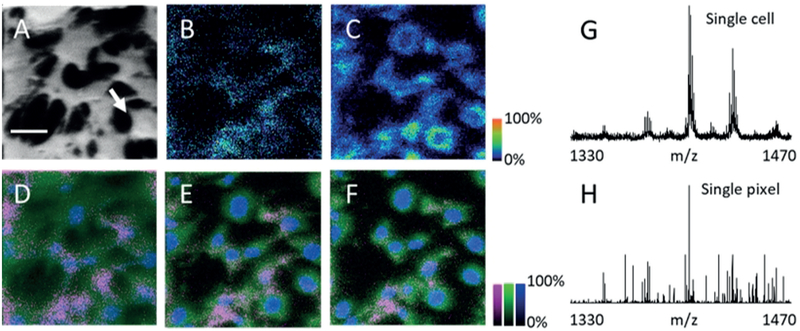
SIMS is promising for exploring single-cell phospholipid changes and depth profiling.[18] The novel GCIB was used to generate lipid images from HT22 cells (a neuronal cell line of hippocampal origin) at a 1 mm pixel size (Figure 2). According to the average cell height and sputter rate, it is estimated that each analysis cycle can detruncate the cell by approximately 200 nm. A total-ion SIMS image of the first layer (surface to subsurface, 0–200 nm) displays the typical HT22 cell-morphology (Figure 2A). Panels 2B and 2C display SIMS second- layer images (200–400 nm) of cellular CL (68:2, m/z 1404.0) and phosphatidylinositol (PI) (38:4, m/z 885.5), respectively. Three successive depth-profiling images display the spatial correlation of organelles, such as nuclei and mitochondria (Figure 2D–F). Overlays of CL (68:2, m/z 1404.0, magenta), PI (38:4, m/z 885.5, green), and a nuclear signal (blue, m/z 257.0 phosphate–deoxyribose–phosphate–H2O or deoxyribose–diphosphate–H2O)[19] show that the PI and CL signals exhibit a perinuclear localization. Previously, only optical techniques have been able to observe this type of mitochondrial distribution (see Figures S15–19 in the Supporting Information for additional species and LC-MS confirmation). Detection of CL at the cellular level is shown by the single- cell spectrum (see Figure 2G and arrow in Figure 2A) displaying three specific mass groupings of CL (66, 68, and 70 fatty-acyl carbons). Furthermore, the main CL signal (m/z 1404.0) could be detected from a single 1 mm pixel (Figure 2H).
Figure 2.
GCIB-SIMS imaging of CL in hippocampal neuronal HT22 cells at 1 μm pixel size. Cells were treated with EDC/PLC immediately prior to analysis to enhance CL signals. A) SIMS first-layer total-ion current image gives the outline of HT22 cells, a representative cell is indicated by an arrow (scale bar=50 μm). B) SIMS second-layer image of cellular CL (68:2, m/z 1404.0) and C) PI (38:4, m/z 885.5) to give subcellular structure. D–F) Depth profiling of HT22 cells. D) First layer, surface to subsurface, (0–200 nm). E) Second (200–400 nm) subsurface and F) third (400–600 nm) subsurface layers. Panels D–F: Green: PI (38:4, m/z 885.6), Blue: deoxyribosediphosphate (m/z 257.0), Magenta: CL (68:2, m/z 1404.0). G) CL region (m/z 1330–1470) SIMS first-layer spectrum of the single cell indicated in (A), and of a representative single pixel (H) in that cell.
Our previous work imaged rat brain using a 20 keV (CO2)3500+ beam to detect CL at resolutions of 9 and 16 mm.[12] CL accounts for 1 to 3 mol% of all phospholipids in brain tissue and PE accounts for 35 to 40 mol%.[20] This low abundance makes CL detection challenging. To maintain sufficient chemical sensitivity, MALDI imaging of CL is often acquired at a spatial resolution of 20–100 μm.[2b] In this study, these technological advances were used to image CL on rat- brain tissue at an unprecedented spatial resolution of 1.6 μm.
