Abstract
Sexual dimorphism of the brain has been well characterized anatomically in Drosophila melanogaster at the single neuron level, yet little is known about the molecular mechanism whereby cellular sex differences are generated except that the neural sex determination gene fruitless (fru) plays a key role. The fru-expressing mAL interneuron cluster is sexually dimorphic in three aspects: the number of cells composing the cluster is 5 in females and 30 in males; the ipsilateral neurite is absent in females and present in males; the contralateral neurite forms Y-shaped branches in the subesophageal ganglion in females while it ends with a simple horsetail-like structure in males. By screens in the compound eye for modifiers of roughness induced by fru+ overexpression, we identified a loss-of-function allele of hunchback (hb) to be a suppressor of this phenotype. Hb was expressed in most of the fru-expressing neurons in the pupal and adult stages. Knocking down hb in mAL MARCM (Mosaic Analysis with a Repressible Cell Marker) clones in the male brain resulted in partial demasculinization of the branching pattern of the contralateral neurites without affecting the cell number and the ipsilateral neurite formation. The present results suggest that Hb is essential for male-typical shaping of the contralateral neurites by Fru.
Introduction
Sexual dimorphism is prevalent in the Drosophila brain (Kondoh et al., 2003; Billeter and Goodwin, 2004; Kimura et al., 2005, 2008; Datta et al., 2008; Sanders and Arbeitman, 2008; Cachero et al., 2010; Rideout et al., 2010; Robinett et al., 2010; Yu et al., 2010), particularly for interneurons that express fruitless (fru), a neural sex determination gene originally identified by its mutant phenotype, which enhances male-to-male courtship (Gill, 1963; Hall, 1978). The best-characterized among them is the interneuronal cluster mAL, which shows three types of sex differences (Kimura et al., 2005; Koganezawa et al., 2010). First, the number of neurons composing the cluster is 30 in males and 5 in females. Second, the male mAL cluster has bilateral projections, in contrast to the female cluster, which has only a contralateral projection. Third, the contralateral projection to the subesophageal ganglion terminates in prominent bifurcating branches in females, whereas the male neurites end with a simple horsetail-like structure.
The Fru proteins play central roles in producing these sex differences, as evidenced by the fact that mAL neurons are feminized in all three ways in fru mutant males (Kimura et al., 2005). The fru gene has multiple promoters, only one of which is active in the nervous system (Ryner et al., 1996; Usui-Aoki et al., 2000). The primary transcript of fru from this promoter is subjected to sex-specific splicing as aided by the female determinant protein Transformer (Ito et al., 1996; Ryner et al., 1996; Du et al., 1998; Demir and Dickson, 2005), resulting in protein-coding mRNAs only in males. Thus, the Fru proteins are male-specifically expressed in the nervous system (Lee et al., 2000; Usui-Aoki et al., 2000).
The difference in the number of mAL neurons between the sexes results from female-specific cell death in a subset of mAL neurons (Kimura et al., 2005). The Fru proteins prevent the male counterparts from dying, allowing the survival of mAL neurons that have an ipsilateral neurite, i.e., male-typical neurons (Kimura et al., 2005). By deleting three major cell-death genes, hid, grim and rpr, those neurons that have an ipsilateral neurite survive through development to the adult stage in the female brain. Interestingly, however, such mAL neurons that escaped from cell death in females had a contralateral neurite with Y-shaped branches in the subesophageal ganglion, i.e., the female-typical structure (Kimura et al., 2005). This observation suggests that the Fru proteins act to shape neurites, apart from their function of inhibiting cell death in the male brain (Kimura et al., 2008). However, nothing is known about the mechanism by which Fru directs neurites to the correct destination in establishing male-typical connections.
In the present work, we performed a genetic screening for modifiers of fru, with the aim of isolating genes that collaborate with it in the formation of neural sexual dimorphism. The screening yielded the gap gene hunchback (hb) as a candidate fru modifier. Our subsequent analysis revealed that hb indeed functions in the elaboration of sex-specific patterning of mAL neurites.
