Abstract
Ecdysis behavior allows insects to shed their old exoskeleton at the end of every molt. It is controlled by a suite of interacting hormones and neuropeptides, and has served as a useful behavior for understanding how bioactive peptides regulate CNS function. Previous findings suggest that crustacean cardioactive peptide (CCAP) activates the ecdysis motor program; the hormone bursicon is believed to then act downstream of CCAP to inflate, pigment, and harden the exoskeleton of the next stage. However, the exact roles of these signaling molecules in regulating ecdysis remain unclear. Here we use a genetic approach to investigate the functions of CCAP and bursicon in Drosophila ecdysis. We show that null mutants in CCAP express no apparent defects in ecdysis and postecdysis, producing normal adults. By contrast, a substantial fraction of flies genetically null for one of the two subunits of bursicon [encoded by the partner of bursicon gene (pburs)] show severe defects in ecdysis, with escaper adults exhibiting the expected failures in wing expansion and exoskeleton pigmentation and hardening. Furthermore, flies lacking both CCAP and bursicon show much more severe defects at ecdysis than do animals null for either neuropeptide alone. Our results show that the functions thought to be subserved by CCAP are partially effected by bursicon, and that bursicon plays an important and heretofore undescribed role in ecdysis behavior itself. These findings have important implications for understanding the regulation of this vital insect behavior and the mechanisms by which hormones and neuropeptides control the physiology and behavior of animals.
Introduction
Neuropeptides are small signaling molecules that regulate animal development, physiology, and behavior (Strand, 1999). The ancient association of neuropeptides with nervous system function is accompanied by a diverse and complex spectrum of actions. In insects, an emblematic case of neuropeptide action is the control of ecdysis, a precisely timed series of behaviors that enables insects to shed the remains of the old exoskeleton at the end of every molt. Research conducted over the last 40 years has revealed that several hormones and neuropeptides regulate the precise order and timing of the different ecdysial behavioral subroutines (for review, see Ewer and Reynolds, 2002; Zitnan and Adams, 2004). The main endocrine signal that commits the animal to executing ecdysis is the phasic release of ecdysis triggering hormone (ETH) that occurs at the end of the molt. Crustacean cardioactive peptide (CCAP) has long been considered the neuropeptide that acts downstream of ETH to turn on the motor program that causes the old exoskeleton to be shed (ecdysis proper). Indeed, adding CCAP peptide to an isolated Manduca CNS activates this motor program and turns off the preparatory motor program of preecdysis (Gammie and Truman, 1997). Also, RNA interference of CCAP signaling in Tribolium causes a failure in ecdysis (Arakane et al., 2008; Li et al., 2011). Finally, Drosophila bearing targeted ablations of CCAP-expressing neurons do not exhibit pupal ecdysis behavior (Park et al., 2003). However, additional studies have implied a more complex model. In Drosophila and other insects, subsets of CCAP neurons express additional neuropeptides (Luo et al., 2005; Kim et al., 2006a,b; Luan et al., 2006; Woodruff et al., 2008), suggesting that some of the functions assigned to CCAP through targeted cell-killing experiments (Park et al., 2003) could be effected by other coexpressed neuropeptides, acting alone or in combination with CCAP. In particular, although bursicon (the so-called tanning hormone) has traditionally been associated with postecdysial functions (Cottrell, 1962; Fraenkel and Hsiao, 1962; Honegger et al., 2008), recent work suggests that it may play a role at ecdysis itself (Loveall and Deitcher, 2010; Veverytsa and Allan, 2011).
To further elucidate the specific role that CCAP plays at ecdysis in Drosophila, we isolated a mutant lacking CCAP function; we also isolated a null allele of pburs, which encodes one of the two subunits of the heteromeric hormone, bursicon, and is expressed in a subset of CCAP neurons. We show that the absence of CCAP causes no detectable defects in ecdysis behavior. By contrast, pburs mutants showed severe failures at pupal ecdysis, in addition to the expected postecdysial defects of adult escapers. Finally, we found that animals lacking both CCAP and pburs function expressed a much more severe phenotype than did flies lacking either hormone, and showed a complete failure to ecdyse. These findings have significant implications for our understanding of the neuroendocrine control of this critical insect behavior. They also show that ecdysis can serve as a tractable model for understanding how neuropeptides and hormones control behavior and physiology in animals.
Materials and Methods
Fly strains and genetics
Fly strains
Fly stocks were maintained at room temperature (22–25°C) on standard agar/cornmeal/yeast media. Unless noted, they were obtained from the Bloomington Drosophila Stock Center (Bloomington, Indiana; http://flystocks.bio.indiana.edu/). The stocks used included:
P-element insertion in the CCAP gene, P{EPgy2}CcapEY15558 [Bloomington stock (BL) #21144];
Two genetic deletions that include the CCAP gene: Df(3R)B+L38, a 40 kb deletion that removes CCAP gene and 14 other genes (Paré et al., 2009), and Df(3R)23D1, ry506/TM3, Sb1 Ser1 [hereafter called Df(3R)23D1; BL#2586; cf., Granderath et al., 1999; breakpoints: 94A3-4; 94D1-4; K. Cook, personal communication to Flybase (http://flybase.bio.indiana.edu/), 2000], ∼800 kb in size, based on information obtained from Flybase;
PiggyBac insertion, PBac{RB}e02061 (stock e02061, Exelixis Harvard Stock Center; insertion in “minus” orientation), 4.2 kb 5′ of the pburs gene, and P-element insertion, P{XP}d02171, (stock d02171, Exelixis Harvard Stock Center; insertion in “minus” orientation) 7.2 kb 3′ of the pburs gene; and
Df(2)Exel6036 (BL#71519; http://flybase.org/reports/FBab0037874.html), an 81 kb deletion that removes the pburs gene and two additional genes.
In addition, the following large deletion uncovering pburs was used: Df(2L)A217, b1, cn1, bw1 (hereafter called Df(2L)A217; BL#6111; http://flybase.org/reports/FBst0006111.html; breakpoints: 34F5-35B3). Finally, the following deletions of the pburs gene region were kindly provided by John Roote (Department of Genetics, University of Cambridge, Cambridge, United Kingdom). All are mutant for pupal (pu; John Roote, personal communication), a gene we show here to be synonymous with pburs: Df(2L)el6D, A379P, b, cn, bw/In(2LR)O, Cy dplvI, pr, cn2 (hereafter abbreviated Df(2)135), and z w11E4; Df(2L)b81a2L, A80R, cn, bw/In(2LR)O, Cy dplvI, b, pr, cn2 [hereafter called Df(2)110]. Stocks bearing homozygous lethal mutations were typically maintained heterozygous with “green” balancer chromosomes, which provide a useful marker for genotyping immature animals via constitutive expression of GFP: CyO, P{w[+mC]=ActGFP}JMR1 (for chromosome 2; from BL#4533) and TM3, P{w[+mC]=ActGFP}JMR2, Ser1 (for chromosome 3; from BL#4534). Other stocks used included y1, w; CyO, H{w[+mC]=PΔ2-3}HoP2.1/Bc1 (BL#2078) and P{ry[+t7.2]=hsFLP}1, w1118; Adv1/CyO (BL#6).
Genetics
Deletion of CCAP gene.
A deletion of the CCAP gene was isolated by imprecise excision of P-element insertion P{EPgy2}CcapEY15558, which occurred within the CCAP gene, 160 bp 3′ of the sequences encoding the CCAP neuropeptide. Flies homozygous for the insertion showed no obvious phenotype but did exhibit decreased CCAP immunoreactivity (data not shown). P{EPgy2}CcapEY15558 was mobilized using a standard scheme involving the “Δ2-3” transposase (Robertson et al., 1988), which was supplied on chromosome 2 by Hobo insertion, H{w[+mC]=PΔ2-3}HoP2.1 on a CyO balancer chromosome (stock BL#2078). Lines were produced using single white-eyed excision males balanced over the P{w[+mC]=ActGFP}JMR2, Ser1 “green balancer.” Candidate deletions were identified by lack of CCAP immunostaining in the CNS of homozygous excision third-instar larva CNS and further characterized molecularly.
Deletion of pburs gene.
Exelixis strains e02061 and d02171, bearing insertions that flank pburs, were used to exclusively delete the pburs gene, using the flippase recombinase/flippase recombinase target (FLP-FRT) system as described by Parks et al. (2004). Briefly, stocks P{ry[+t7.2]=hsFLP}1, w1118; Adv1/CyO (BL#6), e02061, and d02171 were used to obtain F1 larvae bearing both Exelixis elements in trans and a source of FLP under control of the heat-shock promoter. Larval cultures 1–2 d old were heat-shocked daily for 1 h at 37°C for 4 d to induce recombination between the FRT sites within the Exelixis elements. Later, 40 single F2 males were crossed to CyO, P{w[+mC]=ActGFP}JMR1 “green balancer.” From each established line, 3–4 nonbalancer third-instar larvae were screened singly by PCR to identify lines carrying a pburs deletion; these were then further characterized molecularly.