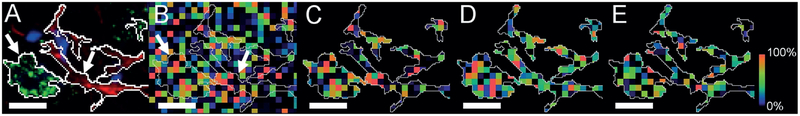
Cellular/subcellular lipid location can be rationalized by combining high-resolution lipid and IHC tissue images. Sections immediately adjacent to those used for lipid imaging were labeled for neuronal (NeuN) markers[21] to define the cells (Figure 3A). The thresholded NeuN signal (Figure 3B) and one of the GCIB-SIMS image signals (Figure 3C) were overlayed (Figure 3D) and cropped (Figure 3E) to allow us to analyze the GCIB-SIMS signal relative to the cellular intensity for NeuN for a given cell (Supporting Information, Figure S20). Importantly, this value is pixel-(GCIB-SIMS) and cell-(NeuN) dependent and can be used to generate quantitative ratios for any phospholipid species in any IHC- labeled intracellular compartment, using constant imaging conditions with a confocal microscope (Supporting Information, Figures S20–S22). Staining for different subcellular entities, including mitochondria, will allow the assessment of the corresponding lipid composition in various cell types.
Figure 3.
Superimposition of immunofluorescence confocal images with SIMS images of coronal rat brain. A) NeuN (green, neurons) and GFAP (red, astroglia) signals are marked with arrows and their thresholded overlays were used to define cell body boundaries (scale bar= 10 μm). B) The PE(36:4p) SIMS signal (m/z 722.5) from the same region is rendered as a pseudocolored heatmapped panel and is intensity-scaled relative to the field of view. The thresholded NeuN and GFAP signals have been overlaid on the SIMS image. C) PE(36:4p) signal is cropped to the NeuN and GFAP thresholds. D) PE(38:4) m/z 766.5, and, E) PE(40:6) m/z 790.5 signals are also cropped to the NeuN and GFAP thresholds.
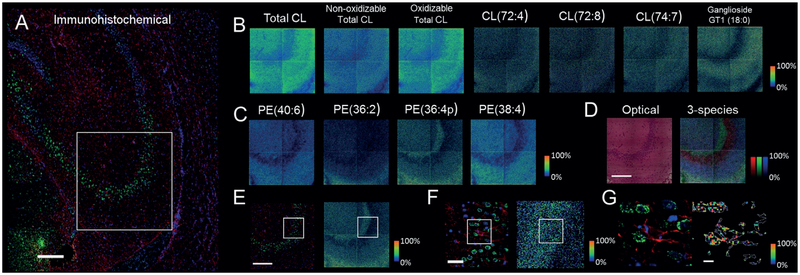
IHC of a rat-brain coronal section of the CA3 region is shown in Figure 4A. SIMS images (1.6 μm pixel size) of total CL, total oxidizable and non-oxidizable CL species (Supporting Information, Figure S7), along with three individual CL species from EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) and PLC (phospholipase C) treated tissue are shown in Figure 4B, with ganglioside GT1(18:0) shown for comparison. PE species (40:6, 36:2, 36:4p, and 38:4) from an untreated tissue section are shown in Figure 4C. Both PE and CL species can be associated with the specific cellular layers of the CA3 region. Figure 4D displays an optical image as well as a 3-color SIMS overlay. PE 36:4p (green) is seen in the outermost layer of the CA3 region, the alveus, followed by PE 40:6 (blue) in the oriens layer. PI 38:5 (red) maps strongly to the pyramidal layer. PE 36:4p (green) also maps to the next narrow layer, the stratum lucidum. Finally, PE(40:6) is seen again in the innermost layer, the stratum radiatum. Successive zoomins of SIMS of PE (36:4p) and IHC images are also shown (Figure 4E–G). Figure 4G (left panel) displays glial fibrillary acidic protein (GFAP)- positive and NeuN-positive cells in the IHC image, which were used for subsequent thresholding of the PE (36:4p) SIMS image (right panel). Any part of the SIMS image can be thresholded in this way (see Figures S5–14 in the Supporting Information for additional species and LC-MS confirmation).
Figure 4.
