Abstract
CO2 sensation represents an interesting example of nervous system and behavioral evolutionary divergence. The underlying molecular mechanisms, however, are not understood. Loss of microRNA-279 in Drosophila melanogaster leads to the formation of a CO2 sensory system partly similar to the one of mosquitoes. Here, we show that a novel allele of the pleiotropic transcription factor Prospero resembles the miR-279 phenotype. We use a combination of genetics and in vitro and in vivo analysis to demonstrate that Pros participates in the regulation of miR-279 expression, and that reexpression of miR-279 rescues the pros CO2 neuron phenotype. We identify common target molecules of miR-279 and Pros in bioinformatics analysis, and show that overexpression of the transcription factors Nerfin-1 and Escargot (Esg) is sufficient to induce formation of CO2 neurons on maxillary palps. Our results suggest that Prospero restricts CO2 neuron formation indirectly via miR-279 and directly by repressing the shared target molecules, Nerfin-1 and Esg, during olfactory system development. Given the important role of Pros in differentiation of the nervous system, we anticipate that miR-mediated signal tuning represents a powerful method for olfactory sensory system diversification during evolution.
Introduction
Sensory systems of animals match distinct ecological niches. Insects use olfactory cues to navigate to food sources and oviposition sites, and to avoid danger. Hence, olfactory organs are highly specialized, and specific environmental cues elicit highly species-specific behaviors. One example is CO2. While attractive to mosquitoes, fruit flies show a robust avoidance behavior to even low levels of CO2 (Gibson, 1996; Suh et al., 2004).
In both fruit flies and mosquitoes, specialized sensory structures on the third segment of the antenna or on the maxillary palp (MP) contain olfactory receptor neurons (ORN) that express receptors tuned to specific odors (Vosshall and Stocker, 2007). In Drosophila, two receptors expressed in the same ORN on the antenna, Gr21a and Gr63a, are necessary and sufficient to detect CO2 and to elicit CO2-mediated escape responses (Jones et al., 2007; Kwon et al., 2007). In contrast to Drosophila, mosquitoes carry CO2 receptor neurons on their MP (Grant et al., 1995; Jones et al., 2007; Lu et al., 2007). In addition to this peripheral difference, CO2 ORN axons innervate different glomeruli in the antennal lobe (AL) in the brain. Drosophila Gr21a/Gr63a-expressing neurons on the antenna innervate a ventral glomerulus, whereas mosquito CO2 ORNs target medial glomeruli, which in both of these species respond to food odors (Suh et al., 2004; Vosshall and Stocker, 2007). Possibly, these peripheral and central changes contributed to the behavioral divergence of mosquito and fruit fly (Cayirlioglu et al., 2008; Jones, 2008; Ramdya and Benton, 2010).
We have recently reported that loss of microRNA-279 leads to formation of ectopic CO2 neurons on MPs of fruit flies, suggesting a potential involvement of miR-279 in CO2 sensory system divergence (Cayirlioglu et al., 2008).
Here, we addressed the mechanistic role of miR-279 in the development of MP CO2 neurons. We show that mutations of the atypical homeobox transcription factor Prospero result in a miR-279 phenotype. Consistent with Prospero's previously reported role in sensory bristles and antenna (Manning and Doe, 1999; Reddy and Rodrigues, 1999; Sen et al., 2003), we find that Pros distinguishes the neuronal pIIb lineage also in MP olfactory sensilla. A new hypomorphic allele of pros revealed that in addition, Pros restricts the number of neurons in specific olfactory sensilla. In mutant MPs, CO2 ORNs are formed as an extra neuron within certain MP sensilla. We apply genetic analysis and in vitro assays to show that Pros is involved in the transcriptional regulation of miR-279, and that reexpression of miR-279 in pros mutants rescues the pros CO2 neuron phenotype. Using RNAi, loss of function analysis, and overexpression, we demonstrate that upregulation of two Pros and miR-279 target genes, nerfin-1 and escargot, is necessary and sufficient for the formation and mistargeting of MP CO2 neurons. Together, our data suggest that Pros regulates miR-279 expression to enhance its own repressor activity, and thereby suppresses a default CO2 neuron program in specific MP olfactory sensilla.
Materials and Methods
Fly genetics.
Fly stocks were maintained in standard medium at 25°C. The genetic screen was performed with EMS (ethyl methanesulfonate) and eyFLP; FRT/FLP mosaic analysis/mosaic analysis with repressible cell marker (MARCM) analysis, as described previously (Cayirlioglu et al., 2008). The screen resulted in four miR-279-like mutants of ∼6000 mutants on the third chromosome that were analyzed. prosIG2227 contains a point mutation at Leu850 to Arg. MARCM analysis was performed on flies of either sex of the following genotype: hsFLP or eyFLP; Or-gal4 UAS-sytGFP/+ (or UAS-mCD8GFP/+); FRT82 mutation/FRT82 Gal80 (E2F). ORN labeling was achieved by fusing the promoter-elements to GAL4 or directly to synaptotagmin-GFP. Mutations included FRT82B miR-279962–7 (Cayirlioglu et al., 2008), FRT82B pros17 (Manning and Doe, 1999), FRT82B prosIG2227 (this study), and FRT82B prosvoila78 (Grosjean et al., 2001). All analyses were done in mosaic animals. Gr21a transcriptional reporter was used for CO2 neurons [i.e., Gr21a-GAL4 driving membrane-bound GFP (UAS-mCD8GFP] to detect cell bodies in the antenna and MP. Elav-GAL4 was used to label neurons. miR-279-GAL4 contains the 2 kb DNA stretch upstream of miR-279 gene. As previously described, this driver rescues the mutant phenotype when used to drive the expression of miR-279 in miR-279 mutants (Cayirlioglu et al., 2008). miR-279-GAL4 was used to label cells and progeny of cells expressing miR-279 in mosaic analysis. Rescue and genetic interaction experiments were performed by using cDNAs fused to UAS transcriptional response elements. UAS-construct expression was under the control of the β-actin promoter, but the expression was restricted to only the mutant tissue in the MP and antenna upon eyFLP expression and mitotic recombination. Transgenic flies carrying RNAi constructs were ordered from the VDRC or Kyoto stock centers. RNAi was expressed in exactly the same manner as the UAS constructs in mutant tissue of antenna and MP only. UAS-Pros constructs were generated and generously provided by F. Matsuzaki (RIKEN Center for Developmental Biology, Kobe, Japan), C. Doe (Institute of Neuroscience, HHMI, University of Oregon, Eugene, OR), and A. Brand (The Gurdon Institute and Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, UK). UAS-dsred-miR-279 was a gift by E. Lai (Sloan-Kettering Institute, Department of Developmental Biology, New York, NY). UAS-nerfin transgenic flies and nerfin-1 loss-of-function alleles were generously provided by Ward Odenwald (Neural Cell-Fate Determinants Section, NINDS, NIH, Bethesda, MD). Escargot loss-of-function and P[lacZ]esg flies were ordered from the Bloomington Stock Center.
Genotypes for analysis.
Control:
eyflp;Gr21a-Gal4,UAS-mCD8GFP/+;FRT82/FRT82Gal80E2F
eyflp;Gr21-sytGFP/+;FRT82/FRT82Gal80E2F
eyflp;miR-279-Gal4,UAS-mCD8GFP/+; FRT82/FRT82Gal80E2F
hsflp;Elav-Gal4,UAS-mCD8GFP/+; FRT82/FRT82Gal80
hsflp;miR-279-Gal4,UAS-mCD8GFP/+; FRT82/FRT82Gal80E2F
eyflp;Gr21a-sytGFP actGal4;FRT82/FRT82ClGal80
eyflp;Gr21a-sytGFP actGal4/RNAipros;FRT82/FRT82ClGal80
eyflp;Gr21a-Gal4 UASmCD8GFP/RNAinerfin-1;FRT82/FRT82ClGal80
eyflp;Gr21a-sytGFP actGal4/RNAispineless;FRT82/FRT82ClGal80 (a.o. RNAi constructs against gcm, senseless, Ptx1)
eyflp;Gr21a-Gal4 UASmCD8GFP/escargotk00606;FRT82 /FRT82ClGal80
eyflp;Gr21a-Gal4 UASmCD8GFP/+;FRT82 hb9kk30/FRT82ClGal80
eyflp;Gr21a-Gal4 UASmCD8GFP/+;FRT82 /FRT82ClGal80
eyflp;OrX-Gal4 UASsytGFP/+;FRT82/FRT82ClGal80
eyflp;OrX-Gal4 UASmCD8GFP/+;FRT82/FRT82ClGal80
eyflp;FRT40A/FRT40A GAL80 or CL;Gr21a-GAL4,UASmCD2
Mutants:
miR-279, prosIG2227, prosVoila78, pros17
eyflp;Gr21a-Gal4,UAS-mCD8GFP/+;FRT82 mutant/FRT82Gal80E2F
eyflp;Gr21-sytGFP/+;FRT82 mutant/FRT82Gal80E2F
eyflp;miR-279-Gal4,UAS-mCD8GFP/+;FRT82 mutant/FRT82Gal80E2F
hsflp;miR-279-Gal4,UAS-mCD8GFP/+;FRT82 mutant/FRT82Gal80E2F
hsflp, Elav-Gal4UASmCD8GFP;FRT82 mutant/FRT82Gal80
eyflp;Gr21a-sytGFP actGal4;FRT82 mutant/FRT82ClGal80
eyflp;Gr21a-sytGFP actGal4/UASmiR-279 or UASpros or UASnerfin-1; FRT82 mutant/FRT82ClGal80
eyflp;Gr21a-sytGFP actGal4/RNAinerfin-1;FRT82 mutant/FRT82ClGal80
eyflp;Gr21a-Gal4 UASmCD8GFP/RNAipros;FRT82 mutant/FRT82ClGal80
eyflp;Gr21a-sytGFP actGal4/RNAispineless;FRT82 mutant/FRT82ClGal80 (a.o. RNAi constructs against gcm, senseless, Ptx1)
eyflp;Gr21a-Gal4 UASmCD8GFP/+;FRT82 mutant hb9kk30/FRT82ClGal80
eyflp;Gr21a-Gal4 UASmCD8GFP/escargotk00606;FRT82 mutant/FRT82ClGal80
eyflp;Gr21a-Gal4 UASmCD8GFP/+;FRT82 mutant nerfin159/FRT82ClGal80
eyflp;OrX-Gal4 UASsytGFP/+;FRT82 mutant/FRT82ClGal80
eyflp;OrX-Gal4 UASmCD8GFP/+;FRT82 mutant/FRT82ClGal80
eyflp;FRT40A escargotk00606/FRT40A GAL80 or CL;Gr21a-GAL4,UASmCD2
eyflp;Gr21a-sytGFP actGal4/UASescargot or UASnerfin-1 or UASescargot, UASnerfin-1;FRT82/FRT82ClGal80
eyflp;Gr21a-sytGFP actGal4/UASp35 or UAScyclinE;FRT82/FRT82ClGal80
Tissue dissection and immunohistochemistry.