Materials and Methods
Fly strains.
Flies were reared on cornmeal-yeast medium at 25°C. The hbB1 allele carrying FRT was a kind gift from Bing Ye (University of California, San Francisco, San Francisco, CA). Canton-S served as a wild-type control. fruNP21 was obtained from the National Institute of Genetics. hb12, UAS-hb+ and UAS-hb RNAi (#29630) stocks were obtained from the Bloomington Stock Center. The UAS-hb RNAi (#107740) line was obtained from the Vienna Drosophila RNAi Center. UAS-pumilio (pum) (full-length) and UAS-pum RNA-binding-domain (pum RBD) flies were a kind gift from Kai Zinn (California Institute of Technology, Pasadena, CA).
Modifier screening.
The flies carrying both GMR-Gal4 and UAS-fru+ transgenes were crossed with the flies from mutant stocks reported to have developmental defects in the nervous system (flybase: http://flybase.bio.indiana.edu/) (supplemental Table S1, available at www.jneurosci.org as supplemental material). To observe the eye structure of hb lethal mutants, hb homozygous clones were generated by the FLP-FRT system using the stock that carried ey-flp on chromosome X and FRT and hb together on chromosome 3. Images of the compound eye surface were obtained with a scanning electron microscope (VE-9800; Keyence Corp.).
Antibodies.
Antibodies used were as follows: guinea pig anti-Hb (Asian Distribution Center for Segmentation Antibodies), rabbit anti-Fru-Male2 (denoted as “anti-Fru” in this study) (Kimura et al., 2008), mouse anti-nc82 (Developmental Studies Hybridoma Bank), rabbit anti-GFP (Invitrogen), rat anti-mCD8 (Caltag), mouse anti-β-tubulin (GE Healthcare), Alexa Fluor 488 anti-rabbit and anti-rat, Alexa Fluor 546 anti-rabbit, Alexa Fluor 647 anti-mouse and anti-guinea pig (Invitrogen).
Dissection, immunohistochemistry, and imaging of the CNS.
The expression of Fru and Hb was examined by staining the neural tissues with the respective antibodies mentioned above according to the method described previously (Kimura et al., 2005). Fru-expressing neurons were also visualized with the Gal4 enhancer-trap fruNP21. In the male brain, fruNP21 labels ∼500 cells per hemibrain, 82% of which are immunopositive to an anti-Fru antibody (Kimura et al., 2008).
The brains of 2- to 7-d-old adult flies were dissected. Fixation and immunohistochemical staining of the flies were performed as described previously (Kimura et al., 2005). Stacks of optical sections at 1 or 2 μm were obtained with a Zeiss LSM 510 confocal microscope and processed with NIH ImageJ (http://rsb.info.nih.gov/ij/) and Adobe Photoshop.
Clonal analysis of mAL neurons.
Somatic clones were produced using the Mosaic Analysis with a Repressible Cell Marker (MARCM) method (Lee and Luo, 1999). The following flies were generated: y hs-flp/Y;G13 UAS-mCD8-GFP/G13 tub-Gal80; fruNP21/+ as the fruNP21/+ control males; and y hs-flp;G13 UAS-mCD8-GFP/G13 tub-Gal80; fruNP21/+ as the fruNP21/+ control females; y w hs-flp UAS-mCD8-GFP/Y;FRT42D UAS-hb RNAi/FRT42D tub-Gal80; fruNP21/UAS-mCD8-GFP and y hs-flp; G13 UAS-mCD8-GFP/G13 tub-Gal80; fruNP21/UAS-hb RNAi as test flies. For the production of neuroblast clones in mAL neurons, embryos at 0–24 h after egg laying (AEL) were heat shocked at 37°C in a water bath for 20 min. For the production of single-cell clones of mAL neurons, larvae at 3–6 d AEL were heat shocked at 37°C for 20 min.
Western blotting assays.
