Abstract
Purpose of Review
This review seeks to provide an informed prospective on the advances in molecular profiling and analysis of colorectal cancer (CRC). The goal is to provide a historical context and current summary on how advances in gene and protein sequencing technology along with computer capabilities led to our current bioinformatic advances in the field.
Recent Findings
An explosion of knowledge has occurred regarding genetic, epigenetic, and biochemical alterations associated with the evolution of colorectal cancer. This has led to the realization that CRC is a heterogeneous disease with molecular alterations often dictating natural history, response to treatment, and outcome. The consensus molecular subtypes (CMS) classification classifies CRC into four molecular subtypes with distinct biological characteristics, which may form the basis for clinical stratification and subtype-based targeted intervention.
Summary
This review summarizes new developments of a field moving “Back to the Future.” CRC molecular subtyping will better identify key subtype specific therapeutic targets and responses to therapy.
Keywords: Consensus molecular subtypes, CMS, Colorectal cancer, RNAseq
Advances Which Have Impacted the Classification of Colorectal Neoplasia
The Data Sets of Life
The central dogma is a biological principle that describes the two-step molecular information transfer involving transcription and translation [1]. This principle involves transcribing DNA information contained in genes that flows through RNA and is translated into proteins: DNA → RNA → protein. Early work on the chemical nature of the substance that transferred the ability to transform pneumococcal subtypes was identified as DNA [2]. The independent functions of DNA versus protein were discovered sometime later [3]. The distribution density and molecular ratios of nucleotide information present in DNA were then revealed [4]. It was unclear how molecular biologic information was maintained until the proposal of the double helix structure of DNA [5•]. It was not until 25 years later that the first sequencing methods emerged [6, 7]. Furthermore, there were no desktop computers to help decipher the complexities of molecular information exchange at that time, and computations were usually done on slide rules.
The informatic focus of those times was on protein. The major data exchange of that day was on 3D X-ray diffraction identification of crystallographic structures. The first of these three-dimensional model of a protein using this technology was the whale myoglobin molecule [8•]. The initial sequencing of proteins involved peptide sequencing [9•], and the first structure elucidated was the amino-acid sequence in the glycyl chain of insulin [10]. Margaret Dayhoff emerged as the founder of bioinformatics by extensively using computational methods in collaboration with Robert S. Ledley, to apply computing resources to biomedical problems that led to the development of COMPROTEIN [11, 12•, 13]. COMPROTEIN was used to elaborate protein primary structure from Edman peptide sequencing data coded on FORTRAN punch cards.
The data sets of life have expanded far beyond the central dogma [14,15] and may have even expanded into the realm of “Personomics” [16], clearly expanding the data sets taken into consideration as part of precision medicine [17•]. We now are considering epigenetic alterations such as DNA methylation defects and aberrant covalent histone modifications that occur in cancer initiation and progression. The extremely high selective pressure brought to bear during natural history of tumor formation are very dynamic. In the case of heterogenous tumors like CRC, tumor heterogeneity [18, 19, 20•, 21–23] and immunity [19, 24–27] add additional selective pressures. These epigenetic changes are likely be considered if detectable throughout the cancer continuum including early onset, progression, and ultimately metastasis, therapeutic resistance, and recurrence [19, 20•, 21, 27–31].
The launching of The Human Genome Project (HGP) on October 1, 1990 involved an international collaborative research program designed to generate a complete map all the genes of human beings. While DNA sequencing [6, 7] and other technologies were initially time consuming and costly, they have progressed over time to be much faster and less expensive [32•]. Then came the early development of RNA sequence-based methods for transcriptome characterization [33]. We now have high-throughput DNA sequencing methodology (next-generation sequencing; NGS) [32•] that has rapidly evolved over recent years with new methods that are continually emerging [34], and we are now entering the third revolution in sequencing technology [35•]. As one example of this progress, digital droplet PCR is advancing our analysis of mutated circulating tumor DNA [36, 37]. As another, single-cell sequencing is helping elaborate clonal tumor evolution and heterogeneity [38·, 39••, 40].
The Internet, World Wide Web, and Bioinformatics
Advances in molecular biology, sequencing, and computer science set the stage for modern bioinformatics. The parallel emergence of these advances is elegantly detailed elsewhere [41]. The Internet began with the concept of wide area networks (WANs) in computer science laboratories in the USA, UK, and France. This enabled the expansion beyond the local area network (LAN) that that interconnected computers within a limited physical site like a laboratory, university campus, or office building. A US Department of Defense contract helped establish Advanced Research Projects Agency Network (ARPANET), an early packet switching network that implemented the protocol suite transmission control protocol/ internet protocol (TCP/IP), forming the foundations of the Internet. This allowed end-to-end data transfer and communication across WANs. ARPANET governance was transferred to the National Science Foundation (NSFNET) and then to commercial network providers of the present day, who are connected to one or more of the network access points (NAPs).
Within this same timeframe, Tim Berners-Lee’s work at the Conseil Europe’en pour la Recherche Nucle’aire (CERN) helped create the World Wide Web as a global information exchange system for interlinked data. This made it possible for bioinformatics to advance further with the ability to exchange numerous forms of data and informatics tools [41]. These included world’s first nucleotide sequence database, the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Data Library, SWISS-PROT, and REBASE [42]. GenBank database also became the responsibility of the National Center for Biotechnology Information (NCBI) and made additional informatics tools publicly available. One of these was the rapid sequence database search tool known as basic local alignment search tool (BLAST) [43]. It was more efficient than the FASTA tool used prior to that. FASTA operated by first rapidly searching for matched data or hash sequence structure followed by applying a dynamic programming algorithm in the same sequence area [44•]. BLAST, by contrast, applied mathematical statistics and the ability to identify structure shared by sequences of high-scoring segment pairs (HSPs) [45]. Current methods using highly sophisticated algorithms and bioinformatic approaches based on NGS are now identifying genomic alterations in human somatic cells, including point mutations, chromosomal rearrangements, gene fusions, epigenetic profiles, and structural variations (SVs) that now provide us with molecular signatures helping to guide precision medicine [46–48] and personalized vaccine development [49].
