Abstract
Era (Escherichia coli Ras-like protein), essential for bacterial cell viability, is composed of an N-terminal GTPase domain and a C-terminal KH domain. In bacteria, it is required for the processing of 16S ribosomal RNA (rRNA) and maturation of 30S (small) ribosomal subunit. Era recognizes 10 nucleotides (1530GAUCACCUCC1539) near the 3’ end of 16S rRNA and interacts with helix 45 (h45, nucleotides 1506–1529). GTP binding enables Era to bind RNA, RNA binding stimulates Era’s GTP-hydrolyzing activity, and GTP hydrolysis releases Era from matured 30S ribosomal subunit. As such, Era controls cell growth rate via regulating the maturation of the 30S ribosomal subunit. Ribosomes manufacture proteins in all living organisms. The GAUCA sequence and h45 are highly conserved in all three kingdoms of life. Homologues of Era are present in eukaryotic cells. Hence, the mechanism of bacterial Era action also sheds light on the cell cycle control of eukaryotes.
Keywords: 16S rRNA, 30S ribosomal subunit, ribosome biogenesis, cell cycle control
INTRODUCTION
I am interested in the structure and function of RNA-processing proteins involved in post-transcriptional gene expression control. This interest was developed when I was studying toward my Ph.D. degree at the University of Oklahoma (1985–1990). Joining the National Cancer Institute (NCI) at Frederick in 1995, I started working on several RNA-processing proteins, including Era (Escherichia coli Ras-like protein). Dr. Alexander Wlodawer is my mentor, supervisor, and also a friend. I am always grateful for his advice, encouragement, and friendship. Alex turns 70 this year. Turning 70 is a major milestone, which I believe is joyous for him as a renowned crystallographer in the world.
Era is the first small GTPase found in bacteria [1]; it is highly conserved [2–5]. Essential for bacterial cell viability, Era plays important roles in cell cycle regulation by coupling cell growth rate with cytokinesis [6,7]. Limiting Era expression or impairing Era’s GTPase activity in E. coli resulted in cells with two or four nucleoids arrested in the cell cycle just before cytokinesis [7,8].
Bacterial ribosome is composed of a large (50S) and a small (30S) ribosomal subunit. The 30S ribosomal subunit contains 16S ribosomal RNA (rRNA) (1542 nucleotides in E. coli) and 21 ribosomal proteins. Era is an RNA-processing protein, required for the processing and maturation of 16S rRNA [9,10]. It has been shown in vitro that Era binds to pre-30S particles at an early stage of assembly, promotes the binding of several late-binding ribosomal proteins to pre-30S, and remains bound to pre-30S throughout several RNA folding and protein binding events [11]. Matured 30S ribosomal subunit contains fully processed 16S rRNA. Required for the processing and maturation of 16S rRNA, Era is naturally essential for the maturation of 30S ribosomal subunit. This review provides a structural view for the functional cycle of Era in the processing of 16S rRNA and maturation of 30S ribosomal subunit.
TWO-DOMAIN ARCHITECTURE
Crystal structures of Era have been determined in complex with a Mg2+ ion, a GNP (non-hydrolyzable GTP analog GDPNP) molecule, and either the 1531AUCACCUCCUUA1542 sequence (Era:MgGNP:RNA, PDB entry 3IEV) [12] or a longer RNA consisted of helix 45 (h45, residues 1506–1529) and the 1530GAUCACCUCCUUA1542 sequence, (Era:MgGNP:RNA301, PDB entry 3R9W; Era:MgGNP:RNA301:KsgA, PDB entry 3R9X) [13]. The RNA (1531–1542) and RNA301 (1506–1542) sequences are taken from A. aeolicus 16S rRNA with original U1506 replaced with a Cyt. The numbering of E. coli 16S rRNA is used for the ease of discussion. KsgA is a universally conserved RNA methyltransferase [14]. It methylates A1518 and A1519 in the tetraloop of h45. These structures show that Era recognizes 10 nucleotides (1530GAUCACCUCC1539) near the 3’ end of 16S rRNA and that Era also interacts with h45.
The Era:MgGNP:RNA301:KsgA structure (PDB entry 3R9X) [13] is illustrated in Figure 1A, showing the Era:MgGNP:RNA301 complex only. The Era molecule has a two-domain arrangement, an N-terminal GTPase domain followed by a C-terminal K homology (KH) domain. The two domains are connected by a flexible linker, 17 amino acid residues in length. The N-terminal GTPase domain resembles p21 Ras [15,16], the G-domain of transducin-α [17], and domain I of EF-Tu [18]. It consists of a central six-stranded β-sheet flanked by five helices (Figure 1B). The C-terminal KH domain is composed of a three-stranded β-sheet and three helices (Figure 1C).
Figure 1.
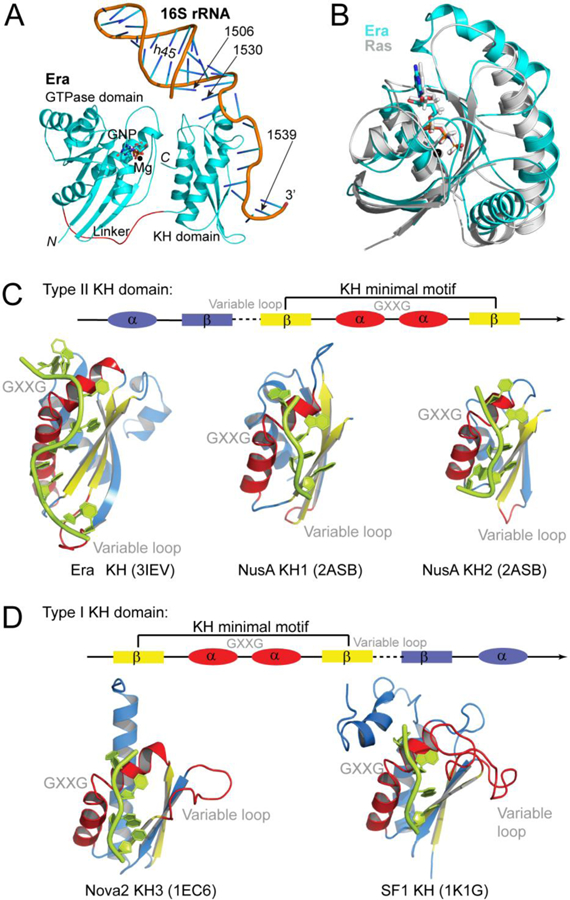
Overall structure of Era. (A) Schematic representation of the Era:MgGNP:RNA301 complex embedded in the Era:MgGNP:RNA301:KsgA structure (PEB entry 3R9X), showing the GTPase domain, KH domain, Mg2+ ion, non-hydrolysable GTP-analog GNP, and RNA301 that contains helix 45 (h45) and the 3’ tail of 16S rRNA. The protein is illustrated as a ribbon diagram (spirals, helices; arrows, strands; and tubes, loops), the Mg2+ ion as a sphere, the GNP molecule as a stick model, and the RNA as a cartoon. Atomic color scheme is used (N, blue; C, cyan; O, red; phosphorus, orange; and Mg, black) except that the linker between the two domains of Era is highlighted in red. (B) The GTPase domain and MgGNP of the Era:MgGNP:RNA301 structure are superimposed with the Ras:MgGNP structure (in grey, PDB entry 4RSG). (C) Type II KH:RNA complexes in the Era:MgGNP:RNA (PDB entry 3IEV) and NusA-RNA (PDB entry 2ASB) structures. (D) Type I KH:RNA complexes in the Nova2:RNA (PDB entry 1EC6) and SF1:RNA (PDB entry 1K1G) structures. The KH domains are aligned on the basis of the helix-turn-helix (HTH) motif, which contains the GXXG loop, and shown separately for clarity. Nucleotide residues that are recognized by the KH domains are 1531AUCACCUCC1539 for Era-KH, 42AGAA45 for NusA-KH1, 48CAAUA52 for NusA-KH2, 12UCAC15 for Nova2, and 6UAAC9 for SF1. Linear representations are used to illustrate the topological differences between the two types of KH folds, in which the dashed line represents the variable loop and the GXXG represents the GXXG loop in the HTH motif. Panels C and D were originally published in Reference [12].