For the analysis of ORN axon targeting in the adult brain, eyFLP mosaic flies were dissected and adult brains were fixed and immunostained. For analysis of cell body number and position, hsFLP and eyFLP were used to create mosaic mutant tissue. Quantifications of ORN number were performed on adult MPs and antennae. Analysis in the developing olfactory neurons was performed using eyFLP. White pupae were selected and incubated at 25°C until the desired stage was reached. MPs and antenna regions were dissected in ice-cold PBS and collected in 4% PFA on ice. Upon fixation for 1 h at room temperature (RT) in 4% PFA, tissues were washed twice for 15 min in PBS-0.5% Triton, following by 1 h incubation in blocking solution (20% donkey serum, 0.5% Triton in PBS). Primary antibody was incubated overnight at 4°C in blocking solution without Triton. After washing twice for 15 min in PBS-0.5% Triton, tissues were incubated with secondary antibody for 1 h at RT in blocking solution without Triton. Tissues were washed twice more for 2× 15 min in PBS-0.5% Triton and mounted with Vectashield mounting medium.
All tissues were analyzed using confocal microscopy with an Olympus FV1000 or a Leica SP2 confocal. Pictures were processed in Adobe Photoshop, Adobe Illustrator, ImageJ, and Microsoft PowerPoint.
Primary and secondary antibodies.
Staining was performed as previously described (Cayirlioglu et al., 2008). The following primary antibodies were used: chick anti-βgal (1:2000; Abcam); mouse anti-NC82 (1:20), mouse anti-disclarge (1:50), mouse anti-pros (1:20), and mouse and rat anti-elav (1:50) (all from Developmental Studies Hybridoma Bank); and rabbit (1:2000) and mouse (1:500) anti-GFP (both from Clontech). Secondary antibodies (anti-mouse-CY5, anti-rat-CY3, and anti-rabbit-488) were ordered from Dianova and used at 1:200.
Electrophysiology.
Extracellular single sensillum recordings from MP were performed according to the procedure described previously (de Bruyne et al., 1999). Briefly, a fly was trapped in a truncated pipette tip with its proboscis protruding out and mounted on a glass slide. The protruding proboscis and MP were secured and stabilized on a coverslip with the help of a tapered glass micropipette. The preparation was visualized with a Leica DM6000 FS microscope at 750× magnification and MP basiconic sensilla were identified by the expression of Gr21a/OrX-GFP in mutant and control flies. A glass reference electrode filled with 0.01 m KCl was inserted into the eye and a recording glass electrode filled with the same solution was used to record from the MP basiconic sensilla. Action potentials were recorded using a CV-7B headstage and MultiClamp 700B amplifier (Molecular Devices). The signals were sampled at 10 kHz, digitized, and fed into a computer by Digidata 1440A. The spikes were visualized and recorded in Clampex 10.2 acquisition software and sorting of spikes were done manually with Clampfit 10.2 software off-line. A continuous and humidified airstream (2000 ml/min) was delivered to the fly throughout the experiment via an 8-mm-diameter glass tube positioned 10 mm away from the preparation. A custom-made odor delivery system was used for stimulation in all experiments (Smartec). For CO2 stimulation, 500 ms pulses of CO2 were delivered into the continuous airstream with the help of mass flow controllers and solenoid valves. For odor stimulation, specific odors were diluted 1:10 in paraffin oil and 300 ml/min odor pulse were delivered into the continuous airstream using headspace method. During the odor stimulation, the continuous airstream flow was maintained always at 2000 ml/min with the help of mass flow controllers and solenoid valves. The spikes in the recorded traces were sorted according to spike amplitude. The spike quantification was done by counting the number of spikes over 500 ms duration immediately after the onset of the response and subtracting the total from the number of spikes counted over a 500 ms window before the stimulation. The obtained number of spikes were doubled and presented as spikes per second.
Bioinformatic binding site predictions.
The presence of the Prospero binding motifs TWAGVYD (Cook et al., 2003) or CWYNNCY (Choksi et al., 2006) in the 2 kb upstream region of the miR-279 gene was tested using the RSA tools software (http://rsat.ulb.ac.be/genome-scale-dna-pattern_form.cgi). The predicted putative binding sites were further evaluated by testing the conservation in six different Drosophila species (D. melanogaster, D. yakuba, D. simulans, D. erecta, D. ananassae, D. pseudoobscura) using the VISTA genome browser (http://pipeline.lbl.gov/cgi-bin/gateway2).
Promoter S2 cell assay.
The genomic 2 kb upstream region of miR-279 was amplified and cloned into the pGL3 vector. This putative enhancer region was further mutagenized at the predicted conserved Prospero binding sites using the Stratagene QuikChange Lightning Multi Site-Directed Mutagenesis Kit. The following primers were used to perform the nucleotide exchange: mutP1 forward: acagttcaaatgtgccgtctaatttctaatgatttaatttc; mutP2 forward: gcgcgtgtgtaagacgttgattgttagtgtacgg; mutP3 forward: cctggtacaatgaagattcgcatttagaataaggca; and mutP4 forward: gggaggaaagcattcacagacaacaacccttctggg.
S2 cells were transfected with a reporter construct carrying either the wild-type enhancer fragment or a construct with mutations at four putative and conserved Prospero binding sites. For normalization, the pTK Renilla vector that expresses the Renilla luciferase was cotransfected. Both vectors were used in a concentration of 100 ng. The cells were seeded in a density of 106 cells and transfected the next day with the constructs described. Approximately 16 h after transfection, the cells were lysed and luciferase expression assayed using the dual luciferase kit (Promega) according to the manufacturer's instructions. For overexpression of Pros, a full-length EST (LD37627; Berkeley Drosophila Genome Project) was subcloned into the pattBUAS vector (donated by the Basler Lab, Institute of Molecular Life Science, University of Zurich, Zurich, Switzerland). The RNAi pros construct was a gift from the VDRC library (Construct ID: 109284). To drive the expression of both constructs, ubiquitinGal4 was cotransfected. All experiments were performed in triplicates.
Electromobility shift assay.
For the electromobility shift assay, the homeodomain of Pros was purified from BL21 cells transfected with the construct pGex-prosL that was kindly provided by Tiffany Cook (Department of Pediatric Ophthalmology, Division of Developmental Biology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH). Oligos containing the predicted Prospero binding sites, as follows, were annealed and radiolabeled with [γ 32P] ATP using T4 polynucleotide kinase (Fermentas): P1 forward: gatgcaagcagcatttacagttcaaatgtgccgtctaattagaaatgatttaatttcaat; mutP1 forward: gatgcaagcagcatttacagttcaaatgtgccgtctaatttctaatgatttaatttcaat; P4 forward: gagggtagcgcaaggaaaggggaggaaagctaagacagacaacaacccttctggg; and mutP4 forward: gagggtagcgcaaggaaagggggaggaaagca ttcacagacaacaacccttctggg.
The labeled oligos were mixed with the purified proteins in binding buffer (20 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm DTT, 5 mm MgCl2) and incubated for 20 min at RT. Subsequently, the mixture was loaded on a 0.5% Tris-borate/EDTA prerun minigel. The gel was dried and exposed to a Phospho screen overnight.
Chromatin immunoprecipitation assay.
Embryos (UAS-Flag-Pros/X; actin-Gal4/+) were collected and fixed as previously described (Sandmann et al., 2006). The embryos were fixed in a cross-linking solution (50 mm HEPES, 1 mm EDTA, 0.5 mm EGTA, 100 mm NaCl, pH 8.0) for 15 min under vigorous shaking. The cross-linking reaction was stopped by adding PBS with 125 mm glycine and 0.1% Triton-X. Embryos were washed twice with PBT (0.1% Triton-X), dried, and frozen in −80°C. For cell lysis, embryos were homogenized in PBS with 0.1% Triton-X and protease inhibitor, centrifuged, and the pellet dissolved in cell lysis buffer (5 mm HEPES, pH 8.0, 85 mm KCl, 0.5% NP-40 and protease inhibitor). After the centrifugation, the pellet was dissolved in nuclear lysis buffer (50 mm HEPES, pH 8.0, 10 mm EDTA, 0.5% N-lauroylsarcosine and protease inhibitor) and incubated at room temperature for 20 min. Lysates were sonicated (Sonicator sonoplus; Bandelin) until an approximate chromatin fragment length of 1000–500 bp was achieved. The sheared chromatin fragments were incubated overnight with magnetic beads and anti-FLAG M2 (Sigma) to detect Flag-Pros or mouse IgG as unspecific binding control. Beads were washed and the bound chromatin was eluted according to the manufacturer's instructions (Magna ChIP G Kit; Millipore). The chromatin was checked for the presence of miR-279 promoter fragments by PCR using the following primer sets: chromatin immunoprecipitation assay (ChIP) P4 forward: gtatataatggacaagaagaagaataagcag; ChIP P4 reverse: catgcggaatttcagttgtttccttttatatc. An input control was included into the PCR.
3′UTR S2 cell assay.
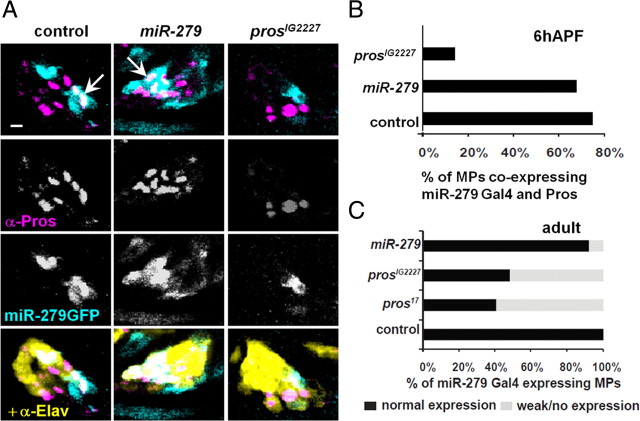
To generate luciferase targets, we amplified a 1.8 kb nerfin fragment (including the entire 3′UTR and 220 bp of downstream sequence), and a 644 bp escargot 3′UTR fragment and cloned these downstream of the renilla luciferase coding region in psiCHECK2; this vector contains an internal firefly luciferase gene that serves as an internal control. For the miR-279 expression construct, we cloned 415 bp of genomic sequence, centered on the miR-279 hairpin, into the 3′UTR of UAS-DsRed vector. We then transfected 100 ng of target, 50 ng of ub-Gal4, and 100 ng of UAS-DsRed-miR-279 plasmids into 1 × 106 S2 cells in 24-well format. Three days later, we lysed the cells and subjected them to dual luciferase assay (Promega) and analyzed these on a plate luminometer (Tecan). Triplicate transfections were performed. Data of four repetitions were pooled (Fig. 9).
Figure 9.
