Abstract
In eukaryotes, genome duplication starts concomitantly at many replication initiation sites termed replication origins. The replication initiation program is spatially and temporally coordinated to ensure accurate, efficient DNA synthesis that duplicates the entire genome while maintaining other chromatin-dependent functions. Unlike in prokaryotes, not all potential replication origins in eukaryotes are needed for complete genome duplication during each cell cycle. Instead, eukaryotic cells vary the use of initiation sites so that only a fraction of potential replication origins initiate replication each cell cycle. Flexibility in origin choice allows each eukaryotic cell type to utilize different initiation sites, corresponding to unique nuclear DNA packaging patterns. These patterns coordinate replication with gene expression and chromatin condensation. Budding yeast replication origins share a consensus sequence that marks potential initiation sites. Metazoan origins, on the other hand, lack a consensus sequence. Rather, they are associated with a collection of structural features, chromatin packaging features, histone modifications, transcription, and DNA-DNA/DNA-protein interactions. These features confer cell type-specific replication and expression and play an essential role in maintaining genomic stability.
Keywords: DNA replication, Cell cycle regulation, Replication origin licensing, Chromatin organization, Histone modification, Replication timing
1. Initiation of DNA Replication
Origins of replication are defined as chromosomal sites where double-stranded DNA unwinds to form single-stranded DNA templates for genome duplication. Genetically, the cis-acting sequences that determine the locations of replication initiation events are termed replicators (Jacob et al. 1963). Replicators can confer the ability to start replication when transferred from their original locations to ectopic sites. Replicators interact with trans-acting factors, termed initiators, to facilitate DNA replication. In eukaryotes, initiators are highly conserved, as all eukaryotes share a group of essential DNA-binding protein complexes that form pre-replication complexes. Pre-replication complexes assemble on chromatin in a process termed “replication licensing,” and the components of pre-replication complexes are orthologous in all eukaryotes (Aladjem 2007; DePamphilis 1999; Fragkos et al. 2015; Masai et al. 2010; Remus and Diffley 2009). Conversely, the chromatin features associated with eukaryotic replicators vary and are often cell type and/or developmental specific (Aladjem 2007; Besnard et al. 2012; Cayrou et al. 2015; Smith and Aladjem 2014; Smith et al. 2016).
The conserved proteins that form pre-replication complexes (Fig. 1) include a DNA-binding origin recognition complex (ORC) that serves as a platform to recruit a conserved group of helicases, polymerases, and accessory proteins that catalyze the initiation of DNA replication. Assembly of the conserved pre-replication and pre-initiation complexes at potential replication origins occurs stepwise during the G1 phase of each the cell cycle (Aladjem 2007; DePamphilis 1999; Fragkos et al. 2015; Remus and Diffley 2009). First, ORC binds to chromatin as cells emerge from mitosis. Then, licensing factors CDC6 and CDT1 bind to ORC, which allows for the binding of the inactive form of the replicative helicase MCM2-7. The resulting pre-replication complex recruits additional proteins required to activate the MCM helicase and initiate DNA replication (Boos et al. 2013; Sansam et al. 2015; Sheu et al. 2016; Tanaka and Araki 2013). The chromatin-bound but inactive MCM2-7 helicase then interacts with additional components, CDC45, MCM10, and GINS (Sld5, Psf1, Psf2, Psf3), to form the complete helicase (CMG) complex. Prompted by cyclin-dependent kinase (CDK)- and Dbf4-dependent kinase (DDK)-mediated phosphorylation, CDC45 interacts with Treslin (Sld3 in yeast), which recruits Sld2/RecQL4 and DPB11/TopBP1 (Bruck et al. 2015; Depamphilis et al. 2012; Masai et al. 2010; Remus and Diffley 2009; Zegerman and Diffley 2007). Chromatin-associated DNA polymerases (Pol-α and pol-δ), replication protein A (RPA), CMG, and Dpb11/TopBP1 then initiate DNA replication (Abid and Costa 2016; Kanemaki and Labib 2006; Takayama et al. 2003).
Fig. 1.