KH domains are conserved RNA-binding motifs [19,20]. They usually occur in tandem repeats and exhibit low sequence similarity. Two types of KH domains, type I and type II, have been characterized. A common βααβ fold, containing a VIGxxGxxIK sequence, is the KH minimal motif shared by the two types (Figure 1, C and D). Type I KH domains, exemplified by the Nova2 KH3 domain [21] and the splicing factor 1 (SF1) KH domain [22], have a topology βααββα [19,20] (Figure 1D). The secondary structural elements of type II KH domains, however, are arranged as αββααβ (Figure 1C). First revealed by the crystal structure of Era [23], other examples of type II KH domains include the KH1 and KH2 of RNA polymerase-associated transcription factor NusA [24] (Figure 1C). The KH domain of Era recognizes 10 nucleotides [13], whereas all other known KH domains recognize 4–5 nucleotides [25].
FOUR REPRESENTATIVE STRUCTURES FROM THREE BACTERIAL SPECIES
To illustrate the functional states of Era, four representative structures are shown in Figure 2. They are the 2.4-Å structure of ligand-free Era from E. coli (apo-Era, PDB entry 1EGA) [23], the 2.8-Å structure of E. coli Era in complex with a GDP molecule (Era:GDP, PDB entry 3IEU) [12], the 1.88-Å structure of Thermus thermophilus Era in complex with a Mg2+ ion and a GNP molecule (Era:MgGNP, PDB entry 1WF3, RIKEN Structural Genomics/Proteomics Initiative), and the 1.9-Å structure of the Era:MgGNP:RNA complex in which the protein is from Aquifex aeolicus (PDB entry 3IEV) [12]. A structure-based sequence alignment reveals that the three Era proteins share ~40% sequence identity; they also share secondary structural elements and functional motifs (Figure 3), suggesting common functions of the protein among genetically distant species. It has been shown that the Era lethal mutants in E. coli can be cross-complemented by Era from other bacterial species [4,26]. Therefore, bacterial species identifiers are not necessary. The amino acid residue numbering of A. aeolicus Era will be used hereafter unless otherwise stated.
Figure 2.
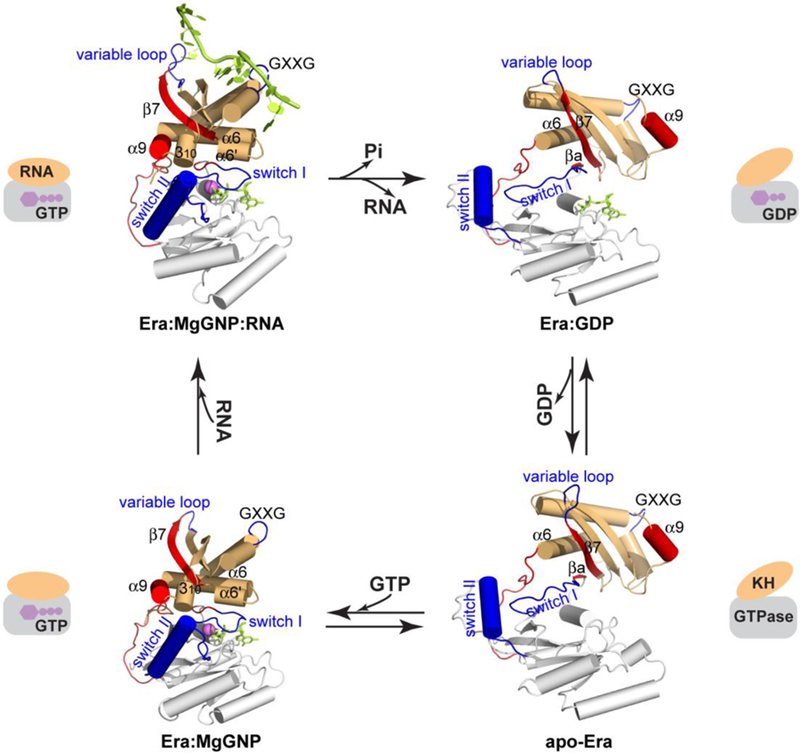
The functional cycle of Era as an essential GTPase. The crystal structures of Era are shown based on superimposed GTPase domains, including Era:GNP:RNA (PDB entry 3IEV) and Era:GNP (PDB entry 1WF3) sharing the closed conformation, and apo-Era (PDB entry 1EGA) and Era:GDP (PDB entry 3IEU) sharing the open conformation. The GTPase domain, the linker and the KH domain are shown in white, red and orange, respectively. Switches I and II and the GXXG and variable loops are highlighted in blue, and the α9 helix, βa and β7 strands, and linker are highlighted in red. The GNP and RNA molecules are colored in green, and the Mg2+ ion in magenta. Highly simplified representation of each structure is composed of a rectangle and an ellipse. This figure was adopted from Reference [12] with revisions and additions.
Figure 3.