Molecular biology
PCR
Screen for pburs deletion.
DNA suitable for PCR screening of candidate lines was obtained from single third-instar larva as described by Gloor et al. (1993), but using 10 μl of “squish buffer” (0.4 μg/μl proteinase K, 10 mm Tris, pH 8, 0.2 mm EDTA, and 25 mm NaCl) per fly larva. One microliter of extract was used for each 20 μl PCR, which was run using the following conditions: 94°C (3 min); then 40 cycles of 94°C (45 s), 58°C (1.5 min), 72°C (1.5 min/kb of product); followed by one cycle at 72°C for 1 min/kb of product. Primer pair used for initial screen for pburs deletion was pburs_F2 + pburs_R1 (see Table 1 and Fig. 1E), which amplifies a 1.1 kb fragment from wild-type DNA. For most reactions, Taq polymerase from Promega was used.
Table 1.
Primers used for PCR amplification
| Name | Use | Sequence |
|---|---|---|
| CCAP F | Fig.1 | AAATCAACTAGGGCACAAATGG |
| CCAP R | Fig.1 | ACAAAAGCAGTGTGGTAACC |
| CCAP_5′ | CCAP rescue | AAATCAACTAGGGCACAAATGG |
| CCAP_3′ | CCAP rescue | TGTCACCAAGAGATAGCATAGGG |
| pburs_F1 | Fig.1 | ACTACTTTCTTTGCTGGCTTGC |
| pburs_F2 | Fig.1 | TTGTTAGCCTTTGGCTTACTGC |
| pburs_R1 | Fig.1 | CAGTCGACATCGTTACTTGTGG |
| pburs R2 | Fig.1 | CCACAATATGTCAAACGAGACC |
| pburs_5′ | pburs rescue | ACTTTGAGTTCCAGCATTGAGC |
| pburs_3′ | pburs rescue | TCAAACACACACGAAGATCC |
| pburs_F3 | In situ probe | GTCCAGGAACTGCTCTTTGTG |
| pburs_R3 | In situ probe | GAGGCATTAACGTGTGAAATCG |
| burs_F1 | In situ probe | CATTCCACGTGAAAGGACACTC |
| burs_R1 | In situ probe | GGCATGGGTATGAGTGCTAAAC |
| P-31 | Fig.1 | CGACGGGACCACCTTATGTTATTTCATCATG |
| X1 | Fig.1 | TACTATTCCTTTCACTCGCACTTATTG |
| X2 | Fig.1 | TCCAAGCGGCGACTGAGATG |
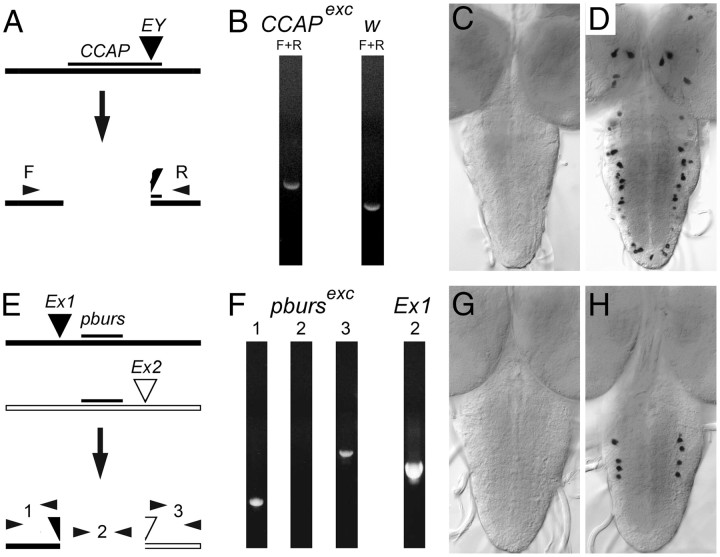
Figure 1.
A–H, Isolation of null mutants in CCAP (A–D) and pburs (E–H) genes. A, Schematic of lesion in CCAP gene caused by the imprecise excision of the EY mobile P-element; F and R: PCR primer pair used for diagnostic PCR, which showed a larger product in excision line (B, CCAPexc) than in controls (B, w) due to retention of some mobile element sequences. C, D, In situ CNS expression of ccap RNA. Prominent expression in CCAP neurons (D) was absent in the CNS of homozygous excision flies (C). E, Schematic of lesion in pburs. Insertions d02171 (Ex1) and e02061 (Ex2), both of which contain FRT sites, were used to create a FLP-induced genetic deletion that exclusively removed the pburs gene. Diagnostic PCR product 2 was absent in homozygous excision flies, whereas DNA distal to Ex1 (PCR product 1) and proximal to Ex2 (PCR product 3) appeared intact in the resulting hybrid element. G, H, In situ CNS expression of pburs RNA. Prominent expression in pburs neurons of controls (H) was absent in the CNS of homozygous excision flies (G). See Table 1 for PCR primer sequences; for E, PCR products 1, 2, and 3 were amplified using primer pairs: pburs_F1 + X1, pburs_F2 + pburs_R1, and X2 + pburs_R2, respectively.
Amplification of genomic DNA.
DNA for genomic rescue constructs was amplified from DNA obtained from the relevant bacterial artificial chromosome (BAC) clone from the RPCI-98 Drosophila melanogaster BAC Library (http://bacpac.chori.org/dromel98.htm) using High Fidelity Expand Long Template PCR system (Roche) following manufacturer's instructions. DNA from BACs BACR23F10 and BACR2L10 were used to amplify CCAP genomic DNA, and DNA from BACR21J17 was used for pburs. PCR products were cloned into pGEM-T Easy vector (Promega), and sequenced for verification. Primers used are listed in Table 1.
Rescue constructs
CCAP genomic DNA (1755 bp), amplified using primers CCAP_5′ + CCAP_3′ (Table 1), included 727 bp 5′ of start ATG and 143 bp 3′ of end of cDNA. The size of the 5′ region was chosen because previous findings (e.g., Park et al., 2003) indicated that 594 bp of 5′ DNA were sufficient to drive gene expression in a pattern that matched CCAP spatial and temporal expression. pburs genomic DNA (1885 bp), amplified using primers pburs_5′ + pburs_3′ (Table 1), included 1086 bp 5′ of start ATG and 315 bp 3′ of TAA stop signal (∼130 bp 3′ of potential polyadenylation site). No prior information was available regarding the suitability of this fragment for rescuing pburs function.
P-elements bearing CCAP and pburs rescue constructs were obtained by first subcloning PCR-amplified genomic DNA into pGEM-T Easy vector (see above). The relevant fragments were then cloned into pGreen H-Pelican P-element vector (Barolo et al., 2000), which was cut with an appropriate restriction enzyme within the multiple cloning site plus SpeI, thereby also excising the EGFP sequences from the vector. Resulting clones were verified by restriction digests and sent to BestGene for germline transformation of a w1118 host. Several (>6) transgenic lines bearing an insertion at different genomic locations were obtained for each construct. Preliminary tests conducted using 2–3 lines showed no qualitative differences in their activity, as expected by the presence of “insulator sequences” that bracket the gene of interest in the pGreen H-Pelican P-element vector.
Immunostaining and in situ hybridization
Immunohistochemistry
Immunostaining was performed as described by Clark et al. (2004). Briefly, late third-instar or early-pupal CNS's were fixed >2 h at room temperature in buffered 4% paraformaldehyde, or 1 h at 4°C in buffered 4% paraformaldehyde plus 7% of a saturated aqueous picric acid solution. Tissues were then rinsed 4 × 10′ in PBS with 0.3% Triton X-100 (Sigma-Aldrich) (PBSTX), and incubated in primary antibody diluted in PBSTX plus 2% normal donkey serum (Jackson ImmunoResearch). The following antisera were used: rabbit anti-CCAP (Clark et al., 2004; 1:5000; generously provided by Hans Agricola); mouse anti-PBURS (Luo et al., 2005; generously provided by Aaron Hsueh and used 1:500), and rabbit anti-BURS (Luan et al., 2006; 1:5000; generously provided by Benjamin White). After overnight incubation on shaker at 4°C, tissues were rinsed 5 × 10′ in PBSTX, and incubated >2 h at room temperature in fluorescently labeled secondary antibodies obtained from Jackson ImmunoResearch and used at 1:200. Preparations were then rinsed, dehydrated, mounted in DPX (Fluka), and viewed under a conventional fluorescent microscope as well as under a confocal microscope (Leica DMR system).