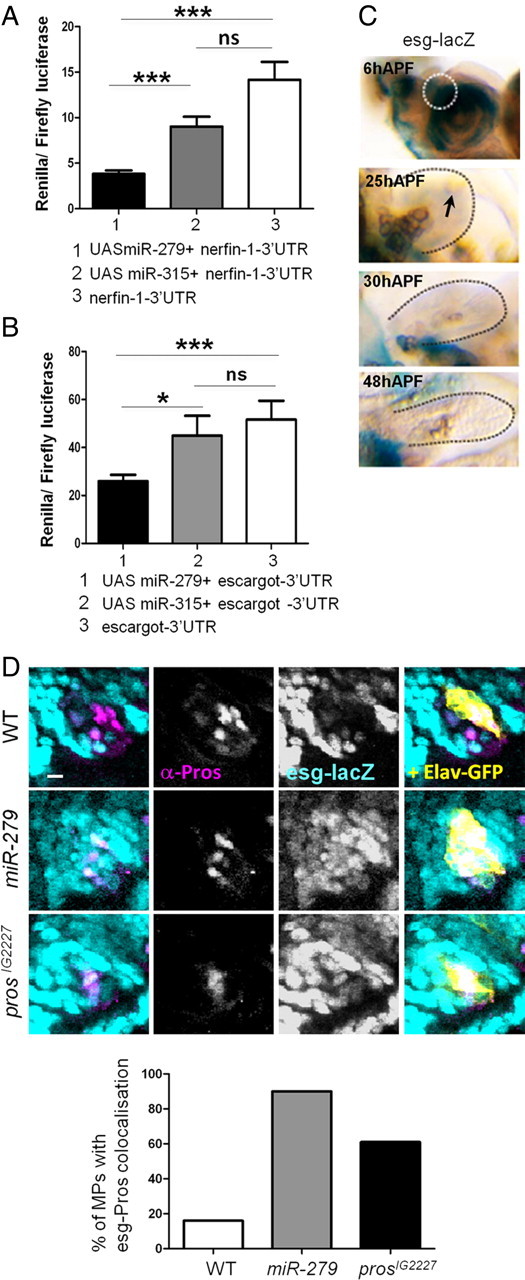
Escargot and Nerfin-1 are shared target of miR-279 and Prospero. A, Quantification of S2 cell assays using nerfin-3′UTR reporter with and without overexpression of miR-279 or miR-315. Experiments were performed in triplicates and data from four repetitions are pooled. B, Quantification of S2 cell assays with esg 3′UTR and overexpression of miR-279 or miR-315. Overexpression of miR-279 reduced esg-3′UTR reporter expression to 51 ± 9%. C, MPs of different developmental stages: LacZ (blue) staining and anti-βGal (turquoise) immunoreactivity reflects expression of esg (P[lacZ]esg). At 6 h APF, esg-lacZ is seen strongly in cells of the antennal disc, including the future MP (circled area). At 25 h APF, esg-lacZ is expressed at much lower levels in the MP. No expression was detected in MPs at 30 or 48 h APF (arrow). D, MPs at 6 h APF from control and mutant flies carrying an esg-lacZ. Esg expression was visualized using anti-βGal (esg-lacZ), expression of Elav was monitored with anti-GFP, and Pros was labeled with anti-Pros. Coexpression of Pros with esg was analyzed in the area of control or mutant clone (Elav-GFP, yellow). In the control case, only 16% of MPs have double-labeled cells (n = 12). In the absence of miR-279 or Pros, repression of esg expression in the developing MP is released. In miR-279 and prosIG2227 mutants, 90% (n = 11) and 61% (n = 13) of the MPs, respectively, counted show cells with Pros-esg coexpression. Scale bars, 5 μm.
Gene ontology term analysis and miR-279 target prediction software.
Prospero in vivo target genes were previously published (Choksi et al., 2006). microRNA-279 target gene predictions were generated with the following online software: TargetScan (http://www.targetscan.org/fly_12/), PicTar (http://pictar.mdc-berlin.de/cgi-bin/new_PicTar_fly.cgi?species=fly), and miRBase (http://www.mirbase.org/). Only targets that appeared in at least two of the predictions were used for the comparison to the list of Prospero target genes. Gene ontology (GO) analysis was performed using GOstat (Beissbarth and Speed, 2004). All predictions were Benjamini-corrected and GO terms with a p > 0.01 were disregarded. Targets for aga-miR-279 were predicted using miRBase. Results can be found at http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/hit_list.pl?genome_id=377&mirna_id=aga-miR-279&external_name=&gene_id=&go_class=function&go_term=&logic=phrase&terms=. aga-nerfin-1 is AGAP002601 and aga-escargot is AGAP008274.
Results
Pros mutants fail to suppress the formation of maxillary palp CO2 neurons
miR-279 activity suppresses the formation of CO2 sensory neurons in the MPs of D. melanogaster (Cayirlioglu et al., 2008). To gain insight into the relationship of miR-279 and regulators of neuron development, we used genetic mosaic screening to identify mutations with a miR-279-like phenotype. We identified a new allele of Pros, prosIG2227, which strongly resembled the phenotype of miR-279 mutants and displayed a similar number of ectopic CO2 neurons on the MP (15 ± 0.9 for prosIG2227 and 16 ± 0.6 for miR-279; Fig. 1A1, B1, C1,F). The number of antennal CO2 neurons in prosIG2227 and miR-279 mutants was not affected and comparable to controls (18 ± 3.2 and 18 ± 3.5 vs 19.4 ± 4, respectively; Fig. 1A2, B2, C2,F). Antennal CO2 ORNs target the V-glomerulus in the AL of the adult fly brain (Fig. 1A3,A4). In prosIG2227 and miR-279 mutants, axons of ectopic MP Gr21a-expressing neurons showed dual targeting specificity. In addition to targeting the V-glomerulus, these neurons innervated a set of medial glomeruli usually innervated by two MP ORN classes, Or42a and Or59c (Fig. 1B3, B4, C3, C4).
Figure 1.

Pros mutants exhibit MP CO2 neurons targeting medial AL glomeruli. A–E, Adult olfactory organs (antenna and MP) and brains stained with anti-GFP (green) or anti-GFP and anti-disc large (magenta) to visualize CO2 neurons or synaptic projections of CO2 neurons (Gr21aGAL4, UAS-sytGFP) in the AL, respectively. Note that only mutant cells are marked by GFP. A1, B1, C1, D1, E1, CO2 neurons in MP labeled with Gr21a-GAL,UAS-mCD8GFP in control and mutant palps. A1, No CO2 neurons were detected in MPs of control flies. B1, C1, D1, E1, Ectopic CO2 neurons were detected in MPs of eyFLP;prosIG2227, eyFLP;prosVoila78, eyFLP;miR-279, and eyFLP;pros17. A2, B2, C2, D2, E2, CO2 neurons in the antenna were labeled with Gr21a-mCD8GFP. While CO2 neuron number appeared normal in prosIG2227, prosVoila78, and miR-279 mutants compared with controls, the number was reduced in eyFLP;pros17 mutants. A3, AL of eyFLP;FRT82B control fly, where CO2 neurons target a single glomerulus, V-glomerulus. B3, C3, D3, E3, CO2 neurons labeled with Gr21a-mCD8GFP innervate medial glomeruli (arrowhead) in addition to the V-glomerulus in brains of eyFLP;prosIG2227, eyFLP;prosVoila78, eyFLP;miR-279, and eyFLP;pros17 flies. Note that CO2 neuron axons project via the labial (arrow) and via the antennal nerve to the V-glomerulus and to medial glomeruli. Axons form contralateral projections. A4, B4, C4, D4, E4, Similar scenario as in A3, B3, C3, D3, and E3; however, here CO2 neurons are labeled with Gr21a-sytGFP to label synaptic connections in the AL. A5, B5, C5, D5, E5, ORNs expressing the Or42a receptor residing on MPs innervate a medial glomerulus (arrowhead) in the AL of eyflp;FRT82B controls (A5). B5, C5, D5, E5, Or42a neurons in eyFLP;prosIG2227, eyFLP;prosVoila78, eyFLP;miR-279, and eyFLP;pros17 target the V-glomerulus (arrow) in addition to the medial glomerulus. A6, B6, C6, D6, E6, Or59c neurons reside on the MP. Note that these neurons show the same behavior as Or42a neurons in control and mutants. F, Quantification of CO2 neurons of antenna and MP for all flies analyzed. Control: n = 15 and 11, pros17: n = 17 and 45 (8 with ectopic CO2 ORNs), prosIG2227: n = 51 and 7, miR-279: n = 37 and 6, ProsVoila78: n = 9 and 10, for antenna and MP, respectively. Control versus prosVoila78 antenna, p = 0.07; control versus pros17 antenna, p = 0.005; prosVoila78 versus prosIG2227 antenna, p = n.s.; miR-279 versus prosIG2227 MP, p = n.s.; pros17 versus prosIG2227 MP, p < 0.001. G, Schematic representation of CO2 neuron projections in wild-type and mutant flies. Ectopic CO2/MP hybrid neurons are detected in the MPs of miR-279 and pros mutants. While the mutant CO2 neurons in the antenna show wild-type innervations of the AL (blue), mutant axons from the MP innervate the V-glomerulus and additional medial glomeruli and express the CO2 receptors and MP ORs (yellow-blue). MP neurons innervate via the labial nerve in both mutants and wild-type flies (yellow). Ant, antenna; pos., positive. Scale bars, 50 μm.
Next, we asked how prosIG2227 relates to previously published alleles of pros. It was previously reported that full loss of pros resulted in a partial conversion of the neural to the non-neural cell lineage and, consequently, in a loss of neurons (Manning and Doe, 1999; Reddy and Rodrigues, 1999; Sen et al., 2003). Consistent with these results, pros17 mutants exhibited ORN number reduction as revealed by the general OR marker Or83b-GFP (Or83b-Gal4,UAS-mCD8GFP) and sparser innervations of a subset of AL glomeruli (Fig. 2A,B). pros17 flies also displayed a loss of antennal CO2 neurons to 51.5% of controls (10 ± 1.3 and 19.4 ± 4; Fig. 1D2,F). Nevertheless, pros17 mutant MPs displayed ectopic CO2 neurons in 17% (4.6 ± 1.4) of flies analyzed (Fig. 1D1,F). These 17% displayed medial mistargeting as miR-279 and prosIG2227 mutants (Fig. 1D3–D4). Another previously published hypomorphic P-element excision, prosVoila78 (Grosjean et al., 2001), showed a phenotype reminiscent of the prosIG2227 phenotype (Fig. 1E1–E4,F).
Figure 2.

Analysis of pros and miR-279 mutants reveals specific and general effects on ORN number and targeting. A, MP ORNs were quantified using the general OR marker Or83b-GFP (Or83b-GAL4, UAS-mCD8GFP). The number of ORNs in prosIG2227 clones was increased compared with control clones. This increase correlated with the number of ectopic CO2 neurons in the mutants. The number of ORNs in pros17 clones was decreased by ∼10 ORNs on average compared with control clones. B, eyFLP mosaic analysis was used to analyze ORN projection patterns in the AL of several classes of ORNs. miR-279 and pros mutants show very mild mistargeting in few classes of non-CO2 receptor ORNs. In general, mistargeting was mild and random. Pros mutants display a reduction of Or22a and Or47a neurons, as shown by the reduced innervations of the AL glomerulus. C, Ectopic CO2 neurons on the MP coexpress Or42a and Or59c. Examples of whole-mount in situ hybridization on control and mutant MP clones expressing the Gr21a-mCD8GFP reporter are shown. Scale bars: B, 50 μm; C, 10 μm.
To address whether the phenotype of the hypomorphic alleles of pros is restricted to the formation of ectopic CO2 ORNs as it is in miR-279 mutants, we next analyzed the projection patterns of several antennal and MP ORNs. prosIG2227 and prosVoila78 again displayed a similar selectivity as miR-279 mutants (Figs. 1, 2, Table 1, and data not shown). As previously reported for miR-279 mutants, two types of MP ORNs, Or59c and Or42a, target the V-glomerulus, in addition to innervating medial glomeruli in all mutants (Fig. 1A5, A6, B5, B6, C5, C6, D5, D6, E5, E6). This mistargeting corresponds to the coexpression of either Or59c or Or42a in ectopic MP CO2 neurons, as shown by in situ hybridization against Or42a or Or59c mRNA in mutant and wild-type MPs (Fig. 2C).