Early Molecular Signature Efforts of Colorectal Cancer
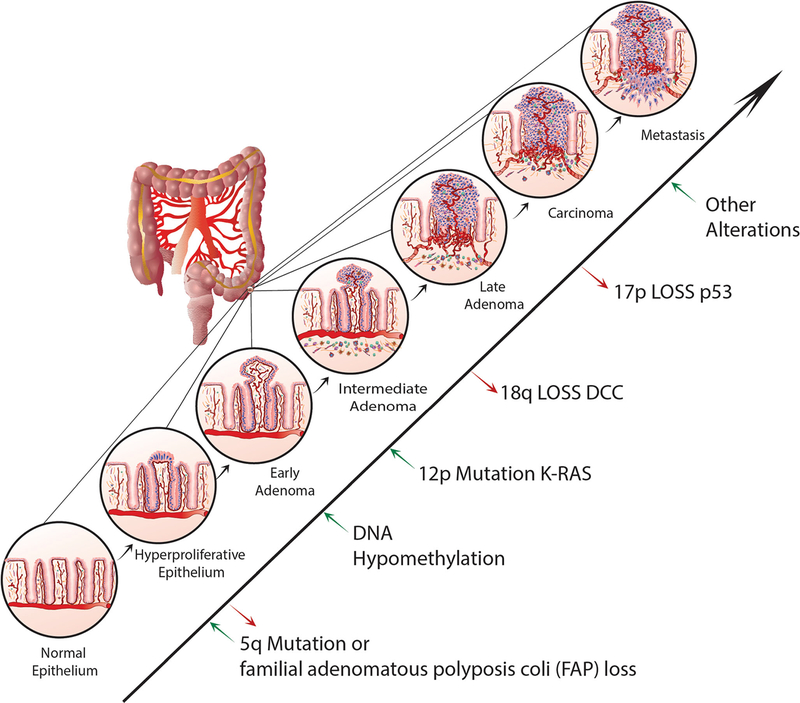
The prevalence of ras gene mutations in human colorectal cancer was first noted by Vogelstein and others [50•]. It was then revealed that the exclusion of the Deleted in Colon Cancer (DCC) gene along with the DCC locus involving a portion of chromosome 18q was accompanied by susceptibility to hereditary nonpolyposis colorectal carcinoma in a kindred analysis [51]. Subsequently, it was revealed that p53 gene mutations were involved in colorectal neoplasia through inactivation of a tumor suppressor function of the wild-type p53 gene [52, 53]. The presence and high prevalence of adenomatous polyposis coli (APC) mutation in familial polyposis coli (FAP) patients and sporadic CRC was also a key early finding [54•, 55]. These cumulative findings were accompanied by the notion of a progression of accumulated molecular lesions leading to colorectal cancer or the early “Vogelgram” [56]. Although many of these original findings hold true, a number of advances have been made with the advent of NGS and bioinformatics (Fig. 1).
Fig. 1.
A genetic model for colorectal tumorigenesis. Adapted from Fearon and Vogelstein Cell, Vol. 61, 759–767, 1990. Tumorigenesis progresses through a series of genetic alterations. These alterations include oncogenes (ras) and tumor suppressor genes (particularly those on chromosomes 5q, 17p, and 18q). Early stages involve 5q mutations or familial adenomatous polyposis coli loss. Alterations in DNA methylation may follow and then K-ras mutations. Loss of 18q or deleted in colorectal cancer (DCC) follow and then 17p loss (p53). Other alterations may precede metastasis
The Cancer Genome Atlas
Colorectal cancer was one of the many tumor types examined within The Cancer Genome Atlas (TCGA) [57]. The TCGA was established under the purview of a collaboration between the National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI). The TCGA was designed to generate comprehensive multi-dimensional data mapping and tools to examine key genomic changes in the analyzed cancer types [58]. The TCGA was scheduled to close, and new genomics initiatives are transitioning to the National Cancer Institute’s Center for Cancer Genomics (CCG).
The colorectal TCGA was designed to characterize somatic alterations in colorectal carcinoma. It was conducted on 276 CRC samples [59••]. It analyzed multiple sets of data, including exome sequence, DNA copy number, promoter methylation, messenger RNA, and microRNA expression.
Ninety-seven of these CRC samples were subjected to using low-depth-of-coverage whole-genome sequencing. Of these colorectal carcinoma samples, 16% were found to be hypermutated and three-quarters of these had high microsatellite instability associated with hypermethylation and MLH1 silencing. One quarter exhibited somatic mismatch-repair gene and polymerase ε (POLE) mutations.
With the exception of hypermutated tumors, CRC displayed consistent subtype signature patterns of genomic alteration. Significantly mutated genes included APC, TP53, SMAD4, PIK3CA, and KRAS as might be expected, along with those in SOX9, FAM123B, and ARID1A. Copy-number changes included amplifications ERBB2 and IGF2. Chromosomal translocation patterns included a fusion between NAV2 and TCF7L1. MYC-directed transcriptional activation or repression was also observed in the initial TCGA report [59••].
A number of subsequent studies identified certain molecularly similar subgroups in CRC. In one study, CRC-intrinsic deficient mismatch repair and epithelial-to-mesenchymal transition subtypes were shown to predict chemotherapy benefit [60]. In another study, CRC gene expression subtypes were identified including surface crypt-like, lower crypt-like, CIMP-H-like, mesenchymal, and mixed [61]. In a separate study, genome-wide mRNA expression subtypes were matched to pharmacologically characterized cell line panels to help determine their potential to develop targeted therapies for defined CRC patient sub-populations [62]. Additional efforts revealed three subtypes with improved disease-free survival (DFS) after surgical resection to potentially spare patients from adverse chemotherapy effects when disease was localized [63]. Within these subtypes, a filamin A expression pattern failed to respond to cetuximab, but based on a cMET subtype, receptor tyrosine kinase inhibitors were proposed for potential efficacy [63]. In separate analyses, chromosomal-instable and microsatelliteinstable cancers were among two molecular subtypes identified, while a third subtype was largely microsatellite stable and contains relatively more CpG island methylator phenotype-positive carcinomas that could not be further separated based on characteristic mutations [64]. Within the same time frame, a classification of CRC into six molecular subtypes was identified, which were hypothesized to arise through distinct biological pathways [65]. Similarly, studies that applied hierarchical clustering identified four robust tumor subtypes with biologically and clinically distinct behavior. These clusters were separated into (1) stromal components, (2) nuclear betacatenin, (3) mucinous histology, and (4) microsatelliteinstability and BRAF mutations [66]. These studies that identified molecular similarities in certain CRC subtypes were ultimately clarified within our large international consortium, formed to resolve reported subtyping inconsistencies [67••].
