Abstract
The pattern of dendritic branching, together with the density of synapses and receptor composition, defines the electrical properties of a neuron. The development of the dendritic arbor and its additional stabilization are highly orchestrated at the molecular level and are guided by intrinsic mechanisms and extracellular information. Although protein translation is known to contribute to these processes, the role of its local component has not been fully explored. For local translation, mRNAs are transported to dendrites in their dormant form as ribonucleoparticles (RNPs). We hypothesized that disturbing spatial mRNA distribution via RNP targeting may result in severe underdevelopment of the dendritic arbor. Zipcode binding protein 1 (ZBP1) controls β-actin mRNA transport and translation in dendrites. We showed that proper cellular levels of ZBP1, its ability to engage in mRNA binding, and Src-dependent release of mRNA cargo from ZBP1 are vital for dendritic arbor development in cultured rat hippocampal neurons. Moreover, β-actin overexpression significantly alleviated the effects of ZBP1 knockdown. These results suggest that ZBP1-dependent dendritic mRNA transport contributes to proper dendritic branching.
Introduction
Neurons have characteristic dendritic arborization patterns (Stuart et al., 2007) that contribute to information processing (Segev and London, 2000). Several cellular processes, including cytoskeleton dynamics and translation, are engaged to support proper dendritic development (Urbanska et al., 2008; Jan and Jan, 2010). Novel protein synthesis in neurons has been observed not only in cell perikarya but also at discrete locations along the dendrites (Aakalu et al., 2001). This local translation is now well known to contribute to proper neuronal connectivity and spine formation (Eom et al., 2003; Goetze et al., 2006; Lebeau et al., 2008; Verpelli et al., 2010).
Local protein synthesis in dendrites requires the repression and transport of dendritically targeted mRNAs. This is enabled by the presence of tagging sequences within mRNA that are recognized and bound by specialized proteins (Kindler et al., 2005; Kiebler and Bassell, 2006; Bramham and Wells, 2007). Zipcode binding protein 1 (ZBP1) is one of several RNA binding proteins found in ribonucleoparticles (RNPs) and dendritic processing bodies (P-bodies) (Barbee et al., 2006; Elvira et al., 2006; Cougot et al., 2008; Zeitelhofer et al., 2008a), which are structures involved in mRNA silencing and transport (Kiebler and Bassell, 2006; Zeitelhofer et al., 2008b). ZBP1 binds a wide variety of transcripts (Jønson et al., 2007), but only its binding to a sequence in the 3′-untranslated region (UTR) of β-actin mRNA has been well characterized (Ross et al., 1997; Oleynikov and Singer, 2003) and confirmed in neurons. The granules that contain ZBP1 are present in dendrites beneath dendritic spines and crucial for cellular β-actin mRNA localization and proper spine growth (Zhang et al., 2001; Eom et al., 2003; Tiruchinapalli et al., 2003).
Much evidence suggests the participation of RNP proteins in dendritic growth (Lee et al., 2003; Barbee et al., 2006; Bestman and Cline, 2008; Vessey et al., 2008), but the precise molecular mechanism and identity of the bound mRNAs that are important for this process remain mostly unknown. The present study addressed these two issues by showing that both knockdown and overexpression of ZBP1 in developing rat hippocampal neurons in culture result in dendritic arbor simplification and β-actin mislocalization. We also provide evidence that localization of ZBP1 to the branching points of dendrites correlates with the need for ZBP1 in proper dendritic arborization. We also show that ZBP1 contributes to dendritogenesis via mRNA binding and Src-dependent release. Finally, the effects of ZBP1 knockdown on dendritic arborization were prevented by β-actin overexpression. Together, these data suggest that ZBP1-dependent dendritic mRNA targeting and unsilencing contribute to the proper formation of dendrites in hippocampal neurons.
Materials and Methods
Antibodies and drugs.
Rabbit anti-ZBP1 antibody has been previously described and was a kind gift from Dr. R. H. Singer (Albert Einstein College of Medicine, Yeshiva University, New York, NY) (Oleynikov and Singer, 2003). The following antibodies and drugs were obtained from commercial sources: goat anti-ZBP1 (Santa Cruz Biotechnology), mouse anti-β-galactosidase (Promega), rabbit anti-β-galactosidase (MP Biomedicals), mouse anti-green fluorescent protein (GFP) (Roche Applied Science), rabbit anti-GFP (Medical and Biological Laboratories), and horseradish peroxidase (HRP)-conjugated sheep anti-digoxigenin (DIG) antibody (Roche). Alexa Fluor 488- and Alexa Fluor 568-conjugated anti-mouse, anti-rabbit, and anti-goat secondary antibodies and HRP-conjugated anti-rabbit, anti-mouse, and anti-goat secondary antibodies were obtained from Invitrogen and Jackson ImmunoResearch, respectively.
DNA constructs.
The following DNA constructs have been previously described: pEGFP-rZBP1 (Eom et al., 2003), pStau1-EYFP, pStau2-EYFP, pSuperior-Stau2sh (Goetze et al., 2006), β-actin-GFP (Jaworski et al., 2005), β-actin-mRFP, FLAG-CaMKIIα (Hoogenraad et al., 2007), β-actin-mCherry (de Vrij et al., 2008), pSuper (Brummelkamp et al., 2002), and pPDGF-EGFP-β-actin (Morales et al., 2000). pcDNA3-c-Myc (Ricci et al., 2004) and pExpress-1-α-actin were acquired from Addgene (plasmid 16011) and ImaGenes (clone IRBPp993E0552D), respectively.
We used pSuper vector for ZBP1, ZBP1-scrambled, and Staufen1 short hairpin RNA (shRNA) expression. The target sequences for shRNAs in the rat ZBP1 (AF541940) and Staufen1 (BC101858) coding sequences were the following: ZBP1sh#1, 400–418, CAAGCCATCATGAAGCTAA; ZBP1sh#2, 650–668, CCATCCGAAACATCACAAA; Stau1sh, 1205–1223, TGAGCAACTGTACTACCT. scZBP1sh#1 (GACAATTAACCACGAGATC) was designed using the online GeneScript tool.
A ZBP1sh#1-resistant mutant of ZBP1, EGFP-ZBP1*1, was generated by introducing five silent mutations in the small interfering RNA target region using the QuikChange Site-Directed Mutagenesis System (Stratagene) with the following primers (mutations are underlined): ZBP1*1, forward (CCGGGAGCAGACCAGGCAGGCAATTATGAAACTCAATGGCCATCAACG); ZBP1*1, reverse (CGTTGATGGCCATTGAGTTTCATAATTGCCTGCCTGGTCTGCTCCCGG).
The substitution of tyrosine with phenylalanine at position 396 of EGFP-ZBP1*1 (ZBP1*1Y396F-EGFP) was performed using the following primers: Y396F, forward (GGGGCTGCTCCCTTCGGCTCCTTCATGC); Y396F, reverse (GCATGAAGGAGCCGAAGGGAGCAGCCCC). ZBP1*1-GXXG, a mutant with decreased RNA binding affinity (Nielsen et al., 2003), was obtained on a basis of EGFP-ZBP1*1 with three subsequent substitutions within the GXXG motif (GKEG, GKKG, GKGG) for the GELG motif in the signature loop of KH1, KH2, and KH3 using the following primers: KH1, forward (GCGCCATCATTGGCGAGCTGGGTGCCACCATCCG); KH1, reverse (CGGATGGTGGCACCCAGCTCGCCAATGATGGCGC); KH2, forward (GTGGGGCGACTCATTGGAGAGCTCGGGCGGAACTTGAAG); KH2, reverse (CTTCAAGTTCCGCCCGAGCTCTCCAATGAGTCGCCCCAC); KH3, forward (GGCGCCATCATCGGCGAGCTGGGCCAGCACATC); KH3, reverse (GATGTGCTGGCCCAGCTCGCCGATGATGGCGCC).
The ZBP1*1ΔC (1-1191 nt) deletion mutant was cloned into the EcoRI/BamHI sites of pEGFP-C1 (Clontech) by PCR using the following primers: ZBP1-EcoRI-1 (GCGAATTCATGAACAAGCTTTACATCGGCAACC); ZBP1-BamHI-1191 (GCGGATCCTCAGCCATAGGGAGCAGCCCCAGTGACG).
Neuron culture and transfection.
Primary hippocampal and cortical cultures were prepared from brains of embryonic day 19 Wistar rats (either sex) (Banker and Goslin, 1988). Cells were plated on coverslips coated with laminin (2 μg/ml) and poly-l-lysine (37.5 μg/ml) at a density of 250/mm2 (hippocampal) or precoated BioCoat plastic plates (BD Biosciences Discovery Labware) at a density of 625/mm2 (cortical). Cultures were grown in Neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen), 0.5 mm glutamine, 12.5 μm glutamate, and penicillin/streptomycin mix. Seven, 14, or 21 d in vitro (DIV) hippocampal neurons were transfected using Lipofectamine 2000 (Invitrogen) according to the procedure described by Jaworski et al. (2009). Briefly, for the transfection of cells growing in the single well of a 24-well format dish, 0.9 μg of DNA was mixed with 1.6 μl of Lipofectamine 2000 in 100 μl of Neurobasal medium and incubated for 30 min. During the incubation time, fresh culture medium was prepared and mixed evenly with the old medium. One-half of the mixed medium was left with the cells for the time of transfection, and the other one-half was saved for medium replacement after transfection. The complexes of DNA and Lipofectamine 2000 were added to the cells for 4 h incubation at 37°C in 5% CO2. Cells were then washed with Neurobasal medium and returned to the saved culture medium. For the cotransfection experiments, plasmids that encoded either the protein of interest or shRNA were used in a 3:1 ratio to β-gal, β-actin-GFP, or β-actin-mRFP plasmids. For the rescue experiments, the ratio of shRNA-encoding plasmids to mutant ZBP1 cDNA or β-actin cDNA was 6:1. Whenever possible, cotransfection was confirmed with double staining for the protein of interest and cotransfected marker (GFP, β-gal, or mRFP). For the ZBP1 distribution experiments, neurons were transfected on DIV7, DIV14, or DIV21 with EGFP-ZBP1 together with mCherry under the control of the β-actin promoter. Cells were fixed 48 h after transfection.
Neuron nucleofection and RNA isolation.
For quantitative real-time PCR (qRT-PCR) validation of ZBP1sh RNA, cortical neurons were transfected on DIV0 using an Amaxa Nucleofector II Device and Amaxa Rat Neuron Nucleofactor Kit (Lonza) according to the adjusted manufacturer's protocol. Two days later, the cells were lysed, and RNA was isolated with the RNeasy Protect Mini Kit (QIAGEN). cDNA was prepared with High Capacity RNA-to-cDNA Master Mix (Applied Biosystems) according to the manufacturer's protocol.