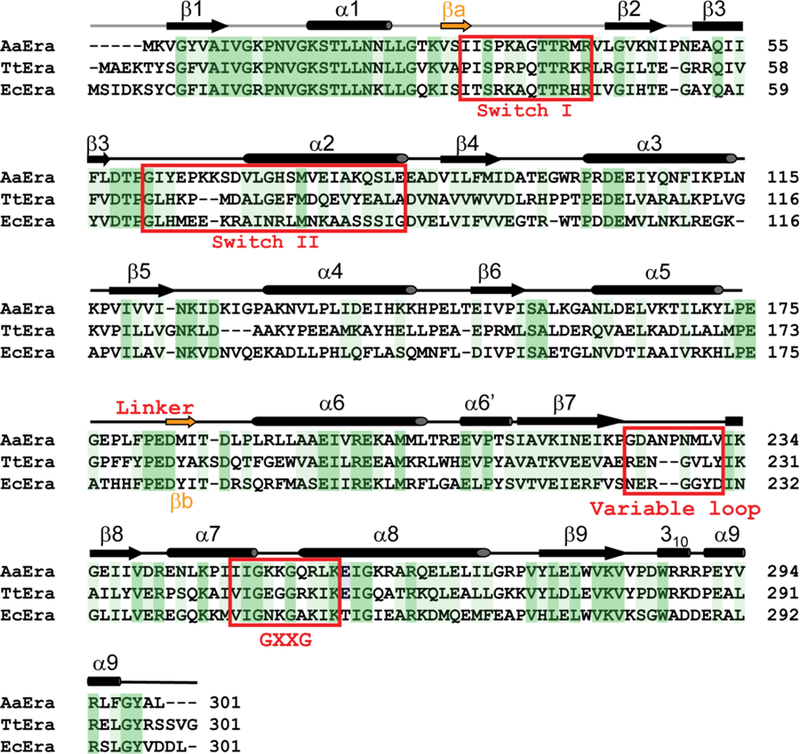
Structure-based sequence alignment of Era proteins from A. aeolicus, T. thermophilus, and E. coli (PDB entries 3IEV, 1WF3, and 1EGA, respectively). Secondary structural elements and the position of the inter-domain 17-residue linker are indicated above the sequences. Switch regions I and II and the GXXG and variable loops are indicated. Identical residues and similar residues are shaded in dark and light green, respectively.
TWO OVERALL CONFORMATIONS FOR FOUR LIGANDED STATES
The 17-residue linker enables dramatic changes to occur in relative positions of the GTPase and KH domains, while the 7-residue connection between β9 and α9 in the KH domain allows large shifts of α9 with respect to the rest of KH (Figures 2 and 3). Nonetheless, the four structures display only two distinct overall conformations. Era:MgGNP:RNA and Era:MgGNP share one conformation in which the GTP-binding site is closed (closed conformation), while Era:GDP and apo-Era share another in which the GTP-binding site is open (open conformation) (Figure 2).
GTP interacts with highly conserved regions of GTPases, including the switch I and switch II regions [27]. In the Era:GDP structure (open conformation), however, GDP binds in a cavity distant from the two switch regions (Figure 2). An inter-domain β-sheet is formed between β7 and βa (Figures 2 and 3). The βa strand and the inter-domain β-sheet exist, however, only in the open conformation. Also distant from the two switch regions is the α9 helix, the last helix of KH and a good marker for conformational changes of the molecule.
In the closed conformation shared by the Era:MgGNP:RNA and Era:MgGNP complexes, the βa strand melts into switch I and the inter-domain β-sheet becomes impossible (Figures 2 and 3). The switch I and switch II regions in the GTPase domain are displaced by ~10 Å and ~15 Å, respectively, when compared with those in the open conformation. While the switch I region serves as a ‘lid’ that covers the entire triphosphate moiety of GTP, the switch II region closes in and interacts with the γ-phosphate (Figure 2). With respect to the GTPase domain, the KH domain repositions itself such that the α9 helix swings from the right- to the left-hand side of the molecule and wedges between β7 of the KH domain and switch II of the GTPase domain (Figure 2).
GTP BINDING AS A PREREQUISITE FOR RNA BINDING
The fact that the four Era structures share two conformations allows us to examine the overall changes of the protein in a systematic manner. That Era:MgGNP:RNA and Era:MgGNP exhibit the same (closed) conformation indicates that the binding of RNA does not elicit significant conformational changes in the protein. Similarly, the observation that Era:GDP and apo-Era assume the same (open) conformation indicates that the release of GDP also does not cause significant conformational changes. In contrast, GTP binding and GTP hydrolysis lead to dramatic changes of Era’s overall conformation, from open to closed and from closed to open, respectively (Figure 2).
GTP binding triggers a chain of conformational changes within and between the two domains, which not only closes the GTP-binding site, but also makes the RNA-binding site accessible to RNA. The KH domain is responsible for the recognition of the 1530GAUCACCUCC1539 sequence in the pre-16S rRNA. However, the RNA-binding site (Figure 4A) on the KH domain is partially shielded by the α9 helix in the open conformation of Era (Figure 4B). It becomes accessible to RNA in the closed conformation of Era where the α9 helix moves away from the RNA-binding site on the KH domain and wedges between the GTPase and KH domains (Figures 2 and 4).
Figure 4.
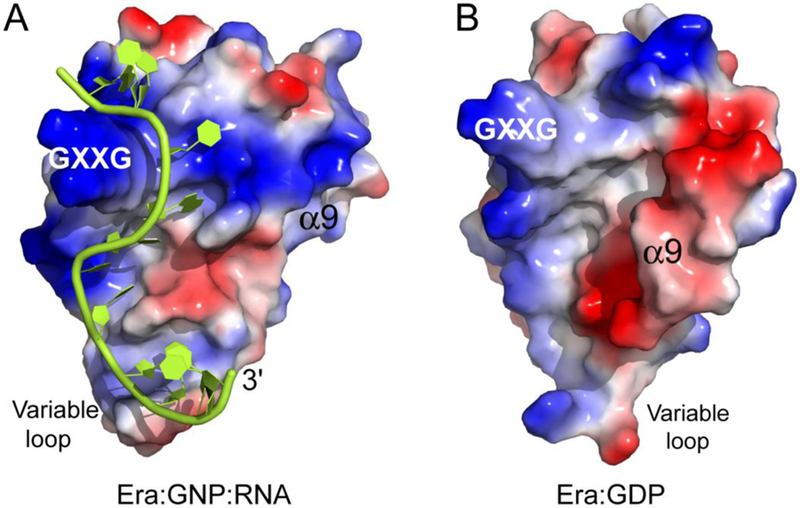
Role of α9 in the regulation of RNA binding by KH. (A) Electrostatic-surface representation of the KH domain and RNA in the Era:GNP:RNA structure (PDB entry 3IEV). (B) Electrostatic-surface representation of the KH domain in the Era:GDP structure (PDB entry 3IEU). On the molecular surfaces, positively charged areas are indicated in blue and negatively charged areas in red. The RNA is illustrated as a cartoon in green. The GXXG motif, variable loop, and α9 helix are indicated. This figure was originally published in Reference [12].
GTP hydrolysis initiates a series of reverse conformational changes, which not only opens the GTP-binding site, but also makes the RNA-binding site inaccessible to RNA by partially shielding the RNA-binding site on the KH domain with the α9 helix (Figure 4B).