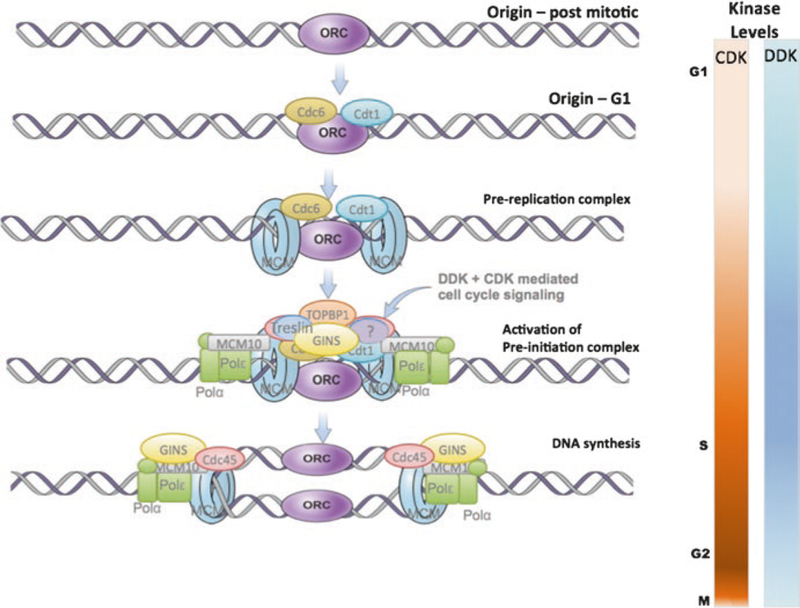
Pre-replication complex proteins bind in a stepwise manner throughout G1. Recruited during the M to G1 transition, the origin recognition complex (ORC) is a platform to recruit a conserved group of helicases, polymerases, and accessory proteins that catalyze the initiation of DNA replication. ORC binds to chromatin as cells emerge from mitosis. Licensing factors Cdc6 and Cdt1 bind to ORC, followed by the inactive form of the replicative helicase MCM2-7. Additional proteins are required to activate the MCM helicase and initiate DNA replication. Specifically, the inactive MCM2-7 helicase then interacts with CDC45, MCM10, and GINS (Sld5, Psf1, Psf2, Psf3) to form the complete helicase (CMG) complex. Cyclin-dependent kinase (CDK)- and Dbf4-dependent kinase (DDK)-mediated phosphorylation activates proteins and allows Cdc45 interacts with Treslin (Sld3 in yeast). Sld2/RecQL4 and DPB11/TopBP1 are then recruited to the complex. Chromatin-associated DNA polymerases (Pol-α and pol-δ), replication protein A (RPA), CMG, and Dpb11/TopBP1 then initiate DNA replication
The assembly of pre-replication complexes and the subsequent initiation are tightly coupled with cell cycle progression by the phosphorylation activities of two kinases, CDK and DDK. Helicase recruitment, in an inactive form, can only occur at low kinase levels (Bell 2002; Remus and Diffley 2009), whereas the activities of DDK and CDK are required at subsequent steps to activate the helicase and initiate replication (Boos et al. 2013; Remus and Diffley 2009; Sansam et al. 2015; Sheu et al. 2016; Tanaka and Araki 2013). The need for low kinase levels in the early stages of pre-replication complex assembly implies that such complexes cannot be assembled once DNA replication has started, insuring orderly cell cycle progression as well as preventing re-replication of cellular DNA, a hallmark of genomic instability (Abbas et al. 2013; Hanlon and Li 2015; Remus and Diffley 2009; Richardson and Li 2014).
Although the events that lead to initiation of DNA replication occur at all potential replication origins, replication initiation occurs with a remarkably consistent order in most cells (Besnard et al. 2012; Cayrou et al. 2011; Martin et al. 2011) to create a coordinated replication timing program (Koren et al. 2014; Mukhopadhyay et al. 2014; Rhind and Gilbert 2013b). The binding patterns of pre-replication complexes do not provide clues to the principles of origin choice as ORC does not exhibit sequence-specific DNA binding (Miotto et al. 2016) and replication origins in most eukaryotes do not share a clear common consensus (Aladjem 2007; Bartholdy et al. 2015; Leonard and Mechali 2013; Masai et al. 2010). Interactions between replication origins and components of pre-replication complexes, therefore, are essential for initiation but cannot intuitively explain the consistent replication patterns observed in most mitotic cell cycles.
2. Genetic Features and Local Determinants of Replication Origins
Replication origins in viruses and in some prokaryotes and eukaryotes exhibit distinct sequence features that facilitate interactions with unique initiator proteins. In DNA tumor viruses, replication origins colocalize with replicator sequences that bind specialized initiators (e.g., SV40 T-antigen and BPV E1 protein) to catalyze DNA unwinding and recruit the host replication machinery (Fanning and Zhao 2009). In budding yeast, replication origins contain an AT-rich, 11 bp consensus ORC binding sequence. High frequency of initiation from yeast replication origins also requires accessory sequences that affect chromatin structure by dictating the efficiency of ORC binding as well as directing initiation in unique chromosomal environments (Hoggard et al. 2013; Marahrens and Stillman 1992; Palacios DeBeer et al. 2003).
In metazoa, replication origin sequences exhibit high heterogeneity (Aladjem 2007; Bartholdy et al. 2015; DePamphilis 1999; Fragkos et al. 2015) and do not share a clear consensus, consistent with the observation that not all potential replication origins initiate replication in all cells each cell cycle. There are two sources of replication origin heterogeneity. First, a large fraction of replication origins exhibit consistent initiation in particular cell types and not in other cells (Besnard et al. 2012; Cayrou et al. 2015; Martin et al. 2011; Smith et al. 2016). For example, less than 50% of all origins identified in a survey of eight cell lines initiated replication in all cells within that cohort, and a large fraction (about 15%) only initiated replication in a single cell line (Smith et al. 2016). Second, within populations of cells of the same type, most replication origins initiate replication stochastically in a fraction of cell cycles. In most somatic metazoan cells, only 10–20% of all potential origins actually initiate replication each cell cycle, suggesting that most origins exhibit flexible initiation patterns (Cayrou et al. 2015). When such flexible origins remain “dormant” and do not initiate replication, they replicate passively from adjacent replication forks (Fig. 2). Notably, however, the presence of excess replication origins plays a role in genome preservation: a marked reduction in the frequency of licensed but dormant origins, achieved either by targeted genetic deletions of origins or by mutating replication licensing factors, increases genomic instability (Abbas et al. 2013; Besnard et al. 2012; Blow et al. 2011; Fragkos et al. 2015; Kawabata et al. 2011; Marks et al. 2016). These observations suggest that flexible or consistent dormant origins, which rarely or never initiate replication during normal cell cycle progression, might serve as backup origins when replication forks stall.
Fig. 2.