Consensus Molecular Subtypes (CMS Classification)
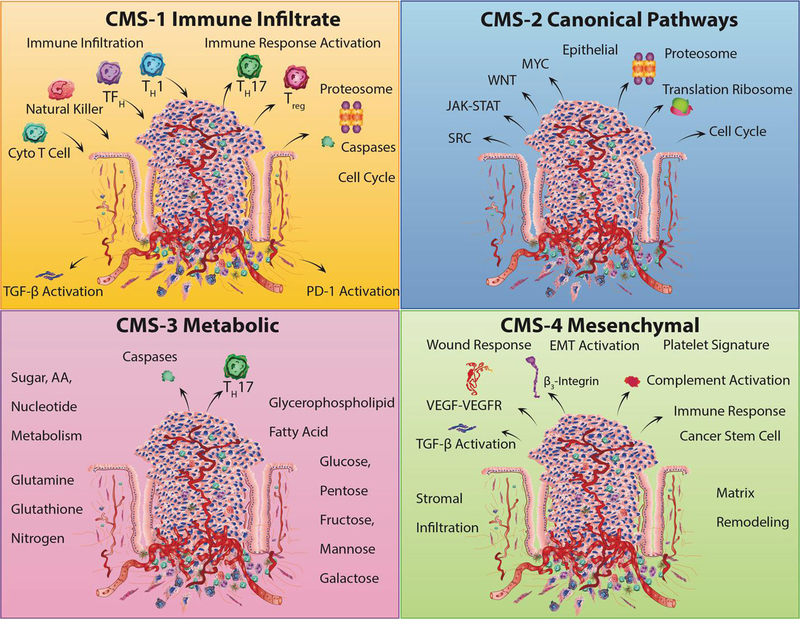
Six groups reporting gene expression-based CRC classifications formed a unique, international consortium that examined shared, large-scale data, and analytics across the expert groups [67••·]. This consortium revealed marked interconnectivity among six independent classification systems that ultimately coalesced into four consensus molecular subtypes (CMSs) with distinguishing mRNA expression along with distinct molecular and clinical features (Fig. 2). Samples were primarily Stage II and III tumors with some normal samples for comparison. Of the four subtypes, CMS2 was named the canonical subtype that made up 37% of all clusters observed and exhibited epithelial characteristics with marked WNT and MYC activated pathway signaling. CMS1 by contrast made up 14% of the categorized molecular clustering and displayed microsatellite instability along with significant immune activation and hypermutated features. The CMS3 subtype made up 13% of the molecular clustering and showed features of epithelial and metabolic dysregulation. The CMS4 subtype was found in 23% of the molecular clusters with epithelial mesenchymal transformation (EMT) characteristics accompanied by prominent stromal invasion and angiogenesis, hallmarked by transforming growth factor-β (TGF-β) activation. The remaining 13% of tumors had molecular features that were mixed and are thought to reflect a transition phenotype or intratumoral heterogeneity, typically with characteristics of multiple CMS. These CMS group classifications are considered the most robust system currently available for CRC that maintain clearly distinct molecular features connected to biological and clinical stratification, which serve as a framework for molecularly targetable interventions. Close collaborations among basic researchers, bioinformaticians, and clinicians may be critical for meeting the challenges of integrating CRC subtyping into routine clinical practice [68], with the goal of maximizing therapeutic response and minimizing adverse side effects for each patient [69].
Fig. 2.
The consensus molecular subtypes. CMS1 displays subtype clusters that involve hypermutation and microsatellite instability along with increased immune cell infiltrates consisting of Th1 lymphocyte, cytotoxic T cell, NK cell infiltration, and upregulated immune checkpoints such as PD-1. CMS2 clustering involves the upregulation of canonical pathways including WNT and MYC downstream targets. CMS3 clustering is defined by dysregulation of metabolic pathways including carbohydrate and fatty acid oxidation and the loss of TH17 cells. CMS4 clustering is referred to as a mesenchymal subtype that involves the upregulation of EMT pathways. CMS4 clustering also shows elevated TGF-β signaling, matrix remodeling, angiogenesis, complement activation as well as integrin-β3 upregulation, stromal infiltration, immune upregulation, and platelet signatures
Continued Molecularly Targeted Mutation and Biomarker Efforts
Since the initial CMS publication, there have been a number of reports focusing on biomarker study designs for individual standard molecular targets such as RAS [70–79] and BRAF [70, 76, 80–82]. Loss of CDX2 has also been paired with BRAF as a criterion for early-stage patients not qualifying for chemotherapy to be reconsidered for such treatment [83]. Separate molecular profile efforts have also focused on EGFR [84–91]. Mutation analysis has also evolved to enable tracking of circulating, free tumor DNA (ctDNA) mutations [84, 89, 92, 93]. The use of patient ctDNA has enabled ~real-time detection of acquired resistance to anti-EGFR therapy in a phase 2 randomized clinical trial [84]. Continued development of molecular tools has enabled additional characterization of ERBB2/ERBB3 in CRC, leading to identification of associations with MSI and co-occurring PIK3CA mutations [93]. These findings may inform therapeutic strategies in the setting of ERBB2/ERBB3 mutations, highlighting the potential clinical impact of molecular tool development.
Continued Consensus Molecular Subtype Efforts
Classically, CRC diagnosis and prognostic stratification are based on histopathologic assessment of cell or nuclear pleomorphism, aberrant mitotic figures, altered glandular architecture, and other phenomic abnormalities [94]. From this standpoint, complexity may involve oncogenic perturbation of spatiotemporal signaling, leading to disruption at multiple levels of tissue organization. Tumor complexity can extend to morphologic plasticity based on a single molecular signature that generates heterogeneous cancer phenotypes. In contrast, morphologically homogeneous tumors can exhibit substantive molecular diversity. When considering a signaling pathway-based or mechanistic interpretation of omics data in a setting of cancer pathology, CRC-CMS stratification is expected to provide clarity for clinical decision making. In one cohort study involving 608 patients, tumor sidedness was evaluated in relationship to CMS [95]. In this study, the prevalence of TP53, KRAS, BRAFV600, PIK3CA, SMAD4, CTNNB1, GNAS, and PTEN mutations differed by side and location. Within this context, transverse colon tumors had mutation profiles that more closely resembled left-sided tumors, suggesting that prognostically, transverse and left-sided tumors should be combined, keeping right-sided tumors separate. CMS prevalence also varied by colon location with CMS1 and CMS3 prevalence decreasing and CMS2 prevalence increasing moving from proximal to distal colon. This study also found that the sigmoid-rectal region appears unique and the transverse colon is distinct from other right-sided locations.
In another study, TGFβ signaling was observed to direct serrated adenomas to the mesenchymal colorectal cancer subtype, CMS4 [96]. This study also showed that TGFβ signaling was elevated in a genetically engineered organoid culture carrying a BRAF(V) (600E) mutation, constituting a model system for sessile serrated adenomas. In a separate study, SMAD4, a key mediator of TGFβ signaling, mutation was found to be independently associated with worse outcomes among patients undergoing resection of colorectal liver metastasis [97]. Another study was able to demonstrate that TGF-beta was a classic EMT inductor, causing upregulation of ANXA2 and internalization of both E-cadherin and ANXA2 in CRC cells [98]. This same study revealed that ANXA2 silencing was able to reduce TGF-beta-induced invasiveness, and inhibitors of the Src/ANXA2/STAT3 pathway were able to reverse EMT. Stromal involvement is significant in CMS4 and in one study of colorectal peritoneal carcinomatosis, primary tumors were found to have high stromal content and CMS4 biology [99]. The authors of this study went on to suggest that patients with colorectal peritoneal carcinomatosis may benefit from therapies targeting tumor-stroma interaction in addition to cytoreductive surgery and hyperthermic intraperitoneal chemotherapy treatments [99]. Furthermore, the additional knowledge of somatic mutations may guide the use of preoperative therapy, extent of surgical margin, and selection for ablation [100].