Quantitative real-time PCR.
Reactions were performed with a 7900HT machine and TaqMan Gene Expression Assays (Applied Biosystems) with the following TaqMan rat probes: GAPDH for endogenous control, IMP1 for ZBP1 detection, KHDRBS1 for IMP2 detection, and IMP3 for IMP3 detection. SDS 2.4 and RQ Manager 1.2.1 programs were used for data acquisition and preliminary analysis. For statistical analysis, Microsoft Excel and GraphPad Prism software were used.
COS-7 cell transfection.
COS-7 cells were transfected with DNA constructs using JetPEI (Polyplus Transfection) and lysed 48 h later for Western blot analysis. For ZBP1 shRNA validation and ZBP1*1 construct validation, cells were cotransfected with pSuper, pSuper-ZBP1sh#1, or pSuper-ZBP1sh#2 together with β-gal and EGFP-ZBP1, pEGFP-C1 (control), or EGFP-ZBP1*1 in a ratio of 10:1:1. For Staufen1 shRNA validation and verification of Staufen1 cross-silencing by Staufen2 shRNA, cells were cotransfected with pSuper, pSuper-Stau1sh, or pSuperior-Stau2sh together with β-gal and Stau1-EYFP or pEGFP-C1 in a ratio of 10:1:1. For Staufen2 shRNA verification and verification of cross-silencing of Staufen2 by Stau1sh, cells were cotransfected with pSuper, pSuper-Stau1sh, or pSuperior-Stau2sh together with β-gal and Stau2-EYFP or pEGFP-C1 in a ratio of 10:1:1.
In situ fluorescent hybridization.
For in situ fluorescent hybridization, cells were fixed in 4% paraformaldehyde for 10 min at room temperature and incubated with 1% H2O2 to minimize background. After washing with PBS containing DEPC (diethylpyrocarbonate), cells were first incubated for 1 h at room temperature with prehybridization buffer [50% (v/v) formamide, 4 μg/ml Escherichia coli tRNA, 2% (v/v) Denhardt's solution (Sigma-Aldrich), 1% (v/v) salmon sperm DNA (Sigma-Aldrich), 2 mm ribonucleoside vanadyl complexes (Sigma-Aldrich)] and then with 50 μl of hybridization buffer [2× SSC, 50% (v/v) formamide, 10% dextran sulfate, 4 μg/ml E. coli tRNA, 2% (v/v) Denhardt's solution, 1% (v/v) salmon sperm DNA, 2% (v/v) ribonucleoside vanadyl complexes] containing 75 ng of heat-denaturated digoxigenin-labeled single-strand sense or antisense RNA probes for 10 h in a hybridization oven set at 56°C. After the hybridization step, cells were rinsed in 2× SSC, washed once in 2× SSC for 1 h at 56°C, washed twice in 0.2× SSC for 30 min at 56°C, and washed four times in 0.2× SSC for 30 min at room temperature. The immunofluorescent detection of digoxigenin-labeled probes was then performed using HRP-conjugated anti-DIG antibody (Roche) and the Tyramide Signal Amplification (TSA) Plus Cyanine 5 System (PerkinElmer Life and Analytical Sciences). For fluorescent in situ hybridization (FISH), we used cRNA probes synthesized and labeled using the Digoxigenin RNA Labeling kit (SP6/T7) (Roche) and T3 polymerase (Roche). The fragment of a sequence of the 5′-UTR of β-actin (nucleotides 6–136) was obtained by PCR using rat hippocampal cDNA and was used as a template. The primers used for the PCR contained promoter sequences for T3 or T7 RNA polymerases on their ends.
Immunocytochemistry.
For staining of transfected proteins, neurons were fixed with cold 4% paraformaldehyde containing 4% sucrose in PBS for 10 min. For β-actin staining, cells were fixed in cold methanol for 10 min at −20°C. After fixation, cells were washed with PBS three times and incubated overnight with primary antibody in GDB buffer (0.2% gelatin, 0.8 m NaCl, 0.5% Triton X-100, 30 mm phosphate buffer, pH 7.4) at 4°C. Secondary antibodies were applied in GDB for 1 h at room temperature. Slides were mounted with VectaShield. For endogenous ZBP1 staining, cells were fixed with −20°C methanol for 10 min at −20°C, air-dried, and incubated in 4% paraformaldehyde at room temperature for an additional 10 min. After fixation, cells were washed with PBS three times, incubated in blocking buffer (10% normal donkey serum in 1× PBS) for 45 min at room temperature, and incubated overnight at 4°C with primary antibody solution in 1× PBS containing 1% BSA and 0.3% Triton X-100. Secondary antibodies were applied for 75 min at room temperature. Slides were mounted with ProLong Gold (Invitrogen).
Image analysis and quantification.
Confocal neuron images were obtained with a Zeiss LSM5 Pascal microscope at 1024 × 1024 pixel resolution for morphological analysis. The 20, 40, and 63× objectives were used. Each image consisted of a series of z-stack images. The resultant stack was flattened into a single image using maximum projection. The confocal settings were constant for all of the scans when fluorescence intensity was compared. Morphometric analysis and quantification were performed with MetaMorph image analysis software (Molecular Devices) for the manual counting of primary dendrites, total number of dendritic tips, and fluorescence intensities and ImageJ software with the NeuronJ plug-in (Meijering et al., 2004) and Sholl plug-in (http://biology.ucsd.edu/labs/ghosh/software) for measurements of total dendrite length and Sholl analysis, respectively. Briefly, we used the tracking function in NeuronJ to manually draw a mask of all dendrites of transfected neuron on a confocal image and measured their length. If more than one transfected cell was present at the particular image, the mask was created only for single cell. The mask was saved as separate image and used in the automated Sholl analysis to avoid background of axons and other cells whose fragments could be present in the original pictures. The original script of Sholl plug-in was modified for more accurate performance with masks instead of raw images. For dendrite counting, we defined a dendrite as a nonaxonal protrusion >10 μm. All dendrites that originated from the cell body were counted as primary. A minimum of 30 cells per experimental point were counted for the morphological studies. The dataset was derived from at least two independent experiments. EGFP-ZBP1 distribution was analyzed using a Zeiss LSM710 NLO laser confocal microscope with a 100×, 1.3, objective. Images were acquired in z-series (six sections, 0.5 μm steps), with identical acquisition parameters for immature and mature neurons. For analysis, z-series were rendered using maximum-intensity projections, exported in TIFF format, and analyzed using MetaMorph software. Profiles of EGFP-ZBP1 fluorescence along the dendrites were made using the Linescan function, with the scan width set to 6 pixels. Branching sites were indicated manually, based on the mCherry imaging of dendrites. The mean intensities of GFP fluorescence at and between the branching sites were calculated using OpenOffice Calc software. Statistical analyses were performed with GraphPad Prism software. For endogenous ZBP1 level analysis, images were acquired using a Zeiss LSM710 NLO laser confocal microscope with a 40×, 1.3, objective and 1.5× zoom in z-series (four sections, 1 μm steps). Fluorescence intensity analysis was performed using MetaMorph with the region measurements function. Statistical analysis was performed with GraphPad Prism. For endogenous β-actin mRNA distribution, images were acquired with a Zeiss LSM5 Pascal microscope with a 63× objective and z-stack (eight sections; 0.3 μm steps). β-Actin mRNA puncta were calculated manually in proximal and distal dendrites and are presented as puncta density per 10 μm.
Statistical analysis.
Unless stated otherwise, statistical analysis was performed with GraphPad Prism using the Kruskal–Wallis test for multiple comparisons, and in case it showed statistical significance, we used Mann–Whitney post test with Bonferroni's adjustment. For comparisons of two groups, the Mann–Whitney or unpaired t test was used. The detailed statistics are provided for all experiments in the supplemental tables (available at www.jneurosci.org as supplemental material).
Results
ZBP1 regulates proper dendritic arbor formation during development
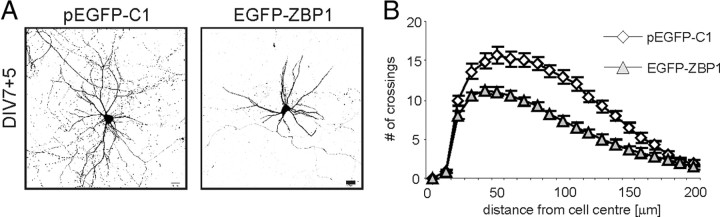
Key regulators of protein synthesis have been previously demonstrated to be important regulators of dendritic arbor growth in developing neurons (Jaworski et al., 2005; Kumar et al., 2005). RNPs are also present in dendrites, often localizing to dendrite branching points, and some of their components are needed for dendritic arbor development (Barbee et al., 2006; Bestman and Cline, 2008; Vessey et al., 2008, 2010). These observations prompted us to determine how selected components of RNPs regulate dendritic arborization. We focused on ZPB1, which regulates the transport and translation of β-actin mRNA and acts as a translational repressor until phosphorylated by Src kinase (Hüttelmaier et al., 2005). We assumed that, if dendritic arbor development depends on dendritic transport and subsequent dendritic translation of mRNA cargo of ZBP1, then the disturbance of these processes, either by overexpression or knockdown of ZBP1, would result in changes in dendritic arbor morphology. In the first case, we expected that ZBP1 mRNA cargo would be properly delivered, but the dendritic arbor would be disturbed because of insufficient cargo release under basal conditions. In contrast, ZBP1 knockdown would impair dendritic growth as a consequence of improper or a lack of mRNA cargo delivery to more distal dendrites. To test this hypothesis, we overexpressed EGFP-ZBP1 in hippocampal neurons cultured in vitro for 5 d beginning on DIV7, a time of very robust dendritic arborization in our preparations (Fig. 1A). Together with EGFP-ZBP1, plasmid encoding β-gal was cotransfected to precisely highlight neuronal morphology and provide unbiased cell filling. We decided to use dissociated cell cultures in our experiments because this is an optimal experimental model for extensive genetic manipulations at any developmental stage of mammalian neurons, which cannot be achieved to such an extent with any other experimental design. The comparison of dendritic arbor morphology between control EGFP-transfected neurons and EGFP-ZBP1-overexpressing neurons revealed a significant 38% decrease in the mean number of dendritic tips per cell in the latter group (pEGFP-C1, 26.57 ± 1.67; EGFP-ZBP1, 16.5 ± 0.72; p < 0.001). However, the mean number of primary dendrites per cell did not change significantly (supplemental Table S1A, available at www.jneurosci.org as supplemental material). Consequently, the dendritic branching index, a quotient of the total number of dendritic ends and primary dendrites, was decreased by 32% in EGFP-ZBP1-overexpressing neurons (pEGFP-C1, 5.014 ± 0.26; EGFP-ZBP1, 3.40 ± 0.14; p < 0.001). The total dendritic length was 39% shorter in ZBP1-overexpressing neurons compared with control neurons (pEGFP-C1, 1922 ± 130 μm; EGFP-ZBP1, 1166 ± 91 μm; p < 0.001).
