Abstract
Gene manipulation and epitope tagging are essential tools for understanding the molecular function of specific genes. The opportunistic human pathogen Candida albicans is a diploid fungus that utilizes a non-canonical genetic code. Since selection markers available in this organism are scarce, several tools based on recyclable markers have been developed for gene disruption, such as the Clox system. This system relies on the Cre recombinase, which recycles selection markers flanked by loxP sites with high efficiency, facilitating single marker or multi-marker recycling. However, PCR-based modules for epitope tagging, such the pFA-modules, mainly use limited non-recyclable auxotrophic markers. To solve this problem, we have used a Gibson assembly strategy to construct a set of new plasmids where the auxotrophic markers of the pFA vectors were swapped with five recyclable marker modules of the Clox system, enhancing the versatility of the pFA plasmids. This new toolkit, named pFA-Clox, is composed of 36 new vectors for gene disruption and epitope tagging (GFP, 3xGFP, mCherry, 3xHA, 5xmyc and TAP). These plasmids contain the dominant NAT1 marker, as well as URA3, HIS1 and ARG4 cassettes, thereby permitting functional analysis of laboratory strains as well as clinical isolates of C. albicans. In summary, we have adapted the Clox system to the pFA-backbone vectors. Thus, the set of primers used for the amplification of previously published pFA modules can also be utilized in this new pFA-Clox system. Therefore, this new toolkit harbors the advantages of both systems, allowing accelerated gene modification strategies that could reduce time and costs in strain construction for C. albicans.
Introduction
Candida albicans is an opportunistic human pathogen that lives as a commensal in the epithelium of the oral cavity, gastrointestinal or urogenital tracts [1]. Due to the particular characteristics of its biology, gene manipulation is a time-consuming process that requires the development of Candida-specific tools. First, C. albicans is mainly a diploid with a particular parasexual life cycle that does not seem to undergo meiosis to complete a standard sexual cycle [2,3]. For that reason, the generation of homozygous deletion mutants requires the successive deletion of both alleles. If more than one locus is going to be deleted, then the genetic markers used to select transformants need to be recycled [4–6]. The second important aspect of its biology is the use of a non-canonical genetic code in which CTG is decoded as Ser instead of Leu [7], which requires the development of codon-optimized tools for this organism (reviewed in [8,9]).
Over the last years, a variety of tools for gene manipulation have been developed for C. albicans, allowing genetic approaches to study function of the genes, including gene disruption [6,10–13], protein tagging [11,14,15] and overexpression [12,15]. In all the cases, these techniques are based on the generation of a cassette by PCR that is integrated in the genome by homologous recombination between sequences in the cassette and the genomic sequences.
The URA blaster method was the classical approach to disrupt genes in C. albicans. This cassette carries two direct repeats of the hisG sequence from Salmonella typhimurium flanking the URA3 gene [4]. After transformation of a ura3/ura3 strain, Uri+ transformants are selected on uracil-deficient medium. Then, spontaneous recombination between two hisG repeats produces the excision of the marker, leaving behind a copy of the hisG sequence in the genome. Ura3- cells can be selected by growth on medium containing 5-fluoroorotic acid (5-FOA), since it is toxic to Uri+ cells. However, 5-FOA is potentially mutagenic and could introduce chromosomal rearrangements in C. albicans [16,17].
To circumvent this problem, strains with additional selectable markers were generated on the SC5314 background, such as BWP17 (Uri- His- Arg-), SN76 (Uri- His- Arg-), SN78 (Uri- His- Leu-) or SN148 (Uri- His- Arg- Leu-) [18,19], and several systems with recyclable markers were also developed. The URA flipper cassette contains two direct repeats of the minimal FLP recombination target (FRT) sequence flanking the URA3 marker. This cassette also contains the FLP recombinase gene under the control of the secreted aspartyl proteinase (SAP2) promoter, which is induced by growth in medium containing bovine serum albumin. The FLP recombinase promotes the homologous recombination between the FRT sites to recycle the URA3 gene for another round of transformation [20]. Similar systems are the SAT1 flipper, which contains the nourseothricin resistance marker SAT1 and the FLP recombinase regulated by the MAL2 promoter [21], and a variant that contains the NAT1 marker, which is a codon-optimized Streptomyces noursei NAT1 gene (nourseothricin acetyltransferase) that also confers nourseothricin resistance regulated by the Ashbya gossypii TEF1 promoter followed by the FLP recombinase gene regulated by the SAP2 promoter [22].
A Cre-loxP system for gene disruption and marker recycling in C. albicans was developed a few years ago [23]. Cre catalyzes the recombination between loxP sequences in P1 bacteriophage [24]. The specificity of sequence recognition has been used to develop different Cre-loxP recombination tools for Saccharomyces cerevisiae and mammalian cells [25,26]. The C. albicans system also involves the use of Cre recombinase to recycle different selection markers flanked by loxP sites [23]. Three different selectable markers flanked by loxP sequences were developed: loxP-ARG4-loxP (LAL), loxP-HIS1-loxP (LHL) and loxP-URA3-loxP (LUL). The initial system used a methionine-regulatable MET3p-cre cassette to induce marker loss that had to be inserted in an additional transformation step after the two target alleles have been disrupted. More recently, an enhanced Cre-loxP toolkit (Clox) was developed [13], which carries a synthetic, intron-containing cre gene under the control of the MET3p promoter and the URA3 or NAT1 selectable markers flanked by loxP sequences (URA3-Clox or NAT1-Clox cassettes). The advantage of these new cassettes is that they facilitate gene disruption using a single marker or multiple markers, with high efficiency of marker loss after induction of the Cre recombinase. The only disadvantage of this toolkit is that the sequences flanking the cassettes used for amplification are different from the commonly used sequences present in pFA plasmids developed for Candida [11,12,14,15], and therefore they require the design of new sets of oligonucleotides for their use.
The development of clustered regularly interspaced short palindromic repeat(s) (CRISPR) genome editing systems has changed genetic engineering in different species (reviewed in [27]). Several groups have reported the development of CRISPR-mediated editing systems for C. albicans [28–30]. These systems generate a Cas9-mediated double-stranded breaks (DSBs) at specific target locus that are repaired by the integration of a donor DNA (dDNA). These new systems are a breakthrough in C. albicans genome manipulation since a homozygous gene deletion mutant is obtained in a single transformation. Recently, an optimized system has been described that allows high-efficiency marker-less homozygous genome editing, rapid cloning-free gRNA generation and marker recycling [31]. In addition, the CRISPR system has also been adapted to regulate gene expression in C. albicans using a Cas9-inactive nuclease [32,33].
Regarding to epitope tagging with recyclable markers, the number of plasmids for epitope tagging that allows marker recycling is more limited. Plasmids containing the 13xmyc epitope or a 6-His/FLAG tandem affinity purification (TAP) tag and the SAT1 flipper cassette have been described [34]. More recently, a collection of vectors for epitope-tagging and overexpression in C. albicans based on the Cre recombinase was also described [35]. Here we describe the development of the pFA-Clox toolkit, a new collection of plasmids for gene deletion and epitope tagging that use the selectable markers of the Cre-loxP system cloned into the pFA vector backbone. This new toolkit combines the advantages of the Clox system (rapid, efficient and flexible gene disruption and marker recycling in C. albicans)[13] with the use of the common flanking regions of the pFA vectors, thereby reducing the need of generating new sets of oligonucleotides. The different cassettes can be amplified with fewer 120-bp primer pairs (100 bp from the gene to be tagged and 20 bp of vector sequences) to tag genes with different epitopes or fluorescent proteins. This PCR strategy is compatible with the pFA-tagging systems previously published [11,12,14]. The plasmids developed here could be used to generate single or multiple deletion strains in auxotrophic laboratory strains and in prototrophic clinical isolates. They also permit simultaneous tagging of different genes with commonly used epitope tags (HA, myc, GFP, mCherry…) followed by marker excision mediated by the Cre recombinase. Consequently, strains generated using this system will have available markers for successive gene manipulations.