When examining miRNA signatures, other CMS4-related studies showed that the miR-200 family, which negatively regulates EMT, was significantly under-expressed in CMS4 tumors, revealing an additional aspect of CMS4 biology [101]. Studies on the CMS paradigm have also evaluated the influence of multiple parameters, such as the origin, developmental route, and micro environmental regulation influence on CMS [102]. One study utilized a multi-omics approach to examine 34 commonly used CRC cell lines, categorizing them into the four CMS subtypes [103•]. This study annotated CMS along with MSI, MSS, POLE, and CpG island methylator phenotype (CIMP) status in this cell line panel. Epigenetics can have additional phenotypic impact on CIMP tumors and comprise 20% of colorectal cancers, and associations have been established with female sex, age, right-sided location, and BRAF mutations [104]. In general, the morphologic appearance of cell lines in CMS1 and CMS4 was mesenchymal, whereas CMS2and CMS3 cell lines were more epithelial-like. In another study, the presence of KRAS mutations was found to be independently associated with a reduction in immune infiltrates and reactivity in CRC, but the extent of this effect varied by CMS [71]. Another study found that CMS might serve as a predictive factor for the efficacy of chemotherapy against mCRCs [105]. Gene expression analyses have also shed further light on distinct CMSs that are differentially distributed between right- and left-sided CRCs [106]. This study also revealed that greater proportions of the “microsatellite unstable/immune” CMS1 and the “metabolic” CMS3 subtypes are found in right-sided colon cancers.
Preclinical CMS Progress
Preclinically, CRC cell lines, primary cultures, and patient-derived xenografts (PDX) were examined and found robustly assigned to one of the four CMSs, independent of the stromal contribution [107]. In this same study, CMS stratification was examined by functional analyses, identifying mesenchymal enrichment in CMS4 and metabolic dysregulation in CMS3. This study also found an association with sensitivity to chemotherapy-induced apoptosis prevalent in CMS2 and CMS4, which correlated with delays in outgrowth of CMS2, but not CMS4 xenografts. A separate preclinical study found that tumor cell proliferation was associated with successful PDX establishment and was able to distinguish patients with poor clinical outcomes within CMS2 [108]. In another preclinical study, a CMS4-like inducible mouse model was also generated based on a Kras mutant allele and conditional null alleles of Apc and Trp53 (iKAP), providing a potential genetically engineered mouse model of CMS4-like CRC [74]. The advantages, disadvantages, and challenges associated with using PDXs in the identification of targets and drug testing were recently summarized in a Nature feature [109].
Improving the CMS Classifier
Other studies have sought to improve upon the CMS classifier for more refined prognosis predictions [110]. There have also been suggestions that CMS be used to guide precision treatment of CRC, necessitating a high degree of confidence in the CMS classification method [111•]. One group retrospectively evaluated CMS as a prognostic factor for stage III CRC patients treated with FOLFOX adjuvant chemotherapy, finding that CMS was predictive in these patients [112]. Another study developed an immunohistochemical-based classifier containing four specific staining profiles involving FRMD6, ZEB1, HTR2B, and CDX2 in combination with cytokeratin [113]. Based on the great potential of CMS clinically and the need for a high degree of confidence in CMS classification, other groups including ours (unpublished results) have continuing, dedicated efforts to improve the CMS classifier, such as CMScaller [114].
CRIS
A separate classifier was recently developed, focusing on a CRC intrinsic signature (CRIS), to cluster samples by patient-of-origin rather than region-of-origin [115•]. This classifier was generated only using PDX tissue, thereby limiting the data to only samples that successfully establish these xenografts from patient samples. The purpose was to emphasize the potential of cancer-cell intrinsic signatures to reliably stratify CRC patients by minimizing the confounding effects of stromal-derived intratumoral heterogeneity (ITH). This study involved 75 RNA transcription profiles, 25 patients’ samples at three regions per sample, namely the invasive front (IF), central tumor (CT), and lymph node (LN). The CRIS were determined using the nearest template prediction (NTP) classifier. Cell-type-specific signatures included epithelial, leukocyte, fibroblast, and endothelial cells. CRIS concordance was broken down into CRIS. A through E and patient clustering for CT and IF was at 92% (22/24 samples). In another study by the same group that compared CMS to CRIS using multiple sampling method approaches, the authors concluded that CRIS provides more spatially and temporally robust classification of molecular subtypes compared to CMS [116]. This group combined CRIS transcriptional subtyping and CD8 immunohistochemistry to identify poor prognosis stage II/III colorectal cancer patients who were able to benefit from adjuvant chemotherapy [117].
Immune-Related Consensus Molecular Subtype
Prior to the CMS report, an immune signature called the co-ordinate immune response cluster (CIRC) was proposed involving 28 genes coordinately regulated across CRC patients [118]. Four patient groups were identified by this method. Group A was heavily enriched for patients with microsatellite instability (MSI-H) and POL mutations, and had high CIRC expression, including several inhibitory molecules: CTLA4, PDL1, PDL2, LAG3, and TIM3. RAS mutation by contrast was enriched in patient groups with lower CIRC expression. RAS mutant tumors predicted a relatively poor immune infiltration and low inhibitory molecule expression.
In the case of CMS-directed studies, by contrast, stratified analyses revealed that chemokine-like factor (CKLF) was a potential prognostic marker in the MSI-immune consensus molecular subtype CMS1 [119]. Heterogeneity in immune function in relation to CMS has also been examined in a study of CRC genome-wide expression datasets, including 1597 tumors and 125 adjacent normal colon tissues [120•]. CRC clusters were identified using a combination of multiple clustering algorithms and multiple validity metrics. The CIBERSORT algorithm was used to compute relative proportions of 22 human leukocyte subpopulations across CRC and normal colon tissue, identifying five clusters of tumor immune infiltrate (COMMUNAL clusters). Distribution of these clusters was then assessed by CMS, finding that four of the clusters overlapped significantly with the four CMSs. Additional analysis identifying differential expression specific to tumor epithelial cells was able to characterize mechanisms of tumor escape from immune surveillance occurring in particular CRC clusters. Common and cluster-specific influx of immune cells into CRCs was found along with several deregulated gene targets to help improve of immunotherapeutic strategies in CRC. In a separate, prospective cohort of 1265 patients with stage II/III cancer, TIL/MMR status and BRAF/KRAS mutations were examined along with CMS status on 142 cases [121]. These authors identified that associations with 5-year disease-free survival (DFS), which were evaluated and validated in an independent cohort of 602 patients and concluded that TIL/MMR subtyping was superior compared with histopathological, genomic, and transcriptomic subtypes [121]. Platelets are derived from immune lineage mega-karyocytes and may also impact tumor CMS [122].
