Abstract
Background/Aims
Gastric cancer is one of the most common malignant tumors worldwide with poor prognosis due to a lack of effective treatment modalities. Recent research showed that a long noncoding RNA named N-BLR modulates the epithelial-to-mesenchymal transition (EMT) process in colorectal cancer. However, the biological role of N-BLR in gastric cancer still remains to be explored. The aim of this study was to investigate the possibility of N-BLR as an EMT modulator in gastric cancer.
Methods
The expression of N-BLR was measured by quantitative polymerase chain reaction in fresh gastric cancer tissue, paired adjacent normal tissues and cell lines. Fresh gastric tissues, paired samples obtained by surgery and clinical data were collected prospectively. Knockdown of N-BLR was induced by small interfering RNA (siRNAs). Cell number and viability were assessed after treatment with siRNAs. The ability of N-BLR to promote metastasis was measured using migration and invasion assays. Additionally, an inverse correlation between N-BLR and miR-200c was measured by TaqMan microRNA assays. Western blotting was performed to detect EMT and apoptosis markers upon knockdown of N-BLR.
Results
N-BLR expression was significantly elevated in gastric cancer cell lines and tissues compared to that in a normal gastric cell line and adjacent normal tissues (p<0.01). Two different siRNAs significantly reduced cell proliferation of gastric cancer cells compared to the siCT. siRNAs for N-BLR significantly suppressed migration and invasion in AGS and MKN28 cells. N-BLR expression was inversely correlated with miR-200c, which is known to regulate EMT.
Conclusions
In this study, we confirmed N-BLR as a regulator of the EMT process in gastric cance
Keywords: RNA, long noncoding, Epithelial-to-mesenchymal transition, Gastric cancer
INTRODUCTION
Gastric cancer is the third leading cause of cancer death worldwide, with no significant biomarkers available for utilization in clinical treatment.1 Recent studies have demonstrated that noncoding RNAs, including miRNA and long noncoding RNA (lncRNA), play important roles in gastric cancer progression and epigenetic gene expression at the transcript and post-transcript levels.2–4 LncRNAs have drawn attention to the new class of noncoding RNA with lengths greater than 200 nucleotides that act in critical roles in biological processes.5–7 Recently, some lncRNAs, such as H19, HOTAIR, and MEG3, have been found to engage in controlling tumorigenesis, tumor progression, and metastasis of diverse cancers.8–10 We previously announced that MALAT1 and HOTAIR can modulate invasion and migration in gastric cancer.10,11 Based on these results, we assumed that other lncRNAs are also involved in gastric carcinogenesis.
N-BLR is a primate-specific lncRNA that has an important contribution to the mechanism of metastasis in colorectal cancer (CRC). In particular, microarray findings, including transcription patterns of 2500+ human genome loci, showed N-BLR abundance in CRC and its role in the epithelial-to-mesenchymal transition (EMT) process.12 However, the functional role of N-BLR in gastric cancer has not yet been discovered.
In this study, we examined the expression of N-BLR in gastric cancer and adjacent normal tissues. We investigated the effects of N-BLR on apoptosis, cell proliferation, colony formation, and cell cycle as a carcinogenic indicator in gastric cancer. Additionally, we analyzed N-BLR’s molecular mechanisms with regard to invasion and metastasis in vitro.
MATERIALS AND METHODS
1. Patients and tissue samples
Fresh gastric cancer tissue and paired adjacent gastric tissue samples were obtained from 38 patients who underwent surgical resection for gastric cancer at Severance Hospital, Yonsei University College of Medicine. All samples were frozen in liquid nitrogen immediately after resection and stored at −80°C until use. In addition, patient and lesion characteristics, comprising age, sex, Helicobacter pylori infection, tumor location, tumor differentiation, depth of tumor invasion, and lymph node metastasis, were obtained (Table 1). This study was approved by the Ethics Committee of Yonsei Hospital. Informed consent was obtained from all patients. Study protocol was reviewed and approved by the Institutional Review Board (IRB number: 4-2011-0753).
Table 1.