GCIB-SIMS shows the localization of different species of CA3-region lipids at 1.6 μm resolution. A) IHC of rat brain coronal section (Red =GFAP, Green= NeuN, Blue =DAPI; scale bar= 250 μm). The corresponding area imaged by SIMS is shown in the square. B) Corresponding SIMS images of CL from an EDC/PLC-treated section are displayed in rainbow intensity scale. Ganglioside GT1(18:0, m/z 2109.9) is shown for comparison. C) Corresponding SIMS images of PE from an untreated tissue section in rainbow scale. D) H&E staining of the untreated imaged section (left) with a 3-color SIMS overlay. (Red: PI(38:5, m/z 883), Blue: PE(40:6, m/z 766), Green: PE(36:4p, m/z 722), scale bar= 250 mm). E–G) Successive zooms of IHC (left) and SIMS (PE(36:4p, m/z 722), right) with regions marked. Scale bar (E)–(G)= 250, 50, and 10 mm, respectively. G) NeuN-positive (green, neurons) and GFAP-positive (red, astroglia) cells in the IHC image (left) used for subsequent thresholding followed by overlaying of the PE(36:4p) SIMS image and cropping, right (intensity scale relative to the field of view).
This brain tissue study displayed the chemical distribution of intact CL, PE, PI, phosphatidylserine (PS), ganglioside, and sterol (ST) lipids in the mass range of m/z 700 to 2500 at a resolution of 1.6 mm, providing vital information for the understanding of lipid function in the CA3 region. More importantly, since CL species can now be mapped at unprecedented resolutions, this facilitates future studies on changes in signal intensity and distribution of CL in brain tissue compromised by TBI.
In summary, by optimizing several GCIB parameters, in combination with chemical/enzymatic treatment methods, we are now able to image low-abundance lipids as intact molecular species at an unprecedented spatial resolution of 1.0–1.6 μm. Aided by IHC/confocal-imaging techniques, different PE species could be further imaged in different neuronal cells on brain tissue. The technical and methodological development in this study have better positioned us to further explore the structure, location, and distribution of critical lipids that function in apoptotic (CL) and ferroptotic (PE) death during injury and disease progression.
Supplementary Material
Acknowledgments
This work was supported by NIH grants 5R01GM113746–21, U54GM103529–08, NS061817, NS076511, P01HL114453, and U19AI068021. HT22 cells were a kind gift from David Schubert of the Salk Institute for Biological Studies.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
“This is the peer reviewed version of the following article: [Secondary ion mass spectrometry images cardiolipins and phosphatidylethanolamines at the subcellular level], which has been published in final form at DOI: 10.1002/anie.201814256 and 10.1002/. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions.”
Contributor Information
Hua Tian, Department of Chemistry, Pennsylvania State University, 209 Chemistry Bldg., University Park, PA 16802 (USA).
Louis J. Sparvero, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health, University of Pittsburgh (USA)
Paul Blenkinsopp, Ionoptika Ltd., Unit B6, Millbrook Cl, Chandler’s Ford, Eastleigh SO53 4BZ (UK).
Andrew A. Amoscato, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health, University of Pittsburgh (USA)
Simon C. Watkins, Department of Cell Biology, University of Pittsburgh (USA)
Hülya Bayır, Department of Chemistry, Pennsylvania State University, 209 Chemistry Bldg., University Park, PA 16802 (USA), Departments of Environmental and Occupational Health, Radiation Oncology, Critical Care Medicine, and Center for Free Radical and Antioxidant Health and Safar Center for Resuscitation Research, University of Pittsburgh (USA) And Children’s Neuroscience Institute, UPMC Children’s Hospital, University of Pittsburgh, 4200 Fifth Ave, Pittsburgh, PA 15260 (USA).
Valerian E. Kagan, Department of Chemistry, Pennsylvania State University, 209 Chemistry Bldg., University Park, PA 16802 (USA), Departments of Environmental and Occupational Health, Chemistry, Radiation Oncology, Center for Free Radical and Antioxidant Health, University of Pittsburgh (USA) and Laboratory of Navigational Redox Lipidomics, IM Sechenov Moscow Medical State University (Russia).