Probes
Templates for pburs and bursicon (burs) probe synthesis were obtained by cloning pburs and burs cDNA using standard techniques. RNA was extracted from third-instar CNS using Trizol (Invitrogen) following manufacturer's instructions. First strand cDNA synthesis was performed using oligo-dT primer. pburs and burs cDNA fragments were then amplified using primer pairs pburs_F4 + pburs_R4 (430 bp product), and burs_F1 + burs_R1 (595 bp product), respectively (see Table 1), cloned into pGEM-T Easy, and sequenced for verification. CCAP template was a cDNA clone (700 bp) described by Park et al. (2003) (also in pGEM-T Easy). DIG-labeled RNA probes were synthesized from linearized cDNA clones following manufacturer's instructions (Roche). After precipitation and resuspension, RNA labeling was verified by spotting a dilution series onto a nylon membrane and processing for DIG immunoreactivity using alkaline-phosphatase-labeled anti-DIG (Roche) used at 1:2000 and visualized using NBT/BCIP (Roche) following manufacturer's recommendations.
RNA in situ hybridization
RNA in situ hybridization was performed using standard methods (e.g., Patel, 1996), using antisense probes at 1:500 dilution. After rinses, tissues were incubated overnight at 4°C in alkaline-phosphatase-labeled anti-DIG (Roche) used at 1:2000, and reacted using NBT/BCIP (Roche) following manufacturer's recommendations. Sense probes (for burs) and CCAP and pburs deletions (Fig. 1) were used as controls, and produced no signal. Tissues labeled for both immunoreactivity and in situ RNA expression were processed sequentially, first for RNA in situ hybridization and reacted with NBT/BCIP (Roche), and then processed for antibody labeling using DAB and H2O2. After final washes in PBS, tissues were mounted on polylysine-covered slides in 80% glycerol.
Behavioral observations
Pupal ecdysis
Flies were crossed in population cages and eggs collected daily on agar/apple juice plates (Wieschaus and Nüsslein-Volhard, 1998). Recently hatched GFP minus first-instar larvae were placed in vials with normal media and kept at 25°C. Animals that had recently pupariated were examined and those containing a bubble in the midregion of the puparium (late stage p4(i); Bainbridge and Bownes, 1981) were selected, placed on their side on a microscope slide, and filmed at room temperature (∼22°C) under dim transmitted light using a Leica DMLB microscope (10× magnification). One experimental and one control animal was filmed simultaneously at one-sixth of the normal speed using a time-lapse video recorder.
Quantification of pupal ecdysis behavior
During pupal ecdysis the animal sheds its larval cuticle and everts its head, thereby completing the transformation, initiated at pupariation, from larva into an adult-shaped pupa; the pupa will then develop into an adult during metamorphosis. In intact wild-type animals, the pupal ecdysis sequence starts with ∼10 min of preecdysis, which consists of slow anterior-directed waves, during which the posterior of the animal separates from the overlying puparium. This period ends with a distinct pullback of the front end of the animal from the puparium, and is immediately followed by 2–3 more rapid anterior-directed waves that sweep the animal, causing the head to evert; also at this time the legs and wings, which everted at pupariation, are extended. Ecdysis is then followed by a protracted postecdysis period, during which the final adult-like shape is attained through progressively smaller body movements.
The timing of the sequences described above could be altered in the various mutants we investigated. In addition to changes to the duration of preecdysis, a lag could occur between the time of occurrence of the pullback of the animal from the front end of the puparium and the start of the anterior-directed peristaltic waves that cause head eversion; finally, the duration and success of this last phase could also be altered. Here we define the end of the preecdysial phase as the moment of pullback from the anterior puparium, and we define ecdysis as the period from the moment this pullback occurs to the moment the head is successfully everted.
In addition to measuring the timing of the different phases of ecdysis, we quantitated the success of ecdysis based on the morphology of the resulting pharate adults. Failures at ecdysis cause defects in head, wing, and leg eversion, resulting in pharate adults with a partial head or no head, and short wings and legs (cf. Park et al., 2003). We therefore scored the fraction of a normal head that was visible, and measured the length of the wings and metathoracic legs, in animals that had reached the end of metamorphosis but were still within the pupal case; one wing and one leg was measured per animal.
Eclosion rhythms
Cultures 6–8 d old were entrained to 12 h light/dark cycle (LD) at 20°C. When most animals had pupated, 1–3-d-old pupae were transferred to Trikinetics eclosion discs, entrained for 2–3 additional days at 20°C, and placed in a Trikinetics eclosion monitoring system. Eclosion was monitored at 20°C either in LD or in constant darkness. Eclosion profiles were analyzed using Matlab analysis programs, kindly provided by Joel Levine (University of Toronto, Canada; Levine et al., 2002).
Statistical analyses
Statistical significance was evaluated using SPSS (PASW Statistics 18). Quantitative results (see Figs. 2, 5, 9C, 10C) were compared by ANOVA followed by Tukey's HSD post hoc analyses. Categorical data based on quantitative measurements (see Fig. 3) were compared by a Kruskal–Wallis test. Following rejection of the null hypothesis, select subsets were analyzed using further Kruskal–Wallis or Mann–Whitney U tests. Remaining categorical data (see Figs. 4, 10B) were analyzed using χ2 tests and, following rejection of the null hypothesis, further χ2 tests were used to analyze select subsets.
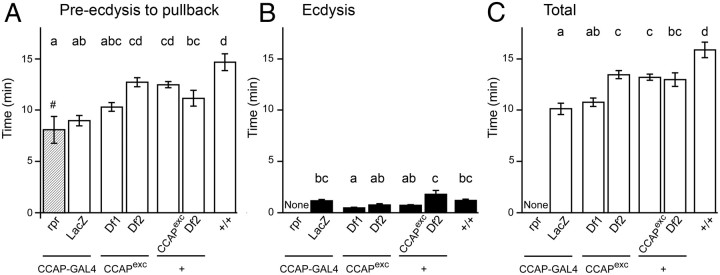
Figure 2.
Pupal ecdysis behavior sequence in the absence of CCAP. A–C, Duration of pupal preecdysis (A), ecdysis (B), and entire ecdysis sequence (preecdysis plus ecdysis) (C) in flies lacking CCAP neurons versus flies mutant for CCAP and controls. Animals bearing targeted ablations of CCAP neurons [expressing cell death gene, reaper, under the control of CCAP-GAL4; (rpr, column 1)] express a weak pullback behavior and then fail to ecdyse (Park et al., 2003). By contrast, controls [flies expressing LacZ under the control of CCAP-GAL4; (LacZ, column 2)] and flies hemizygous for CCAP (columns 3 and 4) express both preecdysis and ecdysis behavior. Although there are differences in the duration of the ecdysial phases among these latter genotypes, these differences do not correlate with the CCAP genotype. Times are averages ± SEM; N = 10–12 per group. # and hatching of column 1 indicate that preecdysis ended with weak pullback behavior. Different letters above columns indicate significantly different timing (p < 0.05). Hemizygous CCAP mutant animals were heterozygous for CCAP excision (CCAPexc) and two different genetic deletions that include CCAP, Df1 [Df(3)23D1] and Df2 [Df(3)B+L38]; see Materials and Methods for more details.
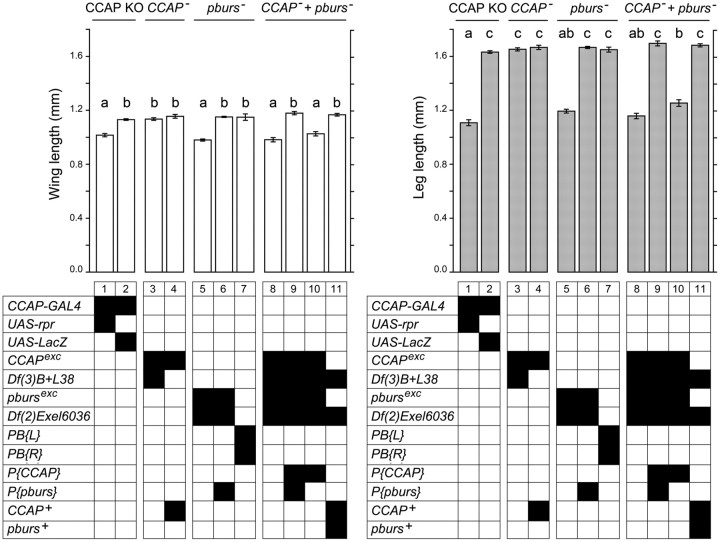
Figure 5.
Role of CCAP and pburs in the regulation of the pharate adult and adult phenotypes: wing (left) and leg (right) length. Groups and genotypes are displayed as described in Figure 3. N > 30 animals per genotype. Different letters above columns indicate significantly different categories (p < 0.05).