THE STIMULATION OF GTP-HYDROLYZING ACTIVITY
The intrinsic GTP-hydrolyzing activity of Ras is low [28], which can be accelerated by a GTPase-activating protein [29,30]. The intrinsic GTP-hydrolyzing activity of Era is also low [31], ranging from 0.01 to 0.02 mmol min−1 mmol-1, which can be stimulated up to 12-fold in the presence of 16S rRNA [32,33], 6-fold in the presence of the 1531AUCACCUCCUUA1542 sequence [12] (Figure 5A), and 10-fold in the presence of RNA301 that contains h45 and downstream sequence (residues 1506–1542) [13] (Figure 5D). Therefore, the 1506–1542 sequence of 16S rRNA is sufficient for the stimulation of Era’s GTPase activity.
Figure 5.
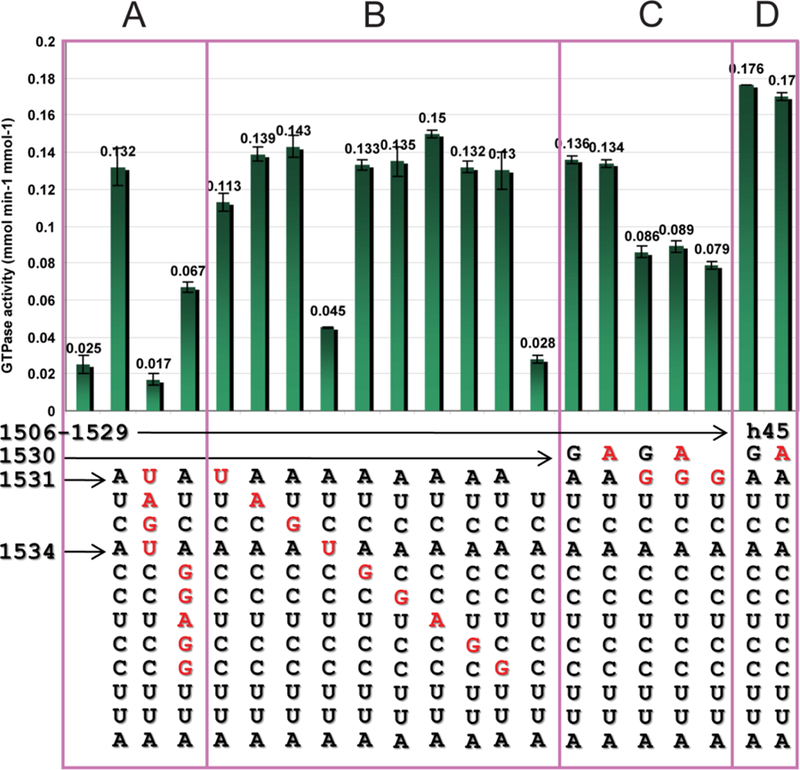
GTP-hydrolyzing activity of Era. (A) GTPase activity of Era in the absence of RNA, in the presence of the 1531AUCACCUCCUUA1542 sequence, in the presence of the sequence with 1531–1534 mutations, and in the presence of the sequence with 1535–1539 mutations. (B) GTPase activity of Era in the presence of the 1531AUCACCUCCUUA1542 sequence with a single mutation at each position from 1531 to 1539 or the deletion of nucleotide 1531. (C) GTPase activity of Era in the presence of the 1530GAUCACCUCCUUA1542 sequence, in the presence of the sequence with a single or double mutation at 1530 and/or 1531, or with the deletion of nucleotide 1530 and a single mutation at 1531. (D) GTPase activity of Era in the presence of longer RNA consisted of h45 (nucleotides 1506–1529) and the 1530GAUCACCUCCUUA1542 sequence, either native (RNA301) or with a G1530A mutation. Data were taken from References [12, 13] and presented graphically here.
Using the transversion mutant of 1531AUCA1534 (1531UAGU1534) and that of 1535CCUCC1539 (1535GGAGG1539), it has been shown that the recognition of AUCA is essential, but the recognition of both AUCA and CCUCC is necessary for optimal stimulation of Era’s GTP-hydrolyzing activity [12] (Figure 5A). Although the CCUCC sequence is necessary for optimal stimulation, the five individual mutations in the CCUCC sequence have no impact on the stimulation (Figure 5B). In contrast, certain mutations in the AUCA sequence have a large impact (Figure 5B). Genetically, A1531 is universally conserved, while A1534 is conserved in RNAs that are recognized by KH domains [12]. The A1531Δ and A1534U mutants exhibit low activities (0.028–0.045 mmol min−1 mmol−1), whereas the U1532A and C1533G mutations do not have any impact on the stimulation (0.139–0.143 mmol min−1 mmol−1). Thus, in the 1531AUCACCUCC1539 sequence, the two Ade nucleotides are essential for stimulation. In contrast, each of the other seven nucleotides, individually, does not contribute to the function.
Although both A1531 and A1534 are important, the transversion mutation of A1531 (A1531U, 0.113 mmol min−1 mmol−1) does not have an impact as dramatic as that of A1534 (A1534U, 0.045 mmol min−1 mmol−1) (Figure 5B). Furthermore, the substitution of A1531 by a Gua (A1531G, 0.079 mmol min−1 mmol−1) also does not have a dramatic impact on stimulation (Figure 5C). The difference between the two Ade nucleotides in their degree of substitution tolerance has well-established structural basis (vide infra).
Surprisingly, the universally conserved G1530 does not further stimulate the GTPase activity (Figure 5C). The specific activities of Era in the absence (0.132 mmol min−1 mmol−1, Figure 5A) or presence (0.136 mmol min−1 mol−1) of G1530 are virtually identical (Figure 5C). Furthermore, the G1530A mutation (0.134 mmol min−1 mmol−1) also does not change the specific activity although the mutation would significantly weaken the interaction (Figure 5C). However, once A1531 is mutated to a Gua with either G1530 or A1530, the specific activity drops to 0.086–0.089 mmol min−1 mmol−1, virtually the same as that of G1530Δ/A1531G (0.79 mmol min−1 mmol−1, Figure 5C). Although G1530 does not contribute to the stimulation of Era’s GTPase activity, its recognition by Era is important for function (vide infra).
Taken together, among the three purines in the 1530GAUCACCUCC1539 sequence, G1530 is not involved in the stimulation of GTPase activity, whereas A1531 and A1534 are determinants for the stimulation. A1531 tolerates mutational substitutions, but A1534 does not. In addition, the h45 sequence upstream from G1530, further stimulates the GTPase activity of Era (Figure 5D). In the presence of h45, the specific activity increases to 0.176 mmol min−1 mmol-1. This increase, however, has nothing to do with G1530 as evidenced by the specific activity (0.17 mmol min−1 mmol−1) for the G1530A mutant of the same RNA (Figure 5D).