CMS and the Microbiome
There have also been reports focused on the microbiome patterns that are associated with CMS [123•]. In this study, CRC subtypes were identified in 34 samples using RNA-sequencing-derived gene expression in concert with the relative abundances of bacterial taxonomic groups using 16S rRNA amplicon metabarcoding. 16S rRNA analysis revealed the enrichment of Fusobacteria and Bacteroidetes, and decreased levels of Firmicutes and Proteobacteria in CMS1. Further analysis of bacterial taxa focused on non-human RNA-sequencing reads uncovered distinct bacterial communities associated with each molecular subtype. The most highly enriched species associated with CMS1 included Fusobacterium hwasookii and Porphyromonas gingivalis. CMS2 was enriched for Selenomas and Prevotella species, while CMS3 had few significant associations. Targeted quantitative PCR validation has also been done, showing an elevation of Fusobacterium nucleatum, Parvimonas micra, and Peptostreptococcus stomatis in CMS1. These results reflected that Fusobacterium was associated with a CRC subtype characterized by CpG island methylation, MSI and inflammatory signatures, and higher prevalence in right-sided tumors. These authors also noted the concept of bacterial biofilms as initiators of CRC that may facilitate microbial invasion of the mucous layer [124].
CMS-Related Drug Development and Treatment: “Back to the Future”
Revealing that there are multiple molecular subtypes in a sense is a game changer for the CRC field. If one considers drug development for example, the assumption that this was a single disease that progressed through a common series of molecular steps or accumulation of mutations is rapidly changing. Take the four CMS described to date for the sake of argument. Recall that we now know that there are four CMSs with identifiable features that separate CMS1 (MSI immune, 14%), hypermutated, microsatellite unstable, and strong immune activation; CMS2 (canonical, 37%), epithelial, marked WNT, and MYC signaling activation; CMS3 (meta-bolic, 13%), epithelial, and evident metabolic dysregulation; and CMS4 (mesenchymal, 23%), prominent TGF-β activation, stromal invasion, and angiogenesis. Suppose we have a drug targeting the metabolic pathways inherent to CMS3 that lead to a response rate of 50% in this subgroup and greatly enhanced progression free and overall survival, this would be considered an unmitigated success. However, absent knowledge of the CMS, had this drug been tried on the entire CRC population, may only have shown a 6.5% response rate assuming that its effects were CMS3 specific, and this drug would likely have been discarded as ineffective for CRC. The identification of molecularly distinct subtypes can provide distinct subsets of patients on which to test new treatments as well as clues for specific biological pathways to target for these subgroups. In a sense, we seem to find our-selves at a “Back to the Future” crossroads that could be expected to result in the reevaluation of many drugs in CMS-related cell lines or PDX models based on molecular changes of a given CMS in response to drug. This would include assessments prior to, following, and in resistance to treatment settings. The same would seem to apply to tumor biomarker and circulating biomarker analysis and assay development. This potential future would enable us to stratify our treatments based on CMS-related analyses, not exposing CRC patients to toxic or targeted drugs from which they will derive no benefit. Such a future would have a higher probability of developing more precise treatments. A future where NGS and other advanced technologies combine with bioinformatics to serve as “Doc’s” time traveling DeLorean. A future where the McFly’s time continuum is selected for the betterment of the CRC field…and Biff’s? ….well you know.
Funding Information
This study was supported by Colorectal Cancer Moon Shot, Duncan Family Institute for Cancer Prevention and Risk Assessment, 1R01CA187238-01, 5R01CA172670-03 and 1R01CA184843-01A1, CA177909, and Cancer Center Support Grant (P30 CA016672).
Abbreviations
- CMS
consensus molecular subtype
- MSI
microsatellite instability
- NGS
bioinformatics, biostatistics, colorectal cancer, targeted therapy, precision medicine
Footnotes
Conflict of Interest The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
Publisher’s Note Springer Nature remains neutral with regard to juris-dictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Crick F Central dogma of molecular biology. Nature. 1970;227(5258):561–3. [DOI] [PubMed] [Google Scholar]
- 2.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79(2):137–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952;36(1):39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamm C, Shapiro HS, Lipshitz R, Chargaff E. Distribution density of nucleotides within a desoxyribonucleic acid chain. J Biol Chem. 1953;203(2):673–88. [PubMed] [Google Scholar]
- 5.•.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356): 737–8. [DOI] [PubMed] [Google Scholar]; The structure of DNA follows that of protein.
- 6.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94(3):441–8. [DOI] [PubMed] [Google Scholar]
- 7.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. [DOI] [PubMed] [Google Scholar]
- 8.•.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181(4610):662–6. [DOI] [PubMed] [Google Scholar]; Bioinformatics beginnings in protein structural analysis.
- 9.•.Edman P A method for the determination of amino acid sequence in peptides. Arch Biochem. 1949;22(3):475. [PubMed] [Google Scholar]; Beginnings of amino acid sequence determination.
- 10.Sanger F, Thompson EO. The amino-acid sequence in the glycyl chain of insulin. I. The identification of lower peptides from partial hydrolysates. Biochem J. 1953;53(3):353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dayhoff MO. Computer aids to protein sequence determination. J TheorBiol. 1965;8(1):97–112. [DOI] [PubMed] [Google Scholar]
- 12.•.Dayhoff MO, Ledley RS. Comprotein: a computer program to aid primary protein structure determination In: Proceedings of the December 4–6, 1962, fall joint computer conference. New York, NY: ACM; 1962. p. 262–74. [Google Scholar]; This is the first computer aided analysis tool that helped establish bionformatic type analyses.
- 13.Ledley RS. Digital electronic computers in biomedical science. Science. 1959;130(3384):1225–34. [DOI] [PubMed] [Google Scholar]
- 14.Zhou W, Dinh HQ, Ramjan Z, Weisenberger DJ, Nicolet CM, Shen H, et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet. 2018;50(4):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesan A Epigenetics: the first 25 centuries. Philos Trans R Soc Lond Ser B Biol Sci. 2018;373(1748):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziegelstein RC. Personomics and precision medicine. Trans Am Clin Climatol Assoc. 2017;128:160–8. [PMC free article] [PubMed] [Google Scholar]
- 17.•.Werner RJ, Kelly AD, Issa JJ. Epigenetics and precision oncology. Cancer J. 2017;23(5):262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an informative treatment of epigenetics in precision medicine.
- 18.Wang Q, Wang G, Liu C, He X. Prognostic value of CpG island methylator phenotype among hepatocellular carcinoma patients: a systematic review and meta-analysis. Int J Surg. 2018;54(Pt a): 92–9. [DOI] [PubMed] [Google Scholar]
- 19.James de Bony E, Bizet M, Van Grembergen O, Hassabi B, Calonne E, Putmans P, et al. Comprehensive identification of long noncoding RNAs in colorectal cancer. Oncotarget. 2018;9(45): 27605–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.•.Mamlouk S, Childs LH, Aust D, Heim D, Melching F, Oliveira C, et al. DNA copy number changes define spatial patterns of heterogeneity in colorectal cancer. Nat Commun. 2017;8:14093. [DOI] [PMC free article] [PubMed] [Google Scholar]; Molecular changes assoiated with tumor heterogeneity.