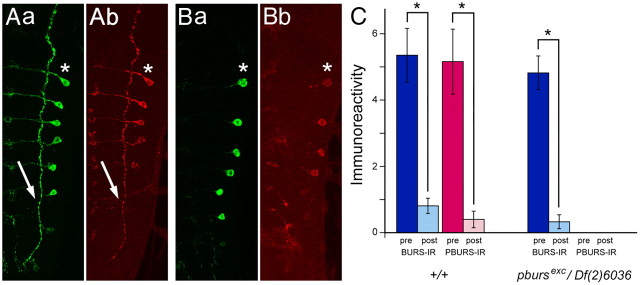
Figure 9.
Bursicon subunits are released at pupal ecdysis. Aa, Ab, Ba, Bb, Pattern of BURS-IR (a) and PBURS-IR (b) before (A) and after (B) pupal ecdysis. Note that prominent immunoreactivity in lateral axon (arrows in Aa and Ab, for BURS-IR and PBURS-IR, respectively) is not visible after ecdysis (Ba and Bb, for BURS-IR and PBURS-IR, respectively). C, Quantitation of immunoreactivity in lateral axon before (pre) and after (post) ecdysis for BURS-IR and PBURS-IR in wild-type (+/+) animals, and for BURS-IR in pburs hemizygous animals [pbursexc/Df(2)Exel6036], showing that bursicon subunits are secreted at pupal ecdysis, and that BURS is released at this time even in the absence of PBURS. Asterisks indicate statistically significant change in immunoreactivity (p < 0.05).
Figure 10.
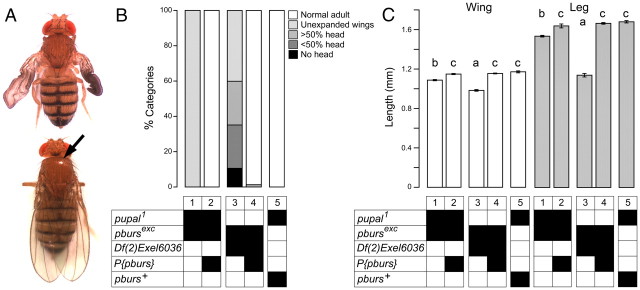
A, pupal is pburs (A) pu1/pbursexc fly (top) showing partially expanded wings and abnormal tanning (e.g., matte exoskeleton). These defects are rescued in pu1/pbursexc fly bearing a P{pburs} transgene (bottom) (black arrow points to reflection indicative of properly sclerotized exoskeleton). B, C, Summary of morphological defects expressed by pu1 hemizygotes, demonstrating allelism with pburs. Wing expansion defects (B, column 1) as well as incomplete wing (C, wing, column 1) and leg (C, leg, column 1) extension are rescued by P{pburs} transgene. Defects expressed by pu1/pbursexc animals (B, C, column 1) are less severe than those expressed by pburs hemizygotes, (B, C, column 3). Genotypes are displayed as described in Figure 3. In C, different letters above wing and leg columns indicate significantly different categories (p < 0.05).
Figure 3.
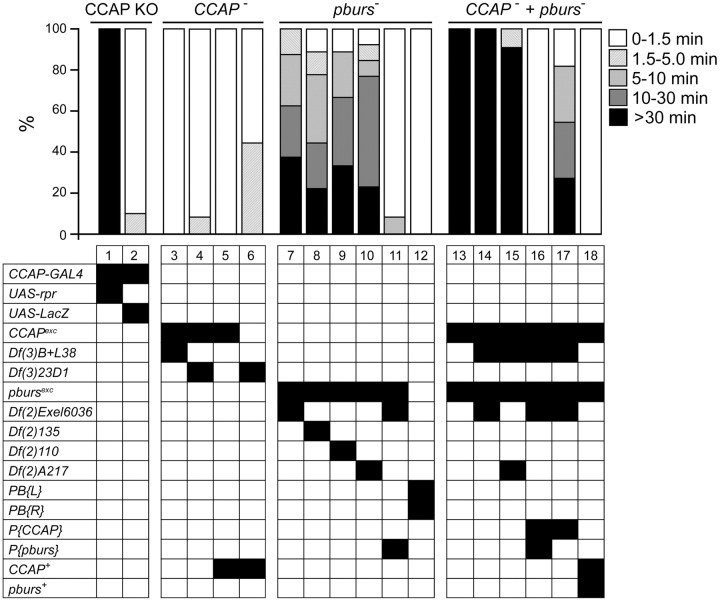
Role of CCAP and pburs in the completion of pupal ecdysis behavior. Top, Success of ecdysis behavior, indicated as the proportion of animals that completed ecdysis within the indicated time intervals, in animals lacking CCAP neurons (CCAP KO group), the CCAP neuropeptide (CCAP− group), the PBURS neurohormone (pburs− group), and both CCAP and PBURS (CCAP− + pburs− group). Genotypes are coded by the combinations of black squares within each table column, and are defined below. Animals lacking PBURS showed severe defects at ecdysis (columns 7–10), which were rescued by a P{pburs} transgene (column 11). Although flies mutant for CCAP completed ecdysis within the normal time (columns 3 and 4), removing CCAP function in animals lacking PBURS greatly potentiated the defects expressed by pburs mutants (columns 13–15 vs columns 7–10). The defects of these CCAP + pburs double mutants were similar to those expressed by flies bearing targeted ablations of CCAP neurons (column 1; cf. Park et al., 2003). Defects expressed by double mutants were fully rescued by P{CCAP} + P{pburs} transgenes (column 16), and were rescued to levels comparable to those of pburs mutants by a P{CCAP} transgene (column 17; columns 7–10 vs 17; p > 0.05). Genotypes, abbreviated in the leftmost column, are as follows: CCAPexc and pbursexc correspond, respectively, to null CCAP and pburs alleles produced in this study; Df(3)B+L38 and Df(3)23D1 are genetic deletions that include the CCAP gene; Df(2)6036, Df(2)135, Df(2)110, and Df(2)A217 are genetic deletions that include the pburs gene; PB{L} and PB{R} represent the two mobile elements flanking pburs that were used to create the pburs excision; P{CCAP} and P{pburs} represent transgenes bearing CCAP and pburs rescue constructs, respectively; CCAP+ and pburs+ represent chromosomes bearing the endogenous, wild-type alleles of the CCAP and pburs genes, respectively. See Materials and Methods for further details. N > 10 animals per group.
Figure 4.
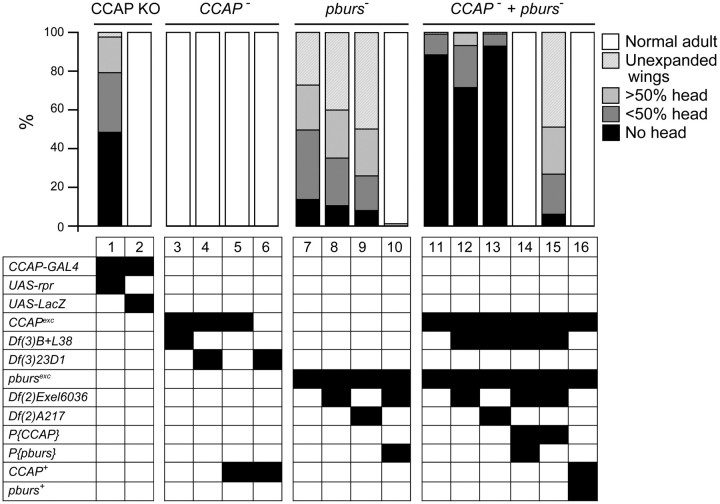
Role of CCAP and pburs in the regulation of head eversion and wing expansion. Morphology of the pharate adult and adult is summarized as the proportion of animals within the indicated categories. Categories and genotypes are displayed as described in Figure 3. The terminal phenotypes expressed by the different genotypes were consistent with the behavioral defects shown in Figure 3; slight differences are likely due to the much greater number of animals examined (N > 50 animals per genotype). Examples of animals in these groups are shown in Figure 6: 6A, normal adult; 6B, unexpanded wings; 6C, no head; 6D, >50% head.
Results
Role of CCAP in pupal ecdysis
Generation of CCAP-null allele
We created a null CCAP allele by excising a P-element inserted within the CCAP gene downstream of the CCAP neuropeptide-encoding sequences and screening for lines that lacked CCAP immunoreactivity (IR) in the CNS. Larvae from a single excision line (of ∼200 single male white-eyed excision lines) were found to lack CCAP-IR (data not shown). Subsequent sequence analyses revealed that this mutant lacked 695 bp of CCAP DNA, starting 29 bp 5′ of transcription start and including all CCAP neuropeptide-encoding sequences (Fig. 1A); it also retained a 1073 bp fragment of the original P-element, thereby explaining the slightly larger size of a diagnostic PCR product (Fig. 1B). As expected, no CCAP RNA could be detected by in situ hybridization of mutant third-instar larval CNS (Fig. 1C; compared with the control, Fig. 1D).