Results
The pFA-Clox kit
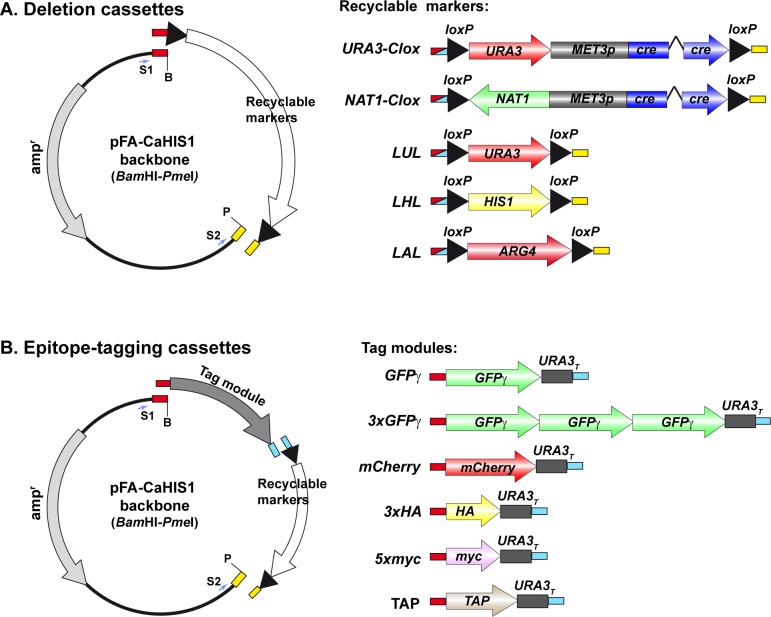
An arsenal of pFA plasmids for PCR-based gene targeting for function analysis in C. albicans has developed since 2003 by Wendland and colleagues [10–12,14]. These pFA series have been generated to delete genes, exchange promoters and tag genes with GFP, myc or HA. An advantage of the pFA series is the minimal set of primers required for the amplification of all modules, reducing the cost of strain construction. However, the use of non-recyclable selection markers diminishes the versatility of these plasmids. Recently, an improved Clox system has been developed that is suitable for gene disruption in C. albicans that allows both multi-marker disruption and single marker recycling strategies with high efficiency [13]. Therefore, we designed an assembly strategy to replace the non-recyclable auxotrophic markers of pFA plasmids for gene disruption and C-terminal tagging with the five recyclables modules of the Clox system (LAL, LHL, LUL, URA3-Clox and NAT1-Clox) (Fig 1A).
Fig 1. Modular strategy used for the construction of pFA plasmids with recyclable markers.
(A) Construction of pFA deletion plasmids. The five recyclable markers flanked by loxP repeats were amplified with oligonucleotides LXL1 and LXL2, which generated overlapping ends with the pFA-CaHIS1 backbone digested with BamHI and PmeI. The amplified fragments were assembled with the vector in 5 independent reactions using NEBuilder HiFi DNA Assembly kit. (B) Construction of epitope-tagging plasmids. The different tagging modules (GFPγ, 3xGFPγ, mCherry, 3xHA, 5xmyc or TAP-TAG) were amplified with oligonucleotides that generated overlapping ends with the pFA-CaHIS1 backbone digested with BamHI (red) and with the 5´end of the recyclable marker module (light blue). The five recyclable markers were independently amplified with oligonucleotides that produced DNA fragments containing overlapping ends with the tagging modules (light blue) and the pFA-CaHIS1 backbone digested with PmeI (yellow). The different modules were assembled with the vector in independent reactions using NEBuilder HiFi DNA Assembly kit.
To generate a new pFA series with these recyclable markers for gene disruption, the pFA-CaHIS1 plasmid [11] was digested with BamHI and PmeI to release the HIS1 sequence and the pFA backbone was assembled with each of the five modules of the Clox system (LAL, LHL, LUL, URA3-Clox and NAT1-Clox) using the NEBuilder HiFI DNA Assembly Cloning Kit as described in Materials and Methods. The resulting plasmids were named pFA-LAL, pFA-LHL, pFA-LUL, pFA-URA3-Clox and pFA-NAT1-Clox (Table 1 and Figure A in S1 File). To amplify the disruption cassettes present in all these plasmids, the same pair of primers (containing S1 and S2 sequences at the ends, Table 2) can be used.
Table 1. The pFA-Clox toolkit.
Plasmids generated and fragment length of the cassettes.
| Plasmid | Accession number | Selectable Marker | Gene | 5´ end | 3´ end | Cassette Size |
|---|---|---|---|---|---|---|
| pFA-LAL | MK652110 | loxP-ARG4-loxP | - | S1 | S2 | 2180 |
| pFA-LHL | MK652111 | loxP-HIS1-loxP | - | S1 | S2 | 1474 |
| pFA-LUL | MK652112 | loxP-URA3-loxP | - | S1 | S2 | 1580 |
| pFA-NAT1-Clox | MK652113 | loxP-NAT1-cre-loxP | - | S1 | S2 | 4576 |
| pFA-URA3-Clox | MK652114 | loxP-URA3-cre-loxP | - | S1 | S2 | 4344 |
| pFA-GFPγ-LAL | MK652105 | loxP-ARG4-loxP | GFPγ | S1-XFP | S2 | 3142 |
| pFA-GFPγ-LHL | MK652106 | loxP-HIS1-loxP | GFPγ | S1-XFP | S2 | 2436 |
| pFA-GFPγ-LUL | MK652107 | loxP-URA3-loxP | GFPγ | S1-XFP | S2 | 2542 |
| pFA-GFPγ-NAT1-Clox | MK652108 | loxP-NAT1-cre-loxP | GFPγ | S1-XFP | S2 | 5538 |
| pFA-GFPγ-URA3-Clox | MK652109 | loxP-URA3-cre-loxP | GFPγ | S1-XFP | S2 | 5306 |
| pFA-3xGFPγ-LAL | MK652100 | loxP-ARG4-loxP | 3xGFPγ | S1-XFP | S2 | 4576 |
| pFA-3xGFPγ-LHL | MK652101 | loxP-HIS1-loxP | 3xGFPγ | S1-XFP | S2 | 3870 |
| pFA-3xGFPγ-LUL | MK652102 | loxP-URA3-loxP | 3xGFPγ | S1-XFP | S2 | 3976 |
| pFA-3xGFPγ-NAT1-Clox | MK652103 | loxP-NAT1-cre-loxP | 3xGFPγ | S1-XFP | S2 | 6972 |
| pFA-3xGFPγ-URA3-Clox | MK652104 | loxP-URA3-cre-loxP | 3xGFPγ | S1-XFP | S2 | 6740 |
| pFA-mCherry-LAL | MK652115 | loxP-ARG4-loxP | yEmCherry | S1-XFP | S2 | 3139 |
| pFA-mCherry-LHL | MK652116 | loxP-HIS1-loxP | yEmCherry | S1-XFP | S2 | 2433 |
| pFA-mCherry-LUL | MK652117 | loxP-URA3-loxP | yEmCherry | S1-XFP | S2 | 2539 |
| pFA-mCherry-NAT1-Clox | MK652118 | loxP-NAT1-cre-loxP | yEmCherry | S1-XFP | S2 | 5535 |
| pFA-mCherry-URA3-Clox | MK652119 | loxP-URA3-cre-loxP | yEmCherry | S1-XFP | S2 | 5303 |
| pFA-HA-LAL | MK652120 | loxP-ARG4-loxP | 3xHA | S1-HA | S2 | 2518 |
| pFA-HA-LHL | MK652121 | loxP-HIS1-loxP | 3xHA | S1-HA | S2 | 1812 |
| pFA-HA-LUL | MK652122 | loxP-URA3-loxP | 3xHA | S1-HA | S2 | 1918 |
| pFA-HA-NAT1-Clox | MK652123 | loxP-NAT1-cre-loxP | 3xHA | S1-HA | S2 | 4914 |
| pFA-HA-URA3-Clox | MK652124 | loxP-URA3-cre-loxP | 3xHA | S1-HA | S2 | 4656 |
| pFA-myc-LAL | MK652125 | loxP-ARG4-loxP | 5xmyc | S1 | S2 | 2614 |
| pFA-myc-LHL | MK652126 | loxP-HIS1-loxP | 5xmyc | S1 | S2 | 1908 |
| pFA-myc-LUL | MK652127 | loxP-URA3-loxP | 5xmyc | S1 | S2 | 2014 |
| pFA-myc-NAT1-Clox | MK652128 | loxP-NAT1-cre-loxP | 5xmyc | S1 | S2 | 5010 |
| pFA-myc-URA3-Clox | MK652129 | loxP-URA3-cre-loxP | 5xmyc | S1 | S2 | 4798 |
| pFA-TAP-LAL | MK652130 | loxP-ARG4-loxP | TAP | S1 | S2 | 2986 |
| pFA-TAP-LHL | MK652131 | loxP-HIS1-loxP | TAP | S1 | S2 | 2280 |
| pFA-TAP-LUL | MK652132 | loxP-URA3-loxP | TAP | S1 | S2 | 2386 |
| pFA-TAP-NAT1-Clox | MK652133 | loxP-NAT1-cre-loxP | TAP | S1 | S2 | 5382 |
| pDIS-NAT1-Clox | MK652134 | loxP-NAT1-cre-loxP | - | N5-up | N5-dw | 5448 |
| pDIS-URA3-Clox | MK652135 | loxP-URA3-cre-loxP | - | N5-up | N5-dw | 5216 |
Table 2. Primers used.