THE RECOGNITION OF NUCLEOTIDES G1530, A1531, AND A1534
The Era:MgGNP:RNA301 and Era:MgGNP:RNA301:KsgA structures (PDB entries 3R9W and 3R9X) [13] show that a pseudo-base pair is formed by the G1530 base and the side chains of E209 and R241 (Figure 6A), which stacks with the first base pair of h45 and plays an important role in Era-RNA interaction. On the basis of structural information, the G1530A mutation would eliminate such pseudo-base pairing: two hydrogen bonds between Era and G1530 could be lost; the mutant Ade base could create an unfavorable stereo clash with the side chain of R241 (Figure 6B). As such, the Era-RNA interaction would be weakened. This structure-based prediction has been validated by isothermal titration calorimetry experiments. The Kd value is 12.6±3.2 nM for RNA301, but is 26.4±4.6 nM for RNA301 with the G1530A mutation, showing a 2-fold decrease in Era-RNA affinity due to the mutation. Therefore, the G1530A mutation affects RNA binding rather than the GTP-hydrolyzing activity of Era [13].
Figure 6.
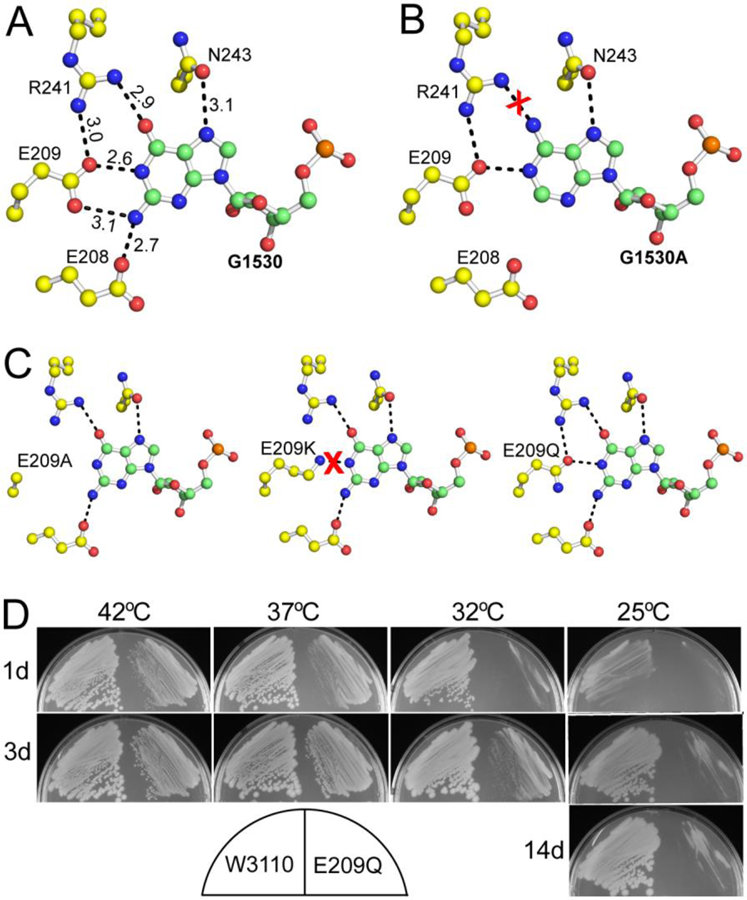
The recognition of G1530 by Era. (A) Details of Era-G1530 recognition. G1530 and the side chains of E208, E209, R241, and N243 are illustrated as ball-and-stick models in atomic colors (C in green or yellow, O in red, N in blue, and P in orange). Dashed lines in black indicate hydrogen bonds and their distances are in Å. (B) Model of the G1530A mutant, suggesting that the mutant is not favorable for the recognition by Era due to the loss of two hydrogen bonds (between E208/E209 and the Gua base, panel A) and the creation of a repulsive interaction (between R241 and the Ade base, indicated with a red cross). (C) Models of the E209A, E209K, and E209Q mutants. Indicated is the impact of each mutation on the pseudo-base paring interaction between the G1530 base and the side chains of residues 209 and 241. (D) The E209Q mutant of Era is cold sensitive. Compared with the wild type (W3110, on the left of each sub-panel), the E209Q mutant (on the right of each sub-panel) grows slower at all temperatures tested. Growth impairment is observed at 37 and 42°C. At 32°C, small colonies appear after three days (3d). At 25°C, however, colonies do not appear even after 14 days (14d). This figure was originally published in Reference [13].
G1530 interacts with four Era side chains (Figure 6A), among which E209 is universally conserved. The impact of three mutations in this residue: E209A, E209 K, and E209Q (Figure 6C) have been tested. The E209A and E209K mutants cannot be made at any growth temperature and are presumed to be lethal, whereas the E209Q mutant can be made but is cold sensitive (Figure 6D). The E209Q mutant severely impacts bacterial growth at 32°C. At 25°C, growth is totally arrested even after 14 days. At higher temperatures (37 or 42°C), the mutation moderately retards cell growth. These data demonstrate that the Era-G1530 interaction is required for the function of Era.
The difference between nucleotide residues A1531 and A1534 in their degree of substitution tolerance can be explained by the structures. As depicted in Figure 7A, the A1531 base fits snugly in a pocket formed by the side chains of T206, R207, E208, E209, V210, I247, and R255. In addition, the base forms two hydrogen bonds with the backbone groups of R207 and the A1531 ribose forms two hydrogen bonds with the guanidinium group of R255. Thus, any mutant of A1531 would still have its base fit in the pocket and its ribose hydrogen bonded to R255. In contrast, although the base of A1534 also forms two hydrogen bonds with the backbone groups of V281, the A1534 ribose does not form hydrogen bonds with the protein (Figure 7B). Moreover, Era does not provide a binding pocket for the A1534 base. Instead, one side of the A1534 base contacts the side chains of I249 and L256, but the other side is perpendicular to and barely contacts the W280 side chain (Figure 7B). Hence, almost an entire side of the A1534 base is exposed to solvent. The impact of the A1534U mutation is therefore two-fold. First, the Uri base cannot form hydrogen bonds with the backbone of V281. Second, the Uri base cannot interact with the W280 side chain, leading to the interrupted recognition of nucleotide residue 1534.
Figure 7.