Table 1.
Quantification of ORN mistargeting phenotypes
| ORN | Wild-type | prosIG2227 | pros17 | prosVoila78 | miR-279 |
|---|---|---|---|---|---|
| 59c | 0/11 (0%) | 12/17 (71%) | 4/35 (11%) | 30/30 (100%) | 10/12 (83%) |
| 42a | 0/9 (0%) | 4/5 (80%) | 7/19 (37%) | 8/11 (73%) | 18/24 (75%) |
| 46a | 0/19 (0%) | 0/4 (0%) | 0/20 (0%) | 2/25 (8%) | 0/25 (0%) |
| 33c | 0/19 (0%) | 0/20 (0%) | 0/14 (0%) | 4/4 (100%) | 0/6 (0%) |
| 47a | 0/12 (0%) | 0/12 (0%) | 0/5 (0%) | 0/10 (0%) | 0/9 (0%) |
| 92a | 0/17 (0%) | 0/12 (0%) | 0/14 (0%) | 0/14 (0%) | 0/33 (0%) |
| 22a | 0/24 (0%) | 0/21 (0%) | 0/20 (0%) | 0/30 (0%) | 0/23 (0%) |
| 67d | 0/11 (0%) | 13/26 (50%) | 0/12 (0%) | 0/22 (0%) | 0/40 (0%) |
| 88a | 0/24 (0%) | 0/19 (0%) | 0/20 (0%) | 18/44 (41%) | 0/11 (0%) |
We tested the functionality of the ectopic CO2/Or59c or Or42a hybrid neurons using single sensillum recordings (Fig. 3). Ectopic MP CO2 neurons on miR-279 and prosIG2227 mutant palps were functional and responded to CO2 (Fig. 3A,B). In addition, sensilla containing ectopic CO2 hybrid neurons also responded to Or42a and Or59c specific odors such as iso-amyl acetate and 3-octanol, in line with their receptor expression profile (Fig. 3C and data not shown) (de Bruyne et al., 1999). Thus, CO2 neurons are formed within Or42a and Or59c sensilla, and coexpress and respond to either of these receptors.
Figure 3.
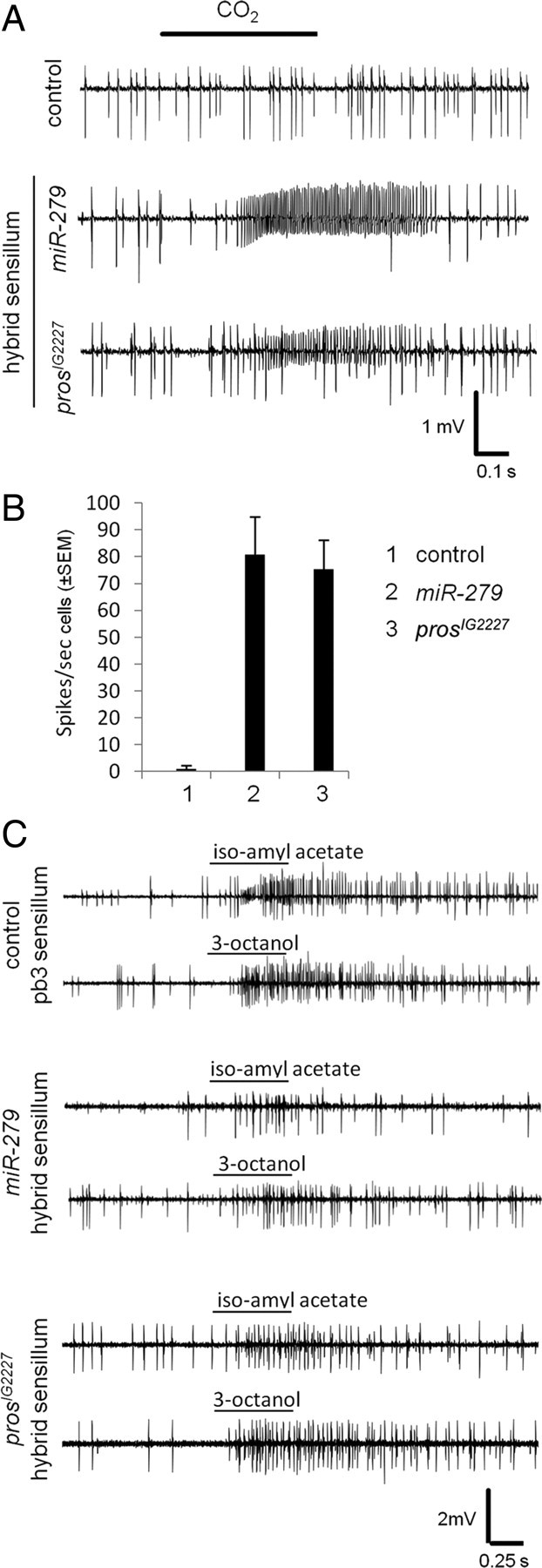
Hybrid neurons respond to CO2 and MP key odors. A, Representative single sensillum recording traces from the MP basiconic sensilla of control, miR-279, and prosIG2227 flies. Control trace (top) shows spikes of two neurons. In both miR-279 (middle) and prosIG2227 (bottom) traces, spikes of an extra neuron are visible. During a 500 ms CO2 stimulation (black bar), the spike frequency of the B neuron in both miR-279 and prosIG2227 increased significantly. No response was observed in control MPs. B, Spike frequency of B neurons to CO2 stimulation in control, miR-279, and prosIG2227 flies are represented in the graph (n = 10 recordings for each genotype, ±SEM). C, Representative single sensillum recording traces from the MP basiconic sensilla of control (n = 5), miR-279 (n = 6), and prosIG2227 (n = 6) flies stimulated with two key ligands of pb3 sensilla, iso-amyl acetate, and 3-octanol.
In summary, mutations in Pros in the olfactory lineage result in the selective formation of ectopic MP CO2 hybrid neurons, as previously reported for miR-279 mutants. Complete lack of Pros also frequently displayed a less ORN-specific and more severe phenotype of neuron loss, including antennal CO2 neurons, comparable to previous reports in ES organs and antenna. These results suggest that Pros, in addition to its early function in pIIb versus pIIa specification, also restricts neuron formation in specific MP olfactory sensilla.
Pros is expressed in the olfactory lineage and maintained in mature sheath cells
The similarity of pros and miR-279 mutant phenotypes suggest that Pros and miR-279 act on the same developmental mechanism and potentially same genetic network. We next asked whether Pros and miR-279 are coexpressed during olfactory system formation. The MP develops during pupal life from a cluster of sensory organ precursors in the antennal part of the eye-antennal imaginal disc (Lebreton et al., 2008). In the mature wild-type MP, every olfactory sensillum contains two ORNs, one sheath, one socket, and one shaft cell (Fig. 4A). In the developing MP, the neural marker Elav was first detected using an anti-Elav antibody at ∼6 h after puparium formation (APF). At this time point, Elav staining overlapped with staining against Pros (Fig. 4B). Two populations stained at different intensities with Pros coexpressed Elav. At 25 h APF, Elav-only cells and Pros-only cells were present next to cells with low levels of Pros that coexpressed high Elav, and vice versa (Fig. 4C). At 45 h APF, Elav and Pros were completely separated, and the final sensillum composition was defined. Two neurons expressed Elav and, conversely, Pros became restricted to the sheath cell (Fig. 4D). These stainings and additional markers are consistent with previous observations (Manning and Doe, 1999), where Elav and Pros are coexpressed early on in the neural lineage pIIb cell (Fig. 4J). The pIIb cell divides and gives rise to a pIIIb and a pIIIb* cell that can be distinguished by their high versus low levels of Pros. Finally, the high Pros-expressing cell divides again and produces a Pros-positive sheath cell and a Pros-negative neuron, whereas the cell with lower Pros levels gives rise to a single neuron (Fig. 4J).
Figure 4.

Prospero and miR-279 are coexpressed in the MP. A, Schematic drawing of a fully differentiated wild-type MP sensillum with two neurons. B, Developing MP at 6 h APF stained with anti-Elav (green, B1) and anti-Pros (pink, B2). Note the almost complete overlap of expression. C, Developing MP at 25 h APF stained with anti-Elav (C1) and anti-Pros (C2). Low and high Pros-expressing cells are indicated with arrow and arrowhead, respectively. Cells with low Pros expression seem to also express Elav. D, MP of elav-GAL4,UAS-mCD8GFP fly at 45 h APF stained with anti-GFP (D1) and anti-Pros (D2). Elav-positive cells (arrow) do not express Pros anymore. Two Elav-expressing cells are located next to one Pros-expressing cell (arrowhead). E, Developing MP of miR-279-Gal4,UAS-nlsGFP at 6 h APF stained with anti-GFP (E1 and E4), anti-Elav (E5), and anti-Pros (E2). miR-279 starts to be expressed and colabels with Pros (arrow and arrowhead). The weaker Pros-positive cell is also Elav-positive (arrow). F, Developing MP of miR-279-GAL4,UAS-mCD8GFP at 30 h APF stained with anti-GFP (F1) and anti-Pros (F2). Many cells express miR-279-GFP, some of which are also Pros-positive (arrowheads) G, MP from miR-279-GAL4,UAS-nlsGFP at 42 h APF stained with anti-GFP (G1) and anti-Pros (G2). miR-279 remains coexpressed with Pros (arrowhead) in some cells. H, MP from miR-279-GAL4,UAS-nlsGFP at 42 h APF stained with anti-GFP (H1) and anti-Elav (H2) miR-279-positive cells (arrowhead) do not colabel with Elav (arrow). I, MP, miR-279-GAL4,UAS-mCD8GFP in an eyFLP control clone at 70 h APF stained with anti-GFP (I1) and anti-Prospero (I2). Arrowhead shows a miR-279-positive cell, which is expressing Pros. J, Model of the divisions of the cells of the inner (pIIb) lineage. Respective expression of Pros, Elav, and miR-279 are indicated. SOP, Sensory organ precursors; n, neuron; sh, sheath cell; L3, larval stage 3. Scale bars, 5 μm.
Next, we used an enhancer element (2 kb upstream of the miR-279 gene), which was sufficient to rescue the phenotype in miR-279 loss-of-function mutants to analyze miR-279 expression (Cayirlioglu et al., 2008). miR-279-GFP (miR-279-GAL4, UAS-mCD8-GFP/-nls-GFP) was first detected in MP precursor cells in the eye–antennal disc at 6 h APF (Fig. 4E1,E4). At this time, both Elav and Pros expression was significantly broader than miR-279-GFP expression, but all miR-279-positive cells were also positive for Pros or Pros/Elav staining (Fig. 4E,E3). By 30 h APF, we detected a large number of miR-279-GFP cells, and the overlap with Pros staining was still high (Fig. 4F). At 42 h APF, expression became more defined and overall dropped to fewer cells compared with 30 h APF (Fig. 4G,H). While the overlap with Pros remained significant, we did not detect any miR-279-cells positive for the neuronal marker Elav (Fig. 4H). In MPs analyzed at 70 h APF, Pros and miR-279 were confined to sheath cells (Fig. 4I).