| For module construction | |
| Primer | Primer sequence |
| LXL1 | TCGTACGCTGCAGGTCGACGGGCGGCCGCTCTAGAACT |
| LXL2 | CGATGAATTCGAGCTCGTTTTTCCTGCAGATTACCCTGTTATCC |
| GFP-LXL1 | TCGTACGCTGCAGGTCGACGGGTGCTGGCGCAGGTGCT |
| GFP-LXL2 | AGCGGCCGCCGGATCTATGCGTCCATCTTTACAGTCCTGTC |
| GFP-LXL3 | GCATAGATCCGGCGGCCGCTCTAGAACT |
| HA1 | TCGTACGCTGCAGGTCGACGGGTCGACGGATCCCCGGG |
| Myc1 | TCGTACGCTGCAGGTCGACGGATCCCCGGGGAACAGAAGCTTATATC |
| TAP1 | TCGTACGCTGCAGGTCGACGGATCCCCGGGTTAATTAATC |
| NEUT7 | TACCGAATTCGAGCTCGTTTGGCGGCCGCTCTAGAACT |
| NEUT8 | GGATCCACTAGTTCTAGAGCTTCCTGCAGATTACCCTGTTATCC |
| PCR primers used to amplify the transformation cassettes | |
| Primer | Primer sequencea |
| S1 | (100 bp target sequence)-GAAGCTTCGTACGCTGCAGGTC |
| S2 | (100 bp target sequence)-TCTGATATCATCGATGAATTCGAG |
| S1–XFP | (100 bp target sequence)-GGT GCT GGC GCA GGT GCT TC |
| S1-HA | (100 bp target sequence)-GGT CGA CGG ATC CCC GGG TAC CCA |
| N5-up | CCTTAACCCACTGAATTCTACATCGAA |
| N5-dw | CAAAGAGAAAGCTCGGAGGAGGCT |
Primer sequences are given in the 5´ to 3´ direction
a The reading frame is indicated by spacing within the sequence; the sequence encoding the (Gly-Ala)3 linker or HA is shown in bold.
Next, we adapted the Clox system for C-terminal tagging with GFP and different epitopes such HA, myc and TAP. To this end, two different DNA fragments were combined in the assembly reaction with the pFA backbone, one containing the fluorescent protein/epitope sequence and the other carrying the recyclable marker (Fig 1B). Two pairs of primers were used to amplify the epitopes and the marker modules with overlapping regions for the assembly reaction (Table 2).
Using this approach, we generated three sets of plasmids for C-terminal tagging with fluorescent proteins. All pFA-Clox modules of these series can be amplified with the same set of primers (containing S1–XFP and S2 sequences at the ends, Table 2). The first group contain the photostable variant of the green fluorescent protein (GFPγ) [36] (pFA-GFPγ-LAL, pFA-GFPγ-LHL, pFA-GFPγ-LUL, pFA-GFPγ-URA3-Clox and pFA-GFPγ-NAT1-Clox) (Table 1 and Figure A in S1 File). The GFPγ protein is codon-optimized for Candida and contains 4 mutations (F64L, S65C, V163A, I167T) that result in a GFP protein with high signal intensity and photostability. In order to tag low abundant proteins, a second set of plasmids was made harboring three in-tandem copies of GFPγ (pFA-3xGFPγ-LAL, pFA-3xGFPγ-LHL, pFA-3xGFPγ-LUL, pFA-3xGFPγ-URA3-Clox and pFA-3xGFPγ-NAT1-Clox, Table 1 and Figure A in S1 File). The 3xGFPγ in tandem fusion was amplified from plasmid pFA-3xGFPγ-URA3, previously described [37]. Finally, the third set of plasmids (pFA-mCherry-LAL, pFA-mCherry-LHL, pFA-mCherry-LUL, pFA-mCherry-URA3-Clox and pFA-mCherry-NAT1-Clox, Table 1 and Figure A in S1 File) was developed to allow double color tagging of different proteins for fluorescence microscopy. The yEmCherry fluorescent protein sequence was obtained from plasmid pFA-yEmCherry-CaURA3 [14].
Next, analogous plasmids for C-terminal epitope tagging were generated. The HA and myc epitopes are highly immunogenic peptides that can then be used for Western blot or immunoprecipitation experiments. The TAP tag was developed to allow purification of protein complexes for mass spectrometry [38]. Plasmids pFA-HA-LAL, pFA-HA-LHL, pFA-HA-LUL, pFA-HA-NAT1-Clox and pFA-HA-URA3-Clox (Table 1 and Figure A in S1 File) were constructed by assembling a PCR fragment that contained 3 copies of the HA epitope followed by the S. cerevisiae URA3 terminator obtained from plasmid pFA-HA-URA3 [15] with the five recyclable markers into the pFA backbone. The integration cassettes can be amplified from these plasmids with primers containing S1-HA and S2 sequences (Table 2). For plasmids pFA-myc-LAL, pFA-myc-LHL, pFA-myc-LUL, pFA-myc-NAT1-Clox and pFA-myc-URA3-Clox (Table 1 and Figure A in S1 File), a PCR fragment containing 5 copies of the myc epitope followed by the S. cerevisiae URA3 terminator obtained from plasmid pFA-myc-URA3 [15] was assembled with the selection markers into the same vector. The integration cassettes can be amplified from this collection of plasmids with primers containing S1 and S2 sequences (Table 2). Finally, plasmids pFA-TAP-LAL, pFA-TAP-LHL, pFA-TAP-LUL and pFA-TAP-NAT1-Clox (Table 1 and Figure A in S1 File) contained the TAP tag followed by the S. cerevisiae URA3 terminator obtained from plasmid pFA-TAP-URA3 [15]. The integration cassettes can be amplified from the four plasmids with primers containing S1 and S2 sequences (Table 2).