Patient and Lesion Characteristics According to the Level of N-BLR Expression
| Variable | N-BLR expression* | p-value | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Age, yr | 0.746 | ||
| <65 | 10 (52.6) | 9 (47.4) | |
| ≥65 | 9 (47.4) | 10 (52.6) | |
| Sex | >0.999 | ||
| Male | 12 (63.2) | 12 (63.2) | |
| Female | 7 (36.8) | 7 (36.8) | |
| Helicobacter pylori infection† | 0.637 | ||
| Absent | 5 (55.6) | 3 (37.5) | |
| Present | 4 (44.4) | 5 (62.5) | |
| Location | 0.798 | ||
| Upper third | 2 (10.5) | 1 (5.3) | |
| Middle third | 8 (42.1) | 7 (36.8) | |
| Lower third | 9 (47.4) | 11 (57.9) | |
| Depth of tumor invasion | 0.461 | ||
| T1&T2 | 4 (21.1) | 6 (37.6) | |
| T3&T4 | 15 (78.9) | 13 (68.4) | |
| Lymph node metastasis | 0.097 | ||
| Absent | 5 (26.3) | 10 (52.6) | |
| Present | 14 (73.7) | 9 (47.4) | |
| Lymphovascular invasion | 0.746 | ||
| Absent | 9 (47.4) | 10 (52.6) | |
| Present | 10 (52.6) | 9 (47.4) | |
| Stage‡ | 0.105 | ||
| I, II | 7 (36.8) | 12 (63.2) | |
| III | 12 (63.2) | 7 (36.8) | |
| Lauren's classification | 0.127 | ||
| Intestinal | 8 (42.1) | 14 (7.37) | |
| Diffuse | 10 (52.6) | 4 (21.1) | |
| Mixed | 1 (5.3) | 1 (5.3) | |
| Differentiation | 0.051 | ||
| AWD or AMD | 9 (47.4) | 11 (57.9) | |
| APD | 3 (15.8) | 7 (36.8) | |
| SRC | 7 (36.8) | 1 (5.3) | |
| Serum CEA value, ng/mL | 0.693 | ||
| ≤5 | 14 (73.7) | 16 (84.2) | |
| >5 | 5 (26.3) | 3 (15.8) | |
| Serum CA19-9 value, U/mL | >0.999 | ||
| ≤37 | 18 (94.7) | 17 (89.5) | |
| >37 | 1 (5.3) | 2 (10.5) | |
Data are presented as number (%).
AWD, well-differentiated adenocarcinoma; AMD, moderately differentiated adenocarcinoma; APD, poorly differentiated adenocarcinoma; SRC, signet ring cell carcinoma; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9.
Patients were classified into two groups according to the median N-BLR expression;
Status of H. pylori infection was missing in 21 patients;
Stage was classified according to the 7th edition of the American Joint Cancer Committee/Union Internationale Contre le Cancer.
2. Cell lines and cell culture
A total of seven normal gastric and cancer cell lines were purchased from the Korean Cell Line Bank (Seoul National University, Seoul, Korea) or the American Type Culture Collection (Rockville, MD, USA). The cells were cultured in RPMI-1640 medium (Thermo Scientific, Rockford, IL, USA) with 10% fetal bovine serum and 1% penicillin/streptomycin (Thermo Scientific). These cells were maintained in a humidified atmosphere of 5% CO2 at 37°C. In addition, whole stomach cancer cells were used up to 30 passages; when cells were used up to 30 passages, they were discarded and new stocks were thawed.
3. Small interfering RNA (siRNA) transfection
For transfection, AGS, MKN 28, SNU719, and KATOIII cells (3×105) were seeded in 6-well culture plates and incubated at 37°C for 24 hours. The cells were transfected with targeted siRNA (lncRNA N-BLR, 50 μM; Invitrogen, Carlsbad, CA, USA) and RNAi negative control (50 μM, siCT; Invitrogen) using Li-pofectamine 2000 reagent (Invitrogen) following the manufacturer’s protocol. The N-BLR siRNA target sequences were as follows: siN-BLR1 sense, UGUGGGAUGGUAGAGAGCAGUCUUU and antisense, AAAGACUGCUCUCUACCAUCCCAGA; siN-BLR2 sense, GCUCAAAUCUACAGGACAAUU and antisense, AAUU GUCCUGUAGAUUUGAGC.
4. Total RNA extraction and quantitative real-time reverse transcription polymerase chain reaction
Total RNA was extracted from gastric cancer tissues and cell lines using TRIzol reagent (Invitrogen). RNA was measured using a Nanodrop (ND-100; NanoDropTechnologies Inc., Wilmington, DE, USA). Purity was confirmed based on the 260/280 nm ratio, and products were loaded on 1% agarose gel. For cDNA synthesis, 2.0 μg of RNA was reverse transcribed using SuperScript™ II (Invitrogen) following the manufacturer’s protocol. The relative level of N-BLR was measured through real-time polymerase chain reaction (PCR) using iQ SYBR Green Supermix (Applied Biosystems Inc., Carlsbad, CA, USA). For miRNA analysis, 10 ng of total RNA were used for cDNA synthesis with RT primers for miR-200c-3p and U6 snRNA via TaqMan MicroRNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. Real-time PCR was carried out as mentioned above, using TaqMan microRNA assays (#002300 and #001973, ThermoFisher Scientific). The Ct value of the sample was normalized to U6 expression, and the 2−ΔΔCt value was calculated. Target sequences for N-BLR and U6 were as follows: N-BLR sense, GTAGCCGAATTGGGGATTTT and antisense, CGGGCTTGATCGTACAATTT; U6 sense, CTCGCTTCG-GCAGCACA and antisense, AACGCTTCAGGAATTTGCGT.