Nicholas Winograd, Department of Chemistry, Pennsylvania State University, 209 Chemistry Bldg., University Park, PA 16802 (USA).
References
- [1].a) Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD, Cell 2017, 171, 273–285; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tyurina YY, Shrivastava I, Tyurin VA, Mao G, Dar HH, Watkins S, Epperly M, Bahar I, Shvedova AA, Pitt B, Wenzel SE, Mallampalli RK, Sadovsky Y, Gabrilovich D, Greenberger JS, Bayir H, Kagan VE, Antioxid. Redox Signaling 2017, 29, 1333–1358; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Neitemeier S, Jelinek A, Laino V, Hoffmann L, Eisenbach I, Eying R, Ganjam GK, Dolga AM, Oppermann S, Culmsee C, Redox Biol 2017, 12, 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Tyurina YY, Tyurin VA, Kaynar AM, Kapralova VI, Wasserloos K, Li J, Mosher M, Wright L, Wipf P, Watkins S, Pitt BR, Kagan VE, Am. J. Physiol.: Lung Cell. Mol. Physiol 2010, 299, L73–L85; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Maguire JJ, Tyurina YY, Moham-madyani D, Kapralov AA, Anthonymuthu TS, Qu F, Amoscato AA, Sparvero LJ, Tyurin VA, Planas-Iglesias J, He R-R, Klein-Seetharaman J, Bayır H, Kagan VE, Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, Okonkwo DO, Cheng JP, Alexander H, Clark RSB, Kochanek PM, Wipf P, Kagan VE, Bayir H, Nat. Neurosci 2012, 15, 1407–1413; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kagan VE, Tyurin VA, Jiang JF, Tyurina YY, Ritov VB, Amoscato AA,Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou MM, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG, Nat. Chem. Biol 2005, 1, 223–232. [DOI] [PubMed] [Google Scholar]
- [4].a) Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, Prokisch H, Trumbach D, Mao G, Qu F, Bayir H, Fullekrug J, Scheel CH, Wurst W, Schick JA, Kagan VE, Angeli JP, Conrad M, Nat. Chem. Biol 2017, 13, 91–98; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kagan VE, Mao G, Qu F, Angeli JP, Doll S, Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, Kapralov AA, Amoscato AA, Jiang J, Anthony-muthu T, Mohammadyani D, Yang Q, Proneth B, Klein-Seetharaman J, Watkins S, Bahar I, Greenberger J, Mallampalli RK, Stockwell BR, Tyurina YY, Conrad M, Bayir H, Nat. Chem. Biol 2017, 13, 81–90; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wenzel SE, Tyurina YY, Zhao J, St Croix CM, Dar HH, Mao G, Tyurin VA, Anthonymuthu TS, Kapralov AA, Amoscato AA, Mikulska-Ruminska K, Shrivastava IH, Kenny EM, Yang Q, Rosenbaum JC, Sparvero LJ, Emlet DR, Wen X, Minami Y, Qu F, Watkins SC, Holman TR, VanDemark AP, Kellum JA, Bahar I, Bayir H, Kagan VE, Cell 2017, 171, 628–641.e626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Amoscato AA, Sparvero LJ, He RR, Watkins S, Bayir H, Kagan VE, Anal. Chem 2014, 86, 6587–6595; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sparvero LJ, Amoscato AA, Fink AB, Anthonymuthu T, New LA, Kochanek PM, Watkins S, Kagan VE, Bayir H, J. Neurochem 2016, 139, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Boughton BA, Hamilton B, Adv. Exp. Med. Biol 2017, 965, 291–321. [DOI] [PubMed] [Google Scholar]
- [7].a) Kompauer M, Heiles S, Spengler B, Nat. Methods 2016, 14, 90; [DOI] [PubMed] [Google Scholar]; b) Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM, J. Mass Spectrom 2012, 47, 1473–1481. [DOI] [PubMed] [Google Scholar]