Finally, we constructed plasmids pDIS-URA3-Clox and pDIS-NAT1-Clox by replacing the NAT1 marker of plasmid pDIS3 [39] with the URA3-Clox or NAT1-Clox cassettes. These plasmids can be used to induce marker loss of any construction generated with the LAL, LHL, or LUL cassettes, allowing the generation of auxotrophic strains.
Testing deletion plasmids
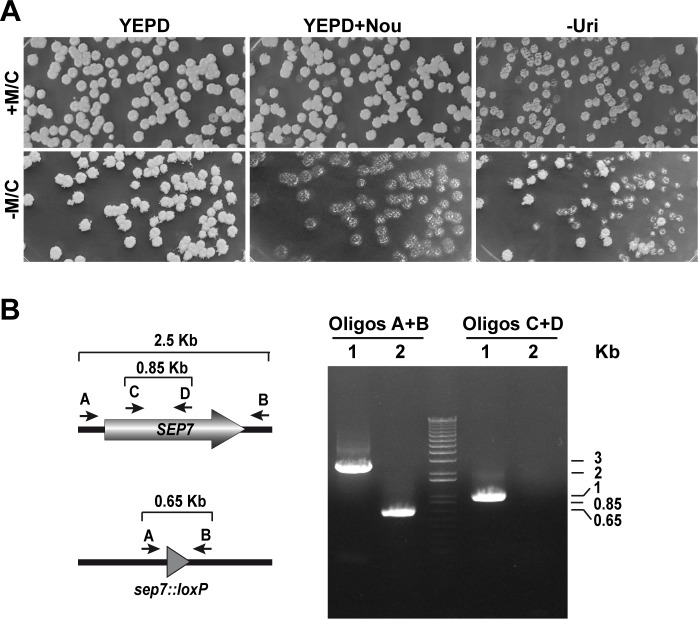
To test the pFA-Clox plasmids for gene disruption, we used them to delete both copies of SEP7, which encodes a non-essential septin subunit that plays a regulatory role for septin function during hyphal growth [40,41]. To delete the first SEP7 allele, BWP17 cells were transformed with a sep7::LUL cassette, amplified using long oligonucleotides carrying S1 and S2 sequences at the 3´ end and 100 nucleotides upstream or downstream of the SEP7 gene, and transformants were selected on SC lacking uridine. To eliminate the second SEP7 allele, the resultant Uri+ strain (SEP7/sep7Δ::LUL) was then transformed with the NAT1-Clox cassette. Nourseothricin resistant (NouR) transformants were selected on YPD supplemented with nourseothricin, methionine and cysteine to repress MET3p-cre expression. To induce the loss of the LUL and NAT1-Clox cassettes, Uri+ NouR isolates (sep7Δ::LUL/sep7Δ::NAT1-Clox) were grown overnight in YPD without supplements and plated onto YPD to obtain single colonies. To identify Uri- NouS cells, the colonies were replica plated onto YPD + nourseothricin and SC -Uri plates (Fig 2A). Interestingly, the NAT1-Clox cassette was resolved in all colonies tested whereas the LUL cassette was recycled in around 66% of the colonies. Growth of the Uri+ NouR isolates overnight in YPD with methionine and cysteine showed that marker resolution due to leaky cre recombinase expression is lower than 5% (Fig 2A). Similar high efficiency was obtained when the URA3-Clox cassette was used to construct a homozygous sep7Δ/sep7Δ null mutant from a sep7::LAL/SEP7 strain (not shown). Functionality of the pDIS-URA3-Clox and pDIS-NAT1-Clox plasmids to induce marker loss after integration of the cassettes at the NEUT5L locus was also tested, and the frequency of marker loss was similar to that obtained with pFA-URA3-Clox or pFA-NAT1-Clox plasmids. The loss of URA3 and NAT1 genes in Uri- NouS isolates was confirmed by diagnostic PCR using two different pairs of oligonucleotides, one annealing at the 5´ and 3´ flanking regions and another internal to the SEP7 coding region (Fig 2B).
Fig 2. Construction of deletion mutants without selection markers.
(A) The sep7::LUL/sep7::NAT1-Clox strain (OL2908) was grown overnight in YPD medium with (+M/C) or without (-M/C) methionine and cysteine to repress or induce the expression of the Cre recombinase respectively, and then plated on YPD medium. After 24 h of incubation at 28ºC, the plate was printed onto YPD+ nourseothricin or SC -Uri plates and incubated another 24 h. (B) PCR analysis of the different strains with specific SEP7 oligonucleotides. Genomic DNA was isolated from the parental strain BWP17 (1) and the sep7::loxP/sep7::loxP (2, OL2909) mutant and analyzed with two different oligonucleotide pairs, one annealing at the 5´ and 3´ flanking regions (A and B) and another internal to the coding region (C and D) to confirm correct integration and excision of the deletion cassettes. The position of the primers in the SEP7 locus and the sep7::loxP allele is depicted to the left.
Tagging multiple genes with fluorescent proteins and recyclable markers
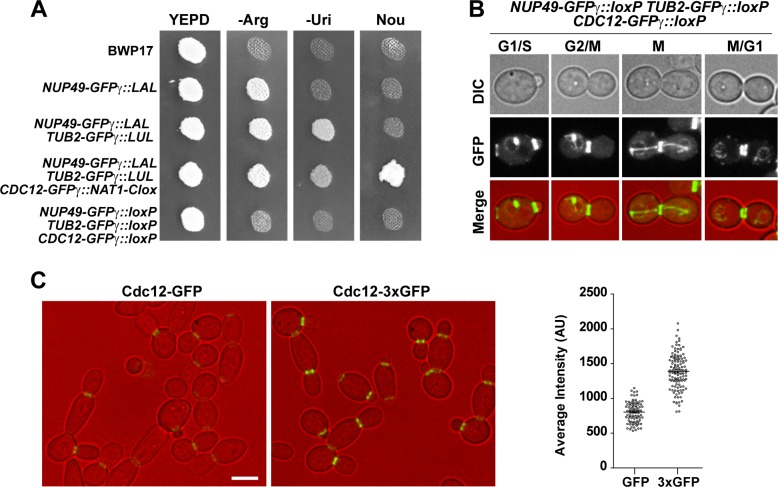
To validate the pFA-Clox plasmids for fluorescence microscopy, we sequentially tagged the nuclear membrane protein Nup49 [42] and the β-tubulin Tub2 [43] with the GFPγ-LAL and GFPγ-LUL cassettes to generate NUP49-GFPγ TUB2-GFPγ cells in a BWP17 background. Then, Arg+ Uri+ cells were transformed with a GFPγ-NAT1-Clox cassette to C-terminal tag the Cdc12 septin subunit [41]. Transformants were selected on YPD plus nourseothricin supplemented with methionine and cysteine to repress the MET3p-cre expression. As expected, cells sequentially transformed with these cassettes were Arg+, Uri+ and NouR (NUP49-GFPγ::LAL TUB2-GFPγ::LUL CDC12-GFPγ::NAT1-Clox). To induce the loss of the three selection markers, Arg+ Uri+ NouR cells were grown overnight at 30ºC in YPD lacking methionine and cysteine. Then, cells were plate on YPD and replica-plated onto selective media. Cells auxotrophic for arginine and uridine and sensitive to nourseothricin were selected (Fig 3A). As expected, the nuclear membrane, microtubules and the septin ring at the bud neck were fluorescence-labeled in the resulting strain (NUP49-GFPγ::loxP TUB2-GFPγ::loxP CDC12-GFPγ::loxP) (Fig 3B).
Fig 3. Generation of auxotroph strains containing three genes tagged with GFPγ.