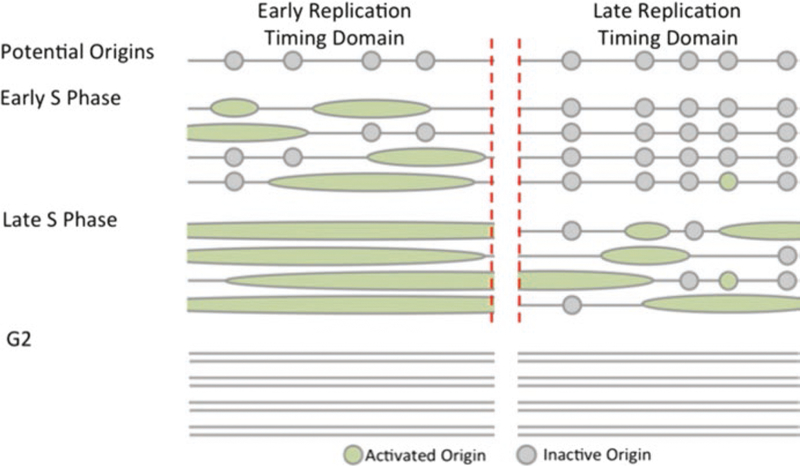
While the profiles of activated replication origins are similar within cell line, specific replication origin chosen by cells within that population varies in location and replication time. Most origins exhibit flexible initiation patterns, where the specific origins activated differ between cells. When such flexible origins remain “dormant” and do not initiate replication, they replicate passively from adjacent replication forks
The exact sequence features that mark replication origins and determine the frequency of initiation at each origin remain to be elucidated. Budding yeast potential replication origins share a consensus sequence that is recognized by the ORC complex. Yeast origins can all initiate replication on plasmids (Marahrens and Stillman 1992; Masai et al. 2010), but chromatin context plays a role in determining the activation rates of particular chromosomal origins (Hoggard et al. 2013; Knott et al. 2012). Metazoan replication origins do not exhibit a prominent single consensus sequence (Aladjem 2007; Fragkos et al. 2015; Mechali et al. 2013; Urban et al. 2015) but share several sequence features (Fig. 3a) including regions that exhibit strand asymmetry, CpG islands, G-quadruplexes, transcription start sites, origin G-rich repeated elements (OGREs), and regions of DNase hypersensitivity (Besnard et al. 2012; Cayrou et al. 2015; Foulk et al. 2015; Martin et al. 2011; Mukhopadhyay et al. 2014; Rao et al. 2014). Of those, genetic association studies on phased genome revealed strong high association with strand asymmetry (Bartholdy et al. 2015).
Fig. 3.
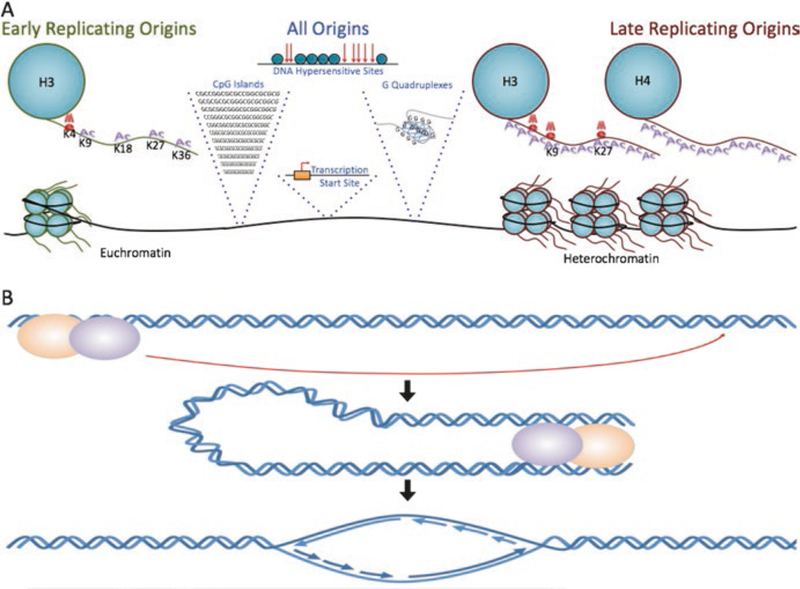
(a) Metazoan replication origins share several sequence features. Origins generally associate with regions that exhibit strand asymmetry, CpG islands, G-quadruplexes, transcription start sites, origin G-rich-repeated elements (OGREs), and regions of DNase hypersensitivity. In agreement, local histone modifications correlate with and can determine replication origin locations and timing. Early replicating regions associate with H3K4me1/2/3, H3K9ac, H3K18ac, H3K36me3, and H3K27ac. These histone modifications also associate with open chromatin and are enriched in moderately active transcription start sites. Late replicating regions tend to associate with H3 and H4 hypoacetylation, H3K9 and H3K27 methylation, and are found in heterochromatic regions. (b) Distal DNA sequences affect origin activity and transcriptional through long-distance interactions. Such interactions can be mediated via protein interaction with enhancers and locus control regions, by chromatin remodeling factors and transcriptional activators that bind enhancers and locus control regions, and by long noncoding RNAs
The primary DNA sequence at replication origins can determine the ability to initiate replication at ectopic sites (Aladjem et al. 1998; Liu et al. 2003), possibly via affecting chromatin modifications (Chen et al. 2013; Conner and Aladjem 2012; Fu et al. 2006; Liu et al. 2003). For example, the replication origin at the human HBB locus can create an open chromatin conformation at ectopic sites (Fu et al. 2006). A group of replication origins colocalizes with ubiquitous chromatin opening elements (UCOES), which maintain open chromatin structure by recruiting transcription factors (Conner and Aladjem 2012; Majocchi et al. 2014) and protecting transcriptional activity despite local repressive epigenetic features (Flickinger 2015). Hence, recruiting chromatin modifiers could allow replication origins to alter the local environment and create a context permissive for both transcriptional activity and replication initiation (Aladjem 2007; Hassan-Zadeh et al. 2012; Huang et al. 2011).