For Western blot analysis of the lysates of flies carrying an hb-knockdown construct (see Fig. 3E), the heads of six adult flies were homogenized in a SDS sample buffer. After fractionation by SDS-PAGE and transfer to nitrocellulose membranes, the blots were reacted with antibodies for guinea pig anti-Hb (1:2000) or mouse anti-β-tubulin (1:2000) followed by horseradish peroxidase-conjugated anti-guinea pig or mouse IgG (Invitrogen). Chemiluminescence was detected with an ECL Plus Western Blotting Detection System (GE Healthcare). Signal intensity was quantified by using NIH ImageJ.
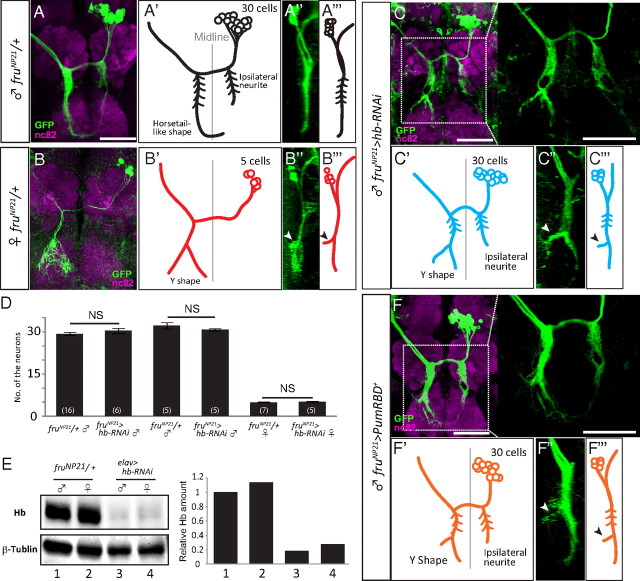
Figure 3.
Effect of hb knockdown on mAL neurons observed in neuroblast MARCM clones. mAL in the male (A, A′, A″, A‴), and female brains (B, B′, B″, B‴). Upon knocking down hb, the male mAL developed side branches (arrowheads) similar to those observed in female neurons in the contralateral neurite (C, C′, C″, C‴). The original images (A–C, frontal views; A″, B″, C″, lateral views) and schematic drawings (A′, B′, C′, frontal views; A‴, B‴, C‴, lateral views) of mAL neurons are shown. Scale bar, 50 μm. D, The mean ± SEM. of the number of mAL neurons. mAL was visualized with the anti-Fru antibody (two left-side bars) or GFP expression under the control of fruNP21 (four right-side bars). Shown in parentheses is the number of hemibrains examined (NS, not significant, p > 0.05 by Student's t test). E, Western blot analysis of the effect of hb knockdown. The numbers shown at the bottom of E indicate the correspondence between the raw data and quantified graphs. F–F‴, The overexpression of Pum RBD demasculinized the male mAL. The original images (F, frontal views; F″, lateral views) and schematic drawings (F′, frontal views; F‴, lateral views) of mAL neurons are shown. The anterior to the left for A‴, B‴, C‴, and F‴. In C and F, enlarged views of the boxed region are also shown. Scale bar, 50 μm.