Pupation behavior of CCAP-null allele
Animals bearing targeted ablations of CCAP neurons show severe behavioral defects at pupal ecdysis (Park et al., 2003; Kim et al., 2006a). Although preecdysis behavior appears normal and the duration of the period between the start of preecdysis and anterior pullback is similar to that of the relevant controls [Fig. 2, preecdysis to pullback, progeny of CCAP-GAL4 × UAS-rpr, column 1 vs that of control, CCAP-GAL4 × UAS-LacZ (column 2); p > 0.05], this anterior pullback is quite weak and is not followed by ecdysis behavior (Fig. 2, ecdysis, column 1, vs control, column 2; Fig. 3, column 1, vs control, column 2); instead, it is followed by progressively weaker preecdysis-like movements (Park et al., 2003). As a result, most animals fail to properly evert their heads and extend their appendages, causing most to have reduced or nonexistent heads (Park et al., 2003; Fig. 4, column 1 vs control, column 2) and shorter than normal legs and wings (Park et al., 2003; Fig. 5, column 1 vs control, column 2, both panels; p < 0.05).
To our surprise, animals bearing the CCAP-null allele, CCAPexc, produced viable, normal-looking and fertile adults, both when homozygous for this mutant allele (Fig. 6A) and when heterozygous with either of two different genetic deletions of the CCAP region [Df(3)23D1 and Df(3)B+L38; see Materials and Methods]. A detailed analysis of their pupal ecdysis behavior did not uncover any abnormality that could specifically be attributed to the lack of CCAP. Indeed, as shown in Figure 2, although the duration of the phases of ecdysis differed among the various genotypes tested, these differences were not due to the lack of CCAP. Thus, for instance, the duration of preecdysis was not significantly different between hemizygous mutant and heterozygous wild-type animals (Fig. 2, preecdysis to pullback, columns 3 and 4, vs controls, columns 5 and 6). Likewise, differences in ecdysis timing did not correlate with the genotype at the CCAP locus. Indeed, hemizygous mutant [CCAPexc/Df(3)23D1 and CCAPexc/Df(3)B+L38, Fig. 2, ecdysis, columns 3 and 4, respectively] grouped with heterozygous wild-type CCAPexc/+ (Fig. 2, ecdysis, column 4), whereas these three genotypes were significantly different from Df(3)B+L38/+ controls (Fig. 2, ecdysis, column 5) (see also, Fig. 3, columns 3 and 4, vs controls, columns 5 and 6). Finally, the resulting adults showed quantitatively normal morphology (Fig. 4, columns 3 and 4 vs controls, columns 5 and 6; and Fig. 5, column 3 vs control, column 4, p > 0.05). Thus, we were unable to detect any defect associated with the absence of CCAP.
Figure 6.
Terminal morphology of flies lacking CCAP or PBURS function. A, Flies mutant for CCAP expressed normal morphology and tanning. B, By contrast, adult flies mutant for pburs failed to inflate their wings and showed abnormal tanning (as evidenced, e.g., by matte exoskeleton). C, D, Examples of pharate adults mutant for pburs that showed extreme (C) and more mild (D) morphological defects caused by abnormal pupal ecdysis, based on the proportion of the head that was everted and the length of the wings and legs (black arrowheads and white arrows, respectively). E, Pharate control fly, showing normal head eversion, and normal wing and leg extension.
Role of CCAP in gating of eclosion
Drosophila adult emergence (eclosion) is regulated by the circadian clock, and is restricted to the early part of the day [dawn and morning in an LD regime; and to the subjective dawn and morning in a dark:dark (DD) regime] (Konopka and Benzer, 1971; Saunders, 2002). In addition, the timing of eclosion can be influenced by environmental stimuli. In particular, a pulse of light delivered at dawn (or subjective dawn, when in DD) triggers the eclosion of the cohort of animals that is developmentally competent to emerge (e.g., McNabb et al., 1997; McNabb and Truman, 2008). It also causes the release of the brain neurohormone eclosion hormone (EH), suggesting that this “lights-on response” may be triggered by the sudden release of EH (McNabb and Truman, 2008).
A small proportion of flies bearing targeted ablations of CCAP neurons is able to eclose and shows normal circadian rhythmicity. However, the pattern of emergence under a LD regime does not show the characteristic lights-on response (Park et al., 2003). This finding suggested that the CCAP neuropeptide, which is believed to act downstream of EH (Ewer and Reynolds, 2002; Zitnan and Adams, 2004), could mediate this surge in eclosion.
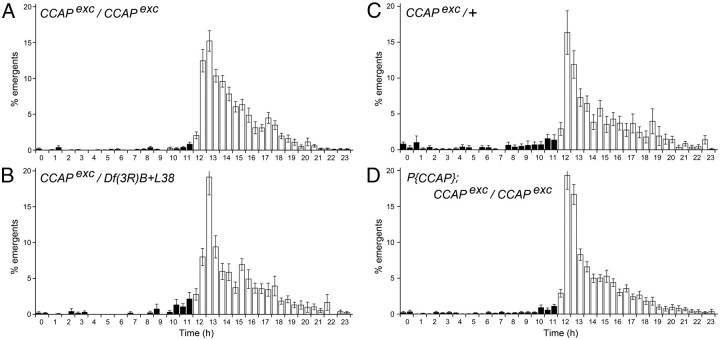
We examined the profile of adult emergence of flies lacking CCAP under an LD regime. As shown in Figure 7, homozygous (Fig. 7A) and hemizygous (Fig. 7B) CCAPexc flies showed an increased eclosion during the first 3 h after lights on, which was similar to that observed in populations of heterozygous control flies (Fig. 7C), homozygous CCAPexc flies rescued with a wild-type CCAP transgene (Fig. 7D), and in wild-type controls (data not shown). Thus, our results suggest that the CCAP neuropeptide is not critical for the lights-on response. The profile of eclosion of CCAP mutants was also normal in DD (data not shown), as suggested by the slight anticipatory increase in the rate of eclosion, which occurred before lights-on (Fig. 7A,B, hours 11 and 12).
Figure 7.
Temporal pattern of adult emergence under LD regime in flies lacking CCAP function. Histogram represents the average percentage (±SEM) of flies that emerged within a 1 h time window. Black and white bars represent dark and light periods of the photoperiod, respectively. A–D, Flies homozygous (A) and hemizygous (B) for CCAPexc showed a profile of eclosion similar to that of controls (C) and of homozygous CCAPexc rescued with a wild-type CCAP transgene (D), including a similar surge of emergence 0–3 h after lights-on. Each panel represents the profile for each population, averaged across different days and for four independent experiments, each including two populations per genotype.
Generation of pburs-null allele
To investigate the role of bursicon at ecdysis, we isolated a null allele of pburs (bursβ), which encodes one of the two subunits of this heterodimeric hormone (Luo et al., 2005; Mendive et al., 2005). This allele was obtained using FRT-bearing mobile elements that flanked the pburs gene, as previously described (Parks et al., 2004). Candidate mutants were identified by the lack of the diagnostic PCR product using primers located between these inserts (Fig. 1E,F, primer pair 2). We then used in situ hybridization to show that hemizygous mutant animals did not show detectable pburs RNA expression in the CNS (Fig. 1G, vs Fig. 1H, controls). Since the bursicon hormone requires the subunits encoded by the pburs and burs (bursα) genes to signal through its receptor (Luo et al., 2005; Mendive et al., 2005), such a mutant would display no bursicon activity.
Phenotypes of pburs-null mutants
Depending on the exact genotype, 30–50% homozygous and hemizygous pburs mutant pupae completed metamorphosis and eclosed. As expected, given the known role of the bursicon neurohormone, 100% of these adults failed to inflate their wings and their exoskeleton did not harden and showed abnormal melanization (Fig. 6B), similar to that caused by burs (Dewey et al., 2004) and rk (Baker and Truman, 2002) mutant alleles. Interestingly however, we noted that the remainder of the pharate adults expressed a spectrum of morphological defects diagnostic of failures at pupal ecdysis, such as partial head eversion, abnormally short legs and wings, and the presence of a persistent space between the end of the abdomen and the overlying puparium (e.g., Fig. 6C,D). To identify the bases of these morphological defects, we next characterized in detail their pupal ecdysis behavior. In wild-type animals, preecdysis ends with the pullback of the front end of the animal from the puparium, which is then immediately followed by ecdysis proper, during which the head is everted. We detected no difference in the duration of preecdysis in pburs homozygous and hemizygous mutants versus controls (data not shown; Fig. 3, ANOVA for genotypes 7–12, p > 0.05). By contrast, we found that pburs homozygotes and hemizygotes expressed a longer and more variable period between pullback and head eversion. Indeed, whereas 90–100% of wild-type animals everted their head within 90 s of pullback (e.g., Fig. 3, columns 2 and 12) the majority of pburs hemizygotes took >90 s to do so, with 20–40% (depending on the exact genotype) failing to do so (defined here as taking >30 min) (Fig. 3, columns 7–10). This defect was rescued to wild-type levels by a single copy of a pburs transgene (Fig. 3, column 11), demonstrating that it was specifically attributable to the absence of pburs function. To obtain an independent measure of ecdysis success, we also quantitated the morphologies of pharate adults. We found that mutations on pburs caused >50% of the animals to show a reduced head, with 20–40% showing a severely reduced or absent head (Fig. 4, columns 7–9); these pharate adults also showed significantly shorter legs and wings (Fig. 5, columns 5 vs 7, both panels; p < 0.05). These defects, which are all attributable to failures of ecdysis, were completely rescued by a single copy of a transgene containing only the pburs gene (head defects: Fig. 4, column 10; wing and leg length: Fig. 5, columns 6 vs 7; p > 0.05). Collectively, these results demonstrate that pburs function contributes significantly to pupal ecdysis behavior.