These data demonstrate that the first miR-279-expressing cells are also positive for Pros. While both Pros and miR-279 initially overlap with the neuronal marker Elav, in midpupal stages when most of the sensory cells have been determined, Pros and miR-279 are excluded from neurons. In the mature MP, Pros and miR-279 are expressed in the sheath cell. While Pros expression is detected early in the pIIb cell, strong miR-279 expression is detectable in the late pIIb and pIIIb cells. We conclude that both miR-279 and Pros are coexpressed during stages of neuronal and sheath cell differentiation.
Pros and miR-279 define neuron number in MP sensilla
pros and miR-279 mutants fail to suppress the formation of CO2 sensory neurons on fly MPs. We next addressed in more detail the development of MP CO2 neurons in the mutants. To selectively label the neuronal lineage in a mutant sensillum, we used MARCM and Elav-GFP (elav-GAL4, UAS-mCD8GFP). In wild-type clones at 45 h APF, Elav-GFP as well as anti-Elav antibody labeled two neurons per single sensillum. Adjacent to each two-neuron cluster, we detected a single Pros-positive cell—the sheath cell (Fig. 5A). In contrast, Elav-GFP labeled three neurons in single sensilla in 100% of the analyzed miR-279 mutants (Fig. 5B). In these three neuron sensilla, all neurons expressed Elav. A Pros-positive cell was found closely associated to the cluster (Fig. 5B). Mutant MP clones of prosIG2227 or pros17, like miR-279 clones, displayed three neurons and one sheath cell per sensillum in 100% of prosIG2227 and 54% of pros17 mutant palps (Figs. 5C,D,G; 6D). Next, we labeled CO2 neurons with Gr21-GAL4,UAS-mCD8GFP in miR-279 and prosIG2227 mutant MPs at 70 h APF. A single Gr21a cell positive for Elav was detected per three neuron sensillum in pros and miR-279 mutant clones (Fig. 5E,F). In 31% of pros17 mutant MPs, Elav-GFP labeled only one single neuron with detectable levels of Elav protein per sensillum (Fig. 6A,B). In 15.5% of pros17 mutant palps, sensilla with up to four cells labeled by Elav-GFP, none of them positive for anti-Elav, were observed, suggesting that these cells are part of the neural lineage but failed to fully differentiate into mature neurons (Fig. 6C,D). In summary, neuron loss in pros-null sensilla shows that in the MP olfactory lineage, Pros helps to establish the neuronal pIIb versus non-neuronal pIIa cell fate (Fig. 6D). The hypomorphic alleles of pros uncovered an additional role of Pros within the olfactory pIIb lineage. Here, Pros restricts the number of neurons, and thereby prevents the formation of MP CO2 neurons without affecting number and differentiation of sheath cells (Fig. 6D). We conclude that Pros and miR-279 are required to prevent the formation of an additional neuron, the CO2 ORN, within Or59c or Or42a ORN-containing MP sensilla of flies.
Figure 5.
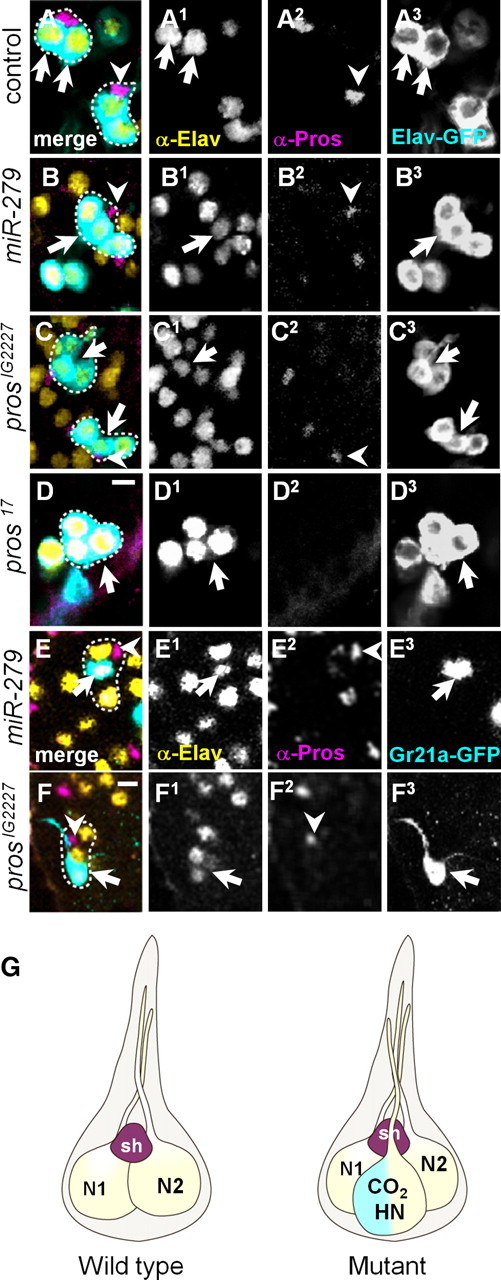
Pros and miR-279 suppress CO2 neuron formation in MP sensilla. A–F, MPs at 45 h APF (A–D) or pharate adults (E, F), stained with anti-GFP (turquoise), anti-Elav (yellow), and anti-Pros (pink). Cells belonging to one sensillum are circled with a dashed line. A, Wild-type showing two Elav-GAL4-positive cells (arrows, A3) costained with anti-Elav (arrows, A1) and one adjacent Pros-expressing cell (arrowhead, A2). B, miR-279 MP clone (circled area) showing three Elav-GFP cells (arrow, B3), which are positive for Elav (arrows, B1). Pros-positive cell is seen next to the cluster (arrowhead). C, Two examples of prosIG2227 clones (circled areas) with three neurons positive for Elav (arrows, C1). Notably, Pros labeling is reduced compared with wild-type clones (arrowhead, C2). D, Similar example of a pros17 clone of three Elav-GAL4-positive cells (arrow). The adjacent neurons outside the circled area belong to different sensilla in close vicinity. Pros expression is not detectable in pros17-null mutant. E, miR-279 clone with single Gr21a-positive cell (arrow, E3), which costains with anti-Elav (arrow, E1) but not anti-Pros (arrowhead, E2). F, Similar example of a prosIG2227 clone with a GR21a-positive cell (arrow, F3), which is positive for anti-Elav (arrow, F1). G, Schematic model of wild-type sensillum housing two neurons (N1 and N2) and one sheath cell (sh). Mutant sensillum with three neurons and one sheath cell is depicted. The additional neuron is a hybrid neuron (HN) and expresses the CO2 receptors and either Or42a or 59c. Scale bars, 5 μm.
Figure 6.

Pros and miR-279 define olfactory neuron number. MPs at 45 h APF stained with anti-GFP (turquoise), anti-Elav (yellow), and anti-Pros (pink). A, In wild-type sensilla, two Elav-GAL4-positive cells costain with anti-Elav (arrows) and one Pros-expressing cell resides next to them (arrowhead). B, pros17 clone with single Elav-positive cells (arrows). Pros expression is not detectable in the pros17-null mutation. C, pros17 clones of Elav-GAL4-positive cells (arrow), which failed to maintain the expression of Elav protein. D, Pros activity is required at distinct steps during lineage formation. pros17 mutants show an early phenotype during pIIb to pIIa specification. The pIIb lineage partly converts to the pIIa lineage in these mutants. This results in a partial or complete loss of ORNs, giving rise to sensilla with only one neuron (31%) or clusters of undifferentiated (undiff.) cells (15.5%). These scenarios are also seen rarely in prosIG2227 (10%) and miR-279 mutants (4%). n(pros17) = 13, n(prosIG2227) = 19, and n(miR-279) = 22 MPs. Hypomorphic alleles of prosIG2227 and prosVoila78 uncover a second phase of Pros requirement during definition of sensilla neuron number. In these mutant palps and in 54% of pros17 mutant palps, a third neuron was formed within a sensillum. This neuron expressed Gr21a receptor (CO2 receptor) starting at late pupal stages. While Pros is involved in pIIb versus pIIa specification, miR-279 is specifically required downstream of Pros within the already determined neuronal lineage to define the number of ORNs in certain MP sensilla. Scale bars, 5 μm.
miR-279 expression suppresses ectopic MP CO2 neurons in Pros mutants
We next addressed the functional relationship between miR-279 and Pros. We inquired whether Pros activity regulates miR-279 expression during MP olfactory sensilla formation. miR-279 expression in miR-279 mutants fully rescued its phenotype (Fig. 7A). The same was observed for Pros expression in prosIG2227 mutants (Fig. 7A). We next expressed miR-279 in eyFLP clones in the developing MP of prosIG2227 mutants. Expression of miR-279 was sufficient to rescue the phenotype of the hypomorphic allele prosIG2227 in 80% of the flies analyzed (Fig. 7A). In contrast, expression of Pros in miR-279-null mutants suppressed ectopic MP CO2 neurons in only 37% of the flies analyzed (Fig. 7A). Thus, miR-279 expression in hypomorphic Pros mutants rescues the prosIG2227 phenotype to a high degree. We propose that, together with the milder Pros-mediated rescue of miR-279-null mutants, Pros acts upstream and also partly in parallel to miR-279 signaling.
Figure 7.