Results
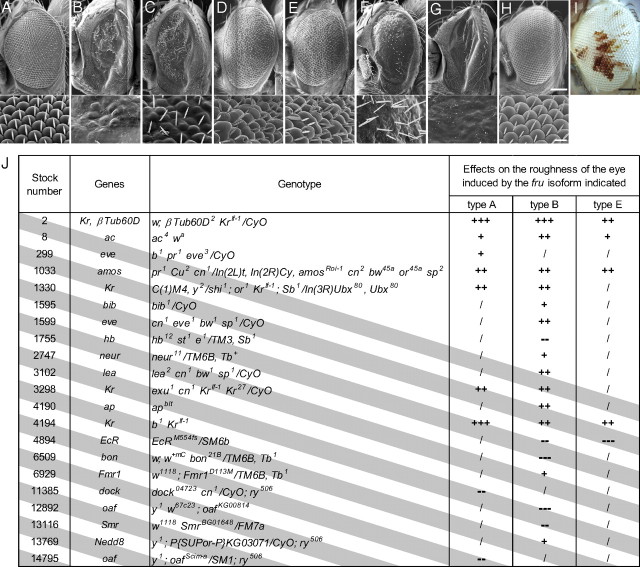
We took advantage of the dominant visible phenotype induced by the overexpression of fru+ in the compound eyes, i.e., the rough-eye phenotype (Fig. 1B), which allowed us to screen many mutations for their ability to enhance or suppress the fru+ effects. By examining 435 heterozygous mutations, we obtained 14 enhancers and 7 suppressors of the fru+-induced rough-eye phenotype (Fig. 1J). Of the 14 enhancers, 4 were Krüppel (Kr) alleles and 1 suppressor was hb (Fig. 1C), a known transcriptional regulator of Kr (Fig. 1J), implicating them as mediators of Fru actions. We therefore decided to investigate the possible involvement of Kr and hb in the fru-dependent sexual differentiation of mAL neurons, leading to the rather unexpected finding that only hb, and not Kr, plays a role in this process. None of the isolated modifier mutants affected the rough-eye phenotype induced by the overexpression of tramtrack (ttk), a gene encoding a member of the BTB-Zn finger protein family, to which Fru belongs (for hb, see Fig. 1D,E). Neither hb− heterozygous nor homozygous tissues exhibited roughness (Fig. 1H,I), supporting the hypothesis that the hb mutant alleles specifically modulated the fru+-induced rough-eye phenotype. Conversely, overexpression of hb+ was found to induce roughness (Fig. 1F), and the simultaneous overexpression of fru+ further disrupted the eye structure (Fig. 1G).
Figure 1.
Genetic interactions between fru and hb in the compound eye. A, The Canton-S wild-type eye. B, The eye with fru+(type B) overexpression under the control of GMR-Gal4. C, The fru+-dominant phenotype alleviated by heterozygous hb12. D, The eye with ttk+ overexpression. E, The ttk+-dominant phenotype in heterozygous hb12. F, The rough eye induced by hb+ overexpression via GMR-Gal4. G, The eye with simultaneous overexpression of fru+ and hb+. H, I, An eye mosaic for hbB observed with scanning electron microscopy (H) or with light microscopy to identify the heterozygous (red) and homozygous (white) tissues (I). The genotype of the test flies was y w ey-flp GMR-lacZ;;FRT82B hbB1/FRT82B w+. The scanning microscopic images are shown at lower (top) and higher (bottom) magnifications. Scale bars: top, 100 μm; bottom, 10 μm. J, The list of modifiers of fru isolated in our screen. The fly lines found to be positive are shown with the Bloomington stock numbers, the names of genes affected, the full description of genotypes and the observed effects on the eye expressing one of the three fru isoforms. The enhancing effects are indicated with “+” marks and the suppressing effects with “−” marks. The strength of the modifying effects is indicated by the number of symbols (lines marked with three symbols are the strongest modifiers). “/”, not tested.