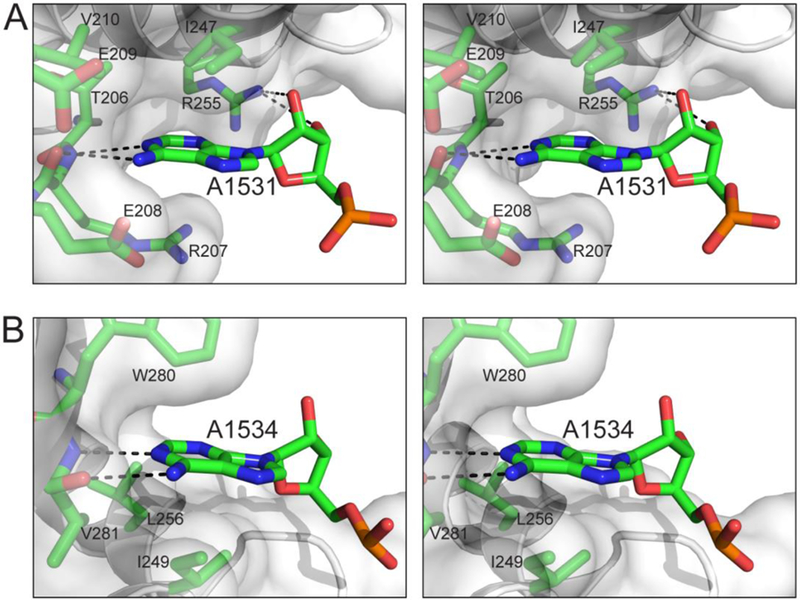
The recognition of nucleotide residues A1531 and A1534 by Era in stereo. (A) Interactions between A1531 and Era. (B) Interactions between A1534 and Era. Protein and nucleotide residues are shown as stick models in atomic colors (C, green; N, blue; O, red; and P, orange). Protein backbone is shown as a cartoon (helices as spirals, strands as arrows, and loops as tubes), and protein surface is shown with 50% transparency. This figure was originally published in Reference [13].
THE FUNCTIONAL CYCLE
The pre-16S rRNA features a 26-basepair stem [34]. Era is required to convert the pre-16S rRNA into 16S rRNA [9,10], during which 115 nucleotides are removed from the 5′ end by RNase E and RNase G, while 33 nucleotides are removed from the 3′ end by an unknown RNase [35,36]. The 1530GAUCACCUCC1539 sequence is only 3 nucleotides away from the cleavage site of the unknown RNase. Therefore, it is also possible that Era binding changes the local conformation of the cleavage site such that the RNA cleaves itself.
Unlike the exchange of the Ras-bound GDP for GTP, which requires a guanine-nucleotide exchange factor [37], the Era-bound GDP can easily exchange with GTP and vise versa [38]. In order words, the GDP- and GTP-bound forms of Era exist in a rapid equilibrium (Figure 2). The recognition of the 10 nucleotide residues and interaction with h45 in the pre-16S rRNA stabilize the Era:MgGTP complex, stimulate the GTP-hydrolyzing activity, and thereby increase the efficiency and/or turnover of GTP hydrolysis by the Era GTPase, which shifts the equilibrium toward GTP-bound form. The functional cycle of Era can be logically described in the context of ribosome biogenesis (Figure 8).
Figure 8.
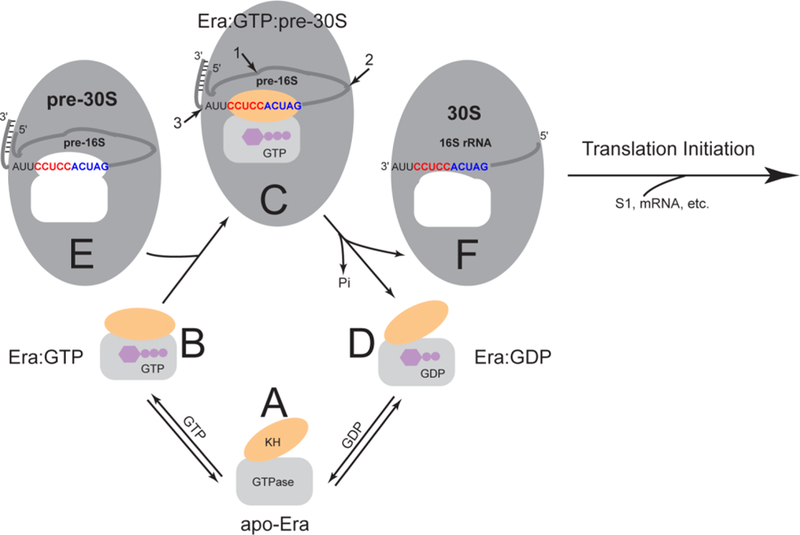
The functional cycle of Era. As also shown in Figure 2, the GTPase domain is represented by a grey rectangle, the KH domain by an orange oval, and the GTP and GDP molecules by purple cartoons. The pre-30S particle and 30S ribosomal subunit are represented by larger grey ovals. The pre-16S rRNA (an RNase III cleavage product with a 26-bp stem and a 2-nt 3′ overhang) and 16S rRNA are represented by a grey line with embedded 1530GAUCACCUCCUUA1542 sequence at the 3′ end. The highly conserved GAUCA sequence is colored in blue. The anti-Shine-Dalgarno (anti-SD) sequence CCUCC, which is unique for bacteria, is in red. The unoccupied Era-binding pocket in the pre-30S particle and that in the 30S ribosomal subunit are indicated in white. Note that the shapes of the pocket are different in the pre-30S particle and the mature 30S subunit. The four functional states, including (A) apo-Era, (B) Era:GTP, (C) Era:GTP:pre-30S and (D) Era:GDP, are represented by the apo-Era (PDB entry 1EGA), Era:GNP (PDB entry 1WF3), Era:GNP:RNA (PDB entry 3IEV), and Era:GDP (PDB entry 3IEU) structures. In panel C, the cleavage sites of RNase E, RNase G and the unknown nuclease are indicated with numbered arrows 1, 2 and 3, respectively. (E) The pre-30S particle contains pre-16S rRNA. (F) The mature 30S ribosomal subunit contains 16S rRNA. This figure was adopted from Reference [12] with updates.
Apo-Era in the open conformation (Figure 8A) can bind either a GDP or GTP molecule. When it binds a GTP molecule, the conformation of Era becomes closed with a closed GTP-binding site in the GTPase domain and an accessible RNA-binding site in the KH domain (Figure 8B). The recognition of the 1530GAUCACCUCC1539 sequence and interaction with h45 of pre-16S rRNA (pre-30S ribosomal particle, Figure 8E) by the Era:GTP complex leads to the formation of the Era:GTP:pre-30S complex (Figure 8C), in which the RNA-Era interactions stimulate the GTP-hydrolyzing activity of the enzyme and also signal the final processing of 16S rRNA, i.e., the removal of the extra nucleotide residues from its 5’ and 3’ ends. In concert with the final processing of 16S rRNA, the completion of GTP hydrolysis triggers the dramatic conformational change of Era from the closed conformation to the open conformation, which is sufficient to facilitate the departure of the Era:GDP complex (Figure 8D) from the mature 30S ribosomal subunit (Figure 8F). The rate of guanine nucleotide release by Era is rapid [38] (Figure 6, D to A). The rate of guanine nucleotide association by the protein is also rapid, especially in the presence of magnesium [38] (Figure 6, A to B). The concentration of GTP in the cell is higher than that of GDP. Therefore, the Ere:MgGTP complex (Figure 6B) is readily available for the next functional cycle of 16S RNA processing and 30S ribosomal subunit assembly and maturation.