5. Cell proliferation analysis
Gastric cancer cells were transfected with 50 μM siN-BLRs from 0 to 72 hours. Cell proliferation was detected using a CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS assay; Promega, Madison, WI, USA) in 96-well culture plates at specified time intervals. The number of viable cells was allowed to react with MTS reagent for 1 hour in the dark. The amount of the reaction was measured with a spectrophotometric plate reader set at 490 nm (Multiskan™ Microplate Photometer; Thermo Scientific).
6. Scratch wound healing assay and invasion assay
AGS cells and MKN28 cells transfected with siN-BLRs or siCT were re-seeded in 6-well culture plates. When cells reached an approximate 60% to 80% confluency, the bottom of each well was scratched using a P-20 tip. The width of the scratched cells was measured at 0 and 24 hours under a bright-field microscope. For the invasion assay, using the same cell line and conditions above, cells were replated on BD BioCoat transwells (BD Biosciences, San Jose, CA, USA) following the manufacturer’s protocol. After 24 hours, non-invading cells within the chamber were removed, and the upper layer of the trans-well was wiped with a cotton swab. The membrane of the bottom part of the upper chamber was fixed with 5% acetaldehyde buffer and stained with crystal violet solution. The invading cells on the membrane were counted under a bright-field microscope.
7. Cell cycle analysis and apoptosis analysis
Transfected AGS cells and MKN28 cells were washed with PBS and fixed with 75% ethanol at −20°C overnight. Cells were resuspended in PBS and treated with RNase for 30 minutes at room temperature. The nuclei of the fixed cells were stained with 50 mg/mL PI (Sigma, St. Louis, MO, USA) in the dark for 20 minutes. Cell cycle phases were determined via flow cytometry (BD Biosciences). For apoptosis analysis, the same two gastric cancer cell lines were washed with PBS and resuspended in 1× binding buffer (BD Biosciences). Fluorescein isothiocyanate (FITC), Annexin V, and propidium iodide staining was conducted with the FITC Annexin V detection kit (BD biosciences) according to the manufacturer’s protocol. The rate of apoptosis was measured using flow cytometry (BD Biosciences).
8. Soft agar colony formation assay
To analyze tumorigenicity in vitro, base and top agarose gels were coated in 96-well culture plates using CytoSelect™ 96-Well Cell Transformation Assay (Cell Biolabs, Inc). Then, 1.5 mL of 2X DMEM containing 1% agarose was poured into each well as a base layer. After 1 hour of solidification, the trans-fected cells were resuspended in 2X DMEM containing 0.7% agarose as a top layer and maintained in 37°C incubator for 2 to 3 weeks. After daily observation, colonies were counted using bright field microscopy.
9. Western blot
Transfected cells with siN-BLRs or siCT were lysed in 1X RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) containing protease inhibitor. Isolated proteins were loaded onto 8% to 15% SDS–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (GE Healthcare, Piscat-away, NJ, USA). The membrane was blocked for 1 hour at room temperature in tris–phosphate buffer containing 0.1% Tween 20 with 5% BSA (BD biosciences), before being incubated with primary antibodies at 4°C overnight. The following primary antibodies were used for Western blot analysis: epithelial marker E-cadherin (1:1,000; BD Biosciences), mesenchymal marker N-cadherin (1:1,000; BD Biosciences), vimentin (1:200; Santa Cruz Biotechnology, Dallas, TX, USA, sc-373717), ZEB1 (1:1,000; Cell Signaling Technology), Snail (1:1,000; Cell Signaling Technology), PARP (1:1,000; Cell Signaling Technology, #9542), bcl-xl (1:1,000; Cell Signaling Technology, #2764), Bax (1:1,000; Santa Cruz Biotechnology, sc-493), cytochrome c (1:1,000; BD Biosciences), and β-actin (1:5,000; Bioworld Technology, St. Louis Park, MN, USA, AP0060). The signal was developed in ECL solution (GenDEPOT, Barker, TX, USA) and exposed to an Image Quant LAS 4000 bio-molecular imager for time intervals between 10 seconds and 6 minutes.