(A) Growth in selective media. Single colonies of strains BWP-17, NUP49-GFPγ::LAL/NUP49 (OL2845), NUP49-GFPγ::LAL/NUP49 TUB2-GFPγ::LUL/TUB2 (OL2847), NUP49-GFPγ::LAL/NUP49 TUB2-GFPγ::LUL/TUB2 CDC12-GFPγ::NAT1-Clox/CDC12 (OL2867) and the resolved strain NUP49-GFPγ::loxP/NUP49 TUB2-GFPγ::loxP/TUB2 CDC12-GFPγ::loxP/CDC12 (OL2866) were grown on YPD plates and replica-printed to -Arg, -Uri and YPD + nourseothricin media. Plates were incubated 2 days at 28ºC. (B) Images of cells carrying Nup49-GFPγ, Tub2-GFPγ and Cdc12-GFPγ during yeast growth. Exponential growing cultures of the NUP49-GFPγ::loxP/NUP49 TUB2-GFPγ::loxP/TUB2 CDC12-GFPγ::loxP/CDC12 strain (OL2866) were imaged. The images are the maximum projection of 5 planes and show the differential interference contrast image (DIC), the GFP channel and the merged image (DIC in red, GFP in green). Scale bar, 2 μm. (C) Comparison of GFPγ and 3xGFPγ intensity. Images of CDC12-GFPγ::LAL/CDC12 or CDC12-3xGFPγ::LAL/CDC12 cells during yeast growth. The images are the maximum projection of 3 planes and show the differential interference contrast image (DIC, in red) and the GFP signal (green). Scale bar, 2 μm. The graph represents the average intensity of the signal ± s.e.m. in both strains. At least 50 single rings were measured in each strain.
To check the pFA-3xGFPγ plasmids, we tagged the CDC12 septin with one or three in-tandem copies of GFPγ to compare the intensity of the fluorescence signal in asynchronous cell cultures. To this end, BWP17 cells were transformed with the corresponding cassettes to generate the CDC12-3xGFPγ::LAL/CDC12 and CDC12-GFPγ::LAL/CDC12 strains. Then, the GFP signal in single septin rings of log-phase cells grown in YPD was quantified (n>50). As expected, fluorescence intensity was higher in CDC12-3xGFPγ::LAL/CDC12 cells (around 2-fold higher) than in cells expressing Cdc12 tagged with a single GFPγ (CDC12-GFPγ::LAL/CDC12) (Fig 3C). Thus, the set of pFA-3xGFPγ-Clox plasmids enhance the signal of the tagged proteins and might be used to study low-expressed proteins in C. albicans.
Dual-color fluorescence labeling with the recyclable marker plasmids
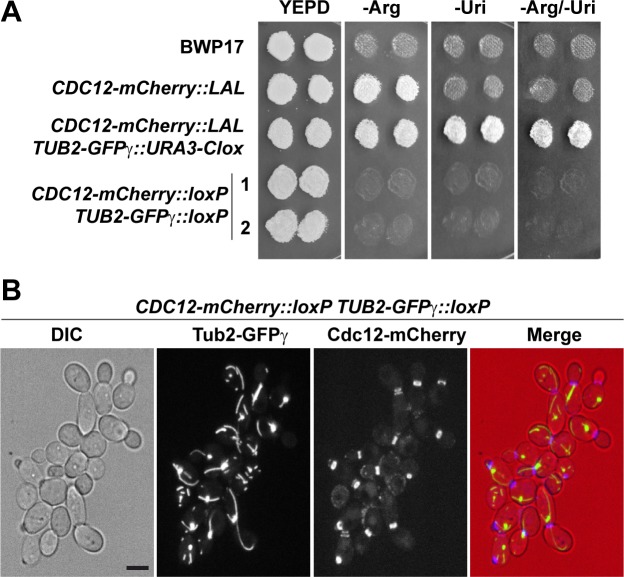
We also tested the utility of the plasmids for tagging proteins with two different fluorescent markers. To this end, the wild-type strain BWP-17 was transformed with a PCR amplified mCherry-LAL cassette to tag the C-terminus of CDC12. Transformants were selected in medium lacking arginine and single colonies were isolated. The CDC12-mCherry::LAL strain was subsequently transformed with a GFPγ-URA3-Clox cassette to be integrated at the C-terminus of the TUB2 gene. Uri+ transformants (CDC12-mCherry::LAL TUB2-GFPγ::URA3-Clox) were selected on SC lacking uridine and containing methionine and cysteine. To recycle both markers, cre expression was induced by growing Arg+ Uri+ cells overnight in YPD. Cells were then plated on YPD and colonies that contained only the loxP sequences were selected by replica-plating to selective media (Fig 4A). As expected, we were able to monitor microtubules and septin rings at the same time in these Arg- Uri- cells (CDC12-mCherry::loxP TUB2-GFPγ::loxP) (Fig 4B). Therefore, this result validates the usefulness of pFA-GFPγ-Clox and pFA-mCherry-Clox plasmids to generate strains for double labelling experiments.
Fig 4. Double fluorescent tagging with GFPγ and mCherry.
(A) Growth in selective media. Single colonies of strains BWP-17, CDC12-mCherry::LAL, CDC12-mCherry::LAL TUB2-GFPγ::URA3-Clox and two independent clones of the resolved strain CDC12-mCherry::loxP TUB2-GFPγ::loxP (1 and 2) were grown on YPD plates and replica-printed to SC -Arg, SC -Uri or SC -Arg/-Uri media. (B) Localization of Cdc12-mCherry and Tub2-GFPγ during yeast growth. Exponential growing cultures of the CDC12-mCherry::loxP TUB2-GFPγ::loxP strain were imaged. The images are the maximum projection of 5 planes and show the differential interference contrast image (DIC), the Tub2-GFPγ and the Cdc12-mCherry channels, and the merged image (DIC in red, Tub2-GFPγ in green and Cdc12-mCherry in blue). Scale bar, 2 μm.
Tagging multiple genes with HA
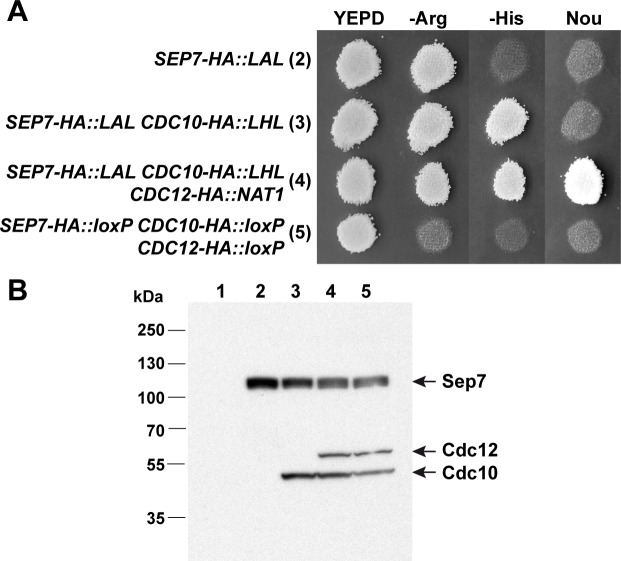
The collection of plasmids carrying HA epitopes and different recyclable markers was also tested. To this end, three of the five septin genes were tagged with the HA epitope at the C-terminus. Integration cassettes were generated using specific oligonucleotides containing 100 bp of the target sequence and either S1-HA or S2 extensions (Table 2), using different plasmids as templates. First, the wild-type strain BWP-17 was transformed with a PCR-amplified HA-LAL cassette to tag the C-terminus of SEP7 to generate the SEP7-HA::LAL/SEP7 strain. Then, positive colonies were transformed with a CDC10-HA::LHL cassette to construct the SEP7-HA::LAL/SEP7 CDC10-HA::LHL/CDC10 strain, and finally, the CDC12 gene was tagged using a CDC12-HA::NAT1-Clox. After marker resolution, colonies carrying loxP sequences were selected by plating colonies in YPD and replica-printing to selective media (Fig 5A). Western blot analysis of the different strains showed that the three septins were correctly tagged (Fig 5B). Similar results were obtained using the plasmids containing the myc epitope.