- 21.Dunne PD, Lawler M. Intratumoural epigenetic heterogeneity in early invasive colorectal cancer: a prognostic imprint? Gastroenterology. 2017;152(6):1622–3. [DOI] [PubMed] [Google Scholar]
- 22.Binder H, Hopp L, Schweiger MR, Hoffmann S, Juhling F, Kerick M, et al. Genomic and transcriptomic heterogeneity of colorectal tumours arising in lynch syndrome. J Pathol. 2017;243(2):242–54. [DOI] [PubMed] [Google Scholar]
- 23.Ogino S, Nishihara R, VanderWeele TJ, Wang M, Nishi A, Lochhead P, et al. Review article: the role of molecular pathological epidemiology in the study of neoplastic and non-neoplastic diseases in the era of precision medicine. Epidemiology. 2016;27(4):602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu XS, Zhao Y, Sun Y, Zhong L, Cheng Y, Zhang Y, et al. The epigenetic modifier PBRM1 restricts the basal activity of the innate immune system by repressing retinoic acid-inducible gene-I-like receptor signalling and is a potential prognostic biomarker for colon cancer. J Pathol. 2018;244(1):36–48. [DOI] [PubMed] [Google Scholar]
- 25.Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA Jr, et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut. 2018;67(6): 1168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamada T, Soong TR, Masugi Y, Kosumi K, Nowak JA, da Silva A, et al. TIME (tumor immunity in the MicroEnvironment) classification based on tumor CD274 (PD-L1) expression status and tumor-infiltrating lymphocytes in colorectal carcinomas. Oncoimmunology. 2018;7(7):e1442999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zewde M, Kiyotani K, Park JH, Fang H, Yap KL, Yew PY, et al. The era of immunogenomics/immunopharmacogenomics. J Hum Genet. 2018;63(8):865–75. [DOI] [PubMed] [Google Scholar]
- 28.Pinto D, Pinto C, Guerra J, Pinheiro M, Santos R, Vedeld HM, et al. Contribution of MLH1 constitutional methylation for lynch syndrome diagnosis in patients with tumor MLH1 downregulation. Cancer Med. 2018;7(2):433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, et al. Colorectal cancer screening: recommendations for physicians and patients from the U.S. multi-society task force on colorectal cancer. Gastroenterology. 2017;153(1):307–23. [DOI] [PubMed] [Google Scholar]
- 30.Verma M The role of epigenomics in the study of cancer biomarkers and in the development of diagnostic tools. Adv Exp Med Biol. 2015;867:59–80. [DOI] [PubMed] [Google Scholar]
- 31.Zeng H, He B, Yi C, Peng J. Liquid biopsies: DNA methylation analyses in circulating cell-free DNA. J Genet Genomics. 2018;45(4):185–92. [DOI] [PubMed] [Google Scholar]
- 32.•.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30(9):418–26. [DOI] [PubMed] [Google Scholar]; An examination of next generation sequencing.
- 33.Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. BioTechniques. 2008;45(1):81–94. [DOI] [PubMed] [Google Scholar]
- 34.Slatko BE, Gardner AF, Ausubel FM. Overview of next-generation sequencing technologies. Curr Protoc Mol Biol. 2018;122(1):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.•.van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34(9): 666–81. [DOI] [PubMed] [Google Scholar]; Highlighting our thrid generation of sequencing technology.
- 36.Mehrotra M, Singh RR, Loghavi S, Duose DY, Barkoh BA, Behrens C, et al. Detection of somatic mutations in cell-free DNA in plasma and correlation with overall survival in patients with solid tumors. Oncotarget. 2018;9(12):10259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohrmann L, Huang HJ, Hong DS, Tsimberidou AM, Fu S, Piha-Paul SA, et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin Cancer Res. 2018;24(1): 181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.•.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tumor evolution revealed by single cell squencing technology.
- 39.••.Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, et al. Punctuated copy number evolution and clonal stasis in triplenegative breast cancer. Nat Genet. 2016;48(10):1119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]; Punctuated evolution of cancer.
- 40.Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, et al. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 2017;27(8):1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gauthier J, Vincent AT, Charette SJ, Derome N. A brief history of bioinformatics. Brief Bioinform. 2018:1–16. [DOI] [PubMed] [Google Scholar]
- 42.Rice CM, Fuchs R, Higgins DG, Stoehr PJ, Cameron GN. The EMBL data library. Nucleic Acids Res. 1993;21(13):2967–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. [DOI] [PubMed] [Google Scholar]
- 44.•.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227(4693):1435–41. [DOI] [PubMed] [Google Scholar]; Bioinformatics tools begin to evolve.
- 45.Kanehisa M, Bork P. Bioinformatics in the post-sequence era. Nat Genet. 2003;33(Suppl):305–10. [DOI] [PubMed] [Google Scholar]
- 46.Yi K, Ju YS. Patterns and mechanisms of structural variations in human cancer. Exp Mol Med. 2018;50(8):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smyth E, Khan K, Valeri N. Translational research and application of basic biology to clinical trial development in GI cancers. Ann TranslMed. 2018;6(9):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nuhn P, De Bono JS, Fizazi K, Freedland SJ, Grilli M, Kantoff PW, et al. Update on systemic prostate Cancer therapies: management of metastatic castration-resistant prostate cancer in the era of precision oncology. EurUrol. 2019;75:88–99. [DOI] [PubMed] [Google Scholar]
- 49.Terbuch A, Lopez J. Next generation Cancer vaccines-make it personal! Vaccines (Basel). 2018;6(3):52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.•.Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327(6120):293–7. [DOI] [PubMed] [Google Scholar]; The importance of ras gene mutations in colorectal cancer.
- 51.Peltomaki P, Sistonen P, Mecklin JP, Pylkkanen L, Jarvinen H, Simons JW, et al. Evidence supporting exclusion of the DCC gene and a portion of chromosome 18q as the locus for susceptibility to hereditary nonpolyposis colorectal carcinoma in five kindreds. Cancer Res. 1991;51(16):4135–40. [PubMed] [Google Scholar]
- 52.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. [DOI] [PubMed] [Google Scholar]
- 53.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244(4901):217–21. [DOI] [PubMed] [Google Scholar]
- 54.•.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359(6392):235–7. [DOI] [PubMed] [Google Scholar]; APC mutations found in colorectal cancer.
- 55.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253(5020):661–5. [DOI] [PubMed] [Google Scholar]
- 56.•.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–67. [DOI] [PubMed] [Google Scholar]; The Vogelgram.
- 57.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296(3):50–7. [PubMed] [Google Scholar]
- 58.Lee H, Palm J, Grimes SM, Ji HP. The cancer genome atlas clinical explorer: a web and mobile interface for identifying clinical-genomic driver associations. Genome Med. 2015;7:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.••.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407): 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; The cancer genome atlas for human colon cancer.