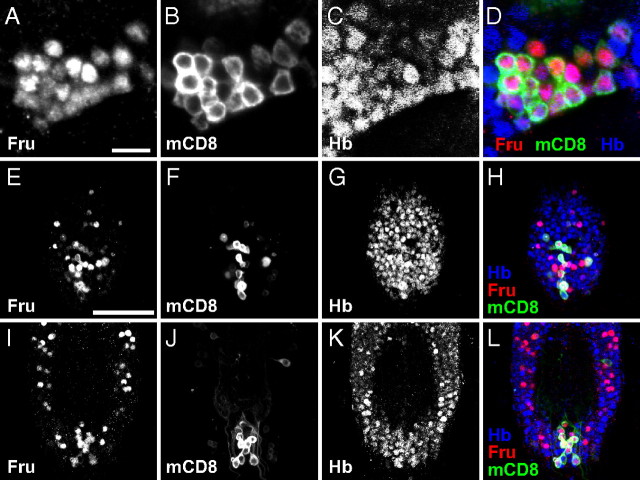
hb is a well known gap gene that participates in the anterior specification of early-stage embryos (Hülskamp et al., 1990; Hoch et al., 1991). Another function of hb is to specify the fate of early-born pioneer neurons in the embryonic nervous system (Novotny et al., 2002; Tran et al., 2010). On the other hand, fru functions primarily in postembryonic stages, particularly the pupal stage, to establish the sexual fate of neurons (Lee et al., 2000; Yamamoto, 2008). To evaluate the possibility that hb might collaborate with fru in the postembryonic stages, we performed double staining of the adult and pupal CNSs with the anti-Fru and anti-Hb antibodies. In keeping with the previous reports (Goodwin et al., 2000; Lee et al., 2000; Billeter and Goodwin, 2004), the anti-Fru antibody was found to label 1000 neurons in the CNS of male pupae and adults. We detected Hb in almost all cells expressing Fru in these stages and other cells without Fru expression (Fig. 2). This result prompted us to further examine possible interactions of Hb with Fru in the sexual differentiation of the postembryonic CNS.
Figure 2.
Coexpression of Hb and Fru in the postembryonic CNS. A–D, Images of mAL neurons in the pupal male brain stained with the anti-Fru antibody (A; red in D), the anti-mCD8 antibody for fruNP21 expression (B; green in D) and the anti-Hb antibody (C; blue in D); and the merged image (D). Scale bar, 10 μm. E–L, The ventral (E–H) and dorsal (I–L) parts of the ventral nerve cord at 24 APF stained with the anti-Fru antibody (E, I; red in H and L), and the anti-mCD8 antibody for fruNP21 expression (F, J; green in H and L) and the anti-Hb antibody (G, K; blue in H and L); the merged images are shown in H and L. Scale bar, 50 μm.
mAL neurons exhibit conspicuous sexual dimorphism that Fru proteins establish (Kimura et al., 2005), thus providing a suitable system for examining the possible role of Hb in sexual differentiation. To examine the effect of hb on the structure of mAL neurons, MARCM clones were generated to selectively label and manipulate them (Lee and Luo, 1999). When the pan-neural driver elav-Gal4 was used to drive UAS-hb RNAi, the Hb protein was markedly decreased (Fig. 3E). This result indicates that hb RNAi effectively knocked down hb expression in neurons.
In accord with previous reports (Kimura et al., 2005), the mAL cluster was composed of 30 cells in males and 5 cells in females in fruNP21 heterozygotes (Fig. 3A,B‴). The male mAL cluster contained neurons that had an ipsilateral neurite, in contrast to the female mAL cluster, which was composed exclusively of neurons without the ipsilateral neurite (Fig. 3A,B‴). The contralateral neurite in the subesophageal ganglion developed Y-shaped branches in females, while the male counterparts exhibited a simple horsetail-like appearance (Fig. 3A,B‴). When hb RNAi was expressed in mAL neuroblast clones under the control of fruNP21, the ensemble of mAL neurons showed intersexual characteristics; although the cluster retained male properties in that it was composed of 30 cells with the ipsilateral neurite (Fig. 3D), it also exhibited a female-like branching pattern of the contralateral neurite in the subesophageal ganglion (Fig. 3C–C‴). The contralateral neurite extending to the subesophageal ganglion is located more anteriorly in females than in males (Fig. 3A‴,B‴). Expression of hb RNAi in male mAL neurons induced an anterior displacement of the contralateral neurite, making it more female-like (Fig. 3C‴).
Pumilio (Pum) is a translational repressor of hb in embryogenesis. To examine whether or not a similar regulation of hb is involved in the sexual differentiation of mAL neurite formation, full-length Pum or the Pum RBD, which is known to recapitulate some of Pum actions (Wharton et al., 1998), was overexpressed in neuroblast clones (Fig. 3F–F‴). Both manipulations phenocopied the hb knockdown (Fig. 3F–F‴), suggesting that the Pum-mediated translational repression of hb contributes to the sexual differentiation of mAL neurite.