Fig 5. Multiple HA tagging.
(A) Growth in selective media. Single colonies of strains SEP7-HA::LAL/SEP7 (2), SEP7-HA::LAL/SEP7 CDC10-HA::LHL/CDC10 (3), SEP7-HA::LAL/SEP7 CDC10-HA::LHL/CDC10 CDC12-HA::NAT1-Clox/CDC12 (4) and SEP7-HA::loxP/SEP7 CDC10-HA::loxP/CDC10 CDC12-HA::lox/CDC12 (5) were grown on YPD plates and replica-printed to SC -Arg, SC -His or YPD+ nourseothricin media. (B) Western blot analysis of the same strains using anti-HA antibody. The BWP17 parental strain (1) was included as an untagged control. Protein extracts were separated on 4–12% Bis-Tris gels, blotted to PDVF membranes and incubated with anti-HA antibody.
Discussion
Gene deletion in C. albicans is a relatively time consuming process due to the particular characteristics of its biology. First, this fungus is diploid, being necessary to delete both chromosomal alleles to generate a homozygous null mutant. Second, it uses a non-canonical genetic code in which the CTG codon is read as Ser instead of Leu [7]. Therefore, specific codon-optimized tools have been developed for generating deletion mutants or epitope-tagged strains [8,9].
A large collection of PCR-based modules for efficient gene disruption has been developed in C. albicans. Most of them are based on the pFA-vector backbone used in the Saccharomyces cerevisiae EUROFAN deletion project [44]. These vectors are used to generate cassettes with flanking regions homologous to the gene of interest that target the module to the desired chromosomal locus [6,11]. In addition, the existing pFA-modules allow promoter-exchange experiments and tagging of genes with epitopes or fluorescent proteins at 5’ or 3’ end to monitor in vivo localization [12,14,15,45,46]. Due to the modular design of the plasmids, an advantage of this pFA system is that a minimal set of primers is required for the amplification of the cassettes, reducing the cost of strain construction.
Molecular techniques for gene manipulation in C. albicans face the problem of the limited range of auxotrophic markers available in laboratory strains (URA3, HIS1, ARG4, LEU2) [4,6,18,47] and the reduced number of dominant antibiotic markers for prototrophic clinical strains (MPAR, SAT1/NAT1, HygB)[21,22,48,49]. To circumvent this problem, strategies that allow the recycling of selectable markers have been used. Currently, the original URA3 blaster method [4] has been replaced by recombinase-mediated systems, such as the FLP recombinase gene disruption system or the Clox toolkit [13,20,21,23]. FLP expression is regulated by inducible promoters to mediate site-specific recombination between the FRT sites that flank the URA3 or SAT1 selection markers. The Clox toolkit also contains several selectable markers that can be recycled after induction of the Cre recombinase, increasing the yields of desired mutants and significantly decreasing the time required to generate these mutants.
In this work, we have replaced the non-recyclable markers of the pFA plasmids with the selectable markers of the Clox system, which facilitate marker recycling during strain construction. We report 36 new plasmids for gene disruption and protein tagging with GFP, mCherry, 3xHA, 5xmyc and TAP tags that facilitate PCR-based genetic modifications of both laboratory strains and clinical isolates of C. albicans. The usefulness of these plasmids is that it combines the advantages of both pFA and Clox systems, since amplification of the modules harboring recyclable markers can be performed with a minimal set of gene-specific primers. Since we have cloned the five cassettes of the Clox system into the pFA backbone, the primer sets already used with published pFA series are valid for these new pFA-Clox plasmids. Recently, a similar vector system for C-terminal and N-terminal epitope-tagging and for inducible and constitutive promoter replacement has been described [35]. However, they were constructed in the pUC19 backbone, and therefore contain different flanking sequences for cassette amplification that are not compatible with the pFA series. In addition, the system cannot be used in prototrophic clinical strains since plasmids do not include a dominant antibiotic marker such SAT1.
We have validated the deletion cassettes (Fig 2) and found that marker resolution occurs with a similar efficiency to that described previously [13]. Importantly, the retrospective genotyping approach used by Shahana and colleagues is also possible with the new plasmids, and PCR diagnosis to confirm the desired homozygous null mutants can be performed after resolution of the two selection markers. Generally, it is enough to analyze several colonies from a few independent transformants to obtain the homozygous mutant.
Plasmids containing fluorescent proteins and recyclable markers have also been constructed. Three sets were generated harboring the photostable variant of the green fluorescent protein known as GFPγ [36], three in-tandem copies of GFPγ [37] or the yEmCherry fluorescent protein [14]. Given all of them are flanked by the same S1-XFP and S2 sequences, the different cassettes can be amplified with the same pair of oligonucleotides. Using these modules, we have generated strains expressing one or several tagged proteins with an efficient resolution of the selectable markers after induction of the Cre recombinase (Figs 3 and 4). For single gene tagging, the fastest approach is to use the plasmids containing either URA3-Clox or NAT1-Clox markers to induce marker resolution after transformation. To tag several genes, plasmids containing the LAL, LHL or LUL should be used for the initial rounds of transformation, leaving the URA3-Clox or NAT1-Clox markers for the final transformation. In this way, all the markers present in a strain can be resolved with a single step, generating the auxotroph strain carrying the tagged proteins.
For epitope tagging, we have constructed plasmids that allow the generation of cassettes to tag proteins with 3xHA, 5xmyc and TAP. Similar to the plasmids containing fluorescent proteins, they can be used individually or in different combinations to construct multiple tagged strains without selection markers in the genome.
Finally, two additional plasmids have been generated, pDIS-URA3-Clox and pDIS-NAT1-Clox. These plasmids target the cassette containing the Cre recombinase to the large intergenic region of NEUT5L locus, which facilitates the integration. It has been shown that integration at NEUT5L has no negative effect on growth and allows expression of ectopic genes, making it ideal for the integration of exogenous DNA in C. albicans [39]. Therefore, they could be useful when it is necessary to resolve the LAL, LHL or LUL markers integrated anywhere in the genome. Since they also induce their own resolution, the resulting stain will only contain a copy of the loxP sequence at the NEUT5L locus. Although we have not validated every plasmid in this study, all cassettes were sequenced to check the absence of mutations. In summary, the new toolkit generated using the recyclable markers of the Clox system represents a significant step forward in the collection of tools available for C. albicans manipulation that should improve the study and characterization of this major human opportunistic pathogen.
Materials and methods
Strains and growth conditions
The strains used in this work are listed in Table A in S1 File. Cells were grown in YPD or in synthetic minimal (SC) medium containing the appropriated supplements at 28ºC. Construction of strains was done using the PCR-mediated procedure previously described [11] using the pFA-derived plasmids generated in this study as template. Transformation of the strains was performed by electroporation [21]. All transformants were checked for correct genome integration by PCR. Correct in-frame insertion of the fluorescent proteins was checked by sequencing or microscopic inspection of the transformants.
Plasmid assembly strategy
The plasmids in the toolkit were constructed using an assembly strategy that allowed the transfer of the deletion cassettes to the pFA backbone as independent modules. Assembly reactions were performed using the NEBuilder HiFI DNA Assembly Cloning Kit (New England Biolabs) following manufacturer’s instructions. Design of the primers required for PCR amplification of the different modules was performed with the NEBuilder Assembly tool (http://nebuilder.neb.com/). The modules were assembled into the pFA backbone obtained by digestion of plasmid pFA-CaHIS1 with BamHI and PmeI [11]. For the deletion plasmids, the selection markers of the Clox system (LAL, LHL, LUL, URA3-Clox and NAT1-Clox) were amplified with primers LXL1 and LXL2 that also carried a common sequence present in the vector, immediately upstream or downstream of the two loxP repeats and generated overlapping regions to the pFA backbone to be used in the assembly reaction.