Figure 1.
Overexpression of EGFP-ZBP1 in hippocampal neurons decreases dendrite branching. A, Representative micrographs of hippocampal neurons transfected on DIV7 for 5 d with control vector or EGFP-ZBP1. Neuronal morphology was visualized by staining for cotransfected β-gal. B, Sholl analysis of neurons transfected with EGFP-ZBP1 or control vector. The horizontal axis of the Sholl plot indicates the distance from the cell soma center. Scale bar, 20 μm.
To analyze the changes in dendritic fields more thoroughly, we used Sholl analysis (Sholl, 1953), which measures the number of dendrites that cross circles at various radial distances from the cell soma. Directional shifts of the Sholl plot describe changes in dendritic arbor complexity and dendritic field area. Specifically, downward and leftward shifts of the plot correspond to decreased complexity and shrinkage of dendritic arbors. Upward and rightward shifts, in contrast, reflect increased complexity and expansion of the dendritic field. In neurons overexpressing EGFP-ZBP1, the maximum number of crossings was reduced by 29% and reached closer to the cell soma compared with control neurons (Fig. 1B; supplemental Table S1A, available at www.jneurosci.org as supplemental material), indicating the simplification of dendritic arbors in ZBP1-overexpressing neurons. The plots for control and ZBP1-transfected neurons diverged between 30 and 170 μm from the center of the cell body, but at distances closer than 30 μm, the dendritic fields of the analyzed neurons were similar.
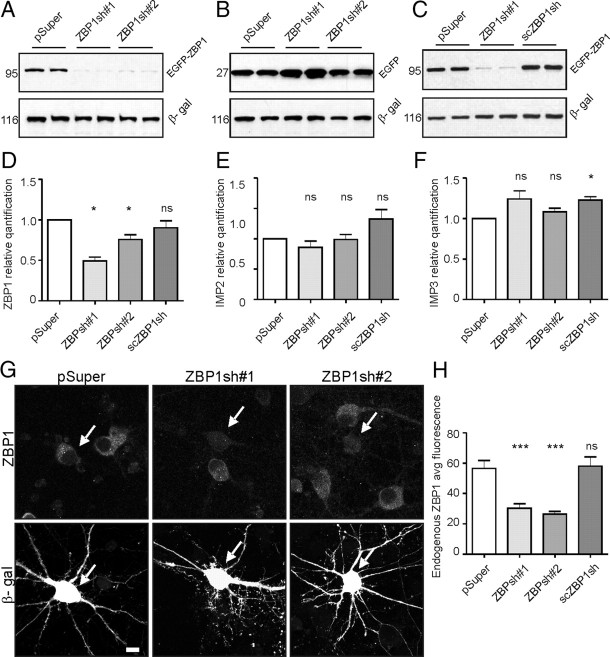
Accordingly to our hypothesis, the lack of ZBP1 should result in changes in dendritic arbor morphology, attributable to inappropriate and insufficient mRNA delivery into dendrites. For ZBP1 knockdown in neurons, we designed two shRNAs against rat ZBP1 (ZBP1sh#1, ZBP1sh#2) and cloned them into pSuper vector (Brummelkamp et al., 2002). We also designed and cloned a scrambled version of ZBP1sh#1 (scZBP1sh) into pSuper as an additional negative control. Both shRNAs against ZBP1 and scZBP1sh were validated with several independent approaches. First, shRNA encoding vectors (i.e., pSuper-ZBP1sh#1, pSuper-ZBP1sh#2, and scZBP1sh) or pSuper were transiently cotransfected with EGFP-ZBP1 or EGFP into COS-7 cells. As shown in Figure 2, both ZBP1sh#1 and ZBPsh#2 markedly decreased the expression of EGFP-ZBP1, with no influence on the levels of EGFP (positive control) or β-gal (control of even transfection). At the same time, scZBP1sh did not affect the expression levels of any of the tested proteins (Fig. 2C). Second, we used qRT-PCR to assess the endogenous levels of ZBP1 mRNA in cortical neurons transfected with ZBP1sh#1, ZBP1sh#2, scZBP1sh, and pSuper at DIV0 (Amaxa nucleofection) for 2 d. Cortical neurons were used in these experiments because of the need for larger amounts of material for analysis. As expected, cells transfected with ZBP1sh#1 and ZBP1sh#2, but not scZBP1sh or pSuper alone, had significantly reduced levels of ZBP1 mRNA (Fig. 2D; supplemental Table S1B, available at www.jneurosci.org as supplemental material). We also tested at this stage the specificity of ZBP1 shRNAs toward other IMP family members. Indeed, none of our constructs silenced the expression of IMP-2 and IMP-3 (Fig. 2E,F; supplemental Table S1B, available at www.jneurosci.org as supplemental material). Third, we tested the effectiveness of ZBP1sh#1 and ZBP1sh#2 toward endogenous ZBP1 protein levels in cultured hippocampal neurons by transfecting them with individual shRNAs together with β-gal as an indicator of transfected cells and by checking for the presence of the target protein with immunofluorescent staining (Fig. 2G,H; supplemental Table S1B, available at www.jneurosci.org as supplemental material).
Figure 2.
Characterization of ZBP1 shRNAs. A–C, shRNA and scrambled RNA validation in COS-7 cell line. Cells were transfected with control or shRNA encoding plasmid together with either EGFP-ZBP1 or pEGFP-C1. Additionally, β-gal was cotransfected as a marker of even transfection. Both ZBP1sh#1 and ZBP1sh#2 strongly reduced EGFP-ZBP1 expression, whereas the scrambled version (scZBP1sh) did not. D–F, Verification of specificity of ZBP1shRNAs using qRT-PCR of cDNAs from neurons transfected on DIV0 with nucleofection with pSuper, ZBP1sh#1, ZBP1sh#2, and scrambled RNA. The graphs show the relative quantification of ZBP1 and two other members of the ZBP1 family: IMP2 and IMP3. *p < 0.05, one-sample t test. G, Representative micrographs of hippocampal neurons transfected on DIV7 with control vector, ZBP1sh#1, or ZBP1sh#2 together with β-gal for identification of transfected cells (indicated with arrowheads). After 3 d of expression, the cells were fixed and immunostained for transfected β-gal (Alexa 568) and endogenous ZBP1 (Alexa 488). The ZBP1 signal was strongly reduced in both the cell body and dendrites of ZBP1sh-transfected neurons compared with untransfected and pSuper-transfected neurons. Scale bar, 20 μm. H, Quantification of ZBP1 silencing in neurons transfected with pSuper, scrambled RNA, ZBP1sh#1, and ZBP1sh#2. Average immunofluorescence was measured in the cell body using MetaMorph software. ***p < 0.0005, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment. Error bars indicate SEM.
ZBP1sh#1 and ZBP1sh#2 efficiently silenced rat ZBP1 expression, and we used them to assess the influence of ZBP1 depletion on dendritic arborization. Developing hippocampal neurons were transfected on DIV7 with pSuper, ZBP1sh#1, or ZBP1sh#2 together with β-gal. Expression proceeded for 3 d. ZBP1 knockdown slightly affected primary dendrite number (ZBP1sh#1, 16% decrease, p < 0.025; ZBP1sh#2, not significant) and significantly decreased the number of dendritic tips compared with control (ZBP1sh#1, 49%; ZBP1sh#2, 35%; p < 0.0005), indicating a strong effect on higher-order branches (Fig. 3B,C; supplemental Table S1C, available at www.jneurosci.org as supplemental material). Indeed, the dendritic branching index was significantly reduced in ZBP1sh#1- and ZBP1sh#2-transfected neurons [69% (p < 0.0005) and 78% (p < 0.001) of control for ZBP1sh#1 and ZBP1sh#2, respectively] (Fig. 3D; supplemental Table S1C, available at www.jneurosci.org as supplemental material). These measurements of dendritic arbor morphology, together with the significant decrease in total dendritic length (ZBP1sh#1, 38%, p < 0.0001; ZBP1sh#2, 27%, p < 0.001) (Fig. 3E; supplemental Table S1C, available at www.jneurosci.org as supplemental material) reflect marked dendritic arbor undergrowth in neurons with ZBP1 knockdown, which was further corroborated by the Sholl analysis. The number of crossings further from the cell soma was significantly lower in ZBP1-knockdown neurons than in controls, indicating a lower branching frequency (Fig. 3F; supplemental Table S1C, available at www.jneurosci.org as supplemental material). Similar to ZBP1-overexpressing neurons, ZBP1-knockdown neurons reached maximum crossing points closer to the cell soma, and their mean maximum number of crossings was significantly lower than control (Fig. 3F; supplemental Table S1C, available at www.jneurosci.org as supplemental material). ZBP1sh#1 and ZBPsh#2 strongly affected dendritic morphology, but scZBP1sh transfection had no significant effects compared with cells transfected with pSuper alone (Fig. 3G,H; supplemental Table S1B, available at www.jneurosci.org as supplemental material). Based on these results, pSuper was used as a valid negative control in the rest of the experiments.
Figure 3.