To determine whether single mAL neurons have the intersexual structure or whether the cluster is a mixture of male-typical and female-typical neurons, single-cell clones were produced by MARCM (Lee and Luo, 1999). Some of the singly stained mAL neurons in which hb was knocked down had side branches near the tip of the contralateral neurite, similar to the case in female mAL neurons (Fig. 4C,C′). There existed mAL neurons that were indistinguishable from the typical male neurons in other single-cell clones where hb was knocked down (Fig. 4A,B′). Approximately 30% of single-cell clones with hb RNAi expression in the male brain exhibited intersexual characteristics (Fig. 4D). In our previous study (Kimura et al., 2005), we reported that 8 types are distinguishable among the male-type mAL neurons although each neurons is not individually identifiable. The mAL neurons with different neurite patterns were demasculinized by hb RNAi expression, suggesting that several distinct mAL neurons are susceptible to this treatment. We conclude that hb knockdown results in the partial demasculinization of a population of mAL neurons that develop female-like extra branches near the tip of the contralateral neurite.
Figure 4.
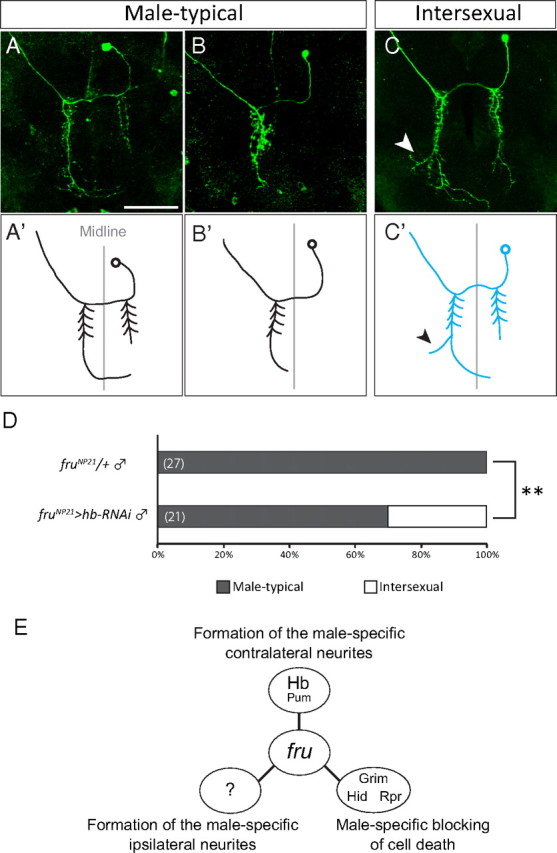
Demasculinization of the contralateral neurite structure of mAL detected in single-cell MARCM clones. A–C′, Three different examples of single-cell clones of mAL are shown. The original images (A–C) and schematic drawings (A′, B′, C′) of mAL neurons are shown. The female-type side branch is highlighted with an arrowhead. Scale bar, 50 μm. D, The proportion of flies that carried single-cell clones of male-type mAL (p < 0.01 by Fisher's exact test). The genotype of the test flies was: y hs-flp/Y;G13 UAS-mCD8-GFP/G13 tub-Gal80;fruNP21 UAS-dicer2/UAS-hb RNAi. E, Three distinct actions of Fru in producing sexual dimorphism in three different characteristics of mAL neurons.
Discussion
In this study, we demonstrated that Hb contributes to the formation of a male-typical neurite structure of fru-expressing interneurons in the postembryonic Drosophila brain. Thus far, the roles of Hb have been explored only during embryogenesis, and virtually nothing has been known about its function in postembryonic development. In early embryogenesis, Hb defines an anterior subdivision of the embryo by its transcriptional regulatory activity on the gap gene network (Schroeder et al., 2004). Hb is distributed along the anteroposterior axis with a concentration gradient (Hülskamp et al., 1990). This gradient of the Hb protein concentration is produced by the translational repression of hb mRNA by nanos (nos) and pum (Wang and Lehmann, 1991; Macdonald, 1992; Wang et al., 1994; Wharton et al., 1998).