In agreement, local histone modifications correlate with, and can determine, replication origin locations (Feng et al. 2006; Leonard and Mechali 2013; Mechali et al. 2013; Rhind and Gilbert 2013b; Smith and Aladjem 2014; Smith et al. 2016; Vogelauer et al. 2002). Comparisons of initiation sites with histone features and replication timing domains identify certain histone modifications as strong indicators of origin utilization (Fig. 3a). For example, early replicating regions associate with H3K4me1/2/3, H3K9ac, H3K18ac, H3K36me3, and H3K27ac (Smith et al. 2016). Origins that associate with those chromatin features also localize in open chromatin and are enriched in moderately active transcription start sites (Besnard et al. 2012; Martin et al. 2011). Replication origins that initiate replication early during S-phase tend to associate with open chromatin features and are often activated in many cell types. In contrast, late replicating regions tend to associate with H3 and H4 hypoacetylation and H3K9 and H3K27 methylation and often initiate replication in a cell type-specific manner (Cayrou et al. 2015; Mechali et al. 2013; Smith et al. 2016). The tendency to initiate replication in most cells or in a particular cell type does not depend on cancer status, as common and cell type-specific initiation activity correlates with cellular lineages rather than with cancer or noncancer (Smith et al. 2016).
Transcriptional activity of local genes and cellular differentiation state can alter the replication timing program. For example, Xenopus early embryos do not exhibit a strong preference for initiation sites, correlated with the absence of transcription (Mechali and Kearsey 1984). In those embryos, induced transcription either through development or by tethering specific transcription factors resulted in increased localized initiation (Fragkos et al. 2015; Mechali et al. 2013). In somatic dividing cells, replication origins often associate with transcription start sites at active genes (Valenzuela et al. 2011) and, in particular, with transcription start sites at moderately active transcribed regions (Martin et al. 2011; Sequeira-Mendes et al. 2009). However, this association is diminished in highly transcribed regions, suggesting that transcription and replication regulate each other to avoid disruptions and polymerase collision events (Martin et al. 2011; Sequeira-Mendes et al. 2009). In agreement, transcriptional activity coordinates with the replication timing program (Rivera-Mulia et al. 2015), and replication delays often accompany gene silencing.
3. Influences of Global Chromatin Organization on Replication Initiation
On a larger scale, replication timing domains, each containing multiple replication origins that replicate concomitantly (Bartholdy et al. 2015; Mukhopadhyay et al. 2014), exhibit high concordance with large-scale chromatin organization units termed topologically associated domains that encompass several hundred kilobases to megabases (Hiratani et al. 2010; Lieberman-Aiden et al. 2009; Mattarocci et al. 2014; Moindrot et al. 2012; Rao et al. 2014; Rhind and Gilbert 2013b; Yaffe et al. 2010). This association suggests that the time of activation of replication origins reflects a fundamental structural property of the nucleus (Hiratani et al. 2010; Moindrot et al. 2012; Pope et al. 2014; Rhind and Gilbert 2013b; Yaffe et al. 2010). In agreement, replication origins are known to associate nuclear structural features such as matrix attachment sites (MARS), scaffold attachment sites (SARs), and stabilizing anti-repressor elements (STARs) (Mechali et al. 2013; Smith and Aladjem 2014) as well as with lamins and cohesins (Cayrou et al. 2015; Smith and Aladjem 2014).
High-resolution whole-genome analyses reveal that replication timing domains often reflect chromatin modifications (Dileep et al. 2015; Pope et al. 2014). Early replicating regions often associate with transcriptionally active topological domains, whereas late replicating origins often associate with heterochromatin. The effects of the primary sequence on replication origin activity and replication timing were assessed using analyses on phased genomes, which permit identification of paternal vs. maternal origins to characterize the effects of specific sequence variations on origin activity (Bartholdy et al. 2015; Mukhopadhyay et al. 2014), and by identifying inherited alleles that affect replication timing following the sequencing of 161 individual proliferating cell samples (Koren et al. 2014). These analyses have demonstrated that cis-acting genetic elements determine, at least in part, the locations of megabase-scale replication timing domains. In cancer cells, the replication of entire chromosomes could be delayed in a sequence-specific manner, and interactions with long noncoding RNAs could alter the timing of replication for entire chromosomes (Donley et al. 2015).
Proteins that catalyze distinct histone modifications can facilitate or modulate initiation in groups of replication origins. HBO1, a histone acetyltransferase that modifies H4K5 and H4K12, binds near origins of replication by associating with ORC1 and Cdt1 (Iizuka et al. 2006; Miotto and Struhl 2010). The chromatin decondensation promoted by HBO1 is enhanced near H3K4me3 and reduced near H3K20me1/2/3 (Huang et al. 2006; Kim et al. 2006; Saksouk et al. 2009). While histone acetyltransferase HBO1 is associated with early replicating origins, histone methyltransferase PR-Set7 and heterochromatin-associated proteins ORC- associated protein (ORCA) and HP1 are associated with late replicating origins. ORCA/LRWD1 stabilizes ORC on origins and promotes late replication by perpetuating histone compaction near the repressive methylation of H3K9, H4K20, and H4K27 (Chakraborty et al. 2011; Giri et al. 2015). HP1 is also known to stabilize ORC on origins via interaction with ORC2 and ORC3 and to bind to methylated H3K9 to establish late replicating domains (Chakraborty et al. 2011; Schwaiger et al. 2010). H4K20me1 serves as a binding domain for other histone modifiers, like Suv4, promoting further chromatin compaction (Tardat et al. 2010). Cells depleted in further methylation of H4K20 are also shown have reduced ORCA and ORC1 binding (Sherstyuk et al. 2014).