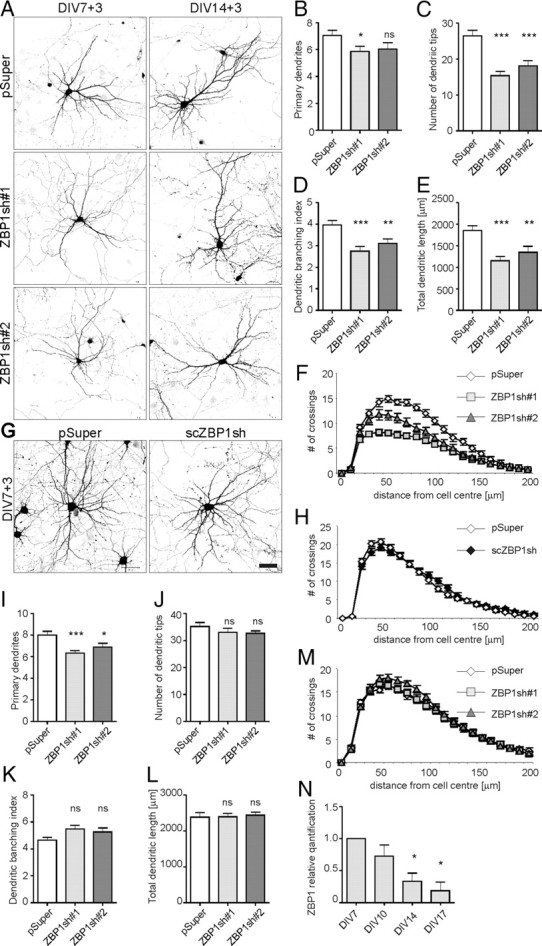
Knockdown of ZBP1 in hippocampal neurons decreases dendritic branching. A, Representative images of hippocampal neurons transfected on DIV7 or DIV14 with pSuper, pSuper-ZBP1sh#1, or pSuper-ZBP1sh#2 for 3 d. Morphology of transfected cells was visualized by staining for cotransfected β-gal. B–F, Mean number of primary dendrites (B), mean total number of dendritic tips (C), dendritic branching index (D), mean total dendritic length (E), and Sholl analysis of neurons transfected on DIV7 (F). G, Representative images of hippocampal neurons transfected on DIV7 with pSuper and scrambled scZBP1sh and fixed 3 d later. Scale bar, 50 μm. H, Comparison of morphologies of dendritic arbors of neurons transfected with pSuper or scZBP1sh. The Sholl analysis revealed no significant difference. I–M, Mean number of primary dendrites (I), mean total number of dendritic tips (J), dendritic branching index (K), mean total dendritic length (L), and Sholl analysis of neurons transfected on DIV14 (M). ***p < 0.0005, **p < 0.005, *p < 0.025, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment. N, qRT-PCR of ZBP1 expression in hippocampal neurons. RNA was isolated from hippocampal neuron cultures collected on DIV7, DIV10, DIV14, and DIV17. The graphs show the relative quantification of ZBP1 normalized to DIV7. *p < 0.05, one-sample t test. Error bars indicate SEM.
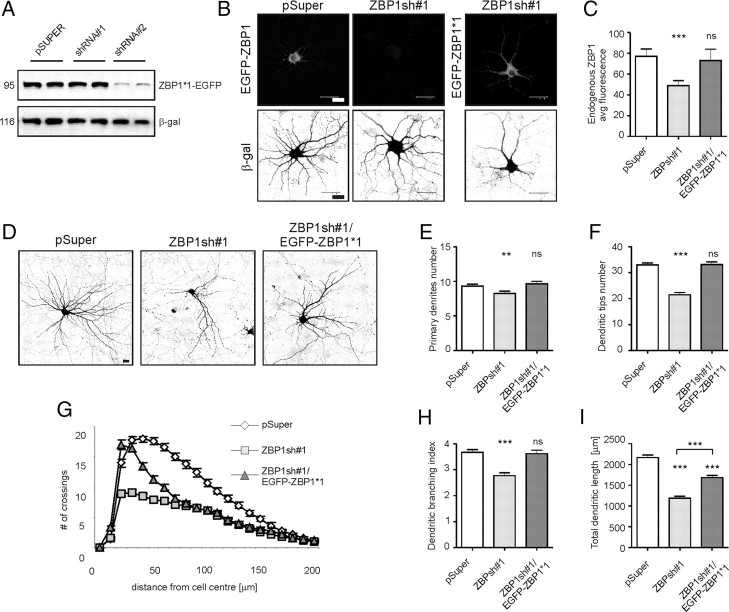
To verify whether the observed effect of shRNA transfection on dendritic arborization is indeed attributable to ZBP1 knockdown, we attempted to rescue the phenotype via ZBP1 replenishment in shRNA-treated cells. We used RNA interference (RNAi) cleavage-resistant EGFP-ZBP1*1, designed specifically to ZBP1sh#1, which was tested in both COS-7 cells and primary hippocampal neurons and proved to be resistant to ZBP1sh#1-driven RNAi (Fig. 4A–C). EGFP-ZBP1*1 significantly rescued the extent of the ZBP1sh#1 effect on dendritic arborization (Fig. 4D–I). Specifically, cotransfection of DIV7 hippocampal neurons with ZBP1*1 and ZBP1sh#1 resulted in the rescue of primary dendrite number, dendritic tips number, and the dendritic branching index, whereas neurons overexpressing ZBP1sh#1 suffered from dendritic loss (Fig. 4D–F,H; supplemental Table S1D, available at www.jneurosci.org as supplemental material), indicating that the ZBP1sh-evoked effects on the number of dendrites and dendritic branching observed in this study were specific to ZBP1 knockdown. However, both knockdown and overexpression of ZBP1 were detrimental for dendritic arborization, and the supplementation of shRNA-resistant ZBP1 in ZBP1-knockdown neurons needed to be precisely fine-tuned to obtain proper levels of total ZBP1 (endogenous plus exogenous) and rescue the phenotype (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). However, even with this, not all parameters of dendritic morphology (e.g., total dendritic length and the coverage of dendritic field) could be fully restored (Fig. 4G,I; supplemental Table S1D, available at www.jneurosci.org as supplemental material).
Figure 4.
RNAi cleavage-resistant ZBP1 rescues ZBP1 knockdown phenotype in hippocampal neurons. A, Western blot analysis of the effect of ZBP1sh#1 and ZBP1sh#2 on the expression of cotransfected EGFP-ZBP1*1 in COS-7 cells. β-gal was cotransfected as a marker of even transfection. B, Representative micrographs of hippocampal neurons transfected on DIV7 with pSuper or pSuper-ZBP1sh#1 together with EGFP-ZBP1 or EGFP-ZBP1*1 for 3 d of expression. Neuronal morphology was visualized by staining for cotransfected β-gal. EGFP-ZBP1*1 was expressed regardless of the presence of ZBP1sh#1. C, Quantification of ZBP1 levels in neurons transfected with pSuper, ZBP1sh#1, and ZBP1sh#1 together with EGFP-ZBP1*1. Average immunofluorescence was measured in the cell body using MetaMorph software. D, Representative micrographs of hippocampal neurons transfected on DIV7 with control vector, ZBP1sh#1, or ZBP1sh#1 together with EGFP-ZBP1*1 for 3 d of expression. Neuronal morphology was visualized by staining for cotransfected β-gal. E–I, Mean number of primary dendrites (E), mean number of dendritic tips (F), Sholl analysis (G), dendritic branching index (H), and mean total dendritic length of transfected neurons (I). ***p < 0.0005, **p < 0.005, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment. Scale bar, 20 μm. Error bars indicate SEM.
ZBP1 is not necessary for stabilization of mature dendritic arbor
To determine whether the presence of ZBP1 is needed for the stabilization of an already developed dendritic pattern, we used DIV14 neurons. In our preparations, intensive dendritic growth is completed by ∼2 weeks in vitro, and we assumed that any detrimental changes to dendritic arborization that occur after this time affect dendritic arbor stabilization rather than growth. Neurons were transfected with pSuper, ZBP1sh#1, or ZBP1sh#2 together with β-gal for 3 d of expression. ZBP1 knockdown did not affect the number of dendritic tips or total dendritic length, and no changes were detectable by the Sholl analysis (Fig. 3A,J,L,M; supplemental Table S1E, available at www.jneurosci.org as supplemental material). However, ZBP1 knockdown slightly decreased the primary dendrite number but did not significantly increase the dendritic branching index (Fig. 3I,K; supplemental Table S1E, available at www.jneurosci.org as supplemental material). The negative effect of ZBP1 knockdown on dendritic arborization was virtually absent and not attributable to the previously suggested lack of ZBP1 in mature neurons. As revealed by the analysis of mRNA levels, although an overall tendency toward decreased ZBP1 expression was observed with the maturation of cultured cortical neurons, ZBP1 mRNA was present in the cells on all days tested (Fig. 3N; supplemental Table S1F, available at www.jneurosci.org as supplemental material). The presence of ZBP1 in cultured mature hippocampal neurons was also confirmed by Western blotting (data not shown). Interestingly, when we analyzed the spatial distribution of EGFP-ZBP1 in hippocampal neurons on DIV9 and DIV16 after 2 d of overexpression, we observed two different distribution patterns. Both in developing and mature neurons, overexpressed ZBP1 formed puncta along the dendrites, but in the young neurons the fluorescence intensity was significantly greater at the dendritic branching points than in the preceding dendritic branches (Fig. 5A–C). A similar spatial distribution was observed for endogenous protein in developing neurons (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). In contrast, in DIV16 neurons, the EGFP-ZBP1 puncta were more evenly distributed, and the differences between branching points and dendrites decreased (Fig. 5D–F). These differences were virtually absent in neurons tested at later time points (DIV23) (data not shown). Thus, we conclude that the particular spatial distribution of ZBP1 seems to be necessary for the proper control of dendritic arborization during the period of intensive dendritogenesis in young neurons but not at the maintenance stage of an already formed dendritic arbor.
Figure 5.