- 60.Roepman P, Schlicker A, Tabernero J, Majewski I, Tian S, Moreno V, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231(1):63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlicker A, Beran G, Chresta CM, McWalter G, Pritchard A, Weston S, et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genet. 2012;5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–8. [DOI] [PubMed] [Google Scholar]
- 65.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez-Villamil B, Romera-Lopez A, Hernandez-Prieto S, Lopez-Campos G, Calles A, Lopez-Asenjo JA, et al. Colon cancer molecular subtypes identified by expression profiling and associated to stroma, mucinous type and different clinical behavior. BMC Cancer. 2012;12:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.••.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Consensus molecular subtypes of colorectal cancer were revealed.
- 68.Wang W, Kandimalla R, Huang H, Zhu L, Li Y, Gao F, et al. Molecular subtyping of colorectal cancer: recent progress, new challenges and emerging opportunities. Semin Cancer Biol. 2018:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wlodarczyk M, Wlodarczyk J, Siwinski P, Fichna J, Sobolewska-Wlodarczyk A. Genetic molecular subtypes in optimizing personalized adjuvant therapy in metastatic colorectal cancer. Curr Drug Targets. 2018;19:1731–7. [DOI] [PubMed] [Google Scholar]
- 70.Smeby J, Sveen A, Merok MA, Danielsen SA, Eilertsen IA, Guren MG, et al. CMS-dependent prognostic impact of KRAS and BRAFV600E mutations in primary colorectal cancer. Ann Oncol. 2018;29(5):1227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lal N, White BS, Goussous G, Pickles O, Mason MJ, Beggs AD, et al. KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin Cancer Res. 2018;24(1):224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Passot G, Denbo JW, Yamashita S, Kopetz SE, Chun YS, Maru D, et al. Is hepatectomy justified for patients with RAS mutant colorectal liver metastases? An analysis of 524 patients undergoing curative liver resection. Surgery. 2017;161(2):332–40. [DOI] [PubMed] [Google Scholar]
- 73.Chun YS, Passot G, Yamashita S, Nusrat M, Katsonis P, Loree JM, et al. Deleterious effect of RAS and evolutionary high-risk TP53 double mutation in colorectal liver metastases. Ann Surg. 2017;1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017;31(4):370–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee MS, Helms TL, Feng N, Gay J, Chang QE, Tian F, et al. Efficacy of the combination of MEK and CDK4/6 inhibitors in vitro and in vivo in KRAS mutant colorectal cancer models. Oncotarget. 2016;7(26):39595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Al-Shamsi HO, Jones J, Fahmawi Y, Dahbour I, Tabash A, Abdel-Wahab R, et al. Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC somatic gene mutations in Arab patients with colorectal cancer: determination of frequency and distribution pattern. J Gastrointest Oncol. 2016;7(6):882–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zimmitti G, Shindoh J, Mise Y, Kopetz S, Loyer EM, Andreou A, et al. RAS mutations predict radiologic and pathologic response in patients treated with chemotherapy before resection of colorectal liver metastases. Ann Surg Oncol. 2015;22(3):834–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira AA, Rego JF, Morris V, Overman MJ, Eng C, Garrett CR, et al. Association between KRAS mutation and lung metastasis in advanced colorectal cancer. Br J Cancer. 2015;112(3):424–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hecht JR, Douillard JY, Schwartzberg L, Grothey A, Kopetz S, Rong A, et al. Extended RAS analysis for anti-epidermal growth factor therapy in patients with metastatic colorectal cancer. Cancer Treat Rev. 2015;41(8):653–9. [DOI] [PubMed] [Google Scholar]
- 80.Jones JC, Renfro LA, Al-Shamsi HO, Schrock AB, Rankin A, Zhang BY, et al. (Non-V600) BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J Clin Oncol. 2017;35(23):2624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cohen R, Svrcek M, Dreyer C, Cervera P, Duval A, Pocard M, et al. New therapeutic opportunities based on DNA mismatch repair and BRAF status in metastatic colorectal cancer. Curr Oncol Rep. 2016;18(3):18. [DOI] [PubMed] [Google Scholar]
- 82.Kopetz S, Desai J, Chan E, Hecht JR, O’Dwyer PJ, Maru D, et al. Phase II pilot study of Vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J Clin Oncol. 2015;33(34): 4032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bruun J, Sveen A, Barros R, Eide PW, Eilertsen I, Kolberg M, et al. Prognostic, predictive, and pharmacogenomic assessments of CDX2 refine stratification of colorectal cancer. Mol Oncol. 2018;12:1639–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Montagut C, Argiles G, Ciardiello F, Poulsen TT, Dienstmann R, Kragh M, et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses: a phase 2 randomized clinical trial. JAMA Oncol. 2018;4(4): e175245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parseghian CM, Parikh NU, Wu JY, Jiang ZQ, Henderson L, Tian F, et al. Dual inhibition of EGFR and c-Src by Cetuximab and Dasatinib combined with FOLFOX chemotherapy in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23(15):4146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Korphaisarn K, Loree JM, Nguyen V, Coulson R, Holla V, Litzenburger BC, et al. Genomic analysis of exceptional responder to regorafenib in treatment-refractory metastatic rectal cancer: a case report and review of the literature. Oncotarget. 2017;8(34): 57882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yiu AJ, Yiu CY. Biomarkers in colorectal cancer. Anticancer Res. 2016;36(3):1093–102. [PubMed] [Google Scholar]
- 88.Sinicrope FA, Okamoto K, Kasi PM, Kawakami H. Molecular biomarkers in the personalized treatment of colorectal cancer. Clin Gastroenterol Hepatol. 2016;14(5):651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morelli MP, Overman MJ, Dasari A, Kazmi SM, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26(4):731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu X, George GC, Tsimberidou AM, Naing A, Wheler JJ, Kopetz S, et al. Retreatment with anti-EGFR based therapies in metastatic colorectal cancer: impact of intervening time interval and prior anti-EGFR response. BMC Cancer. 2015;15:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee MS, Kopetz S. Current and future approaches to target the epidermal growth factor receptor and its downstream signaling in metastatic colorectal cancer. Clin Colorectal Cancer. 2015;14(4): 203–18. [DOI] [PubMed] [Google Scholar]
- 92.Mehrotra M, Singh RR, Chen W, Huang RSP, Almohammedsalim AA, Barkoh BA, et al. Study of preanalytic and analytic variables for clinical next-generation sequencing of circulating cell-free nucleic acid. JMol Diagn. 2017;19(4):514–24. [DOI] [PubMed] [Google Scholar]