For the assembly of plasmids containing fluorescent proteins or epitope tags, the Clox selection markers were amplified with primers GFP-LXL3 and LXL2 that generated overlapping regions between the epitope modules and the 3´ end of the pFA backbone. The different modules containing fluorescent proteins or epitope tags were amplified with a pair of primers that also included overlapping regions with the 5´ end of the pFA backbone and the selection module. For pFA-GFPγ-Clox plasmids, the GFPγ sequence, along with the ScURA3 terminator was amplified using primers GFP-LXL1 and GFP-LXL2 and plasmid pFA-GFPγ-URA3 [36] as template. The 3xGFPγ sequence with the ScURA3 terminator present in pFA-3xGFPγ-Clox plasmids and the yEmCherry-ScURA3 terminator module were amplified using the same primers (GFP-LXL1 and GFP-LXL2) and plasmids pFA-3xGFPγ-URA3 [37] or pFA-yEmCherry-CaURA3 [14] as templates, respectively. To amplify the module containing 3 in-tandem copies of the HA epitope with the S. cerevisiae URA3 terminator, primers HA1 and GFP-LXL2 were used, with pFA-HA-URA3 as template [15]. For pFA-myc-Clox plasmids, a PCR fragment containing 5 copies of the myc epitope and the S. cerevisiae URA3 terminator was amplified from plasmid pFA-myc-URA3 [15] using primers Myc1 and GFP-LXL2. Finally, the TAP-ScURA3 terminator module was amplified using primers TAP1 and GFP-LXL2 from plasmid pFA-TAP-URA3 [15]. The modules containing the epitopes or fluorescent proteins were mixed with the recyclable markers and the pFA backbone in independent assembly reactions and transformed in E. coli. Confirmation of the different plasmids was performed by PCR amplification with internal oligonucleotides and/or restriction digestion. Sequencing of the plasmids to verify the correct sequence was performed at the DNA Sequencing Facility-NUCLEUS of the University of Salamanca.
To construct plasmids pDIS-URA3-Clox and pDIS-NAT1-Clox, plasmid pDIS3 [39] was digested with NotI and PmeI to release the NAT1 marker and assembled with the URA3-Clox or NAT1-Clox modules respectively, amplified with oligonucleotides NEUT7 and NEUT8 that generated overlapping regions for the assembly reaction. To integrate these cassettes at the NEUT5L locus, the recyclable markers flanked by NEUT5L sequences can be amplified using oligonucleotides N5-up and N5-dw (Table 2). Alternatively, the whole cassette can be isolated by digestion with SfiI (Figure A in S1 File).
The sequences and annotations of all plasmids are available in GenBank. Accession numbers are indicated on Table 1. The plasmids are available from the authors.
Cre-mediated marker resolution
Elimination of the auxotrophic or resistance markers was performed using a variation of the previously described method [13]. In brief, single colonies of C. albicans transformants were grown overnight in YPD medium without methionine and cysteine (to induce MET3p-cre expression). Cells were plated onto YPD plates and replica-printed onto selective media to select colonies that had lost the auxotrophic and/or resistance markers. The genotype of the resolved mutants was confirmed by diagnostic PCR.
Western blot
Protein extracts were prepared as previously described [50]. In brief, cells from 5- to 10-ml mid-log cultures were harvested and resuspended in 1 ml of 20% trichloroacetic acid (TCA). The supernatant was removed after centrifugation, and the pellet was resuspended in 100 μl of 20% TCA. Cells were broken using a FastPrep FP120 cell disrupter (BIO 101 ThermoSavant) and the lysate was recovered. Lysates were centrifuged at 1000×g for 3 min, and the pellet was thoroughly resuspended in 100 μl of 2×Laemmli buffer and 50 μl of 2 M Tris base. After boiling for 5 min, 10–20 μl were loaded in the gels. Proteins were resolved by 10% SDS-PAGE, transferred to Hybond-P (Amersham Bioscience) membranes and probed with anti-HA High Affinity antibody (3F10, Roche). Horseradish peroxidase-conjugated anti-rat antibody (NA935V, GE Healthcare) was used as secondary antibody. Detection of proteins was performed using the Luminata Forte Western HRP substrate (Merck Millipore).
Microscopy
Fluorescence microscopy was performed with an Olympus IX81 microscope equipped with a spinning-disc confocal system (Roper Scientific). Three to five z-planes were collected with step sizes of 0.4 μm. Average fluorescence intensity was quantified with ImageJ (http://rsb.info.nih.gov/ij/).
Supporting information
Maps and nomenclature of modules cloned into the pFA vector background. LAL, LHL, LUL, URA3-Clox and NAT1-Clox were used as marker genes. The GFPγ, 3xGFPγ and yEmCherry served as fluorescent protein-encoding ORFs. HA, myc and TAP tags are also shown. The annealing sites of the oligonucleotides used for cassette amplification are indicated in blue. Only unique restriction sites are show.
(PDF)
Acknowledgments
We thank Alistair Brown for generously providing the Clox system, James Konopka for the photostable version of the GFP, Jeong-Yoon Kim for her kind gift of the pFA-3xGFPγ-URA3 plasmid, J. Wendland for the pFA-yEmCherry-CaURA3 plasmid, and J. Berman for the pDIS3 plasmid. We thank the DNA Sequencing Facility-NUCLEUS of the University of Salamanca for plasmid sequencing.
Data Availability
All the sequences from the vectors are available from the GenBank database (accession number(s) MK652110, MK652111, MK652112, MK652113, MK652114, MK652105, MK652106, MK652107, MK652108, MK652109, MK652100, MK652101, MK652102, MK652103, MK652104, MK652115, MK652116, MK652117, MK652118, MK652119, MK652120, MK652121, MK652122, MK652123, MK652124, MK652125, MK652126, MK652127, MK652128, MK652129, MK652130, MK652131, MK652132, MK652133, MK652134 and MK652135).