The mature 30S r-subunit (Figure 8F) is ready for translation initiation. However, it may also bind apo-Era to form an Era:30S complex, locking the 30S ribosomal subunit in a conformation that is not favorable for association with the 50S ribosomal subunit [39]. According to a cryo-electron microscopy (cryo-EM) structure (PDB entry 1X1L), neither the closed conformation (Era:MgGTP, Figure 8B), nor the open conformation (apo-Era, Figure 8A; Era:GDP, Figure 8D) is compatible with the cavity left behind by Era in the mature 30S ribosomal subunit (Figure 8F). Docking of Era into this cavity requires dramatic conformational changes that obscure Era’s GTP-binding pocket [12]. Furthermore, it has been observed that addition of GTP or GDP precludes Era binding to the mature 30S r-subunit [9], demonstrating that guanine nucleotide-bound Era cannot be bound by the mature 30S ribosomal subunit.
CHAPERON FOR 16S PROCESSING AND CHECKPOINT FOR 30S MATURATION
Era binds to pre-16S when it is complexed with GTP and releases mature 16S upon the completion of GTP hydrolysis. It recognizes the 1530GAUCACCUCC1539 sequence that is very close to the mature 3′ end of 16S rRNA, suggesting that Era binding protects the GAUCACCUCC sequence from accidental damage during processing by, for example, the unknown RNase (Figure 8C). In addition, Era binding may change the conformation of pre-16S rRNA to assist RNases in processing and facilitate the activity of other factors, such as KsgA. KsgA methylates two adjacent adenosine residues in h45 [40–42]. The methylation occurs at a later stage of processing and maturation of 16S rRNA but before the 30S ribosomal subunit becomes competent to initiate translation [40–42]. Facilitating multiple steps during the process, Era acts as a chaperone for the processing and maturation of 16S rRNA.
The selection of correct initiation codon and translational reading frame in an mRNA depends on base pairing between the Shine-Dalgarno (SD) sequence, GGAGG, located upstream of the initiator codon and the anti-SD sequence CCUCC [43, 44]. Therefore, the binding of the GAUCACCUCC sequence by Era prevents base pairing between the anti-SD and SD sequences, thereby preventing mRNA recruitment to the pre-30S particle. The Era-binding site on the 30S largely overlaps with that of the S1 ribosomal protein [39]. Hence, the binding of 16S rRNA by Era would also occlude the binding of S1 (Figure 8). The S1 protein is known to directly affect the SD/anti-SD interaction [45,46] and so the initiation of mRNA translation by the 30S ribosomal subunit becomes possible only after Era is released. As such, Era functions as a checkpoint for ribosome assembly and final activation of the mature 30S ribosomal subunit.
UNIQUE PROTEIN FUNCTION IN BACTERIA
Era functions as an RNA chaperone for the processing and maturation of 16S rRNA and a checkpoint for the assembly of 30S ribosomal subunit [11–13]. It recognizes the 1530GAUCACCUCC1539 sequence near the 3′ end of 16S rRNA, the significance of which is four-fold. First, Era binding protects the GAUCACCUCC sequence from accidental damage during 16S rRNA processing and maturation. Second, the binding prevents base pairing between the anti-SD and SD sequences, thereby preventing mRNA recruitment. Third, the binding of Era occludes the binding of S1 that directly affects the association of the SD and anti-SD sequences. Fourth, conformational changes of the 16S rRNA precursor induced by the binding of Era may facilitate the activity of other ribosome biogenesis factors, including RNases that remove extra nucleotide residues from both ends of pre-16S rRNA [39] and KsgA that methylates A1518 and A1519 in the tetraloop of h45 [47].
The structural and functional data suggest a common mechanism for a highly conserved Era function in all forms of life by recognizing the GAUCA sequence with a “twist” for non-eukaryotic Era proteins by also recognizing the CCUCC. Era is present in nearly every bacterial species and is essential for cell growth and division, which is unique among all other known protein functions of bacteria. Inhibition of bacterial Era function will likely stop the synthesis of bacterial ribosome. Hence, Era is an attractive target for the development of novel antibiotics to fight the worldwide crisis of antibiotic resistance. The eukaryotic homologue of Era (EARL1) is a candidate for a tumor suppressor [48]. It is located in the small subunit of mitochondrial ribosome and interacts with the 12S rRNA, playing important roles in mitochondrial ribosome assembly and cell viability [49]. Currently, structure-based inhibitor design and structure-and-function studies of human and mouse ERAL1 proteins are undertaken in my laboratory.
Acknowledgements:
I would like to thank all my coworkers who contribued to the work cited from this laboratory. I am particularly indebted to Drs. Xin Chen and Chao Tu. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations:
- Era
Escherichia coli Ras-like protein
- GNP
non-hydrolyzable GTP analog GDPNP
- KH domain
K homology domain
- SD
Shine-Dalgarno
LITERATURE
- [1].Ahnn J, March PE, Takiff HE, Inouye M (1986) A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc Natl Acad Sci USA 83: 8849–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].March PE, Lerner CG, Ahnn J, Cui X, Inouye M (1988) The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene 2: 539–544 [PubMed] [Google Scholar]
- [3].Inada T, Kawakami K, Chen SM, Takiff HE, Court DL, Nakamura Y (1989) Temperature-sensitive lethal mutant of ERA, a G protein in Escherichia coli. Journal of Bacteriology 171: 5017–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zuber M, Hoover TA, Dertzbaugh MT, Court DL (1997) A Francisella tularensis DNA clone complements Escherichia coli defective for the production of Era, an essential Ras-like GTP-binding protein. Gene 189: 31–34 [DOI] [PubMed] [Google Scholar]
- [5].Eltsov M, Zuber B (2006) Transmission electron microscopy of the bacterial nucleoid. J Struct Biol 156: 246–254 [DOI] [PubMed] [Google Scholar]
- [6].Britton RA, Powell BS, Dasgupta S, Sun Q, Margolin W, Lupski JR, Court DL (1998) Cell cycle arrest in Era GTPase mutants: A potential growth rate-regulated checkpoint in Escherichia coli. Mol Microbiol 27: 739–750 [DOI] [PubMed] [Google Scholar]
- [7].Gollop N, March PE (1991) A GTP-binding protein (Era) has an essential role in growth rate and cell cycle control in Escherichia coli. Journal of Bacteriology 173: 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Britton RA, Powell BS, Court DL, Lupski JR (1997) Characterization of mutations affecting the Escherichia coli essential GTPase era that suppress two temperature-sensitive dnaG alleles. J Bacteriol 179: 4575–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sayed A, Matsuyama S, Inouye M (1999) Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem Biophys Res Commun 264: 51–54 [DOI] [PubMed] [Google Scholar]
- [10].Inoue K, Alsina J, Chen J, Inouye M (2003) Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol Microbiol 48: 1005–1016 [DOI] [PubMed] [Google Scholar]