10. Statistical analysis
All analytical data for continuous variables are presented as the mean±standard error. Categorical variables were demonstrated using proportions. Statistical tests comprised the t-test, chi-square test, and Fisher exact test. Based on the median of N-BLR expression, the expressions of N-BLR on gastric cancer were classified into low and high. The Kaplan-Meier method and log-rank test were used for survival analysis. A value of p<0.05 was regarded as a statistically significant difference for comparisons between groups. All statistical processes were accomplished using the statistical software SPSS for Windows version 18.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
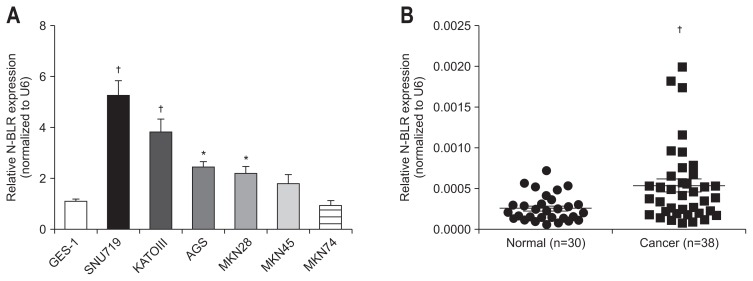
1. N-BLR is upregulated in gastric cancer cell lines and gastric cancer tissues
To further investigate the effect of N-BLR on gastric cancer, we observed the expression of N-BLR in normal epithelial gastric GES-1 and six gastric cancer cell lines, SNU719, KATOIII, AGS, MKN28, MKN 45, and MKN74. The six gastric cancer cell lines revealed statistically significant elevation of N-BLR expression compared to GES-1 (Fig. 1A). The expression of N-BLR was significantly higher in cancer tissues than adjacent non-cancer tissues from 38 patients with gastric cancer (p<0.01) (Fig. 1B).
Fig. 1.
N-BLR was overexpressed in gastric cell lines and tissues. (A) The relative expression of N-BLR was upregulated in gastric cancer cells including SNU719, KATOIII, AGS, MKN28, MKN45, and MKN74, compared to GES-1, a normal gastric cell line. (B) N-BLR expression was estimated in gastric cancer tissues (n=38) and paired adjacent noncancerous tissues (n=30). The expression of N-BLR was calculated by the 2–ΔΔCt method using U6 levels for normalization. These data represent the mean±SEM. *p<0.05 and †p<0.01 show a statistically significant difference compared with a scrambled control. N-BLR was not detected in eight adjacent noncancerous tissue samples by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR).
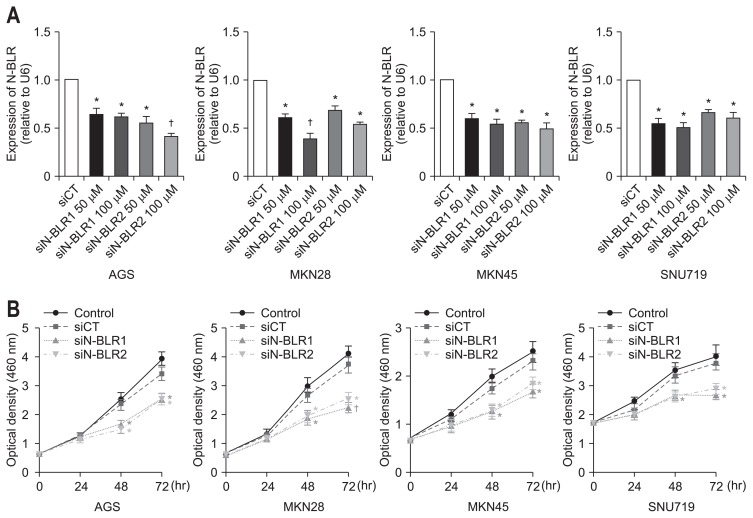
2. Inhibition of N-BLRs by siRNAs down-regulated cell proliferation of gastric cancer cell lines
We employed two different siRNA for N-BLR in AGS, MKN28, SNU719, and MKN45 cells to explore mechanisms of carcinogenesis. After treatment with siN-BLR1 or siN-BLR2 in gastric cancer cell lines, significantly reduced expression of N-BLR was seen in AGS, MKN28, SNU719, and MKN45 compared to that of siCT in a dose-dependent manner (Fig. 2A). Both siN-BLR1 and siN-BLR2 showed almost 50% knock-down at a concentration of 50 μM in gastric cancer cell lines.
Fig. 2.
Knockdown of N-BLR expression suppressed gastric cancer cell proliferation. AGS, MKN28, MKN45, and SNU719 cells were transfected with siN-BLR1, siN-BLR2, or scrambled RNA (siCT, 50 μM), and expression of N-BLR (A) and cell viability were measured by MTS assay in gastric cancer cell lines (B). The data shown in the figures are from three independent experiments and represent the mean±SEM. *p<0.05 and †p<0.01 show a statistically significant difference compared with a scrambled control.