Interestingly, it has been reported that the overexpression of pum or of nos-tub3′ UTR reduced the number of higher-order dendritic branches of class III and class IV dendritic arborization neurons, yet this action of nos and pum did not require hb (Ye et al., 2004). Our experiment (Fig. 3F–F‴) showed that overexpression of the full-length Pum or Pum RBD in MARCM clones phenocopies the effect of hb knockdown. This suggests that, unlike in the case of sensory neurons, the sex-specific shaping of dendritic branches in mAL neurons involves Hb, the translation of which may be regulated by the Nos-Pum complex. In the Drosophila neuromuscular junction, Pum shapes presynaptic terminals by presumably repressing translation of the transcription factor elF-4E (Menon et al., 2004). In this system, Pum RBD failed to replace full-length Pum, in contrast to the embryonic abdominal segmentation (Wharton et al., 1998) and mAL neurite morphogenesis (this study).
The synergy between fru and hb forcibly expressed via Gal4 was observed in the production of the rough-eye phenotype, and this finding favors the idea that their protein products collaborate in the transcriptional regulation of downstream genes, although our experiments do not provide any further insights about the mode of action of these proteins in such processes. No direct target for Fru has been identified (Goldman and Arbeitman, 2007). On the other hand, several genes have been demonstrated to be direct targets of Hb. For example, Hb directly regulates pdm in the embryonic CNS (Kambadur et al., 1998) and hb itself in the blastoderm (Treisman and Desplan, 1989). Other downstream genes include Krüppel (Kr), which is activated directly (Hoch et al., 1991), and zfh2, cut, runt, and cas, which are repressed, although it is not known whether Hb regulates these genes directly or indirectly (Kambadur et al., 1998; Isshiki et al., 2001; Grosskortenhaus et al., 2006). These potential targets of Hb are obvious candidates for mediating the role of Hb in the sexual differentiation of mAL neurons.
In a separate study, we found a different mode of sexual switching, in which Fru determines the sexual fate of mAL neurons in an all-or-none manner; when the chromatin regulator HDAC1 or HP1 is knocked down in the fru hypomorphic male brain, the proportion of female-typical mAL neurons increases or decreases without producing any neurons that bear intersexual characteristics (H. Ito, unpublished data). These chromatin regulators presumably mediate the Fru's action as the primary sex determinant by coordinating distinct downstream mechanisms (Fig. 4E). One of such downstream mechanisms is the male-specific blocking of cell death by Fru (Kimura et al., 2005). In the present study, the number of mAL neurons was never affected by knocking down hb, indicating that hb regulates fru-dependent sex differentiation in mAL neurons by a mechanism distinctly different from the sex-specific cell death. Also, hb knockdown had no effect on the male-specific ipsilateral neurite, implying yet another mechanism regulating this process. Thus Fru appears to orchestrate at least three different mechanisms for neural sexual differentiation (Fig. 4E).
Footnotes
This work was supported in part by Specially Promoted Research Grant 1802012 from the Ministry of Education, Culture, Sport, Science and Technology to D.Y., a grant from the Strategic Japanese-French Cooperative Program from the Japan Science and Technology Agency to D.Y., a grant from the Tohoku Neuroscience Global COE (Center of Excellence) program to D.Y. and a Grant-in-Aid for JSPS (Japan Society for the Promotion of Science) Fellows (22-6778) to J.G. We thank the Bloomington Stock Center, the National Institute of Genetics, Kyoto Genetic Resource Center, and the Vienna Drosophila RNAi Center for numerous stocks, the Developmental Studies Hybridoma Bank and Asian Distribution Center for segmentation antibodies for the antibodies, and K. Sawaguchi and H. Sato for secretarial assistance.