- [8].Alberts B, Johnson A, Lewis J, Walter P, Raff M, Roberts K, Molecular Biology of the Cell, 4th ed., Routledge, Abingdon-on-Thames, 2002. [Google Scholar]
- [9].Vickerman JC, Briggs D, ToF-SIMS: Surface Analysis by Mass Spectrometry, IM Publications, Chichester, 2001. [Google Scholar]
- [10].a) Touboul D, Kollmer F, Niehuis E, Brunelle A, Laprévote O, J. Am. Soc. Mass Spectrom 2005, 16, 1608–1618; [DOI] [PubMed] [Google Scholar]; b) Steele AV, Schwarzkopf A, McClelland JJ, Knuffman B, Nano Futures 2017, 1, 015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fletcher JS, Sadia R, Barber AM, Lockyer NP, Vickerman JC, Surf. Interface Anal 2013, 45, 273–276. [Google Scholar]
- [12].Tian H, Sparvero LJ, Amoscato AA, Bloom A, Bayır H, Kagan VE, Winograd N, Anal. Chem 2017, 89, 4611–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Angerer TB, Blenkinsopp P, Fletcher JS, Int. J. Mass Spectrom 2015, 377, 591–598. [Google Scholar]
- [14].a) Angerer TB, Mohammadi AS, Fletcher JS, Biointer-phases 2016, 11, 02A319; [DOI] [PubMed] [Google Scholar]; b) Wehrli PM, Angerer TB, Farewell A, Fletcher JS, Gottfries J, Anal. Chem 2016, 88, 8680–8688; [DOI] [PubMed] [Google Scholar]; c) Sämfors S, Ståhlman M, Klevstig M, Borén J, Fletcher JS, Int. J. Mass Spectrom 2019, 437, 77–86; [Google Scholar]; d) Phan NTN, Fletcher JS, Ewing AG, Anal. Chem 2015, 87, 4063–4071. [DOI] [PubMed] [Google Scholar]
- [15].Matsuo J, Torii S, Yamauchi K, Wakamoto K, Kusakari M, Nakagawa S, Fujii M, Aoki T, Seki T, Appl. Phys. Express 2014, 7, 056602. [Google Scholar]
- [16].Passarelli MK, Pirkl A, Moellers R, Grinfeld D, Kollmer F, Havelund R, Newman CF, Marshall PS, Arlinghaus H, Alexander MR, West A, Horning S, Niehuis E, Makarov A, Dollery CT, Gilmore IS, Nat. Methods 2017, 14, 1175–1183. [DOI] [PubMed] [Google Scholar]
- [17].Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, Vickerman JC, Anal. Chem 2008, 80, 9058–9064. [DOI] [PubMed] [Google Scholar]
- [18].a) Do TD, Comi TJ, Dunham SJB, Rubakhin SS, Sweedler JV, Anal. Chem 2017, 89, 3078–3086; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Passarelli MK, Ewing AG, Winograd N, Anal. Chem 2013, 85, 2231–2238; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Fletcher JS, Vickerman JC, Winograd N, Curr. Opin. Chem. Biol 2011, 15, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Strzelecka D, Chmielinski S, Bednarek S, Jemielity J, Kowalska J, Sci. Rep 2017, 7, 8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].a) Bayir H, Tyurin VA, Tyurina YY, Viner R, Ritov V, Amoscato AA, Zhao Q, Zhang XJ, Janesko-Feldman KL, Alexander H, Basova LV, Clark RSB, Kochanek PM, Kagan VE, Ann. Neurol 2007, 62, 154–169; [DOI] [PubMed] [Google Scholar]; b) Samhan-Arias AK, Ji J, Demidova OM, Sparvero LJ, Feng W, Tyurin V, Tyurina YY, Epperly MW, Shvedova AA, Greenberger JS, Bayir H, Kagan VE, Amoscato AA, Biochim. Biophys. Acta Biomembr 2012, 1818, 2413–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Lavezzi AM, Corna MF, Matturri L, J. Neurol. Sci 2013, 329, 45–50; [DOI] [PubMed] [Google Scholar]; b) Sarnat HB, Nochlin D, Born DE, Brain and Development 1998, 20, 88–94; [DOI] [PubMed] [Google Scholar]; c) Mullen RJ, Buck CR, Smith AM, Development 1992, 116, 201–211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.