Role of CCAP in pupal ecdysis in the absence of pburs
Although our findings reveal that bursicon plays a role in the control of pupal ecdysis, flies bearing targeted ablations of CCAP neurons express much more severe defects at pupation. For example, most pburs homozygotes and hemizygotes everted their head within 30 min of pullback (only 30–40% took longer; Fig. 3, pburs, columns 7–10), whereas almost all flies lacking CCAP neurons failed to do so within 30 min (Fig. 3, CCAP KO, column 1). The morphology of the resulting pharate adults was also less abnormal for pburs mutants than for CCAP KO animals. For example, only 5–10% of pburs mutants showed no head at the pharate adult stage (Fig. 4, pburs, columns 7–9), compared with ∼50% when CCAP neurons were ablated (Fig. 4, CCAP KO, column 1). This suggests that other neuropeptides expressed in CCAP neurons may be involved in controlling pupation behavior.
This observation together with the known role of CCAP in the control of ecdysis of other insects prompted us to examine the phenotype of flies lacking both CCAP and bursicon function. Pharate pupae mutant for both genes were readily obtained, suggesting normal viability during the larval stages. Nevertheless, we observed that they expressed very severe defects at pupation. Indeed, although these animals expressed preecdysis behavior, this phase rarely ended with a distinct pullback of the front end of the animal from the puparium. Instead, pullback was typically weak or absent, and preecdysis transitioned into a weaker preecdysis-like behavior, which later resembled postecdysis behaviors. These behaviors were variable and were not characterized in detail. Yet, it was clear that the vast majority of animals failed to express any head eversion behaviors within 30 min (Fig. 3, CCAP− + pburs−, columns 13–15), and the morphology of the resulting pharate adults was also severely affected, with 70–90% showing no head (Fig. 4, CCAP− + pburs−, columns 11–13). Both defects were rescued by transgenes bearing wild-type copies of the CCAP gene and the pburs gene (ecdysis: Fig. 3, CCAP− + pburs−, column 16; morphology: Fig. 4, CCAP− + pburs−, column 14). Furthermore, the phenotype of CCAP, pburs double mutants, was similar to that of pburs single mutants when the double mutant was rescued with only the CCAP-bearing transgene (ecdysis: Fig. 3, CCAP− + pburs−, column 17, compare with columns 7–10, p > 0.05; morphology: Fig. 4, CCAP−+ pburs−, column 15, compare with columns 7–9). Thus, our results suggest that CCAP and bursicon both participate in the control of ecdysis, even though a function for CCAP could only be uncovered in the absence of bursicon. The similarities between the severe defects expressed by animals lacking CCAP neurons (Figs. 3, 4, CCAP KO, column 1) and those of flies lacking both CCAP and bursicon (Figs. 3, 4, CCAP− + pburs−, columns 13–15 and 11–13, respectively), suggest that these two molecules mediate the majority of the actions subserved by these neurons at this stage.
Timing of bursicon release
We used immunohistochemistry to verify that bursicon is released at pupal ecdysis. We first determined the pattern of burs and pburs expression. In the third-instar larval CNS, BURS and PBURS-IR is strictly confined to CCAP neurons (Dewey et al., 2004; Luo et al., 2005; Zhao et al., 2008). However, PBURS has a much more limited expression: whereas BURS is expressed in most CCAP neurons of the abdominal (A) ganglia, PBURS immunoreactivity is restricted to just one of the two pairs of CCAP neurons in segments A1–4 (Luo et al., 2005; Peabody et al., 2008). By contrast, both subunits show coincident expression in pharate adults (Luan et al., 2006; Peabody et al., 2008).
We used anti-BURS immunohistochemistry in combination with in situ hybridization to burs and pburs to determine the pattern of expression of the two bursicon subunits in pharate pupae. In our hands, PBURS-IR was not always robust enough to reliably label the full complement of PBURS neurons. Thus, we used pburs in situ RNA expression in combination with BURS-IR to aid in assessing colocalization. We also performed BURS-IR plus burs in situ hybridization to validate this method. As shown in Figure 8, the spatial pattern of expression of the bursicon subunits in the CNS of pharate pupae was similar to that for third-instar larvae. Thus, BURS-IR occurred in 1–2 pairs of neurons per segment, from thoracic (T) T1 to A8, and was largely coincident with the in situ pattern of gene expression (Fig. 8A–C); the only exceptions were some serial homologs (e.g., those in segments A5–8) that showed weak staining using one method but a weak or no signal with another, which likely reflects an overall low level of expression in these neurons. As occurs in larvae (Fig. 1H), pburs in situ RNA expression was limited to one of the two CCAP neurons in A1–4 (Fig. 8A,D), all of which also expressed burs mRNA (Fig. 8C) and BURS-IR (Fig. 8A,B). Thus, as occurs in the larval CNS, the pharate pupal CNS expresses BURS and pburs in subsets of CCAP neurons, and BURS is more widely expressed than is pburs. The lack of complete overlap in the pattern of expression of these two genes is intriguing because the homodimeric hormones (BURS+BURS and PBURS+PBURS) are inactive in in vivo tanning assays and do not activate the rk receptor (Luo et al., 2005). This raises the possibility that BURS may regulate processes that are independent of PBURS and that would not be mediated by RK. A similar lack of strict colocalization has also been described for other insects (e.g., Manduca sexta; Dai et al., 2008).
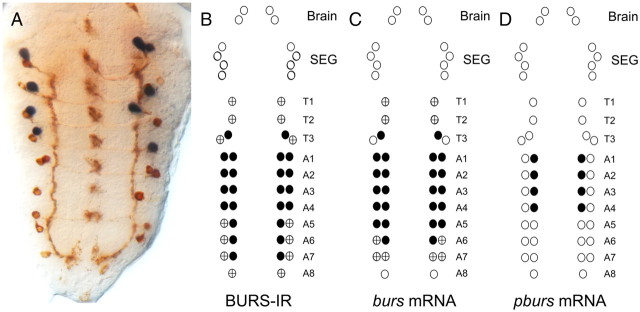
Figure 8.
Spatial pattern of expression of bursicon subunits in the ventral CNS of pharate pupae. A, Pattern of BURS-IR (brown) and pburs in situ RNA expression (blue), showing that pburs is expressed in a subset of four pairs of BURS-IR neurons. B–D, Summary of pattern of BURS-IR (B), and burs (C) and pburs in situ RNA expression (D). Circles represent the complement of CCAP neurons in the brain, SEG, and ventral nervous system. Filled circles, strong staining; +, weak staining. For each stain, 8–10 preparations were scored.
Recently, Loveall and Deitcher (2010) showed that BURS-IR decreases at pupal ecdysis from peripheral synaptic terminals. Here we examined the changes in BURS-IR and PBURS-IR in the CNS at this time. For this, animals before ecdysis (late stage p4(i); Bainbridge and Bownes, 1981) and 30 min after ecdysis were selected, and their CNS processed for BURS-IR and PBURS-IR. We found a clear and significant decrease following ecdysis in BURS-IR and PBURS-IR from central axons (Fig. 9Aa vs Ba for BURS-IR and Fig. 9Ab vs Bb for PBURS-IR, and quantitated in Fig. 9C, +/+) (p < 0.05), suggesting that the bursicon heterodimer is released at this time. These results are consistent with previous findings that BURS-IR is reduced from synaptic terminals at pupation (Loveall and Deitcher, 2010). Interestingly, we found that BURS is released at pupation even in the absence of PBURS. Indeed, we detected a normal fall in BURS-IR following the pupation of pburs hemizygous mutant animals (Fig. 9C, pbursexc/Df(2)6036; p < 0.05). Thus, BURS, either in the form of a monomer or as homodimer, is correctly packaged and secreted at this time.