- 93.Loree JM, Bailey AM, Johnson AM, Yu Y, Wu W, Bristow CA, et al. Molecular landscape of ERBB2/ERBB3 mutated colorectal cancer. J Natl Cancer Inst. 2018;110:1409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Campbell FC, Loughrey MB, McClements J, Deevi RK, Javadi A, Rainey L. Mechanistic insights into colorectal cancer phenomics from fundamental and organotypic model studies. Am J Pathol. 2018;188(9):1936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loree JM, Pereira AAL, Lam M, Willauer AN, Raghav K, Dasari A, et al. Classifying colorectal cancer by tumor location rather than sidedness highlights a continuum in mutation profiles and consensus molecular subtypes. Clin Cancer Res. 2018;24(5): 1062–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fessler E, Drost J, van Hooff SR, Linnekamp JF, Wang X, Jansen M, et al. TGFbeta signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med. 2016;8(7): 745–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mizuno T, Cloyd JM, Vicente D, Omichi K, Chun YS, Kopetz SE, et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol. 2018;44(5):684–92. [DOI] [PubMed] [Google Scholar]
- 98.Rocha MR, Barcellos-de-Souza P, Sousa-Squiavinato ACM, Fernandes PV, de Oliveira IM, Boroni M, et al. Annexin A2 over-expression associates with colorectal cancer invasiveness and TGF-ss induced epithelial mesenchymal transition via Src/ANXA2/STAT3. Sci Rep. 2018;8(1):11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ubink I, van Eden WJ, Snaebjornsson P, Kok NFM, van Kuik J, van Grevenstein WMU, et al. Histopathological and molecular classification of colorectal cancer and corresponding peritoneal metastases. Br J Surg. 2018;105(2):e204–11. [DOI] [PubMed] [Google Scholar]
- 100.Yamashita S, Chun YS, Kopetz SE, Vauthey JN. Biomarkers in colorectal liver metastases. Br J Surg. 2018;105(6):618–27. [DOI] [PubMed] [Google Scholar]
- 101.Fessler E, Jansen M, De Sousa EMF, Zhao L, Prasetyanti PR, Rodermond H, et al. A multidimensional network approach reveals microRNAs as determinants of the mesenchymal colorectal cancer subtype. Oncogene. 2016;35(46):6026–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fessler E, MedemaJP. Colorectal cancer subtypes: developmental origin and microenvironmental regulation. Trends Cancer. 2016;2(9):505–18. [DOI] [PubMed] [Google Scholar]
- 103.•.Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, et al. Multi-omics of 34 colorectal cancer cell lines—a resource for biomedical studies. Mol Cancer. 2017;16(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Consensus molecular subtypes revealed in colorectal cancer cell lines.
- 104.Advani SM, Advani P, DeSantis SM, Brown D, VonVille HM, Lam M, et al. Clinical, pathological, and molecular characteristics of CpG island methylator phenotype in colorectal cancer: a systematic review and meta-analysis. Transl Oncol. 2018;11(5): 1188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Okita A, Takahashi S, Ouchi K, Inoue M, Watanabe M, Endo M, et al. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9(27):18698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee MS, Menter DG, Kopetz S. Right versus left colon cancer biology: integrating the consensus molecular subtypes. J Natl Compr Cancer Netw. 2017;15(3):411–9. [DOI] [PubMed] [Google Scholar]
- 107.Linnekamp JF, Hooff SRV, Prasetyanti PR, Kandimalla R, Buikhuisen JY, Fessler E, et al. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ. 2018;25(3):616–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prasetyanti PR, van Hooff SR, van Herwaarden T, de Vries N, Kalloe K, Rodermond H, et al. Capturing colorectal cancer inter-tumor heterogeneity in patient-derived xenograft (PDX) models. Int J Cancer. 2019;144:366–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Willyard C The mice with human tumours: growing pains for a popular cancer model. Nature. 2018;560(7717):156–7. [DOI] [PubMed] [Google Scholar]
- 110.Bramsen JB, Rasmussen MH, Ongen H, Mattesen TB, Orntoft MW, Arnadottir SS, et al. Molecular-subtype-specific biomarkers improve prediction of prognosis in colorectal cancer. Cell Rep. 2017;19(6):1268–80. [DOI] [PubMed] [Google Scholar]
- 111.•.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17(4):268. [DOI] [PubMed] [Google Scholar]; Consensus molecular subtypes in precision medicine.
- 112.Kwon Y, Park M, Jang M, Yun S, Kim WK, Kim S, et al. Prognosis of stage III colorectal carcinomas with FOLFOX adjuvant chemotherapy can be predicted by molecular subtype. Oncotarget. 2017;8(24):39367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ten Hoorn S, Trinh A, de Jong J, Koens L, Vermeulen L. Classification of colorectal cancer in molecular subtypes by immunohistochemistry. Methods Mol Biol. 2018;1765:179–91. [DOI] [PubMed] [Google Scholar]
- 114.Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer preclinical models. Sci Rep. 2017;7(1):16618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.•.Dunne PD, Alderdice M, O’Reilly PG, Roddy AC, McCorry AMB, Richman S, et al. Cancer-cell intrinsic gene expression signatures overcome intratumoural heterogeneity bias in colorectal cancer patient classification. Nat Commun. 2017;8:15657. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cancer intrinsic gene expression signatures found in colorectal cancer.
- 116.Alderdice M, Richman SD, Gollins S, Stewart JP, Hurt C, Adams R, et al. Prospective patient stratification into robust cancer-cell intrinsic subtypes from colorectal cancer biopsies. J Pathol. 2018;245(1):19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Allen WL, Dunne PD, McDade S, Scanlon E, Loughrey M, Coleman H, et al. Transcriptional subtyping and CD8 immunohistochemistry identifies poor prognosis stage II/III colorectal cancer patients who benefit from adjuvant chemotherapy. JCO Precis Oncol. 2018:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lal N, Beggs AD, Willcox BE, Middleton GW. An immunogenomic stratification of colorectal cancer: implications for development of targeted immunotherapy. Oncoimmunology. 2015;4(3):e976052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dunne PD, O’Reilly PG, Coleman HG, Gray RT, Longley DB, Johnston PG, et al. Stratified analysis reveals chemokine-like factor (CKLF) as a potential prognostic marker in the MSI-immune consensus molecular subtype CMS1 of colorectal cancer. Oncotarget. 2016;7(24):36632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.•.Karpinski P, Rossowska J, Sasiadek MM. Immunological landscape of consensus clusters in colorectal cancer. Oncotarget. 2017;8(62):105299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]; Consensus immunologic clusters in colorectal cancer.
- 121.Williams DS, Mouradov D, Jorissen RN, Newman MR, Amini E, Nickless DK, et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2018:1–10. [DOI] [PubMed] [Google Scholar]
- 122.Lam M, Roszik J, Kanikarla-Marie P, Davis JS, Morris J, Kopetz S, et al. The potential role of platelets in the consensus molecular subtypes of colorectal cancer. Cancer Metastasis Rev. 2017;36(2): 273–88. [DOI] [PubMed] [Google Scholar]
- 123.•.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7(1):11590. [DOI] [PMC free article] [PubMed] [Google Scholar]; Microbiome associated with consensus molecular subtypes.
- 124.Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014;111(51):18321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]