Next, we performed an MTS assay to assess viable cells after transfection with siRNAs for N-BLR. In four gastric cancer cell lines, both siN-BLR1 and siN-BLR2 significantly decreased cell viability at both 48 and 72 hours (Fig. 2B).
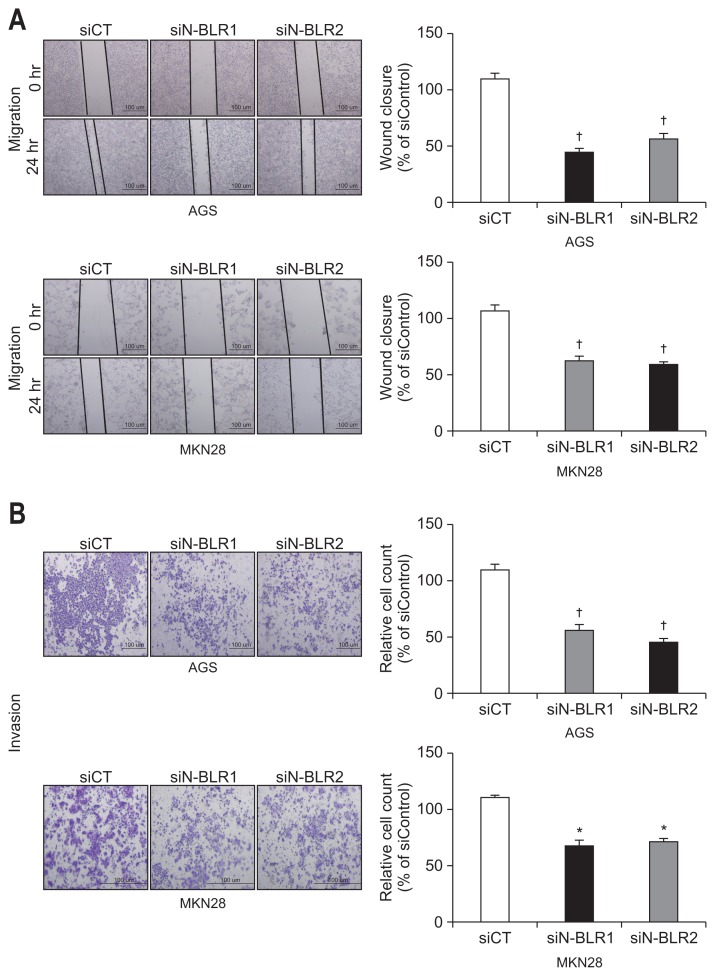
3. Inhibition of N-BLR decreased migration and invasion of MKN28 and AGS cells
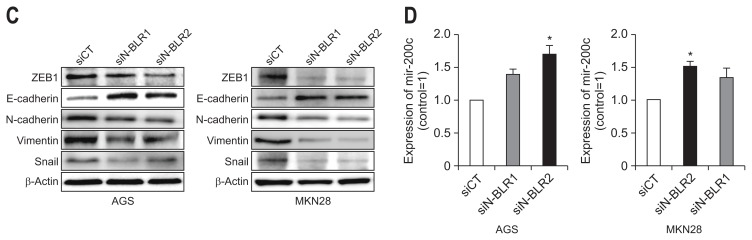
We explored the invasion and migration of gastric cancer cells using siRNAs for N-BLR. In the wound-healing assays, the migration of MKN28 and AGS cells was significantly inhibited by all siN-BLRs compared to that of the siCT (Fig. 3A). In a Matrigel invasion assay, all siRNAs significantly reduced invasion, which is consistent with the wound healing assay results (Fig. 3B). After treatment of siN-BLRs, the expressions of ZEB1, Snail, N-cadherin, and Vimentin were reduced; however, E-cadherin increased in both MKN28 and AGS cells (Fig. 3C). Treating siN-BLRs increased the level of miR-200c-3p (Fig. 3D) in both MKN28 and AGS cells.
Fig. 3.
siN-BLRs suppressed cell migration and invasion. (A) Wound healing assay was observed by microscopy at 0 and 24 hours. Scale bar refers to 100 μm. (B) Matrigel invasion assay was performed using an invasion chamber after knockdown of N-BLR expression. After treatment with siRNAs, AGS and MKN28 cells were stained with 0.1% coomassie brilliant blue. Scale bar refers to 100 μm. (C) EMT markers were detected by immunoblotting in transfected AGS and MKN28 cells. (D) The influence of transient transfection with siN-BLR on the miR-200 family. MiR-200c-3p was increased in both N-BLR small interfering RNA (siRNA)-transfected cell lines compared with cells transfect-ed with the scramble control. The data shown in the figures are from three independent experiments and represent the mean±SEM. *p<0.05 and †p<0.01 show a statistically significant difference compared with a scrambled control.