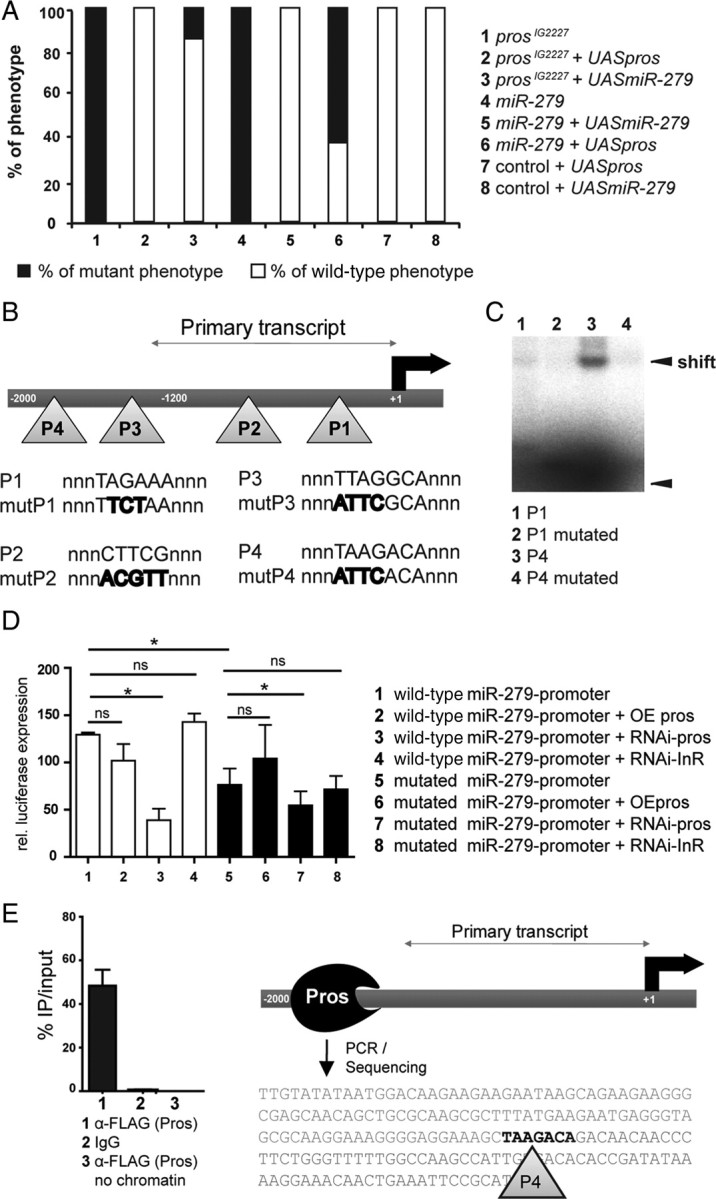
miR-279 and Pros interact in vitro and in vivo. A, Expression of UAS-pros as well as UAS-miR-279 in the eyFLP;prosIG2227 mutant background rescued the phenotype almost fully (100%, n = 25 and 80%, n = 81, respectively), while expression of UAS-pros in the eyFLP;miR-279 mutant background only led to a partial rescue of 37% (n = 25). Overexpression of Pros or miR-279 in wild-type clones did not result in a detectable phenotype in Gr21a-expressing neurons (n = 20). B, Scheme of the putative enhancer region of miR-279 depicting the relative positions of the predicted Pros binding sites (P1–P4). The binding sites include either the consensus motif TWAGVYD or CWYNNCY. The primary transcript starts ∼1200 bp upstream of miR-279. Sequences of putative binding sites and sequences of mutated sites within the consensus sequence are shown. C, Gel shift assay using purified homeodomain of Pros. Radiolabeled oligos carrying either P1 or P4 sites were assayed for direct Pros binding. Pros homeodomain binds to P4, which results in a strong shift (lane 3). The shift is strongly reduced when mutated P4 oligos were tested (lane 4). D, S2 cell miR-279 promoter reporter assays with wild-type and mutated versions of the promoter. Note that RNAi against Pros strongly reduced miR-279 reporter expression, while Pros overexpression did not significantly enhance reporter expression of the wild-type and mutated promoter constructs (*p < 0.05). E, Quantification of percentage PCR product of immunoprecipitated (IP) chromatin compared with the input. Anti-FLAG to pull down Pros-FLAG was compared with mouse IgG plus chromatin and anti-FLAG antibody without chromatin. The PCR was conducted with a primer set amplifying the region around P4. The sequence of the PCR product is shown.
Pros might act as a direct transcriptional activator of miR-279 expression. In bioinformatics predictions, we detected 18 putative Pros-binding sites in the 2 kb upstream region of miR-279, of which we focused on the four most conserved sites. miRs are first transcribed as longer pri-miRs, before they get exported and processed into pre-miR and mature miR by Dicer (Lee et al., 2003). To define the length of the primary miR-279 transcript, we used 5′RACE, and determined the transcriptional start site at 1.2 kb upstream of the miR-279 gene (Fig. 7B). We used electromobility shift assays (EMSA) to demonstrate direct Pros binding to the miR-279 enhancer. Purified Pros homeodomain bound strongly only to oligos containing the P4 site as shown by the gel shift (Fig. 7C). The shift was significantly reduced when the oligos were mutated to a nonconsensus motif. We conclude that Pros protein can bind the miR-279 upstream putative enhancer regions in vitro.
To test whether Pros induces expression of the miR-279 promoter in a cellular assay, we transfected S2 cells with a miR-279 promoter construct fused to a firefly luciferase gene (Fig. 7D and data not shown). miR-279 wild-type reporter was strongly expressed in S2 cells, consistent with endogenous miR-279 expression in these cells (Fig. 7D) (Cayirlioglu et al., 2008). To test whether promoter activity was due to endogenous Pros expression in S2 cells, we manipulated Pros levels using RNAi and overexpression. While Pros overexpression did not further increase reporter expression, RNAi against Pros reduced reporter activity by 70 ± 6.2% (Fig. 7D). Next, we mutated up to four putative Pros binding-sites in the promoter region (Fig. 7D). We observed 42 ± 10% reduced expression of the mutated versus the wild-type promoter construct (Fig. 7D). We tested the effect of Pros RNAi and Pros overexpression on the mutated promoter. Similar to the wild-type construct, Pros overexpression did not significantly increase enhancer activity, while Pros RNAi reduced activity levels to 49 ± 7%, similar to the wild-type reporter (Fig. 7D). These data suggest that Pros is required for expression of miR-279 in S2 cells. Nevertheless, they also indicate that additional sites of the 18 predicted Pros binding-sites might be involved.
To test the binding of Pros to the miR-279 promoter in vivo, we performed ChIP. We analyzed chromatin from embryos that overexpress FLAG-tagged Pros, and immunoprecipitated Pros-FLAG with α-FLAG antibody. PCR was used to amplify the region flanking the P4 binding site. Forty-eight percent of the input DNA was immunoprecipitated using α-FLAG, whereas control IgG and α-FLAG without input did not produce a detectable PCR product (Fig. 7E). The resulting PCR product was sequenced to confirm that it contained P4 Pros binding site in the promoter of miR-279 (Fig. 7E).
We then investigated the effect of loss of Pros on the expression of miR-279 in vivo. To this aim, we used the miR-279-GAL4 driver line, described above, to monitor miR-279 expression in control, miR-279, and pros mutant MP clones during development and adulthood. At 6 h APF, miR-279 expression is first detected in Pros-positive cells in the developing MP. At this time point, miR-279 expression in Pros-expressing cells was detected in only 16% of the prosIG2227 MPs compared with in 75% of the wild-type and 69% of the miR-279 mutant MPs (Fig. 8A,B). Similar results were obtained in adult MPs (Fig. 8C). The reduction in miR-279 expression in pros mutants correlated well with the proportion of prosIG2227 MPs that showed a high number of MP CO2 neurons (43%: >15; 54%: >12; 70%: >10 CO2 neurons; compare Fig. 1). These data show that Pros activity contributes to the expression of miR-279 in MPs.
Figure 8.
miR-279 expression is strongly reduced in Pros mutant MPs. A, B, Representative expression patterns of miR-279-GAL4 (turquoise) combined with anti-Pros (pink) and anti-Elav (yellow) staining in control and mutant clones in the developing MP at 6 h APF. Pros and miR-279 coexpressing cells are indicated with arrows. We detected colabeled cells in 73% (n = 15) and 66% (n = 18) of control and miR-279 MPs, respectively. In prosIG2227 mutant clones, only 14% (n = 14) of the MPs showed cells coexpressing miR-279 and Pros. C, Expression of miR-279-GAL4, UAS-mCD8GFP in adult MPs of wild-type and mutant clones. miR-279-GFP expression is strongly reduced in 59% and 52% of MP of pros17 and prosIG2227 mutants compared with controls, respectively (n = 117, 37, 112). miR-279-GFP expression is only slightly reduced in miR-279 MP (n = 107, 92% compared with controls). Note that efficiency of FLP recombination was not different among the genotypes (data not shown). Scale bars, 5 μm.
We conclude that Pros directly binds the miR-279 enhancer and contributes to its expression to suppress the formation of ectopic CO2 neurons in the fly MP.
Common targets Escargot and Nerfin-1 are sufficient to induce MP CO2 neurons
Previously, in vivo Pros/DNA-interaction sites were mapped in the fly embryo (Choksi et al., 2006). Identification of putative miR targets has been quite successful using several bioinformatic analysis tools. We combined the predicted miR-279 targets from three different databases and compared them to the list of in vivo Pros targets. Twenty Pros target genes were predicted to be miR-279 targets (Table 2). Next, we used GO analysis to assess the putative role of the common targets of Pros and miR-279. Shared targets fell into six categories, with 12 of 20 targets falling into two GO terms—cell fate determination and nervous system development (data not shown). Thus, they appeared highly relevant for the described phenotype and were the focus of our further analysis. These predictions suggested that miR-279 might act on Pros target genes, being a Pros-target itself.
Table 2.
Predicted common targets of Pros and miR-279
| Common target | Gene | Molecular function |
|---|---|---|
| CG13906 | nerfin-1 | Transcription factor |
| CG3758 | escargot | Transcription factor |
| CG11988 | neuralized | Ubiquitin ligase |
| CG3827 | scute | Transcription factor |
| CG9626 | unknown | Unknown |
| CG13521 | robo | Receptor |
| CG9239 | B4 | Unknown |
| CG10580 | fringe | UDP-glycosyltransferase |
| CG9739 | frizzled-2 | Wnt receptor |
| CG17835 | invected | Transcription factor |
| CG12245 | gcm | Transcription factor |
| CG1447 | Ptx1 | Transcription factor |
| CG6993 | spineless | Transcription factor |
| CG1072 | Arrowhead | Transcription factor |
| CG18408 | CAP | Vinculin binding |
| CG2679 | goliath | Transcription factor |
| CG9623 | inflated | Receptor cell adhesion |
| CG1004 | rhomboid | Serine-type peptidase, cleaves EGF |
| CG5507 | Transcript 48 | Unknown |
| CG31048 | unknown | Unknown |
List of the twenty predicted target genes shared between Pros and miR-279. Genes include transcription factors and enzymes implicated in neuronal development.
Two high-ranking miR-279 targets and in vivo targets of Pros are the snail-family transcription factor Escargot (Esg) and the neurogenic zinc-finger transcription factor Nerfin-1 (Kuzin et al., 2005; Choksi et al., 2006). We used a P[lacZ] insertion into the esg locus to analyze Esg expression in vivo. While at 6 h APF, Esg was expressed in the developing MP and was low or excluded from Pros-expressing pIIb cells. At later stages (25, 30, and 48 h APF), Esg expression appeared overall lower and went down to finally undetectable levels (Fig. 9C). Similar to Esg, Nerfin-1 is also expressed in the developing MP at low levels (Cayirlioglu et al., 2008). Therefore, Esg is expressed at the right time and place to act as a downstream target of Pros and miR-279 during MP olfactory sensilla development.
To test whether miR-279 can silence the expression of Esg via the 3′UTR in vitro, we assayed 3′UTR-luciferase reporter activity in S2 cells. These reporter constructs carried the coding region of the renilla luciferase followed by the 3′UTR of the respective target gene. miR-279 overexpression significantly silenced the expression of the esg 3′UTR and the nerfin-1 3′UTR reporter constructs (Fig. 9A,B). The expression of the esg 3′UTR and the nerfin-1 3′UTR were reduced to 51 ± 9% and to 27 ± 10%, respectively (Fig. 9A,B). Therefore, Esg and Nerfin-1 are direct targets of miR-279 in vitro.
To test the impact of miR-279 and prosIG2227 on Esg expression in vivo, we crossed the esg-P[lacZ] into the mutant background of miR-279 and prosIG2227 (Fig. 9D). We analyzed MPs at 6 h APF. In control palps, a low percentage (16%) of MPs displayed coexpression of Pros and β-gal (esg), while in miR-279 and prosIG2227 mutant clones, the colocalization of Pros and esg[lacZ] occurred in up to 90% and 61% of all palps analyzed, respectively. We conclude that miR-279 as well as Pros can repress the expression of Esg in vitro and in vivo.