In addition to modifying histones near origins of replication, some trans-acting factors, like Rif1, Taz1, and FKH1/2, facilitate the recruitment of replication factors to origins. Taz1 and Rif1 help promote replication initiation in heterochromatic, telomeric regions (Cornacchia et al. 2012; Hayano et al. 2012; Tazumi et al. 2012; Yamazaki et al. 2012). Both Taz1, which prevents early-S replication activation, and Rif1, which recruits protein phosphatase 1 (PP1) and modulates the chromatin binding of pre-initiation complex components (Dave et al. 2014; Foti et al. 2016; Hiraga et al. 2014; Kanoh et al. 2015), are associated with late replication. Taz1 and Rif1 might delay replication by interfering with DDK phosphorylation of Mcm2-7 and associating with nuclear architectures that anchor heterochromatin (Foti et al. 2016; Tazumi et al. 2012). Inhibition of MCM phosphorylation subsequently interferes with CDC45 and Sld3 loading (Francis et al. 2009; Tazumi et al. 2012). The replication and DNA repair features regulated by Rif1 are conserved across eukaryotes (Mattarocci et al. 2014). Rif1 organizes replication timing domains by associating with G-quadruplexes to suppress replication (Foti et al. 2016; Kanoh et al. 2015; Mattarocci et al. 2014).
Unlike Taz1 and Rif1, FKH1/2 can promote early replication by recruiting replication factors to early replicating DNA (Knott et al. 2012). In addition, FKH1/2 overexpression advances the replication time of late replicating origins (Knott et al. 2012). These proteins help either activate or repress replication by facilitating inter-chromosomal interactions (Musialek and Rybaczek 2015). FKH1/2 advance the time of initiation by acting during the late G1 phase of the cell cycle (Peace et al. 2016), indicating that replication timing can be reset subsequently to origin licensing.
Distal DNA sequences affect transcriptional activity and origin activity through long-distance interactions (Aladjem et al. 1998; Gerhardt et al. 2014; Norio et al. 2005). Such interactions (Fig. 3b) can be mediated via protein interaction with enhancers and locus control regions (Huang et al. 2011) or by chromatin remodeling factors and transcriptional activators that bind enhancers and locus control regions (Aladjem 2007; Fragkos et al. 2015). RepID, a protein that interacts with a group of replication origins, is associated with an origin-activating chromatin loop between the origin and the locus control region at the human HBB locus (Zhang et al. 2016). Long-distance interactions that modulate replication timing can also be mediated by long noncoding RNAs (lncRNAs) such as Xist and HOTAIR, which guide histone and chromatin remodeling proteins to specific DNA sequences and facilitate chromatin interactions (Fragkos et al. 2015; Nagano and Fraser 2011). lncRNAs can stabilize ORC to origins in viruses (Fragkos et al. 2015; Nagano and Fraser 2011) and can affect the timing of replication for entire chromosomes in cancer cells (Donley et al. 2015).
4. Role of Replication Origins
The apparent excess of potential replication origins and the absence of sequence- specific initiation during early embryogenesis both suggest that a consistent replication initiation program is not a mechanistic requirement for genome duplication. Replication also proceeds stochastically with no apparent replication timing domains within the human inactive X chromosome (Koren et al. 2014), again suggesting that a consistent replication timing program is not required merely to insure genome duplication. The replication program could be established to coordinate replication with other chromatin transactions, primarily transcription. Consistent replication initiation sites could facilitate genome integrity by coordinating replication with transcription and chromatin assembly on the shared chromatin template. The consistent replication timing programs establish regions that replicate late during S-phase and might serve to establish and maintain specific nuclear compartments, such as heterochromatin. Late replication of heterochromatin can be required to preserve the structural integrity of the nucleus by preventing rapid, massive chromatin decondensation and re-condensation that could and lead to DNA damage (Bustin and Misteli 2016).
Although the severe effects of changes in replication timing support a critical role for replication timing regulation in maintaining genomic stability, recent mathematical models suggest that the relative efficiencies of initiation at replication origins are sufficient to determine the organization of replication timing domains. A mathematical model can predict replication timing with high accuracy in human cells without assuming any “replication timing factor,” using two variables: the known distribution of replication origins as correlates of DNase hypersensitive sites and the assumption that replication initiation is restricted by the availability of a single rate-limiting activator (Gindin et al. 2014). A second model (Lob et al. 2016) was also able to predict the general progression of DNA replication and in addition predicted the three-dimensional spatial organization of replication events based on higher chromatin organization, assuming spontaneous stochastic initiation within euchromatin and facultative heterochromatin. Again, this model did not assume a replication timing factor, and replication timing could be deduced without such a factor assuming concomitant inhibition of replication initiation at distances below the size of chromatin loops and a domino-like effect by which replication at a particular origin would induce initiation from adjacent origins. A third model was able to predict replication timing in yeast with high accuracy relying on the density of the MCM replicative helicase, assuming a high level of MCMs at early origins (Das et al. 2015). Together, all models suggest that the spatial distribution of replication origins determines the temporal organization of replication.