EMT, epithelial-to-mesenchymal transition.
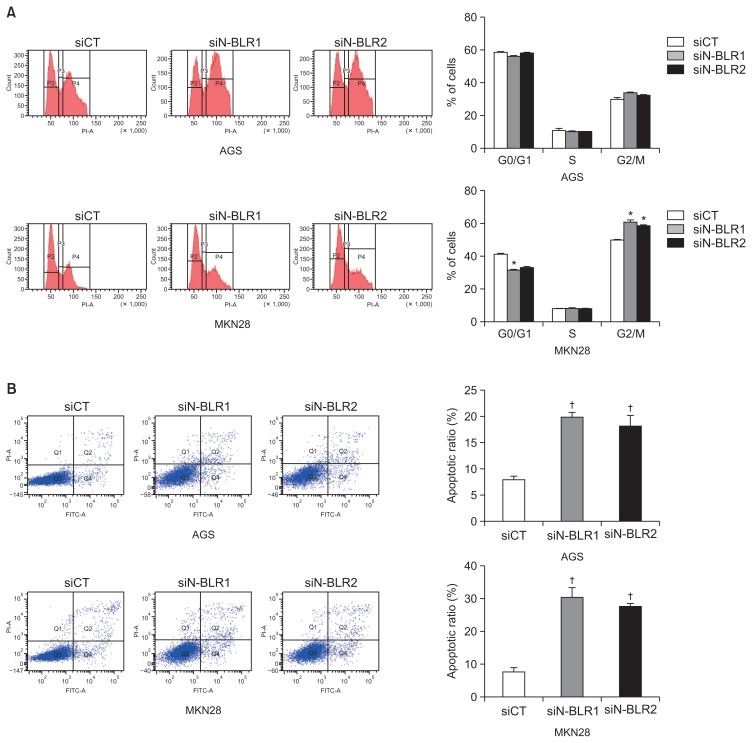
4. Suppression of N-BLR expression induces cell cycle arrest and apoptosis
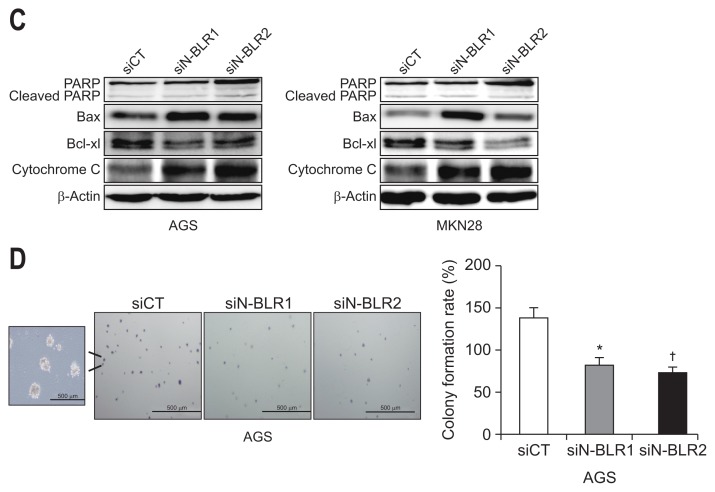
To follow up on our cell proliferation results, we evaluated the role of N-BLR in cell cycle progression and apoptosis. Treatment with siN-BLRs promoted cell cycle arrest in the G2/M phase and decreases the number of MKN28 cells in G0/G1 (Fig. 4A). The same phenomenon was not observed in AGS cells. Furthermore, staining the cells with PI/Annexin V and siN-BLRs significantly increased apoptosis in AGS and MKN28 cells (Fig. 4B). Both early and late apoptosis was increased by siN-BLRs. siN-BLRs increased cytochrome C, Bax, and PARP expression and decreased Bcl-XL expression in both cell lines (Fig. 4C). Next, we examined a soft agar colony formation assay to verify cellular anchorage-independent growth in vitro. The capability for colony formation decreased significantly in both siN-BLRs compared to siCT (Fig. 4D). Based on these results, we concluded that N-BLR induced cell cycle arrest and apoptosis and decreased colony formation correlated to cell proliferation in gastric cancer.
Fig. 4.