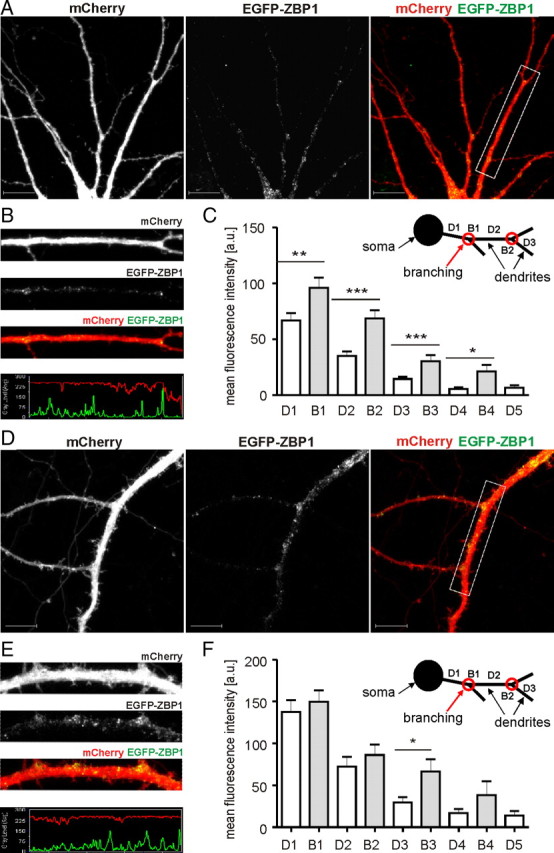
EGFP-ZBP1 is enriched in dendritic branching points of developing but not mature neurons. A, D, Micrographs of hippocampal neurons transfected on DIV7 (A) and DIV14 (D) with EGFP-ZBP1. Expression proceeded for 2 d. Neuronal morphology was visualized by cotransfected mCherry. B, E, Representative micrographs of single dendrites with corresponding profiles of fluorescence. C, F, Mean intensities of fluorescence measured in dendrites. ***p < 0.001, **p < 0.01, *p < 0.05, Mann–Whitney test. Scale bar, 10 μm. Error bars indicate SEM.
mRNA binding is pivotal for ZBP1 function in dendritogenesis
ZBP1 contains two RNA recognition motifs and four RNA-binding domains (KH domains), which enable its interaction with the β-actin zipcode. Specifically, the KH3 and KH4 domains were shown to be necessary and sufficient for zipcode binding (Farina et al., 2003). To determine whether ZBP1 RNA binding capacity is required for the regulation of dendritogenesis, we expressed truncated and site-mutated forms of ZBP1*1 (ZBP1*1ΔC and ZBP1*1-GXXG; schematic in Fig. 6A; distribution in neurons in supplemental Fig. S3, available at www.jneurosci.org as supplemental material) in wild-type neurons or neurons with endogenous ZBP1 knockdown. ZBP1*1ΔC is unable to bind RNA and form RNP granules because of the removal of the KH3 and KH4 domains (Farina et al., 2003). The ZBP1*1-GXXG mutant has a 50-fold decreased RNA binding capacity as a consequence of three double-amino acid substitutions in the key regions of three KH domains (Nielsen et al., 2003). Five day overexpression of these mutants in developing neurons transfected on DIV7 did not affect the number of dendritic tips, dendritic branching index, or total dendritic length, in contrast to EGFP-ZBP1*1 overexpression, which decreased these parameters (Fig. 6B,D,F,G; supplemental Table S2A, available at www.jneurosci.org as supplemental material). The Sholl analysis did not show any significant changes in dendritic arborization in the neurons transfected with ZBP1*1ΔC or ZBP1*1-GXXG mutants compared with controls, whereas ZBP1*1 overexpression resulted in a shrinkage of the dendritic field and a decrease in dendritic arbor complexity (Fig. 6E; supplemental Table S2A, available at www.jneurosci.org as supplemental material). The observed differences in dendritic arbor morphology were not attributable to the different expression levels of ZBP1*1 and its mutants, which were even (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). These data indicate that the inhibitory effect of ZBP1 overexpression on dendritic arbor complexity depends on its RNA binding capacity.
Figure 6.
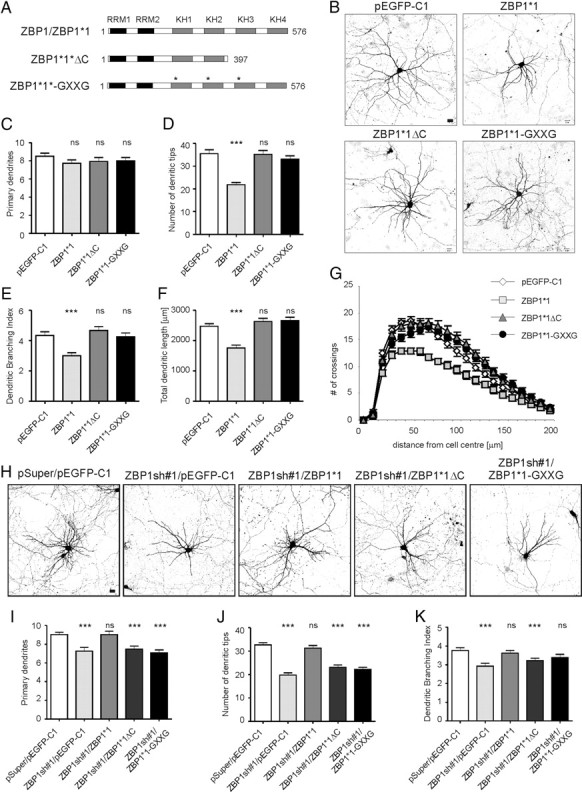
ZBP1 mutants with impaired RNA binding do not affect dendritic branching when overexpressed and are not sufficient for phenotype rescue in neurons with endogenous ZBP1 knockdown. A, Schematic representations of mutants used in this set of experiments. B, Representative micrographs of hippocampal neurons transfected on DIV7 with control vector, EGFP-ZBP1*1, EGFP-ZBP1*1ΔC, or EGFP-ZBP1*1-GXXG. Neuronal morphology was visualized by staining for cotransfected β-gal. Scale bar, 20 μm. C–G, Mean number of primary dendrites (C), mean total number of dendritic tips (D), dendritic branching index (E), total length of dendrites (F), and Sholl analysis of neurons overexpressing EGFP, EGFP-ZBP1*1, EGFP-ZBP1*1ΔC, or EGFP-ZBP1*1-GXXG (G). H, Representative micrographs of hippocampal neurons transfected on DIV7 with pEGFP-C1 together with empty pSuper vector as a control, ZBP1sh#1, or ZBP1sh#1 together with EGFP-ZBP1*1, EGFP-ZBP1*1ΔC, or EGFP-ZBP1*1-GXXG. Expression proceeded for 3 d. Neuronal morphology was visualized by staining for cotransfected β-gal. Scale bar, 20 μm. I–K, Mean number of primary dendrites (I), mean total number of dendritic tips (J), and dendritic branching index of transfected neurons (K). ***p < 0.00025, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment. ns, Not significant. Error bars indicate SEM.
We also tested the ability of ZBP1*1ΔC and ZBP1*1-GXXG to rescue the phenotype of ZBP1-knockdown neurons. DIV7 neurons were transfected with ZBP1sh#1, together with ZBP1*1, ZBP1*1ΔC, or ZBP1*1-GXXG. ZBP1*1 fully rescued the number of primary dendrites, total number of dendritic tips, and dendritic branching index, which were significantly decreased by ZBP1sh#1, but neither ZBP1*1ΔC nor ZBP1*1-GXXG reversed the effects of ZBP1 knockdown and enabled normal dendritogenesis in the presence of ZBP1sh#1 (Fig. 6H–K; supplemental Table S2B, available at www.jneurosci.org as supplemental material). The removal of RNA binding capacity is sufficient to nullify the negative effect of ZBP1 overexpression on dendritic arborization, and ZBP1 that cannot bind mRNA is not able to rescue dendritic arbor growth in neurons with ZBP1 knockdown. Therefore, we conclude that RNA binding is essential for ZBP1 function during dendritogenesis.
Tyrosine 396 phosphorylation of ZBP1 restricts dendritic arborization
To further investigate the mechanisms by which ZBP1 controls dendritogenesis, we focused on the step of de-repression of translation of ZBP1-bound transcripts, which has been suggested to be triggered by Src phosphorylation on ZBP1 tyrosine 396 (Hüttelmaier et al., 2005). If the local translation of ZBP1 cargo is involved in dendritic arborization, then this particular phosphorylation would be vital for it. To examine this possibility, we used a ZBP1*1-Y396F mutant that cannot de-repress bound transcripts because of a disabled Src phosphorylation site (Hüttelmaier et al., 2005) and is resistant to ZBP1sh#1-driven RNAi. We used EGFP-ZBP1*1-Y396F, in addition to EGFP-ZBP1*1 and pEGFP-C1, to transfect neurons on DIV7, together with β-gal to reveal neuronal morphology. Similar to the previous experiments, 5 d overexpression of EGFP-ZBP1*1 severely affected the dendritic arbors of transfected neurons, and EGFP-ZBP1*1-Y396F evoked a similar effect (Fig. 7A). EGFP-ZBP1*1-Y396F overexpression decreased the number of dendritic tips, dendritic branching index, and total dendritic length by 43, 38, and 32%, respectively. The Sholl analysis revealed downward and leftward shifts of the plot for EGFP-ZBP1*1-Y396F-overexpressing neurons (Fig. 7C,E–G; supplemental Table S3A, available at www.jneurosci.org as supplemental material). The number of dendritic tips and dendritic branching index were lower in EGFP-ZBP1*1-Y396F- than EGFP-ZBP1*1-overexpressing neurons, although the decrease was not statistically significant (supplemental Table S3A, available at www.jneurosci.org as supplemental material). However, after 5 d of overexpression, the extent of dendrite loss was so pronounced in both cases that discriminating between them was hindered. Therefore, we overexpressed EGFP-ZBP1*1 and EGFP-ZBP1*1-Y396F for a shorter period of time (3 d) to assess the speed of detrimental changes evoked by overexpression of these proteins. At this time point, neurons that overexpressed EGFP-ZBP1*1-Y396F had more severely retarded dendritic arborization than EGFP-ZBP1*1-transfected neurons (Fig. 7D), reflected by an additional decrease in the number of dendritic tips (13% vs ZBP1*1, p < 0.025) (Fig. 7I; supplemental Table S3B, available at www.jneurosci.org as supplemental material), which showed that the detrimental changes occurred faster in neurons overexpressing ZBP1*1 with a disabled Src phosphorylation site. To further corroborate the importance of the Src-dependent release of mRNA cargo from ZBP1 for dendritic growth, we assessed the ability of ZBP1*1-Y396F to rescue the phenotypical effects of ZBP1 knockdown. As shown in Figure 7K–N and supplemental Table S3C (available at www.jneurosci.org as supplemental material), the EGFP-ZBP1*1-Y396F mutant was unable to neutralize the dendritic arbor deterioration caused by ZBP1sh#1. In summary, these results indicate that ZBP1 RNA binding capacity and the regulation of the translational competence of ZBP1-bound transcripts are involved in dendritic arbor development.
Figure 7.