The ability to modify the spatial and temporal initiation profile also allows a cell to accommodate its specific transcription program. A large fraction of the human genome exhibits changes in the order of replication during nuclear reorganization associated with differentiation and development (Pope et al. 2014; Rhind and Gilbert 2013a; Rivera-Mulia and Gilbert 2016). Consistent with the need to activate initiation at distinct times to accommodate changes in transcription, cell type- specific replication origins are often located in regions that exhibit differentiation-specific and tissue-specific gene expression (Gerhardt et al. 2014; Norio et al. 2005). Origins that are activated in a narrow set of distinct cell types tend to initiate replication late in S-phase in regions that contain few and sparse origins, whereas origins that are commonly activated in many cell type initiate replication throughout S-phase (Smith et al. 2016). Since chromatin and histone modifications influence transcription and replication patterns, varying recruitment of modifiers and other proteins by transcription factors can markedly influence the replication program and vice versa (Bar-Ziv et al. 2016). Massive alterations in replication initiation patterns can be programmed, associated with activation of a differentiation cascade leading to changes in gene expression patterns (Gerhardt et al. 2014; Norio et al. 2005). Conversely, since transcription can hinder initiation of DNA replication on the common chromatin template (Martin et al. 2011), regions that exhibit massive differentiation-induced transcription might contain fewer and sparser replication origins due to the paucity of genetic elements that can support initiation. The precise timing of origin activation within replication timing domains is determined anew after each mitotic cell division (Wu and Gilbert 1996), facilitating dynamic and flexible changes in replication order (Rhind and Gilbert 2013a).
The excess of replication origins might also play a regulatory role to facilitate genomic stability by allowing for timely, accurate replication under stress. Activation of stress responses in actively proliferating cells, including changes in the rate of replication fork progression, can signal for changes in the utilization of certain origins. Conversely, alterations in the frequency of initiation can affect the rate of DNA synthesis. In yeast, replication can proceed upon depletion of the most or all replication origins in specific chromosomes, but those cells exhibit elevated chromosome loss rates (Dershowitz et al. 2007). In addition, a lower number of potential origins can increase the abundance of DNase hypersensitive regions, chromosome fragility, and chromosomal rearrangements (Huang and Koshland 2003; Lengronne and Schwob 2002).
Mammalian cells often exhibit increased frequency of replication initiation events (activation of “dormant origins”) in response to events that slow the progression of replication forks, including changes in nucleotide pool levels (Anglana et al. 2003), exposure to histone deacetylase inhibitors (Conti et al. 2010) diminished homologous recombination (Daboussi et al. 2008) and dysfunctional DNA modifying enzymes such as topoisomerase I (Tuduri et al. 2009) and Mus81 endonuclease (Fu et al. 2015). Since the enhanced frequency of initiation in those cases associates with a mild decrease in replication fork progression rates, it is unclear whether the overall increase in initiation frequency indicates a global compensatory mechanism linking replication fork rates and origin activity or reflects local changes in a group of loci (e.g., fragile sites) that are particularly prone to potentially genotoxic lesions under conditions of slow replication fork progression.
5. Conclusions and Future Questions
Cells duplicate their genomes starting from many origins and proceeding along a well-established program that sequentially replicate the entire genome. Consistent replication origins are evident in most cells, although they are not essential for complete genome duplication. Origin activation dynamics might therefore primarily play a role in establishing local and global chromatin structure and facilitate the cellular response to adverse events that perturb the replication process.
Although the ability to initiate DNA replication can partially be conferred by the primary DNA sequence, most replication origins exhibit flexible initiation, as their activation in a fraction of cells is affected by the chromatin environment and by interactions with distal DNA elements. This flexible initiation program facilitates coordination between replication and transcription and preserves genome stability by maintaining a group of “reserve” potential replication origins that can be activated if replication at adjacent replicons stalls.
Understanding the molecular interactions at replication origins is critical for establishing a complete picture of how cells coordinate chromatin transactions, including transcription, chromatin decondensation and compaction, and DNA synthesis. Despite rapid progress in mapping the locations and timing of replication initiation events, we have yet to identify the corresponding molecular interactions that dictate initiation of DNA replication at particular sites. Future progress in addressing these issues will be achieved by identifying the exact combination of DNA-binding proteins or chromatin modifiers that activate DNA replication. The evident flexibility of the replication program also necessitates studies that characterize cell cycle signaling pathways that repress replication from origins that remain “dormant” during particular cell cycles and modulate replication initiation to coordinate with changes in the transcription program. Finally, future studies characterizing how cell cycle checkpoint pathways affect molecular interactions at replication origins could lead to a better understanding of cellular responses to potentially genotoxic stress.