pupal1 is a pburs allele
The genetic region around pburs includes pu, an unmapped gene. Weak alleles of pu (e.g., pu1) cause failures in wing inflation and cuticle tanning (Natzle et al., 2008). Unpublished postings on Flybase [e.g., Davis, T. (2001.4.23); see http://flybase.org/reports/FBrf0138570.html], which predate the discovery that CG15284 encodes pburs, discuss the possibility that pu mutants are alleles of CG15284. Given the similarity between the adult phenotype of pupal mutants and that of pburs and rk mutants, we performed complementation tests between pupal and pburs mutant alleles. As shown in Figure 10 using our pburs-null allele, the pu1 mutant allele, deletions known to uncover pburs or pu, and flies bearing a transgenic pburs rescue construct, we showed that pu is an allele of pburs. Indeed, pu1/pbursexc expressed a phenotype similar to that of pu1/pu1 flies (Fig. 10A, top), with 100% of such animals failing to inflate their wings normally (Fig. 10B, column 1). Importantly, these defects were completely rescued by a pburs transgene (Fig. 10A, bottom; Fig. 10B, column 2), as were those of pupal1 homozygotes (data not shown). The pu1 allele is a weak hypomorphic allele, since it causes the expression of a much milder phenotype than is seen in pbursexc homozygous or hemizygous animals (Fig. 10B, column 3). Defects in wing (Fig. 10C, wing) and leg (Fig. 10C, leg) length showed a similar pattern of complementation. Thus, mutations in pupal are alleles of pburs; we suggest renaming pupal mutant alleles pburspupal (e.g., pburspupal1 for pupal1).
Discussion
Our fragmentary understanding of the regulation of ecdysis behavior indicates that it is controlled by a suite of neuropeptides and hormones that show complex hierarchical and reciprocal relationships, and in which a given neuropeptide (or hormone) may act on different targets or act in a combinatorial manner on a specific target with other neuropeptides (or hormones). Here we isolated mutants null for CCAP and pburs to better define the functions of these genes and to investigate possible synergistic actions. We were surprised to find that animals lacking CCAP expressed normal pupal ecdysis behavior because CCAP is believed to be the key neuropeptide that controls ecdysis. For instance, application of CCAP to an isolated Manduca CNS will turn on the ecdysis motor program (Gammie and Truman, 1997). Also, RNA interference (RNAi) of CCAP or its receptor, CCAPR-2, by injection of double-stranded RNA, causes arrest at ecdysis in Tribolium (Arakane et al., 2008; Li et al., 2011). Furthermore, because bursicon has previously only been associated with the regulation of postecdysis events following adult eclosion (e.g., Dewey et al., 2004), we were also surprised to discover that pburs-null mutants showed severe defects at pupation. Nevertheless, RNAi of both bursicon subunits as well as of its receptor (rickets, rk) cause a quantitative weakening of preecdysis behavior in Tribolium (Arakane et al., 2008), and release of bursicon during Drosophila pupal preecdysis has recently been reported (Loveall and Deitcher, 2010), suggesting a role in the control of early phases of the ecdysis sequence. Furthermore, Loveall and Deitcher (2010) reported that interference of rk function in Drosophila causes defects at pupation, although the range of additional nonecdysial defects observed suggest that such manipulations interfered with other pathways, rendering the interpretation of their findings more difficult. In this regard, our results using animals mutant for pburs indicate that PBURS plays an important role, and that this role is primarily restricted to the correct execution of ecdysis behaviors (a role in postecdysis has not been investigated at this stage). A role for bursicon specifically at pupal ecdysis was recently uncovered by showing that defects at pupation, caused by the elimination of the retrograde signal needed for CCAP and PBURS expression in the CNS, could be partially rescued by specifically restoring pburs expression in the relevant neurons (Veverytsa and Allan, 2011).
Although flies lacking CCAP were ostensibly entirely normal (compare Figs. 2–5, 6A), we were able to uncover a critical function for this peptide at ecdysis by examining pupation in animals lacking pburs function. Indeed, in this mutant background, eliminating CCAP caused an almost complete failure of ecdysis. This suggests that both CCAP and PBURS regulate ecdysis, with CCAP playing a minor role and PBURS playing a major role. The bases of CCAP and bursicon actions, however, remains unclear. The CCAP-expressing neurons in the ventral CNS consist of a pair of efferent neurons (CCAPE) in segments T3-A4 (Zhao et al., 2008; homologous to cell 27s in other insects; Honegger et al., 2008), and a pair of interneurons (CCAPIN) in segments T1-A9 (Zhao et al., 2008; homologous to IN704 in other insects; Honegger et al., 2008). In Manduca, addition of CCAP to an isolated CNS can activate and sustain the ecdysis motor program (Gammie and Truman, 1997). The limited arborization of CCAPE within the CNS would imply that this activational role would be subserved by CCAPIN, and is consistent with the type of role that these neurons play after adult emergence (Luan et al., 2006); by contrast, CCAPE neurons release bursicon into the hemolymph to first plasticize then harden and melanize the wings and exoskeleton, and play no behavioral role (Peabody et al., 2008). Alternatively, the activation of ecdysis could be mediated by a pair of CCAP neurons in the subesophageal ganglion (SEG). At least at adult emergence, it is these neurons that command postecdysial behaviors, such as air-swallowing and wing inflation (Peabody et al., 2008). However, both CCAP neurons in the SEG and the CCAPINs express CCAP but not PBURS, and we were unable to detect any ecdysial defects in CCAP-null mutants. By contrast, recent findings show that reducing CCAP and bursicon expression from CCAPEs causes severe defects in pupation (Veverytsa and Allan, 2011), implying that these neurons may be key for the activation of ecdysis behavior. Since CCAPEs have a sparse arborization within the CNS, these results would also imply that the activational roles of CCAP and bursicon could be indirect.
While pupation requires activation of motor programs, changes in hemolymph pressure may also be essential for correct eversion of the head and the proper extension of legs and wings. Thus, it is also possible that CCAP's role in Drosophila ecdysis is at least in part based on its cardioactive function (cf. Zitnan and Adams, 2004), instead of or in addition to a neural activation role. Indeed, CCAP is cardioactive in insects (cf. Dircksen, 1998) including Drosophila (Nichols et al., 1999; Dulcis et al., 2005) and, in addition to serving to better disperse coreleased neuropeptides and neurohormones, including bursicon, this cardioactive function may be necessary for the successful transformation into a pupa. The identification of the direct targets of CCAP and bursicon coupled with functional studies will be needed for the full understanding of the exact roles that these peptides play at pupation.
Independent of the exact nature of their functions, the actions of CCAP and bursicon show the hallmark complexity of neuropeptide control of physiology and behavior. In addition to each of these molecules acting on different targets (e.g., bursicon, which activates ecdysis and also causes wing inflation and the hardening and pigmentation of the adult exoskeleton), we show that CCAP and bursicon act synergistically to control ecdysis behavior. Such convergence is seen in a number of peptide systems, and appears to be the basis for the integration of multiple signals and many time-independent signals. Such a situation occurs, for example, in the control of arousal, which depends on inputs related to food intake and satiation, as well as from inputs from the circadian clock (Adamantidis and de Lecea, 2008). Another role for multiple peptidergic inputs may be to increase the precision and robustness of a response. For example, mammalian circadian clocks cause daily rhythms of locomotor activity to be expressed with a precision of ∼1 min/d (King and Takahashi, 2000). The basis for this precision is not entirely understood, but is likely mediated by the action of multiple clock output neuropeptides (Dibner et al., 2010), all of which can affect the pattern of activity/inactivity. Ecdysis likewise shows a very precise timing, and naturally occurring failures are extremely rare (Reynolds, 1980). Although we have shown that CCAP is not essential for ecdysis in the laboratory, it may nevertheless provide a signal that, under particular conditions, is essential for the successful and seamless execution of the behavior. This signal may also vary in different insects, reflecting a bias toward one of several possible actions in organisms with different body plans. Thus, for instance, CCAP may primarily play a cardioactive role in some insects, whereas for others it may play a critical role in activating a motor program itself. We hope that future comparative work using insects with different developmental and anatomical constraints will help elucidate the logic behind such biases. In addition to such an approach, work in Drosophila (e.g., Park et al., 2002) and Tribolium (e.g., Arakane et al., 2008; Li et al., 2011) has clearly shown that molecular genetics provides a unique tool to understand the essential as well as the redundant functions of every ecdysis neuropeptide and hormone. The combination of both approaches will shed light on the mechanism that enables insects to flawlessly complete a complex behavioral sequence almost regardless of conditions. It will also provide a useful model for understanding how neuropeptides control the physiology and behavior of all animals.