- [11].Bunner AE, Nord S, Wikstrom PM, Williamson JR (2010) The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J Mol Biol 398: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tu C, Zhou X, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X (2009) Structure of ERA in complex with the 3’ end of 16S rRNA: implications for ribosome biogenesis. Proc Natl Acad Sci USA 106: 14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tu C, Zhou X, Tarasov SG, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X (2011) The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3’ end of 16S rRNA. Proc Natl Acad Sci USA 108: 10156–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xu Z, O’Farrell HC, Rife JP, Culver GM (2008) A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol 15: 534–536 [DOI] [PubMed] [Google Scholar]
- [15].Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A (1990) Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO Journal 9: 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tong LA, de Vos AM, Milburn MV, Kim SH (1991) Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J Mol Biol 217: 503–516 [DOI] [PubMed] [Google Scholar]
- [17].Noel JP, Hamm HE, Sigler PB (1993) The 2.2 A crystal structure of transducin-a complexed with GTP gamma S. Nature 366: 654–663 [DOI] [PubMed] [Google Scholar]
- [18].Jurnak F (1985) Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science 230: 32–36 [DOI] [PubMed] [Google Scholar]
- [19].Musco G, Stier G, Joseph C, Castiglione Morelli MA, Nilges M, Gibson TJ, Pastore A (1996) Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell 85: 237–245 [DOI] [PubMed] [Google Scholar]
- [20].Musco G, Kharrat A, Stier G, Faternali F, Gibson TJ, Nilges M, Pastore A (1997) The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat Struct Biol 4: 712–716 [DOI] [PubMed] [Google Scholar]
- [21].Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323–332 [DOI] [PubMed] [Google Scholar]
- [22].Liu ZH, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M (2001) Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102 [DOI] [PubMed] [Google Scholar]
- [23].Chen X, Court DL, Ji X (1999) Crystal structure of ERA: A GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc Natl Acad Sci USA 96: 8396–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA (2005) Structure of a Mycobacterium tuberculosis NusA-RNA complex. Embo Journal 24: 3576–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Valverde R, Edwards L, Regan L (2008) Structure and function of KH domains. FEBS J 275: 2712–2726 [DOI] [PubMed] [Google Scholar]
- [26].Pillutla RC, Sharer JD, Gulati PS, Wu E, Yamashita Y, Lerner CG, Inouye M, March PE (1995) Cross-species complementation of the indispensable Escherichia coli era gene highlights amino acid regions essential for activity. Journal of Bacteriology 177: 2194–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sprang SR (1997) G protein mechanisms: insights from structural analysis. Annu Rev Biochem 66: 639–678 [DOI] [PubMed] [Google Scholar]
- [28].Bollag G, McCormick F (1995) Intrinsic and GTPase-activating protein-stimulated Ras GTPase assays. Methods Enzymol 255: 161–170 [DOI] [PubMed] [Google Scholar]
- [29].Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A (1997) Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry 36: 4535–4541 [DOI] [PubMed] [Google Scholar]
- [30].Gideon P, John J, Frech M, Lautwein A, Clark R, Scheffler JE, Wittinghofer A (1992) Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol Cell Biol 12: 2050–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen SM, Takiff HE, Barber AM, Dubois GC, Bardwell JC, Court DL (1990) Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J Biol Chem 265: 2888–2895 [PubMed] [Google Scholar]
- [32].Meier TI, Peery RB, McAllister KA, Zhao G (2000) Era GTPase of Escherichia coli: binding to 16S rRNA and modulation of GTPase activity by RNA and carbohydrates. Microbiology 146: 1071–1083. [DOI] [PubMed] [Google Scholar]
- [33].Meier TI, Peery RB, Jaskunas SR, Zhao G (1999) 16S rRNA is bound to era of Streptococcus pneumoniae. J Bacteriol 181: 5242–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Srivastava AK, Schlessinger D (1990) Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol 44: 105–129 [DOI] [PubMed] [Google Scholar]
- [35].Li Z, Pandit S, Deutscher MP (1999) RNase G (CafA protein) and RNase E are both required for the 5’ maturation of 16S ribosomal RNA. EMBO J 18: 2878–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kaczanowska M, Ryden-Aulin M (2007) Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J (1998) The structural basis of the activation of Ras by Sos. Nature 394: 337–343 [DOI] [PubMed] [Google Scholar]
- [38].Sullivan SM, Mishra R, Neubig RR, Maddock JR (2000) Analysis of guanine nucleotide binding and exchange kinetics of the Escherichia coli GTPase Era. Journal of Bacteriology 182: 3460–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sharma MR, Barat C, Wilson DN, Booth TM, Kawazoe M, Hori-Takemoto C, Shirouzu M, Yokoyama S, Fucini P, Agrawal RK (2005) Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell 18: 319–329 [DOI] [PubMed] [Google Scholar]
- [40].Thammana P, Held WA (1974) Methylation of 16S RNA during ribosome assembly in vitro. Nature 251: 682–686 [DOI] [PubMed] [Google Scholar]
- [41].Connolly K, Rife JP, Culver G (2008) Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol 70: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Desai PM, Rife JP (2006) The adenosine dimethyltransferase KsgA recognizes a specific conformational state of the 30S ribosomal subunit. Arch Biochem Biophys 449: 57–63 [DOI] [PubMed] [Google Scholar]
- [43].Gold L, Pribnow D, Schneider T, Shinedling S, Singer BS, Stormo G (1981) Translational initiation in prokaryotes. Annu Rev Microbiol 35: 365–403 [DOI] [PubMed] [Google Scholar]
- [44].Shine J, Dalgarno L (1974) The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA 71: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Komarova AV, Tchufistova LS, Supina EV, Boni IV (2002) Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 8: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wilson DN, Nierhaus KH (2005) Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol 40: 243–267 [DOI] [PubMed] [Google Scholar]
- [47].Lu Q, Inouye M (1998) The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of era, an essential RAS-like GTP-binding protein in Escherichia coli. Journal of Bacteriology 180: 5243–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Britton RA, Chen SM, Wallis D, Koeuth T, Powell BS, Shaffer LG, Largaespada D, Jenkins NA, Copeland NG, Court DL, Lupski JR (2000) Isolation and preliminary characterization of the human and mouse homologues of the bacterial cell cycle gene era. Genomics 67: 78–82 [DOI] [PubMed] [Google Scholar]
- [49].Uchiumi T, Ohgaki K, Yagi M, Aoki Y, Sakai A, Matsumoto S, Kang D (2010) ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res 38: 5554–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]