References
- Abbas T, Keaton MA, Dutta A (2013) Genomic instability in cancer. Cold Spring Harb Perspect Biol 5:a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abid A, Costa A (2016) The MCM helicase motor of the eukaryotic replisome. J Mol Biol 428:10. [DOI] [PubMed] [Google Scholar]
- Aladjem MI (2007) Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8:588–600 [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Rodewald LW, Kolman JL, Wahl GM (1998) Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005–1009 [DOI] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114:385–394 [DOI] [PubMed] [Google Scholar]
- Bartholdy B, Mukhopadhyay R, Lajugie J, Aladjem MI, Bouhassira EE (2015) Allele-specific analysis of DNA replication origins in mammalian cells. Nat Commun 6:7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ziv R, Voichek Y, Barkai N (2016) Chromatin dynamics during DNA replication. Genome Res 26:1245–1256. 10.1101/gr.201244.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev 16:659–672 [DOI] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM (2012) Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19:837–844 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA (2011) How dormant origins promote complete genome replication. Trends Biochem Sci 36:405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D, Yekezare M, Diffley JF (2013) Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 340:981–984 [DOI] [PubMed] [Google Scholar]
- Bruck I, Perez-Arnaiz P, Colbert MK, Kaplan DL (2015) Insights into the initiation of eukaryotic DNA replication. Nucleus 6:449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Misteli T (2016) Nongenetic functions of the genome. Science 352:aad6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R et al. (2011) Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res 21:1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau JC, van Helden J, Mechali M (2015) The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res 25:1873–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Shen Z, Prasanth SG (2011) “ORCanization” on heterochromatin: linking DNA replication initiation to chromatin organization. Epigenetics 6:665–670 [DOI] [PubMed] [Google Scholar]
- Chen X, Liu G, Leffak M (2013) Activation of a human chromosomal replication origin by protein tethering. Nucleic Acids Res 41:6460–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner AL, Aladjem MI (2012) The chromatin backdrop of DNA replication: lessons from genetics and genome-scale analyses. Biochim Biophys Acta 1819:794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, Aladjem MI, Pommier Y (2010) Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res 70:4470–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchia D, Dileep V, Quivy JP, Foti R, Tili F, Santarella-Mellwig R, Antony C, Almouzni G, Gilbert DM, Buonomo SB (2012) Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J 31:3678–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi F, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M, Lopez BS (2008) A homologous recombination defect affects replication-fork progression in mammalian cells. J Cell Sci 121:162–166 [DOI] [PubMed] [Google Scholar]
- Das SP, Borrman T, Liu VW, Yang SC, Bechhoefer J, Rhind N (2015) Replication timing is regulated by the number of MCMs loaded at origins. Genome Res 25:1886–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A, Cooley C, Garg M, Bianchi A (2014) Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep 7:53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML (1999) Replication origins in metazoan chromosomes: fact or fiction? BioEssays 21:5–16 [DOI] [PubMed] [Google Scholar]
- Depamphilis ML, de Renty CM, Ullah Z, Lee CY (2012) “The Octet”: eight protein kinases that control mammalian DNA replication. Front Physiol 3:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A, Snyder M, Sbia M, Skurnick JH, Ong LY, Newlon CS (2007) Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Mol Cell Biol 27:4652–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM (2015) Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res 25:1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N, Smith L, Thayer MJ (2015) ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet 11:e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Zhao K (2009) SV40 DNA replication: from the A gene to a nanomachine. Virology 384:352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YQ, Desprat R, Fu H, Olivier E, Lin CM, Lobell A, Gowda SN, Aladjem MI, Bouhassira EE (2006) DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet 2:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger RA (2015) Possible role of H1 histone in replication timing. Develop Growth Differ 57:1–9 [DOI] [PubMed] [Google Scholar]
- Foti R, Gnan S, Cornacchia D, Dileep V, Bulut-Karslioglu A, Diehl S, Buness A, Klein FA, Huber W, Johnstone E et al. (2016) Nuclear architecture organized by Rif1 underpins the replication-timing program. Mol Cell 61:260–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulk MS, Urban JM, Casella C, Gerbi SA (2015) Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res 25:725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkos M, Ganier O, Coulombe P, Mechali M (2015) DNA replication origin activation in space and time. Nat Rev Mol Cell Biol 16:360–374 [DOI] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP (2009) Incorporation into the prereplicative complex activates the Mcm2–7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev 23:643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Wang L, Lin CM, Singhania S, Bouhassira EE, Aladjem MI (2006) Preventing gene silencing with human replicators. Nat Biotechnol 24:572–576 [DOI] [PubMed] [Google Scholar]
- Fu H, Martin MM, Regairaz M, Huang L, You Y, Lin CM, Ryan M, Kim R, Shimura T, Pommier Y et al. (2015) The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat Commun 6:6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL (2014) The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell 53:19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S (2014) A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol Syst Biol 10:722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri S, Aggarwal V, Pontis J, Shen Z, Chakraborty A, Khan A, Mizzen C, Prasanth KV, Ait-Si-Ali S, Ha T et al. (2015). The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon SL, Li JJ (2015) Re-replication of a centromere induces chromosomal instability and aneuploidy. PLoS Genet 11:e1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan-Zadeh V, Chilaka S, Cadoret JC, Ma MK, Boggetto N, West AG, Prioleau MN (2012) USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol 10:e1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H (2012) Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26:137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Alvino GM, Chang F, Lian HY, Sridhar A, Kubota T, Brewer BJ, Weinreich M, Raghuraman MK, Donaldson AD (2014) Rif1 controls DNA replication by directing protein phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev 28:372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S et al. (2010) Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20:155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard T, Shor E, Muller CA, Nieduszynski CA, Fox CA (2013) A link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast. PLoS Genet 9:e1003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Koshland D (2003) Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev 17:1741–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM (2006) Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312:748–751 [DOI] [PubMed] [Google Scholar]
- Huang L, Fu H, Lin CM, Conner AL, Zhang Y, Aladjem MI (2011) Prevention of transcriptional silencing by a replicator-binding complex consisting of SWI/SNF, MeCP1, and hnRNP C1/C2. Mol Cell Biol 31:3472–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Matsui T, Takisawa H, Smith MM (2006) Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 26:1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Brenner J, Cuzin F (1963) On the regulation of DNA replication in bacteria. Cold Spring Harbor Dymp Quant Biol 28:329 [Google Scholar]