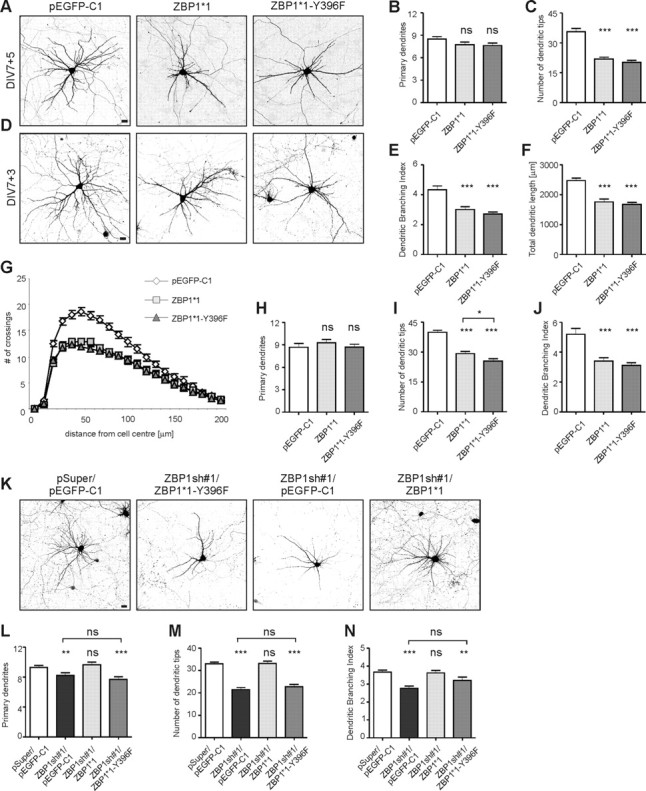
ZBP1 mutant resistant to Src phosphorylation decreases dendritic branching when overexpressed and is not sufficient for phenotype rescue in neurons with endogenous ZBP1 knockdown. A, D, Representative micrographs of hippocampal neurons transfected on DIV7 for 5 d (A) or 3 d (D) with control vector, EGFP-ZBP1*1, or EGFP-ZBP1*1-Y396F. Neuronal morphology was visualized by staining for cotransfected β-gal. B, C, E–G, Mean number of primary dendrites (B), mean total number of dendritic tips (C), dendritic branching index (E), total length of dendrites (F), and Sholl analysis of neurons overexpressing EGFP, EGFP-ZBP1*1, or EGFP-ZBP1*1-Y396F for 5 d (G). H–J, Mean number of primary dendrites (H), mean total number of dendritic tips (I), and dendritic branching index of neurons overexpressing EGFP, EGFP-ZBP1*1, or EGFP-ZBP1*1-Y396F for 3 d (J). ***p < 0.0005, *p < 0.025, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment; ns, not significant. K, Representative micrographs of hippocampal neurons transfected on DIV7 with pEGFP-C1 together with empty pSuper vector as a control, ZBP1sh#1, or ZBP1sh#1 together with EGFP-ZBP1*1 or EGFP-ZBP1*1-Y396F. Expression proceeded for 3 d. Neuronal morphology was visualized by staining for cotransfected β-gal. L–N, Mean number of primary dendrites (L), mean total number of dendritic tips (M), and dendritic branching index of neurons transfected with control vector, ZBP1sh#1, ZBP1sh#1 together with EGFP-ZBP1*1, or ZBP1sh#1 together with EGFP-ZBP1*1-Y396F (N). ***p < 0.00033, **p < 0.0033, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment; ns, not significant. Scale bar, 20 μm. Error bars indicate SEM.
ZBP1 levels influence dendritic distribution of β-actin
The original ZBP1 cargo identified in dendrites is β-actin mRNA. Therefore, the explanation for the effect of ZBP1 depletion on dendritic arborization could be that it leads to disturbances in the cytoskeleton via impediments to the local translation of actin, originating from impairment of dendritic targeting of β-actin mRNA. Thus, we verified whether the distribution of β-actin mRNA and protein is disturbed under our experimental conditions. Fluorescence in situ hybridization with probes specific to β-actin mRNA revealed its presence in puncta in cell soma and in dendrites of control cells transfected with pSuper on DIV7 (Fig. 8A). However, on knockdown of ZBP1 with ZBP1sh#1, the FISH signal was still present in primary dendrites but diminished in more distal dendritic branches (Fig. 8B,B′,B″,E; supplemental Table S4A, available at www.jneurosci.org as supplemental material). EGFP-ZBP1 overexpression did not visibly alter the dendritic distribution of β-actin mRNA puncta (data not shown).
Figure 8.
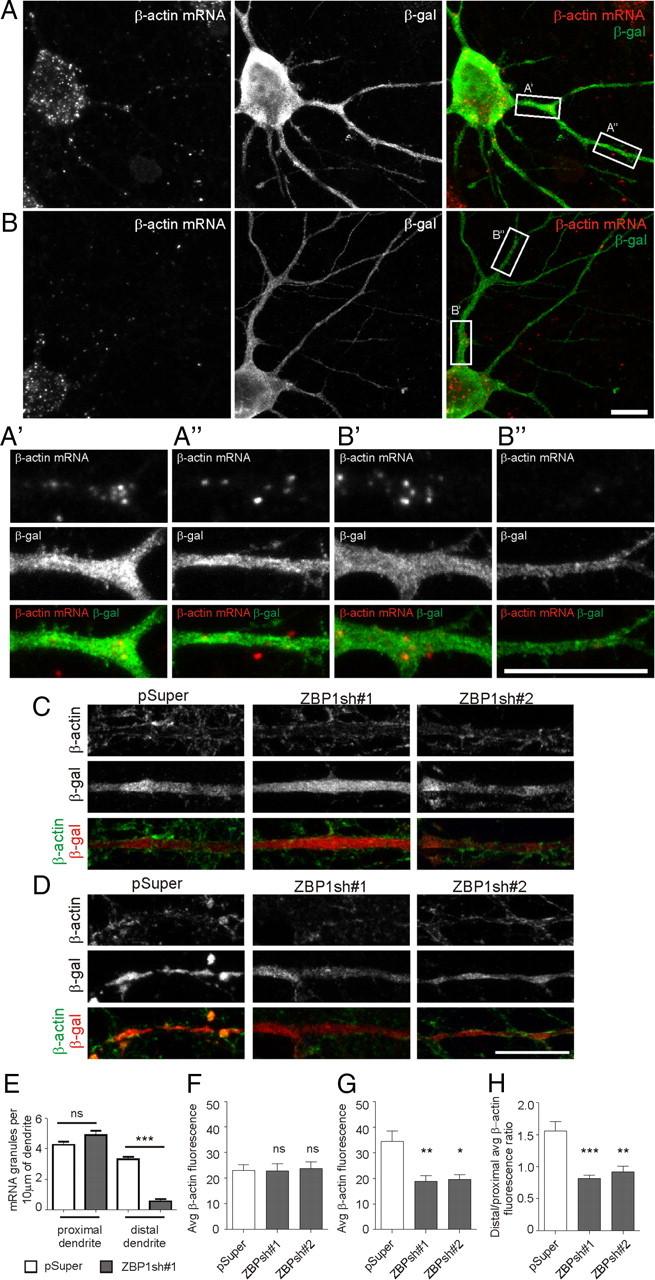
β-Actin mRNA and protein localization are disturbed in ZBP1-knockdown neurons. A, B, Representative micrographs of fluorescent β-actin in situ hybridization (red) in hippocampal neurons that were transfected on DIV7 with pSuper (A) or ZBP1sh#1 (B) for 36 h. Neuronal morphology is visualized by staining for cotransfected β-gal (green). Scale bar, 10 μm. A′, A″, B′, and B″ show in greater magnification fragments of proximal (A′, B′) and distal (A″, B″) dendrites of transfected neurons. Scale bar, 10 μm. C, D, Hippocampal neurons were transfected at DIV7 for 48 h with pSuper, ZBP1sh#1, or ZBP1sh#2. Pictures show β-actin staining in proximal (C) and distal (D) dendrites of transfected cells. Scale bar, 10 μm. E, Mean number of β-actin mRNA granules per 10 μm of dendrite measured in proximal and distal dendrites (after branching). ***p < 0.001; ns, not significant; unpaired t test with Welch's correction. F–H, Average intensity of fluorescence of β-actin protein in proximal (F) and distal (G) dendrites, and ratio of average fluorescence intensities in distal and proximal dendrites (H). ***p < 0.001; **p < 0.01; *p < 0.05; ns, not significant; Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment. Error bars indicate SEM.
We then verified by immunofluorescence the levels of β-actin protein in proximal and distal dendrites. As shown in Figure 8, C, D, and F–H, and supplemental Table S4A (available at www.jneurosci.org as supplemental material), in control neurons transfected with pSuper on DIV7, immunofluorescence was stronger in distal dendrites than in proximal dendrites. However, transfection of ZBP1sh#1 or ZBP1sh#2 resulted in a significant decrease in β-actin levels in distal dendrites but not in proximal dendrites (Fig. 8C,D,F–H; supplemental Table S4A, available at www.jneurosci.org as supplemental material). Consequently, the ratio of β-actin immunofluorescence in distal dendrites to proximal dendrites dropped (Fig. 8H; supplemental Table S4A, available at www.jneurosci.org as supplemental material). A similar change was observed in neurons overexpressing EGFP-ZBP1 compared with EGFP-C1-transfected neurons (distal/proximal immunofluorescence ratio: EGFP-C1, 1.56 ± 0.15; EGFP-ZBP1, 0.64 ± 0.08; p < 0.001) (supplemental Table S4A, available at www.jneurosci.org as supplemental material), which is consistent with ZBP1 being a translational repressor (Hüttelmaier et al., 2005).
β-Actin is crucial for ZBP1-dependent dendritic arborization
To determine whether β-actin abundance and protein diffusion are sufficient for overcoming the negative effect of ZBP1 knockdown, we overexpressed the β-actin coding sequence under the control of an SV40 late polyadenylation signal (Morales et al., 2000) in developing neurons cotransfected with ZBP1sh#1. GFP-β-actin was efficiently expressed and present in dendrites of transfected neurons (supplemental Fig. S4A,B, available at www.jneurosci.org as supplemental material). We found that β-actin overexpression nullified the negative effect of ZBP1sh#1 on the primary dendrite number, dendritic tip number, and dendritic branching index (Fig. 9A–D; supplemental Table S4B, available at www.jneurosci.org as supplemental material). Similar to ZBP1*1, β-actin overexpression also significantly inhibited, but did not fully prevent, the ZBP1sh#1-induced decrease in total dendritic length (30% increase vs ZBP1sh#1, p < 0.001; 19% decrease vs control, p < 0.001) (Fig. 9E; supplemental Table S4B, available at www.jneurosci.org as supplemental material). Additionally, the maximum number of crossings in the Sholl analysis was restored in ZBP1sh#1- and β-actin-cotransfected neurons. However, their Sholl plots remained shifted leftward and downward compared with control but upward compared with ZBP1sh#1 (Fig. 9F; supplemental Table S4B, available at www.jneurosci.org as supplemental material). Coexpression of β-actin had a protective effect in ZBP1sh#1-transfected neurons, regardless of the fact that its expression in wild-type neurons transfected with the same amount of β-actin-coding plasmid did not increase dendritic arbor growth (supplemental Fig. S4C, Table S4C, available at www.jneurosci.org as supplemental material).