N-BLR silencing induced apoptosis and cell cycle arrest. AGS and MKN28 cells were transfected with siCT or siN-BLRs, followed by cell-cycle analysis using propidium iodide (PI) staining (A) and an apoptosis assay using PI/Annexin-V staining (B). (C) Western blotting analysis showed apoptotic and cell cycle markers in transfected AGS and MKN28 cells. (D) Soft agar colony formation was carried out with siN-BLRs stained with 0.5% crystal violet. Scale bar refers to 500 μm. The bar graph reveals the proportion of colony formation. The data represent the mean±SEM. *p<0.05 and †p<0.01 show a statistically significant difference compared with a scrambled control.
5. The relationships between N-BLR expression and clinico-pathologic features in gastric cancer
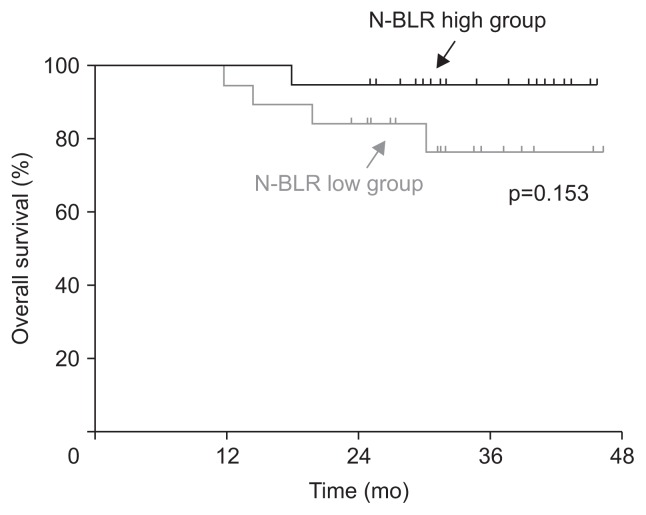
Age, sex, tumor location, depth of invasion, and lymph node metastasis did not differ between the N-BLR-low and N-BLR-high groups. Histological differentiation tended to be different between the N-BLR-low and N-BLR-high groups, although the difference was not statistically significant (p=0.051). The proportion of well- to moderately-differentiated adenocarcinomas was similar between the groups (N-BLR-low vs high: 45.0% vs 55.0%). However, poorly differentiated adenocarcinomas were mainly observed in the N-BLR-high group (N-BLR-low vs high: 30.0% vs 70.0%), while signet ring cell carcinoma was predominantly identified in the N-BLR-low group (N-BLR-low vs high: 87.5% vs 12.5%). In the subgroup analysis of tumors with undifferentiated histology (either poorly differentiated adenocarcinoma or signet ring cell carcinoma), the histology did significantly differ between the N-BLR-low and N-BLR-high groups (p=0.025) (Table 1). In the survival analysis, disease-free and overall survival did not differ between the groups (disease-free survival, p=0.905; overall survival, p=0.153) (Fig. 5).
Fig. 5.
Kaplan-Meier plots represent disease-free survival and overall survival depending on the level of N-BLR in patients who underwent surgery.
DISCUSSION
Increasing evidence has shown that lncRNAs have vital roles in regulating biological processes.13 Many new lncRNAs have been found to be involved in the development and progression of cancer.14,15 Our previous work has aided in the discovery of a new lncRNA called N-BLR, which has been shown to be associated with tumor progression in CRC.12 In this study, we found that N-BLR was significantly involved in the development and progression of gastric cancer. N-BLR was elevated in gastric cancer compared to normal tissue. Knock-down of N-BLR inhibited the proliferation of four different gastric cancer cell lines, and this finding was supported by the observation that N-BLR inhibited apoptosis in AGS and MKN28 cell lines in our study. N-BLR promoted the EMT transition through regulation of E-cadherin, N-cadherin, ZEB1, and Snail. Knock-down of N-BLR significantly increased the expression of miR-200c, which is known to inhibit EMT. These results suggest that N-BLR may be involved in cancer development and progression not only in CRC, but also in stomach cancer.
N-BLR is a pyknon-containing, primate-specific lncRNA located in the cytoplasm. In 2006, Rigoutsos et al.16 identified pyknons, which are a class of short DNA sequence motifs, in the human genome. A core property of pyknons is that they have multiple exact copies in the intergenic and intronic regions of the genome and in at least one mRNA. LncRNA containing pyknons was expected to play a role in regulation of biological processes.12,16 In fact, N-BLR plays a role in CRC according to a microarray targeting pyknon motifs.12
N-BLR was reported as a novel modulator of the EMT process and apoptotic pathway in CRC.12 N-BLR localizes to the cytoplasm where it directly interacts with miR-141-3p and miR-200c-3p, two members of the highly conserved miR-200 family known to inhibit EMT in CRC. Our observations showed that N-BLR suppresses miR-200c and regulates EMT in gastric cancer. In addition, knock-down of N-BLR reduced markers involved in EMT, such as E-cadherin, N-cadherin, ZEB1, and Snail.