Footnotes
Funding was provided by National Science Foundation Grant IBN-0343699000 and Fondo Nacional de Desarollo Cientifico y Tecnólogico Grants 1071079 and 1111023. We thank Gunisha Kauer, Tashana Williams, Chloe Ota, and Bianca Chiang for assistance during various stages of this work. We are grateful to Hans Agricola for anti-CCAP antiserum, Aaron Hsueh for anti-PBURS antiserum, and Benjamin White for anti-BURS antiserum. We thank John Roote for pupal alleles. We appreciate comments on the manuscript from Ralph Greenspan and Hans-Willi Honegger. This project was initiated by J.E. in the Department of Entomology, Cornell University, Ithaca, NY 14853.
References
- Adamantidis A, de Lecea L. Sleep and metabolism: shared circuits, new connections. Trends Endocrinol Metab. 2008;19:362–370. doi: 10.1016/j.tem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, Park Y. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech Dev. 2008;125:984–995. doi: 10.1016/j.mod.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- Baker JD, Truman JW. Mutations in the Drosophila glycoprotein hormone receptor, rickets, eliminate neuropeptide-induced tanning and selectively block a stereotyped behavioral program. J Exp Biol. 2002;205:2555–2565. doi: 10.1242/jeb.205.17.2555. [DOI] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726, 728–730, 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Clark AC, del Campo ML, Ewer J. Neuroendocrine control of larval ecdysis behavior in Drosophila: complex regulation by partially redundant neuropeptides. J Neurosci. 2004;24:4283–4292. doi: 10.1523/JNEUROSCI.4938-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell CB. The imaginal ecdysis of blowflies. Detection of the blood-borne darkening factor and determination of some of its properties. J Exp Biol. 1962;39:413–430. [Google Scholar]
- Dai L, Dewey EM, Zitnan D, Luo CW, Honegger HW, Adams ME. Identification, developmental expression, and functions of bursicon in the tobacco hawkmoth, Manduca sexta. J Comp Neurol. 2008;506:759–774. doi: 10.1002/cne.21575. [DOI] [PubMed] [Google Scholar]
- Dewey EM, McNabb SL, Ewer J, Kuo GR, Takanishi CL, Truman JW, Honegger HW. Identification of the gene encoding bursicon, an insect neuropeptide responsible for cuticle sclerotization and wing spreading. Curr Biol. 2004;14:1208–1213. doi: 10.1016/j.cub.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dircksen H. Conserved crustacean cardioactive peptide (CCAP) neuronal networks and functions in arthropod evolution. In: Coast GM, Webster SG, editors. Recent advances in arthropod endocrinology. Cambridge: Cambridge UP; 1998. pp. 302–333. [Google Scholar]
- Dulcis D, Levine RB, Ewer J. Role of the neuropeptide CCAP in Drosophila cardiac function. J Neurobiol. 2005;64:259–274. doi: 10.1002/neu.20136. [DOI] [PubMed] [Google Scholar]
- Ewer J, Reynolds S. Neuropeptide control of molting in insects. In: Pfaff DW, Arnold AP, Fahrbach SE, Etgen AM, Rubin RT, editors. Hormones, brain and behavior. San Diego: Academic; 2002. pp. 1–92. [Google Scholar]
- Fraenkel G, Hsiao C. Hormonal and nervous control of tanning in the fly. Science. 1962;138:27–29. doi: 10.1126/science.138.3536.27. [DOI] [PubMed] [Google Scholar]
- Gammie SC, Truman JW. Neuropeptide hierarchies and the activation of sequential motor behaviors in the hawkmoth, Manduca sexta. J Neurosci. 1997;17:4389–4397. doi: 10.1523/JNEUROSCI.17-11-04389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR. Type I repressors of P element mobility. Genetics. 1993;135:81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granderath S, Stollewerk A, Greig S, Goodman CS, O'Kane CJ, Klämbt C. loco encodes an RGS protein required for Drosophila glial differentiation. Development. 1999;126:1781–1791. doi: 10.1242/dev.126.8.1781. [DOI] [PubMed] [Google Scholar]
- Honegger HW, Dewey EM, Ewer J. Bursicon, the tanning hormone of insects: recent advances following the discovery of its molecular identity. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194:989–1005. doi: 10.1007/s00359-008-0386-3. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zitnan D, Galizia CG, Cho KH, Adams ME. A command chemical triggers an innate behavior by sequential activation of multiple peptidergic ensembles. Curr Biol. 2006a;16:1395–1407. doi: 10.1016/j.cub.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zitnan D, Cho KH, Schooley DA, Mizoguchi A, Adams ME. Central peptidergic ensembles associated with organization of innate behavior. Proc Natl Acad Sci U S A. 2006b;103:14211–14216. doi: 10.1073/pnas.0603459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1–26. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Beeman RW, Park Y. Functions of duplicated genes encoding CCAP receptors in the red flour beetle, Tribolium castaneum. J Insect Physiol. 2011;57:1190–1197. doi: 10.1016/j.jinsphys.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Loveall BJ, Deitcher DL. The essential role of bursicon during Drosophila development. BMC Dev Biol. 2010;10:92. doi: 10.1186/1471-213X-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, Hsueh AJ. Bursicon, the insect cuticle hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci U S A. 2005;102:2820–2825. doi: 10.1073/pnas.0409916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb SL, Truman JW. Light and peptidergic eclosion hormone neurons stimulate a rapid eclosion response that masks circadian emergence in Drosophila. J Exp Biol. 2008;211:2263–2274. doi: 10.1242/jeb.015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb SL, Baker JD, Agapite J, Steller H, Riddiford LM, Truman JW. Disruption of behavioral sequence by targeted death of peptidergic neurons in Drosophila. Neuron. 1997;19:813–823. doi: 10.1016/s0896-6273(00)80963-0. [DOI] [PubMed] [Google Scholar]
- Mendive FM, Van Loy T, Claeysen S, Poels J, Williamson M, Hauser F, Grimmelikhuijzen CJ, Vassart G, Vanden Broeck J. Drosophila molting neurohormone bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. FEBS Lett. 2005;579:2171–2176. doi: 10.1016/j.febslet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Natzle JE, Kiger JA, Jr, Green MM. Bursicon signaling mutations separate the epithelial-mesenchymal transition from programmed cell death during Drosophila melanogaster wing maturation. Genetics. 2008;180:885–893. doi: 10.1534/genetics.108.092908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R, Kaminski S, Walling E, Zornik E. Regulating the activity of a cardioacceleratory peptide. Peptides. 1999;20:1153–1158. doi: 10.1016/s0196-9781(99)00118-7. [DOI] [PubMed] [Google Scholar]
- Paré AC, Dean DM, Ewer J. Construction and characterization of deletions with defined end points in Drosophila using P elements in trans. Genetics. 2009;181:53–63. doi: 10.1534/genetics.108.094193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Schroeder AJ, Helfrich-Förster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–2656. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- Park Y, Filippov V, Gill SS, Adams ME. Deletion of the ecdysis-triggering hormone gene leads to a lethal ecdysis deficiency. Development. 2002;129:493–503. doi: 10.1242/dev.129.2.493. [DOI] [PubMed] [Google Scholar]
- Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Patel NH. In situ hybridization to whole-mount Drosophila embryos. In: Krieg PA, editor. A laboratory guide to RNA: isolation, analysis and synthesis. Wiley; 1996. pp. 357–369. [Google Scholar]
- Peabody NC, Diao F, Luan H, Wang H, Dewey EM, Honegger HW, White BH. Bursicon functions within the Drosophila CNS to modulate wing expansion behavior, hormone secretion, and cell death. J Neurosci. 2008;28:14379–14391. doi: 10.1523/JNEUROSCI.2842-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SE. Integration of behaviour and physiology at ecdysis. Adv Insect Physiol. 1980;15:475–595. [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders DS. Insect clocks, third edition. Amsterdam: Pergamon; 2002. [Google Scholar]
- Strand FL. Neuropeptides: regulators of physiological processes. Cambridge, MA: MIT; 1999. [Google Scholar]
- Veverytsa L, Allan DW. Retrograde BMP signaling controls Drosophila behavior through regulation of a peptide hormone battery. Development. 2011;138:3147–3157. doi: 10.1242/dev.064105. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila: a practical approach. Oxford: IRL; 1998. pp. 179–214. [Google Scholar]
- Woodruff EA, 3rd, Broadie K, Honegger HW. Two peptide transmitters copackaged in a single neurosecretory vesicle. Peptides. 2008;29:2276–2280. doi: 10.1016/j.peptides.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Gu T, Rice HC, McAdams KL, Roark KM, Lawson K, Gauthier SA, Reagan KL, Hewes RS. A Drosophila gain-of-function screen for candidate genes involved in steroid-dependent neuroendocrine cell remodeling. Genetics. 2008;178:883–901. doi: 10.1534/genetics.107.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitnan D, Adams M. Neuroendocrine regulation of insect ecdysis. In: Gilbert L, Kostas I, Gill S, editors. comprehensive molecular insect science. Amsterdam: Elsevier; 2004. pp. 1–60. [Google Scholar]