Figure 9.
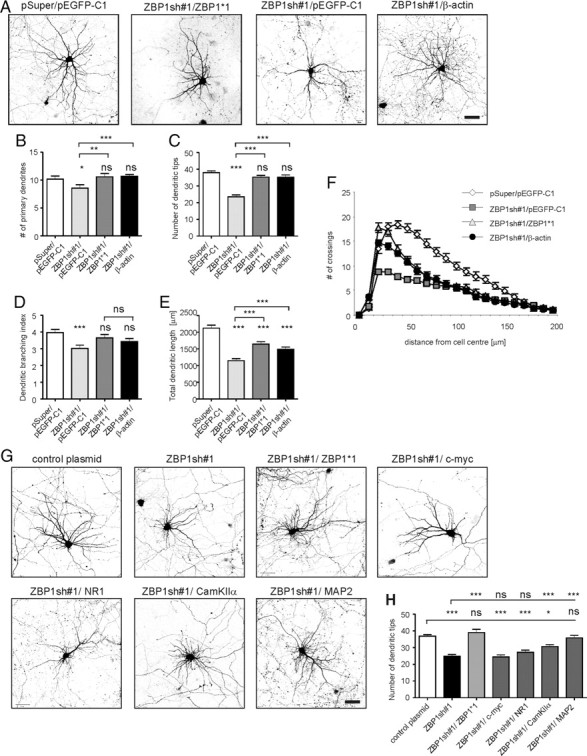
β-Actin overexpression in hippocampal neurons with ZBP1 knockdown is sufficient for partial phenotype rescue. A, Micrographs of hippocampal neurons transfected on DIV7 with pEGFP-C1 together with pSuper vector as a control, ZBP1sh#1, or ZBP1sh#1 together with EGFP-ZBP1*1 or EGFP-β-actin. Expression proceeded for 3 d. Neuronal morphology was visualized by staining for cotransfected β-gal. B–F, Mean number of primary dendrites (B), mean number of dendritic tips (C), dendritic branching index (D), mean total dendritic length (E), and Sholl analysis of transfected neurons (F). ***p < 0.00033, **p < 0.0033, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment; ns, not significant. Scale bar, 50 μm. G, Phenotype rescue attempted with other possible ZBP1 targets and unrelated cDNAs. The figure shows representative images of neurons transfected with control plasmid, ZBP1sh#1, ZBP1sh#1/EGFP-ZBP1*1, ZBP1sh#1/c-Myc, ZBP1sh#1/GW1-NR1, ZBP1sh#1/GFP-CaMKIIα, and ZBP1sh#1/MAP2. H, Mean number of dendritic tips of transfected neurons. ***p < 0.00016, *p < 0.008, Kruskal–Wallis test followed by Mann–Whitney post hoc test with Bonferroni's adjustment; ns, not significant. Scale bar, 50 μm. Error bars indicate SEM.
As a next step, we tested whether β-actin overexpression rescue effect is specific for ZBP1 knockdown. We first developed and validated shRNA against Staufen1 that did not recognize its homolog Staufen2 in a COS-7 cell line (supplemental Fig. S5, available at www.jneurosci.org as supplemental material). Staufen1 is a mammalian homolog of Drosophila Staufen, which plays a role in the stage of early RNA sorting, binds numerous transcripts in neurons, and regulates dendritic branching (Monshausen et al., 2001; Mallardo et al., 2003; Vessey et al., 2008; Tübing et al., 2010). Staufen1, however, was not shown to bind β-actin mRNA directly. Consistent with a previous experimental design, we then tested the effects of 3 d of Staufen1 knockdown in neurons transfected on DIV7 in the presence or absence of β-actin coding plasmid. As expected, transfection with Stau1sh severely impaired dendritic arbor morphology (supplemental Fig. S5C–F, Table S4D, available at www.jneurosci.org as supplemental material). β-Actin overexpression, however, was not able to reverse this effect (supplemental Fig. S5C–F, Table S4D, available at www.jneurosci.org as supplemental material).
Finally, we investigated whether the ZBP1 knockdown phenotype can be rescued by overexpression of either c-Myc (another ZBP1 target) or the NMDA receptor subunit 1 (NR1), Ca2+/calmodulin-dependent protein kinase (CaMKIIα), microtubule-associated protein 2 (MAP2), three locally translated proteins [the mRNA of which was reported to be the target of fragile X mental retardation protein (FMRP), Staufen1, and MARTA1, respectively] (Rehbein et al., 2002; Mallardo et al., 2003; Schütt et al., 2009). DIV7 neurons were transfected with ZBP1sh#1 together with plasmid encoding cDNAs encoding these proteins. As shown in Figure 9, c-Myc, NR1, and CaMKIIα did not fully reverse the decrease in the number of dendritic tips induced by ZBP1sh#1 (supplemental Table S4E, available at www.jneurosci.org as supplemental material), although overexpression of CaMKIIα was partially effective. Intriguingly, overexpression of MAP2, a microtubule-stabilizing protein, fully prevented the phenotypic effects of ZBP1 knockdown on the number of dendritic tips (Fig. 9H; supplemental Table S4E, available at www.jneurosci.org as supplemental material).
Discussion
The present study describes the essential contribution of a protein component of RNP, ZBP1, to the development of proper dendritic morphology in hippocampal neurons. It also shows that ZBP1 is not required for the stabilization of already developed dendritic arbors. This difference correlates with the lower abundance and more even dendritic distribution of ZBP1 in mature neurons than developing neurons. A detailed analysis of the contribution of ZBP1 to dendritic development showed that the ability of ZBP1 to bind, transport, and release its cargo was necessary to sustain a correct branching pattern of dendritic arbors. Moreover, the negative effects of ZBP1 knockdown were significantly, but not completely, alleviated by ZBP1-independent overexpression of β-actin, a well known target of ZBP1. Thus, β-actin mRNA transport and translational control contribute to the role of ZBP1 in dendritogenesis.
The above conclusion is supported by several studies, in which dendritic arborization was shown to rely on local translation. First, dendritic growth depends on crucial regulators of protein synthesis (Jaworski et al., 2005). Second, the mRNA and proteins of transport RNPs, as well as dendritic processing bodies (P-bodies) (i.e., other structures important for transport and mRNA metabolism), are present in dendrites (Tang et al., 2001; Eom et al., 2003; Tiruchinapalli et al., 2003; Barbee et al., 2006; Zeitelhofer et al., 2008a). Third, the roles of some of these proteins have been demonstrated for proper dendritic arbor morphology (Lee et al., 2003; Pan et al., 2004; Ye et al., 2004; Restivo et al., 2005; Barbee et al., 2006; Bestman and Cline, 2008, 2009; Vessey et al., 2008, 2010; Fiore et al., 2009). The present study adds to this evidence. ZBP1, a protein that participates in mRNA transport and local translation, was shown to contribute to the regulation of dendritic arbor morphology.
However, ZBP1 regulates various aspects of cellular mRNA distribution and metabolism and is a component of different types of RNPs (Zeitelhofer et al., 2008b; Buchan and Parker, 2009). Although ZBP1 is widely considered a component of dendritic transport RNPs (Kindler et al., 2005; Kiebler and Bassell, 2006), it also shuttles between the cytoplasm and nucleus (Nielsen et al., 2003), can be recruited to P-bodies and stress granules (Barbee et al., 2006; Stöhr et al., 2006; Cougot et al., 2008) (but see Zeitelhofer et al., 2008a), and regulates not only translational competence but also the stability of bound transcripts (Hüttelmaier et al., 2005; Stöhr et al., 2006). Thus, caution should be taken when interpreting the role of ZBP1 in cellular physiology at the molecular level. For example, the fact that both ZBP1 overexpression and knockdown led to the simplification of dendritic arbors may appear contradictory. However, considering the complex role of ZBP1 in mRNA transport and its translational silencing, these findings support the hypothesis of the contribution of ZBP1-dependent translational control of transported mRNA to dendrite development. Each of our genetic manipulations should result in a disturbance of the discrete arrangement of translational events in dendrites, either by preventing mRNA transport (knockdown) or increasing the threshold for its local release (overexpression). The first conclusion is supported by the following observations: (1) RNA binding-deficient ZBP1 mutants are insufficient for the rescue of the ZBP1sh phenotype, (2) ZBP1 knockdown leads to a disturbed spatial distribution of β-actin mRNA and protein, and (3) an increase in β-actin levels, presumably without its natural spatial distribution, only partially rescues the more complex parameters of dendritic arbors in ZBP1-deficient neurons. The second conclusion is supported by the finding that the simplification of dendritic arborization caused by ZBP1 knockdown could not be rescued with ZBP1*1-Y396F overexpression. This ZBP1 mutant normally binds to and transports target mRNA to its destination, but it cannot release it for translation (Hüttelmaier et al., 2005). Additionally, short-term overexpression of this mutant (3 d instead of 5 d) revealed that it affected dendritic arborization more robustly than wild-type ZBP1. Finally, overexpression of ZBP1 affected the dendritic levels of β-actin protein. However, considering various cellular roles of ZBP-1, future experiments with translational reporters (Leung et al., 2006) are needed to directly visualize a role for ZBP1 in local protein synthesis during dendritogenesis.
Our studies also revealed a correlation between the localization of ZBP1 to the branching points of dendrites and the contribution of ZBP1 localization to dendritic arbor development and stability. In developing neurons, overexpressed ZBP1 preferentially localizes to branching points, and ZBP1 knockdown restricts dendritic arborization. In mature neurons, in contrast, ZBP1 becomes distributed more evenly along dendrites, and its knockdown no longer influences dendritic arbors. These results led us to question how preferential localization of ZBP1 to branching points regulates dendrite development and stabilization. Preferential localization to branching points and an involvement in the regulation of dendritic branching have been reported for at least two other proteins: Me31B (a component of dendritic P-bodies) and cytoplasmic polyadenylation element binding protein (a protein responsible for dendritic mRNA delivery) (Barbee et al., 2006; Bestman and Cline, 2008, 2009). Intriguingly, dendritic P-bodies disperse or exchange their content more dynamically in response to increased neuronal activity, which has been interpreted as a release of stored mRNA for induced translation (Cougot et al., 2008; Zeitelhofer et al., 2008a). Bestman and Cline (2009) suggested that such release at a branching point could actually be a part of a mechanism of dendritic arbor development. Therefore, a reasonable speculation is that ZBP1 accumulates at branching points in the form of autonomous RNPs or as a component of P-bodies and regulate mRNA storage and release to support the process of dendritic arborization. But places of actual translation need to be further investigated with use of translational reporters (Leung et al., 2006).