- Kanemaki M, Labib K (2006) Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J 25:1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh Y, Matsumoto S, Fukatsu R, Kakusho N, Kono N, Renard-Guillet C, Masuda K, Iida K, Nagasawa K, Shirahige K et al. (2015) Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat Struct Mol Biol 22:889–897 [DOI] [PubMed] [Google Scholar]
- Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, Shima N (2011) Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell 41:543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM (2012) Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148:99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Handsaker RE, Kamitaki N, Karlic R, Ghosh S, Polak P, Eggan K, McCarroll SA (2014) Genetic variation in human DNA replication timing. Cell 159:1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Schwob E (2002) The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol Cell 9:1067–1078 [DOI] [PubMed] [Google Scholar]
- Leonard AC, Mechali M (2013) DNA replication origins. Cold Spring Harb Perspect Biol 5:a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Malott M, Leffak M (2003) Multiple functional elements comprise a mammalian chromosomal replicator. Mol Cell Biol 23:1832–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lob D, Lengert N, Chagin VO, Reinhart M, Casas-Delucchi CS, Cardoso MC, Drossel B (2016) 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat Commun 7:11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majocchi S, Aritonovska E, Mermod N (2014) Epigenetic regulatory elements associate with specific histone modifications to prevent silencing of telomeric genes. Nucleic Acids Res 42:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817–823 [DOI] [PubMed] [Google Scholar]
- Marks AB, Smith OK, Aladjem MI (2016) Replication origins: determinants or consequences of nuclear organization? Curr Opin Genet Dev 37:67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, Ryan M, Kim R, Zakas AL, Fu H, Lin CM, Reinhold WC, Davis SR, Bilke S, Liu H et al. (2011) Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res 21:1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem 79:89–130 [DOI] [PubMed] [Google Scholar]
- Mattarocci S, Shyian M, Lemmens L, Damay P, Altintas DM, Shi T, Bartholomew CR, Thoma NH, Hardy CF, Shore D (2014) Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep 7:62–69 [DOI] [PubMed] [Google Scholar]
- Mechali M, Kearsey S (1984) Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 38:55–64 [DOI] [PubMed] [Google Scholar]
- Mechali M, Yoshida K, Coulombe P, Pasero P (2013) Genetic and epigenetic determinants of DNA replication origins, position and activation. Curr Opin Genet Dev 23:124–131 [DOI] [PubMed] [Google Scholar]
- Miotto B, Struhl K (2010) HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Ji Z, Struhl K (2016) Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci U S A 113:E4810–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Audit B, Klous P, Baker A, Thermes C, de Laat W, Bouvet P, Mongelard F, Arneodo A (2012) 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res 40:9470–9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Lajugie J, Fourel N, Selzer A, Schizas M, Bartholdy B, Mar J, Lin CM, Martin MM, Ryan M et al. (2014) Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet 10:e1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musialek MW, Rybaczek D (2015) Behavior of replication origins in Eukaryota – spatio-temporal dynamics of licensing and firing. Cell Cycle 14:2251–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Fraser P (2011) No-nonsense functions for long noncoding RNAs. Cell 145:178–181 [DOI] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL (2005) Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20:575–587 [DOI] [PubMed] [Google Scholar]
- Palacios DeBeer MA, Muller U, Fox CA (2003) Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev 17:1817–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peace JM, Villwock SK, Zeytounian JL, Gan Y, Aparicio OM (2016) Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res 26:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK et al. (2014) Topologically associating domains are stable units of replication-timing regulation. Nature 515:402–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159:1665–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Diffley JF (2009) Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol 21:771–777 [DOI] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM (2013a) DNA replication timing. Cold Spring Harb Perspect Med 3:1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM (2013b) DNA replication timing. Cold Spring Harb Perspect Biol 5:a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Li JJ (2014) Regulatory mechanisms that prevent re-initiation of DNA replication can be locally modulated at origins by nearby sequence elements. PLoS Genet 10:e1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Gilbert DM (2016) Replication timing and transcriptional control: beyond cause and effect-part III. Curr Opin Cell Biol 40:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ et al. (2015) Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res 25:1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Côté V et al. (2009) HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 33:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CG, Goins D, Siefert JC, Clowdus EA, Sansam CL (2015) Cyclin-dependent kinase regulates the length of S phase through TICRR/TRESLIN phosphorylation. Genes Dev 29:555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Kohler H, Oakeley EJ, Stadler MB, Schubeler D (2010) Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res 20:771–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M (2009) Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5:e1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstyuk VV, Shevchenko AI, Zakian SM (2014) Epigenetic landscape for initiation of DNA replication. Chromosoma 123:183–199 [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Kinney JB, Stillman B (2016) Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Res 26:315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith OK, Aladjem MI (2014) Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J Mol Biol 426:3330–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith OK, Kim R, Fu H, Martin MM, Lin CM, Utani K, Zhang Y, Marks AB, Lalande M, Chamberlain S et al. (2016) Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17:1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H (2013) Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol 5:a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E (2010) The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12:1086–1093 [DOI] [PubMed] [Google Scholar]
- Tazumi A, Fukuura M, Nakato R, Kishimoto A, Takenaka T, Ogawa S, Song JH, Takahashi TS, Nakagawa T, Shirahige K et al. (2012) Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev 26:2050–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C et al. (2009) Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11:1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JM, Foulk MS, Casella C, Gerbi SA (2015) The hunt for origins of DNA replication in multicellular eukaryotes. F1000prime Rep 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela MS, Chen Y, Davis S, Yang F, Walker RL, Bilke S, Lueders J, Martin MM, Aladjem MI, Massion PP et al. (2011) Preferential localization of human origins of DNA replication at the 5′-ends of expressed genes and at evolutionarily conserved DNA sequences. Plos One 6:e17308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M (2002) Histone acetylation regulates the time of replication origin firing. Mol Cell 10:1223–1233 [DOI] [PubMed] [Google Scholar]
- Wu JR, Gilbert DM (1996) A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science 271:1270–1272 [DOI] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I (2010) Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6:e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Ishii A, Kanoh Y, Oda M, Nishito Y, Masai H (2012) Rif1 regulates the replication timing domains on the human genome. EMBO J 31:3667–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445:281–285 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Fu H, Smith OK, Lin CM, Utani K, Rao M, Reinhold WC, Redon CE, Ryan M et al. (2016) A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells. Nat Commun 7:11748. [DOI] [PMC free article] [PubMed] [Google Scholar]