References
- Billeter JC, Goodwin SF. Characterization of Drosophila fruitless-gal4 transgenes reveals expression in male-specific fruitless neurons and innervation of male reproductive structures. J Comp Neurol. 2004;475:270–287. doi: 10.1002/cne.20177. [DOI] [PubMed] [Google Scholar]
- Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GS. Sexual dimorphism in the fly brain. Curr Biol. 2010;20:1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, Flores J, Balonze K, Dickson BJ, Axel R. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452:473–477. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Du C, McGuffin ME, Dauwalder B, Rabinow L, Mattox W. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol Cell. 1998;2:741–750. doi: 10.1016/s1097-2765(00)80289-0. [DOI] [PubMed] [Google Scholar]
- Gill KS. A mutation causing abnormal courtship and mating behavior in males of Drosophila melanogaster. Am Zool. 1963;3:507. [Google Scholar]
- Goldman TD, Arbeitman MN. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 2007;3:e216. doi: 10.1371/journal.pgen.0030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SF, Taylor BJ, Villella A, Foss M, Ryner LC, Baker BS, Hall JC. Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics. 2000;154:725–745. doi: 10.1093/genetics/154.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 2006;20:2618–2627. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JC. Courtship among male due to a male-sterile mutation in Drosophila melanogaster. Behav Genet. 1978;8:125–141. doi: 10.1007/BF01066870. [DOI] [PubMed] [Google Scholar]
- Hoch M, Seifert E, Jäckle H. Gene expression mediated by cis-acting sequences of the Krüppel gene in response to the Drosophila morphogens bicoid and hunchback. EMBO J. 1991;10:2267–2278. doi: 10.1002/j.1460-2075.1991.tb07763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Pfeifle C, Tautz D. A morphogenetic gradient of Hunchback protein organizes the expression of the gap genes Krüppel and knirps in the early Drosophila embryo. Nature. 1990;346:577–580. doi: 10.1038/346577a0. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcriptional factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci U S A. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by Castor and Hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Ote M, Tazawa T, Yamamoto D. fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Koganezawa M, Haba D, Matsuo T, Yamamoto D. The shaping of male courtship posture by lateralized gustatory inputs to male-specific interneurons. Curr Biol. 2010;20:1–8. doi: 10.1016/j.cub.2009.11.038. [DOI] [PubMed] [Google Scholar]
- Kondoh Y, Kaneshiro KY, Kimura K, Yamamoto D. Evolution of sexual dimorphism in the olfactory brain of Hawaiian Drosophila. Proc Biol Sci. 2003;270:1005–1013. doi: 10.1098/rspb.2003.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Foss M, Goodwin SF, Carlo T, Taylor BJ, Hall JC. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol. 2000;43:404–426. doi: 10.1002/1097-4695(20000615)43:4<404::aid-neu8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Macdonald PM. The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development. 1992;114:221–232. doi: 10.1242/dev.114.1.221. [DOI] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Sanders LE, Arbeitman MN. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev Biol. 2008;320:378–390. doi: 10.1016/j.ydbio.2008.05.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ. Recombineering hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identify. Development. 2010;137:1421–1430. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J, Desplan C. The products of the Drosophila gap genes hunchback and Krüppel bind to the hunchback promoters. Nature. 1989;341:335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Usui-Aoki K, Ito H, Ui-Tei K, Takahashi K, Lukacsovich T, Awano W, Nakata H, Piao ZF, Nilsson EE, Tomida J, Yamamoto D. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat Cell Biol. 2000;2:500–506. doi: 10.1038/35019537. [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Dev Dyn. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. The pumilio RNA-binding domain is also a translational regulator. Mol Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- Yamamoto D. Brain sex differences and function of the fruitless gene in Drosophila. J Neurogenet. 2008;22:309–332. doi: 10.1080/01677060802298491. [DOI] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. nanos and pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Yu JY, Kanai MI, Demir E, Jefferis GS, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]