We showed previously that reduction of Nerfin-1 expression by removing one allele of nerfin-1 in the background of miR-279 mutants ameliorated the ectopic CO2 neuron phenotype (Cayirlioglu et al., 2008). Using the same experimental set-up for prosIG2227 mutants, we found that reduction of Nerfin-1 reduced the number of ectopic CO2 neurons significantly by 55% compared with prosIG2227 mutant palps only (Fig. 10A). In MPs of miR-279 and prosIG2227 flies, reduction of esg reduced the number of ectopic CO2 neurons by 29% and 49%, respectively (Fig. 10A,B). Removal of one copy of both nerfin-1 and esg in the background of prosIG2227 did not further reduce the number of ectopic CO2 neurons (Fig. 10A). Hb9 (exex), a predicted target of miR-279, but not a target of Pros, did not show a genetic interaction with miR-279 (Fig. 10B). We conclude that shared miR-279 and Pros target genes are required for CO2 neuron formation on MPs.
Figure 10.
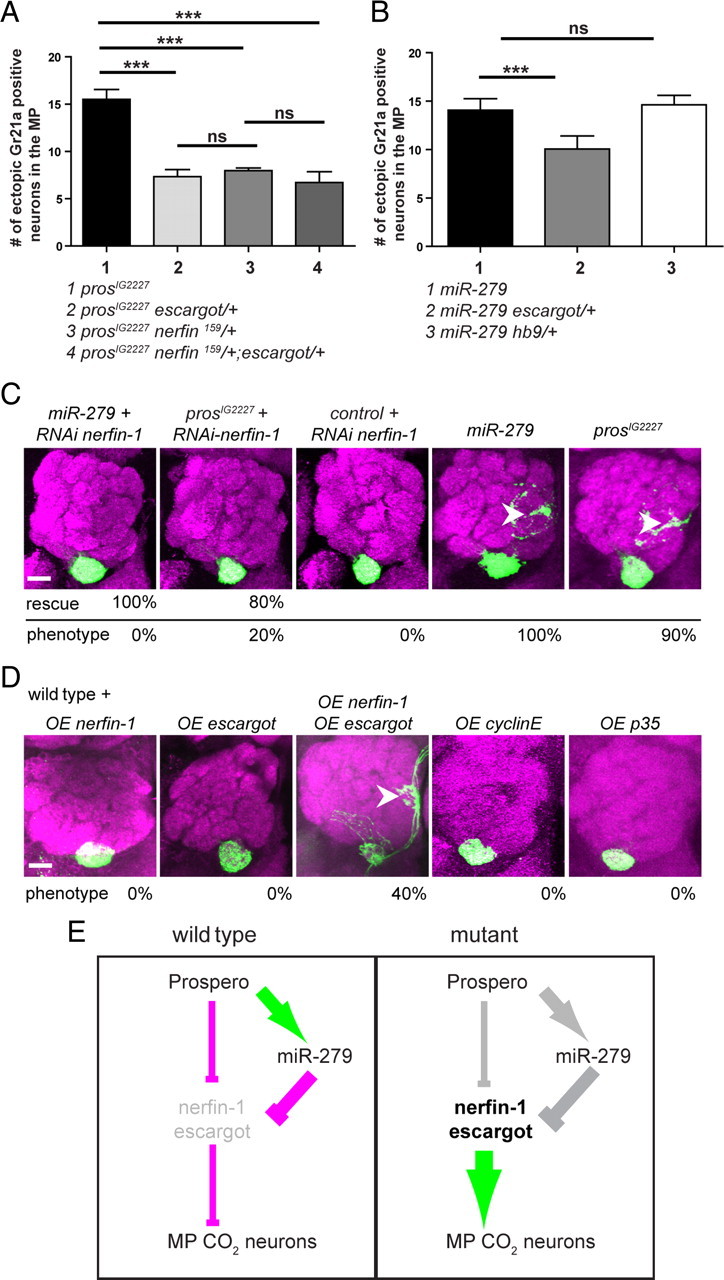
Increased expression of Nerfin-1 and Escargot is required and sufficient to induce CO2 neuron formation and mistargeting on the MP. A, Genetic interaction of prosIG2227 and two target genes nerfin-1 and esg. The number of ectopic Gr21a-positive cells in prosIG2227 mutant MPs missing either one allele of esg, nerfin-1, or both esg and nerfin-1 is reduced by 55% (n = 22), 49% (n = 30), and 40% (n = 20) compared with controls (n = 37), respectively. B, Quantification of the genetic interaction of miR-279 and esg showing a reduction by 44% of the ectopic Gr21a neurons after removal of one copy of esg in the miR-279 mutant background (n = 30) compared with miR-279 mutant only (n = 30). The removal of one copy hb9 (exex) did not result in a reduced number of ectopic CO2 neurons (n = 24). C, nerfin-1-RNAi expression in the background of the miR-279 and the prosIG2227 background rescues the phenotype by 100% (n = 35) and 80% (n = 29), respectively. Nerfin-1 RNAi in control flies induced no phenotype (n = 20). D, Overexpression of targets in the wild-type background. Single overexpression of Nerfin-1 and Esg did not result in Gr21a medial mistargeting. In contrast, overexpression of both Esg plus Nerfin-1 phenocopied the phenotype of miR-279 and prosIG2227 in 40% (n = 24) of all fly brains analyzed. Overexpression of the apoptosis inhibitor p35 (n = 20) and the cell cycle gene CyclinE (n = 25) did not result in a miR-279-like phenotype. E, Mechanistic model of a network of Pros, miR-279, Nerfin-1, and Escargot: In wild-type MPs, Pros directly binds and represses the expression of at least two target genes, nerfin-1 and escargot. In a parallel pathway, Pros directly activates the expression of miR-279, which in turn represses nerfin-1 and esg on a posttranscriptional level. Our genetic data indicate that indirect repression via miR-279 post-transcriptionally is more powerful compared with direct Pros repression on the genomic level (indicated by line thickness). This regulation represses the formation of MP CO2 neurons. In pros or miR-279 mutants, the repression of Nerfin-1 and Esg expression directly and/or indirectly is lost. As a consequence, an excess of Nerfin-1 and Esg leads to the formation of ectopic CO2 neurons within Or42a and Or59c olfactory sensilla on the MP. Scale bars, 20 μm.
To address the role of Nerfin-1 during axon targeting, we expressed an RNAi construct against nerfin-1 in miR-279, pros, and control palps. Nerfin-1 RNAi was able to rescue the miR-279- and prosIG2227-targeting phenotype in 100% and 80% of all flies analyzed, respectively, but had no effect on CO2 neurons in wild-type clones (Fig. 10C). A similar experiment with esg was not possible in lieu of a working RNAi construct. We tested four additional potential downstream factors of miR-279 and Pros for interaction with pros or miR-279 mutants by means of in vivo RNAi: spineless, senseless, gcm, and Ptx-1. None of them rescued the targeting defect of miR-279 or pros mutants (data not shown).
To test whether elevated levels of Nerfin-1 and Esg are sufficient to induce the formation and mistargeting of CO2 neurons, we overexpressed Esg and Nerfin-1 separately or together in wild-type MP clones. Overexpression of only Nerfin-1 or Esg did not result in an altered targeting pattern of CO2 neurons (Fig. 10D). In contrast, overexpression of both targets together phenocopied the miR-279 and pros phenotype in 40% of all flies analyzed (Fig. 10D). Overexpression of p35 and cyclinE in the wild-type background did not induce ectopic CO2 neurons and mistargeting, suggesting that lack of apoptosis or altered cell cycle progression are not sufficient to induce ectopic CO2 neurons (Fig. 10D).
These experiments led us to propose a genetic network including the activities of Pros, miR-279, and two shared downstream target genes. We provide evidence that Pros is a positive regulator of miR-279 expression. miR-279 in turn represses at least two of Prospero's targets. Upregulation of Esg and Nerfin-1 is sufficient for the formation and mistargeting of MP CO2 neurons. In conclusion, we suggest that Pros uses its transcriptional activator activity to enhance its own repressor function via the induction and activity of a single microRNA, miR-279 (Fig. 10E).
Discussion
Here, we show that the transcription factor Pros prevents the formation of MP CO2 neurons by inducing miR-279 to enhance its own repressor activity on two shared target genes, nerfin-1 and esg (Fig. 10). We first describe the expression of Pros and miR-279 during the formation of two neurons in the MP wild-type neural lineage. Second, we demonstrate that Pros and miR-279 are required to prevent the formation of a third ORN within certain MP sensilla. Third, we present evidence that Pros regulates miR-279 expression in vivo, possibly by direct binding to the miR-279 enhancer. Fourth, we show that re-expression of miR-279 in pros mutant clones rescues the pros phenotype. Finally, we find that increased levels of two Pros and miR-279 target genes, Nerfin-1 and Escargot, induces the formation and mistargeting of MP CO2 neurons. We propose a genetic network in which Pros uses two activity modes—one direct and one indirect via miR-279 activation—to repress target genes Nerfin-1 and Esg (Fig. 10). This fine-tuning of Pros activity defines the number of neurons in two specific olfactory sensilla and thereby prevents the expression of a CO2 neuron genetic program on MPs.
Pros together with miR-279 defines neuron numbers in specific olfactory sensilla
Our analysis of both wild-type and mutant olfactory lineages suggests that the development of MP olfactory sensilla follows the canonical lineage model (Lai and Orgogozo, 2004). Previous analysis of the role of Pros in ES and antennal olfactory sensilla suggested that complete loss of Pros results in an early phenotype in the sensory organ lineage. It affects the proper establishment of the pIIb cell that ultimately gives rise to all neurons and the sheath cell in each sensillum (Manning and Doe, 1999; Reddy and Rodrigues, 1999; Sen et al., 2003). In agreement with this work, we also find that pros-null mutants showed neuron loss in several classes of ORNs, including antennal CO2 neurons. Interestingly, we describe a new hypomorphic allele of pros and reveal that Pros also has a later role during the definition of neuron number in specific olfactory sensilla. This function of Pros is similar to its described role in the embryonic CNS, where Pros presumably uses its repressor activity to prevent mitosis (Choksi et al., 2006). miR-279-null mutants, in contrast to pros-null mutants, do not display neuron loss but, like pros hypomorphic mutants, fail to repress the formation of an extra CO2 neuron in two classes of MP sensilla. Thus, miR-279 is selectively required during the second Pros phase uncovered in the pros hypomorphic alleles. Using this mechanism, miR-279-mediated repression downstream of Pros appears to impose functional specificity on this pleiotropic neural development regulator.
Despite the expression of Pros and miR-279 in all olfactory sensilla, Pros' and miR-279's function in CO2 neuron repression appears restricted to the MP. Although we currently cannot answer why Pros and miR-279 affect MP sensilla differently than antennal sensilla, we speculate that the reason might be linked to the different developmental and evolutionary origin of these organs that depend on two different proneural genes, Atonal and Amos (Jhaveri et al., 2000; Markitantova et al., 2003). Similar to the three ORN mutant MP sensillum in the fly, the wild-type CO2 sensory neuron of mosquito also belongs to the Ato lineage and resides in a three-neuron sensillum on the MP (Lu et al., 2007).