N-BLR acts as an inhibitor of apoptosis in CRC.12 Our analytical results show that knock-down of N-BLR significantly induced apoptosis in AGS and MKN28 cells. Knock-down of N-BLR induced both early and late apoptosis, which subsequently reduced the proliferation of AGS and MKN28 cells. Our results also confirmed that knock-down of N-BLR can reduce tumorigenesis by reducing colony formation. There are many studies that involve lncRNAs in gastric cancer apoptosis and contribute to the development of gastric cancer. Recently, studies have found that lncRNA expression, including that of HOXA11-AS, HOTAIR, LINC00673, TINCR, TUSC7, and MALAT1, significantly regulates apoptosis.10,11,17–20 However, the precise mechanism by which these lncRNAs are involved in apoptosis is still unclear.
N-BLR was reported to mediate the switch from an epithelial to a mesenchymal cell phenotype by sequestering miR-141-3p and miR-200c-3p.12 In this, knock-down of N-BLR also induced a mesenchymal cell phenotype. siN-BLR reduced the expression of typical mesenchymal markers including ZEB1, Snail, N-cadherin, and Vimentin. However, siN-BLR was found to increase E-cadherin in both MKN28 and AGS cells.
In CRC, the expression of N-BLR significantly affected the stage and prognosis.12 In the current study, the expression of N-BLR was significantly up-regulated in cancer tissues compared to adjacent non-cancer tissues. However, we did not find a correlation between degree of N-BLR expression and prognosis in gastric cancer patients in survival and multivariate analysis. These differences can be explained with several reasons. First, the colon cancer study included many patients from two cohorts, whereas our study included only 38 patients with gastric cancer. We think that the number of patients included in our study with gastric cancer was too small to have statistically significant results. As a result, further study with a larger sample size of patients is needed. Second, stomach cancer has relatively high tumor heterogeneity compared to CRC.21
In conclusion, we found that N-BLR was related to apoptosis in gastric cancer carcinogenesis and promoted the EMT transition. Furthermore, this study also introduced the new N-BLR, which acts on the development and metastasis of gastric cancer. Based on the results of this study, we expect further studies to examine the use of N-BLR in the diagnosis and treatment of gastric cancer.
ACKNOWLEDGEMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Ko-rea (NRF) funded by the Ministry of Science and ICT (NRF-2015R1C1A1A01054352).
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Study concept and design: Y.H.Y. Data acquisition, data analysis and interpretation: H.J.B. Critical revision of the manuscript for important intellectual content: J.H.Y. Statistical analysis: C.H.P. Drafting of the manuscript and study supervision: S.K.L.
REFERENCES
- 1.Nobili S, Bruno L, Landini I, et al. Genomic and genetic alterations influence the progression of gastric cancer. World J Gastroenterol. 2011;17:290–299. doi: 10.3748/wjg.v17.i3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8:81572–81582. doi: 10.18632/oncotarget.19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658–3664. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niland CN, Merry CR, Khalil AM. Emerging roles for long non-coding RNAs in cancer and neurological disorders. Front Genet. 2012;3:25. doi: 10.3389/fgene.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nana-Sinkam SP, Croce CM. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Mol Oncol. 2011;5:483–491. doi: 10.1016/j.molonc.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 9.Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee NK, Lee JH, Park CH, et al. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451:171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 11.Lee NK, Lee JH, Ivan C, et al. MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer. 2017;17:46. doi: 10.1186/s12885-016-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigoutsos I, Lee SK, Nam SY, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. 2017;18:98. doi: 10.1186/s13059-017-1224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Liu S, Hu X. Long non-coding RNAs: crucial regulators of gastrointestinal cancer cell proliferation. Cell Death Discov. 2018;4:50. doi: 10.1038/s41420-018-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigoutsos I, Huynh T, Miranda K, Tsirigos A, McHardy A, Platt D. Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc Natl Acad Sci U S A. 2006;103:6605–6610. doi: 10.1073/pnas.0601688103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 18.Ba MC, Long H, Cui SZ, et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8:95542–95553. doi: 10.18632/oncotarget.20980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 20.Qi P, Xu MD, Shen XH, et al. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer. 2015;137:1269–1278. doi: 10.1002/ijc.29516. [DOI] [PubMed] [Google Scholar]
- 21.Hudler P. Challenges of deciphering gastric cancer heterogeneity. World J Gastroenterol. 2015;21:10510–10527. doi: 10.3748/wjg.v21.i37.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]