An intriguing question is which specific mRNAs are transported and translationally regulated in dendrites to support dendritic arbor development. Scores of mRNAs that encode proteins that fall into different functional groups have been shown to localize to dendrites (Moccia et al., 2003; Schratt et al., 2004; Poon et al., 2006; Zhong et al., 2006). According to Zhong et al. (2006), cytoskeleton elements and regulators are among the mRNAs abundantly present in dendrites. Our studies primarily focused on β-actin as a potential ZBP1 mRNA target important for dendritic growth because it is the best described ZBP1 substrate, which fits well into a molecular mechanism of dendritic growth. The importance of the actin cytoskeleton for the development and stability of dendritic arbors is well established (Threadgill et al., 1997; Nakayama et al., 2000; Urbanska et al., 2008). However, no studies have indicated that local actin production plays an important role in these processes. In the present study, we provide supporting evidence showing that ZBP1 knockdown led to local changes in dendritic β-actin and to the deterioration of dendritic arborization, which was rescued by β-actin overexpression. The rescue effect was specific to ZBP1-β-actin because (1) other proteins known to be locally translated (CaMKIIα, NR1) could not reverse the ZBP1 knockdown phenotype and (2) dendritic deterioration attributable to Staufen1 knockdown was not prevented by β-actin overexpression. However, in non-neuronal cells, ZBP1 was shown to bind hundreds of transcripts (Jønson et al., 2007; Furic et al., 2008; Hafner et al., 2010) so other factors may contribute to its role in dendritogenesis. We show that another canonical ZBP1 target, c-myc, is unable to rescue the ZBP1sh phenotype in hippocampal neurons. However, among the ZBP1-bound transcripts, mRNAs encoding proteins involved in protein secretion and ubiquitin-dependent protein degradation are enriched (Jønson et al., 2007), and Guo et al. (2001) provided evidence for proteasome-dependent dendritic growth. Additional studies are needed to verify whether ZBP1 exerts its effects on dendritic growth through the transport of these particular mRNAs. Intriguingly, we found that MAP2 overexpression, in addition to β-actin, rescued the ZBP1 knockdown phenotype. Indeed, MAP2 is crucial for proper dendritic arbor morphology (Teng et al., 2001; Urbanska et al., 2008) but does not contain a consensus sequence for ZBP1. Thus, this result cannot be explained similarly to β-actin, and the mechanism of the importance of MAP2 for ZBP1-dependent dendritic arbor development requires additional investigation. ZBP1 can be recruited to mRNA also by alternative mechanisms, including (1) binding to non-3′-UTR sequences, (2) the recognition of a particular RNA spatial structure, or (3) an interaction with other mRNA binding proteins (Rackham and Brown, 2004; Patel and Bag, 2006; Atlas et al., 2007). MAP2 dendritic transport requires the rat homolog of ZBP2 (Rehbein et al., 2002), a protein that is also needed for the binding of ZBP1 to β-actin mRNA (Pan et al., 2007).
Very few studies have investigated the identity of the dendritic mRNA species regulated by RNP proteins that would play a role in dendritogenesis. The data presented herein add ZBP1-β-actin mRNA to the previously described FMRP-Rac1 mRNA in this task (Lee et al., 2003). Notably, Rac1 regulates the spatial organization of microfilament networks. Therefore, these examples suggest that the local translation of cytoskeleton elements may be involved in the control of dendritic growth.
Footnotes
This work was supported by the Polish Ministry of Science and Higher Education Grant NN301 3147 33, Polish–Norwegian Research Fund Grant PNRF-96-AI-1/07, and FP7-HealthProt Grant 229676. We thank Dr. Gary Bassell for the EGFP-ZBP1 plasmid, Dr. Paolo Macchi for the Staufen1-EYFP and Staufen2-EYFP plasmids and pSuperior-Stau2sh, Dr. Casper Hoogenraad for β-actin-mRFP, β-actin-mCherry, FLAG-CaMKIIα, and EGFP-MAP2c, Dr. Robert Pawlak for pPDGF-GFP-β-actin, Dr. Anthone Dunah for GW1-NR1, and Dr. Robert Singer for ZBP1 antibody. We are thankful to Jakub Mieczkowski and Magda Dziembowska for consultation on statistics and experimental design, Malgorzata Urbanska and Anna Malik for help with the experiments, and Monika Dudek for technical assistance. We also express our gratitude to Clive Bramham, Leszek Kaczmarek, Anna Malik, Malgorzata Urbanska, and Lukasz Swiech for fruitful discussions and their comments on our work.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Atlas R, Behar L, Sapoznik S, Ginzburg I. Dynamic association with polysomes during P19 neuronal differentiation and an untranslated-region-dependent translation regulation of the tau mRNA by the tau mRNA-associated proteins IMP1, HuD, and G3BP1. J Neurosci Res. 2007;85:173–183. doi: 10.1002/jnr.21099. [DOI] [PubMed] [Google Scholar]
- Banker G, Goslin K. Developments in neuronal cell culture. Nature. 1988;336:185–186. doi: 10.1038/336185a0. [DOI] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci U S A. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The relationship between dendritic branch dynamics and CPEB-labeled RNP granules captured in vivo. Front Neural Circuits. 2009;3:10. doi: 10.3389/neuro.04.010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Bhattacharyya SN, Tapia-Arancibia L, Bordonné R, Filipowicz W, Bertrand E, Rage F. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28:13793–13804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrij FM, Levenga J, van der Linde HC, Koekkoek SK, De Zeeuw CI, Nelson DL, Oostra BA, Willemsen R. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol Dis. 2008;31:127–132. doi: 10.1016/j.nbd.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sánchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille JC, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a β-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze B, Tuebing F, Xie Y, Dorostkar MM, Thomas S, Pehl U, Boehm S, Macchi P, Kiebler MA. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J Cell Biol. 2006;172:221–231. doi: 10.1083/jcb.200509035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Lin Y, Horbinski C, Drahushuk KM, Kim IJ, Kaplan PL, Lein P, Wang T, Higgins D. Dendritic growth induced by BMP-7 requires Smad1 and proteasome activity. J Neurobiol. 2001;48:120–130. [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Feliu-Mojer MI, Spangler SA, Milstein AD, Dunah AW, Hung AY, Sheng M. Liprinalpha1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev Cell. 2007;12:587–602. doi: 10.1016/j.devcel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, Krugers H, Defilippi P, Akhmanova A, Hoogenraad CC. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Jønson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau G, Maher-Laporte M, Topolnik L, Laurent CE, Sossin W, Desgroseillers L, Lacaille JC. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol Cell Biol. 2008;28:2896–2907. doi: 10.1128/MCB.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Müller J, Goetze B, Macchi P, Peters C, Kiebler MA. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc Natl Acad Sci U S A. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Moccia R, Chen D, Lyles V, Kapuya E, E Y, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, Martin KC. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen M, Putz U, Rehbein M, Schweizer M, DesGroseillers L, Kuhl D, Richter D, Kindler S. Two rat brain staufen isoforms differentially bind RNA. J Neurochem. 2001;76:155–165. doi: 10.1046/j.1471-4159.2001.00061.x. [DOI] [PubMed] [Google Scholar]
- Morales M, Colicos MA, Goda Y. Actin-dependent regulation of neurotransmitter release at central synapses. Neuron. 2000;27:539–550. doi: 10.1016/s0896-6273(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Adolph SK, Rajpert-De Meyts E, Lykke-Andersen J, Koch G, Christiansen J, Nielsen FC. Nuclear transit of human zipcode-binding protein IMP1. Biochem J. 2003;376:383–391. doi: 10.1042/BJ20030943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13:199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Hüttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol Cell Biol. 2007;27:8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr Biol. 2004;14:1863–1870. doi: 10.1016/j.cub.2004.09.085. [DOI] [PubMed] [Google Scholar]
- Patel GP, Bag J. IMP1 interacts with poly(A)-binding protein (PABP) and the autoregulatory translational control element of PABP-mRNA through the KH III–IV domain. FEBS J. 2006;273:5678–5690. doi: 10.1111/j.1742-4658.2006.05556.x. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Brown CM. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 2004;23:3346–3355. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehbein M, Wege K, Buck F, Schweizer M, Richter D, Kindler S. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J Neurochem. 2002;82:1039–1046. doi: 10.1046/j.1471-4159.2002.01058.x. [DOI] [PubMed] [Google Scholar]
- Restivo L, Ferrari F, Passino E, Sgobio C, Bock J, Oostra BA, Bagni C, Ammassari-Teule M. Enriched environment promotes behavioral and morphological recovery in a mouse model for the fragile X syndrome. Proc Natl Acad Sci U S A. 2005;102:11557–11562. doi: 10.1073/pnas.0504984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci MS, Jin Z, Dews M, Yu D, Thomas-Tikhonenko A, Dicker DT, El-Deiry WS. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol. 2004;24:8541–8555. doi: 10.1128/MCB.24.19.8541-8555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütt J, Falley K, Richter D, Kreienkamp HJ, Kindler S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J Biol Chem. 2009;284:25479–25487. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev I, London M. Untangling dendrites with quantitative models. Science. 2000;290:744–750. doi: 10.1126/science.290.5492.744. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Hüttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart G, Spruston N, Häusser M. Dendrites. Ed 2. Oxford: Oxford UP; 2007. [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Teng J, Takei Y, Harada A, Nakata T, Chen J, Hirokawa N. Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J Cell Biol. 2001;155:65–76. doi: 10.1083/jcb.200106025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tübing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci. 2010;30:4160–4170. doi: 10.1523/JNEUROSCI.3537-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska M, Blazejczyk M, Jaworski J. Molecular basis of dendritic arborization. Acta Neurobiol Exp (Wars) 2008;68:264–288. doi: 10.55782/ane-2008-1695. [DOI] [PubMed] [Google Scholar]
- Verpelli C, Piccoli G, Zanchi A, Gardoni F, Huang K, Brambilla D, Di Luca M, Battaglioli E, Sala C. Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J Neurosci. 2010;30:5830–5842. doi: 10.1523/JNEUROSCI.0119-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tübing F, Aparicio SA, Abel T, Kiebler MA. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci U S A. 2008;105:16374–16379. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci U S A. 2010;107:3222–3227. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M, Dahm R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008a;28:7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitelhofer M, Macchi P, Dahm R. Perplexing bodies: the putative roles of P-bodies in neurons. RNA Biol. 2008b;5:244–248. doi: 10.4161/rna.6948. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]