Funding Statement
This work was supported by grants from the Spanish Ministry of Science and Innovation to CRV (BIO2015-70195-C2-1-R) and to JC-B (BIO2015-70195-C2-2R), the Junta de Extremadura to JC-B (GR15008). All Spanish funding is co-sponsored by the European Union FEDER programme. The IBFG is supported by Programa "Escalera de Excelencia" from Junta de Castilla y León (CLU-2017-03, P.O. FEDER de Castilla y León 14-20). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kumamoto CA. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14: 386–391. 10.1016/j.mib.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett RJ. The parasexual lifestyle of Candida albicans. Curr Opin Microbiol. 2015;28: 10–17. 10.1016/j.mib.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid J, Magee PT, Holland BR, Zhang N, Cannon RD, Magee BB. Last hope for the doomed? Thoughts on the importance of a parasexual cycle for the yeast Candida albicans. Curr Genet. 2016;62: 81–85. 10.1007/s00294-015-0516-8 [DOI] [PubMed] [Google Scholar]
- 4.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Backer MD, Magee PT, Pla J. Recent developments in molecular genetics of Candida albicans. Annu Rev Microbiol. 2000;54: 463–498. 10.1146/annurev.micro.54.1.463 [DOI] [PubMed] [Google Scholar]
- 6.Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16: 65–70. [DOI] [PubMed] [Google Scholar]
- 7.Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23: 1481–1486. 10.1093/nar/23.9.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu QR, Yan L, Lv QZ, Zhou M, Sui X, Cao YB, et al. Molecular genetic techniques for gene manipulation in Candida albicans. Virulence. 2014;5: 507–520. 10.4161/viru.28893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samaranayake DP, Hanes SD. Milestones in Candida albicans gene manipulation. Fungal Genet Biol. 2011;48: 858–865. 10.1016/j.fgb.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walther A, Wendland J. PCR-based gene targeting in Candida albicans. Nat Protoc. 2008;3: 1414–1421. 10.1038/nprot.2008.137 [DOI] [PubMed] [Google Scholar]
- 11.Gola S, Martin R, Walther A, Dunkler A, Wendland J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20: 1339–1347. 10.1002/yea.1044 [DOI] [PubMed] [Google Scholar]
- 12.Schaub Y, Dunkler A, Walther A, Wendland J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J Basic Microbiol. 2006;46: 416–429. 10.1002/jobm.200510133 [DOI] [PubMed] [Google Scholar]
- 13.Shahana S, Childers DS, Ballou ER, Bohovych I, Odds FC, Gow NA, et al. New Clox Systems for rapid and efficient gene disruption in Candida albicans. PLoS One. 2014;9: e100390 10.1371/journal.pone.0100390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reijnst P, Walther A, Wendland J. Dual-colour fluorescence microscopy using yEmCherry-/GFP-tagging of eisosome components Pil1 and Lsp1 in Candida albicans. Yeast. 2011;28: 331–338. 10.1002/yea.1841 [DOI] [PubMed] [Google Scholar]
- 15.Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9: 578 10.1186/1471-2164-9-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5´-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197: 345–346. [DOI] [PubMed] [Google Scholar]
- 17.Wellington M, Rustchenko E. 5-Fluoro-orotic acid induces chromosome alterations in Candida albicans. Yeast. 2005;22: 57–70. 10.1002/yea.1191 [DOI] [PubMed] [Google Scholar]
- 18.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4: 298–309. 10.1128/EC.4.2.298-309.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181: 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morschhauser J, Michel S, Staib P. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol. 1999;32: 547–556. 10.1046/j.1365-2958.1999.01393.x [DOI] [PubMed] [Google Scholar]
- 21.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341: 119–127. 10.1016/j.gene.2004.06.021 [DOI] [PubMed] [Google Scholar]
- 22.Shen J, Guo W, Kohler JR. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun. 2005;73: 1239–1242. 10.1128/IAI.73.2.1239-1242.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennison PM, Ramsdale M, Manson CL, Brown AJ. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol. 2005;42: 737–748. 10.1016/j.fgb.2005.05.006 [DOI] [PubMed] [Google Scholar]
- 24.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150: 467–486. 10.1016/0022-2836(81)90375-2 [DOI] [PubMed] [Google Scholar]
- 25.Sauer B. Functional expression of the Cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7: 2087–2096. 10.1128/mcb.7.6.2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85: 5166–5170. 10.1073/pnas.85.14.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro RS, Chavez A, Collins JJ. CRISPR-based genomic tools for the manipulation of genetically intractable microorganisms. Nat Rev Microbiol. 2018;16: 333–339. 10.1038/s41579-018-0002-7 [DOI] [PubMed] [Google Scholar]
- 28.Min K, Ichikawa Y, Woolford CA, Mitchell AP. Candida albicans Gene Deletion with a Transient CRISPR-Cas9 System. mSphere. 2016;1 10.1128/mSphere.00130-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vyas VK, Barrasa MI, Fink GR. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv. 2015;1: e1500248 10.1126/sciadv.1500248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang MY, Mitchell AP. Marker Recycling in Candida albicans through CRISPR-Cas9-Induced Marker Excision. mSphere. 2017;2 10.1128/mSphere.00050-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen N, Quail MMF, Hernday AD. An Efficient, Rapid, and Recyclable System for CRISPR-Mediated Genome Editing in Candida albicans. mSphere. 2017;2 10.1128/mSphereDirect.00149-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roman E, Coman I, Prieto D, Alonso-Monge R, Plá J. Implementation of a CRISPR-Based System for Gene Regulation in Candida albicans. mSphere. 2019;4 10.1128/mSphere.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wensing L, Sharma J, Uthayakumar D, Proteau Y, Chavez A, Shapiro RS. A CRISPR Interference Platform for Efficient Genetic Repression in Candida albicans. mSphere. 2019;4 10.1128/mSphere.00002-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernday AD, Noble SM, Mitrovich QM, Johnson AD. Genetics and molecular biology in Candida albicans. Methods Enzymol. 2010;470: 737–758. 10.1016/S0076-6879(10)70031-8 [DOI] [PubMed] [Google Scholar]
- 35.Chang P, Wang W, Igarashi Y, Luo F, Chen J. Efficient vector systems for economical and rapid epitope-tagging and overexpression in Candida albicans. J Microbiol Methods. 2018;149: 14–19. 10.1016/j.mimet.2018.04.016 [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Konopka JB. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot Cell. 2010;9: 224–226. 10.1128/EC.00327-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HJ, Kim JM, Kang WK, Yang H, Kim JY. The NDR Kinase Cbk1 Downregulates the Transcriptional Repressor Nrg1 through the mRNA-Binding Protein Ssd1 in Candida albicans. Eukaryot Cell. 2015;14: 671–683. 10.1128/EC.00016-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17: 1030–1032. 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- 39.Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J. Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology. 2013;159: 565–579. 10.1099/mic.0.064097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Novo A, Correa-Bordes J, Labrador L, Sánchez M, Vázquez de Aldana CR, Jiménez J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol Biol Cell. 2008;19: 1509–1518. 10.1091/mbc.E07-09-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol Biol Cell. 2002;13: 2732–2746. 10.1091/mbc.E02-01-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finley KR, Berman J. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot Cell. 2005;4: 1697–1711. 10.1128/EC.4.10.1697-1711.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith HA, Allaudeen HS, Whitman MH, Koltin Y, Gorman JA. Isolation and characterization of a beta-tubulin gene from Candida albicans. Gene. 1988;63: 53–63. 10.1016/0378-1119(88)90545-8 [DOI] [PubMed] [Google Scholar]
- 44.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10: 1793–1808. 10.1002/yea.320101310 [DOI] [PubMed] [Google Scholar]
- 45.Gerami-Nejad M, Dulmage K, Berman J. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast. 2009;26: 399–406. 10.1002/yea.1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerami-Nejad M, Hausauer D, McClellan M, Berman J, Gale C. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast. 2004;21: 429–436. 10.1002/yea.1080 [DOI] [PubMed] [Google Scholar]
- 47.Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182: 5730–5736. 10.1128/jb.182.20.5730-5736.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohler GA, White TC, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol. 1997;179: 2331–2338. 10.1128/jb.179.7.2331-2338.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basso LR Jr., Bartiss A, Mao Y, Gast CE, Coelho PS, Snyder M, et al. Transformation of Candida albicans with a synthetic hygromycin B resistance gene. Yeast. 2010;27: 1039–1048. 10.1002/yea.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Acosta I, Ontoso D, San-Segundo PA. The budding yeast polo-like kinase Cdc5 regulates the Ndt80 branch of the meiotic recombination checkpoint pathway. Mol Biol Cell. 2011;22: 3478–3490. 10.1091/mbc.E11-06-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maps and nomenclature of modules cloned into the pFA vector background. LAL, LHL, LUL, URA3-Clox and NAT1-Clox were used as marker genes. The GFPγ, 3xGFPγ and yEmCherry served as fluorescent protein-encoding ORFs. HA, myc and TAP tags are also shown. The annealing sites of the oligonucleotides used for cassette amplification are indicated in blue. Only unique restriction sites are show.
(PDF)
Data Availability Statement
All the sequences from the vectors are available from the GenBank database (accession number(s) MK652110, MK652111, MK652112, MK652113, MK652114, MK652105, MK652106, MK652107, MK652108, MK652109, MK652100, MK652101, MK652102, MK652103, MK652104, MK652115, MK652116, MK652117, MK652118, MK652119, MK652120, MK652121, MK652122, MK652123, MK652124, MK652125, MK652126, MK652127, MK652128, MK652129, MK652130, MK652131, MK652132, MK652133, MK652134 and MK652135).