It is possible that some olfactory sensilla are especially sensitive to Pros signaling. Work in the ES lineage has shown that Notch and Pros can cooperate to prevent additional mitosis of pIIIb cells in the lineage (Simon et al., 2009). Notch is also involved in regulation of neuron type and targeting in the olfactory system (Endo et al., 2007). Given the variety of olfactory bristles with different neuron numbers and types, differential sensitivity to levels of lineage determinants such as Pros, possibly due to different Notch activities, could have contributed to the formation of subsets of this large repertoire.
microRNAs and Prospero in cell fate decisions
miRs have been implicated in stabilizing gene expression by engaging in different modes of feedback regulation (Herranz and Cohen, 2010). They are posttranscriptional repressors of gene expression, which makes them perfectly suited for fine-tuning or tightly controlling expression levels of genes (Biryukova et al., 2009; Coolen and Bally-Cuif, 2009; Liu et al., 2009). Here, we provide evidence that Pros regulates miR-279 activity to suppress its own target genes. Why does Prospero engage a microRNA? We envision three scenarios: (1) Pros repressor activity on the genomic level is simply too weak, (2) Pros acting on the transcriptional level is not fast enough, and (3) Pros uses miR-279 not only to enhance and speed up its activity but also to temporally define expression levels. The observation that re-expression of Pros in miR-279 mutants rescued the miR-279 phenotype to a certain degree is consistent with a role of miR-279 as enhancer of repressor activity. miR-279 mutants phenocopy only a specific aspect of the pros phenotype. This argues for a role of miR-279 in temporal control of Pros activity.
We showed that overexpression of two shared targets of Pros and miR-279 Nerfin-1 together with Esg was sufficient to induce MP CO2 neurons. Nerfin-1 and Esg reduction lowers the numbers of ectopic CO2 neurons in miR-279 and pros mutants. Thus, Nerfin-1 and Esg are essential and sufficient to trigger CO2 neuron formation in MP, suggesting that they are part of a MP CO2 neuron program normally suppressed in flies. Consistent with a role as possible axon targeting factor, Nerfin-1 was shown previously to be essential for axon guidance decisions in the developing embryonic CNS. nerfin-1 mutants failed to properly express guidance factors such as robo2, wnt5, derailed, G-oalpha47A, Lar, and futsch (Kuzin et al., 2005). Our data, together with previous analysis of esg mutants (Yang et al., 2010), points to a role in neurogenesis. However, single mutants of esg did not show a defect in antennal CO2 neuron development (data not shown). Simple interference with cell cycle progression or apoptosis was not sufficient to induce ectopic CO2 neurons, arguing for a more complex biology controlled by both Nerfin-1 and Esg. Future analysis will address the relationship and exact function of Nerfin-1 and Esg.
Divergence of CO2 sensory systems of fruit flies and mosquitoes
MP ORN axons in fruit flies and mosquitoes target medial glomeruli in the AL. This is also the case for the axons of the ectopic CO2 neurons in the mutant fly MP. It is possible that certain developmental constraints on the wiring of the olfactory neurons to the AL make it difficult for MP neurons not to innervate food-related glomeruli in the AL (Ghysen, 1992; Sweeney et al., 2007). Thus, it might have been more feasible to relocate the neurons and take advantage of an existing wiring program than to leave the neurons in the same sensillum and redirect them to a new glomerulus. The pathway we describe here might have contributed to the differential localization of CO2 neurons in insects with opposite CO2 preferences. pros, miR-279, as well as nerfin-1 and escargot genes are present in the mosquito genome. We speculate that the genetic and functional interaction of Pros, miR-279, Nerfin-1, Escargot, and likely additional factors was either lost in mosquito or acquired in the fly to allow or prevent the formation of MP CO2 neurons, respectively. In support of this, we analyzed putative target genes for Anopheles gambiae miR-279 (aga-miR-279) using miRBase prediction. Notably, both aga-escargot and aga-nerfin-1 are not among the first 150 predicted targets of aga-miR-279 (see Materials and Methods, above). Given that overexpression of Nerfin-1 and Esg are sufficient to induce MP CO2 neurons, it is possible that Nerfin-1 and Esg are key factors in a CO2 neuron program actively suppressed by both Pros and miR-279 in fly MPs. Hence, although the relocation of CO2 neurons from antenna to MP might be a rather unusual or extreme case, it is conceivable that this pathway or similar miR-mediated mechanisms could have been used to produce sensilla, including nonolfactory sensilla, of different neuronal numbers in different species.
Is MP location and medial mistargeting sufficient to convert repulsion into attraction? MP CO2 neurons in the mutants are functional. Mutant flies with antennae still avoid CO2, while antennaless mutants appear indifferent to CO2 (data not shown). One of many interpretations of these data is that MP CO2 ORNs activate attraction and repulsion glomeruli simultaneously, resulting in behavioral indifference. Nevertheless, it has been reported that individual glomeruli mediate innate attraction versus repulsion (Semmelhack and Wang, 2009).
In conclusion, our study identifies the first miR regulated by transcriptional activity of Pros that represses Pros target molecules. We propose that miR-mediated regulation of pleiotropic neural genes played a role in the divergence of sensory systems. Failure of miR-mediated fine-tuning of intrinsically noisy genetic pathways could have created opportunities for nervous system divergence. Thus, this mode of activity restriction presents an attractive alternative to enhancer-mediated divergence of protein expression during evolution.
Footnotes
This work was supported by an Emmy-Noether grant of the German Research Foundation and the career development Award of the Human Frontiers Science Organization to I.G.K. The Max Planck Society provided further support through funds to I.G.K. We are very grateful to L. Zipursky and P. Cayirlioglu Volkan for discussions and sharing data in the earlier stages of this work. We thank J. F. Ferveur, A. Brand, C. Doe, T. Cook, S. Cohen, T. Suzuki, and F. Matsuzuki for providing important reagents and fly stocks. We acknowledge the Bloomington, Kyoto, and VDRC stock centers for providing fly stocks. We thank R. Klein, T. Suzuki, F. Schnorrer, and members of the laboratories for helpful discussions and comments on this manuscript.
The prosIG2227 mutant was discovered as part of a screen that was performed by I.C.G.K. and P. Cayirlioglu Volkan while they were postdoctoral fellows in the Zipursky lab.
The authors declare no financial conflicts interest.
References
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Biryukova I, Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 2009;327:487–496. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GS, Gunning D, Lai EC, Zipursky SL. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 2008;319:1256–1260. doi: 10.1126/science.1149483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Clyne PJ, Carlson JR. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Aoki T, Yoda Y, Kimura K, Hama C. Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat Neurosci. 2007;10:153–160. doi: 10.1038/nn1832. [DOI] [PubMed] [Google Scholar]
- Ghysen A. The developmental biology of neural connectivity. Int J Dev Biol. 1992;36:47–58. [PubMed] [Google Scholar]
- Gibson G. Genetics, ecology and behaviour of anophelines. Ciba Found Symp. 1996;200:22–37; discussion 37–47. [PubMed] [Google Scholar]
- Grant AJ, Wigton BE, Aghajanian JG, O'Connell RJ. Electrophysiological responses of receptor neurons in mosquito maxillary palp sensilla to carbon dioxide. J Comp Physiol A. 1995;177:389–396. doi: 10.1007/BF00187475. [DOI] [PubMed] [Google Scholar]
- Grosjean Y, Balakireva M, Dartevelle L, Ferveur JF. PGal4 excision reveals the pleiotropic effects of Voila, a Drosophila locus that affects development and courtship behaviour. Genet Res. 2001;77:239–250. doi: 10.1017/s0016672301005006. [DOI] [PubMed] [Google Scholar]
- Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri D, Sen A, Reddy GV, Rodrigues V. Sense organ identity in the Drosophila antenna is specified by the expression of the proneural gene atonal. Mech Dev. 2000;99:101–111. doi: 10.1016/s0925-4773(00)00487-1. [DOI] [PubMed] [Google Scholar]
- Jones WD. MicroRNA mutant turns back the evolutionary clock for fly olfaction. Bioessays. 2008;30:621–623. doi: 10.1002/bies.20780. [DOI] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Kuzin A, Brody T, Moore AW, Odenwald WF. Nerfin-1 is required for early axon guidance decisions in the developing Drosophila CNS. Dev Biol. 2005;277:347–365. doi: 10.1016/j.ydbio.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Orgogozo V. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev Biol. 2004;269:1–17. doi: 10.1016/j.ydbio.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Lebreton G, Faucher C, Cribbs DL, Benassayag C. Timing of Wingless signalling distinguishes maxillary and antennal identities in Drosophila melanogaster. Development. 2008;135:2301–2309. doi: 10.1242/dev.017053. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Liu SP, Fu RH, Yu HH, Li KW, Tsai CH, Shyu WC, Lin SZ. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplant. 2009;18:1039–1045. doi: 10.3727/096368909X471224. [DOI] [PubMed] [Google Scholar]
- Lu T, Qiu YT, Wang G, Kwon JY, Rutzler M, Kwon HW, Pitts RJ, van Loon JJ, Takken W, Carlson JR, Zwiebel LJ. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr Biol. 2007;17:1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning L, Doe CQ. Prospero distinguishes sibling cell fate without asymmetric localization in the Drosophila adult external sense organ lineage. Development. 1999;126:2063–2071. doi: 10.1242/dev.126.10.2063. [DOI] [PubMed] [Google Scholar]
- Markitantova YV, Makariev EO, Pavlova GV, Zinovieva RD, Mitashov VI. Location of the Prox1 gene expression during newt lens and retina regeneration. Dokl Biol Sci. 2003;391:361–364. doi: 10.1023/a:1025119121348. [DOI] [PubMed] [Google Scholar]
- Ramdya P, Benton R. Evolving olfactory systems on the fly. Trends Genet. 2010;26:307–316. doi: 10.1016/j.tig.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Rodrigues V. Sibling cell fate in the Drosophila adult external sense organ lineage is specified by prospero function, which is regulated by Numb and Notch. Development. 1999;126:2083–2092. doi: 10.1242/dev.126.10.2083. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Semmelhack JL, Wang JW. Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature. 2009;459:218–223. doi: 10.1038/nature07983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Reddy GV, Rodrigues V. Combinatorial expression of Prospero, Seven-up, and Elav identifies progenitor cell types during sense-organ differentiation in the Drosophila antenna. Dev Biol. 2003;254:79–92. doi: 10.1016/s0012-1606(02)00021-0. [DOI] [PubMed] [Google Scholar]
- Simon F, Fichelson P, Gho M, Audibert A. Notch and Prospero repress proliferation following cyclin E overexpression in the Drosophila bristle lineage. PLoS Genet. 2009;5:e1000594. doi: 10.1371/journal.pgen.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh GS, Wong AM, Hergarden AC, Wang JW, Simon AF, Benzer S, Axel R, Anderson DJ. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431:854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- Sweeney LB, Couto A, Chou YH, Berdnik D, Dickson BJ, Luo L, Komiyama T. Temporal target restriction of olfactory receptor neurons by Semaphorin-1a/PlexinA-mediated axon-axon interactions. Neuron. 2007;53:185–200. doi: 10.1016/j.neuron.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Yang DJ, Chung YJ, Lee SJ, Park SY, Pyo JH, Ha NC, Yoo MA, Park BJ. Slug, mammalian homologue gene of Drosophila escargot promotes neuronal differentiation through suppression of HEB/daughterless. Cell Cycle. 2010;9:2789–2802. [PubMed] [Google Scholar]