Abstract
Sleep is a unique physiological state, which is behaviorally defined, and is broadly conserved across species from mammals to invertebrates such as insects. Because of the experimental accessibility provided by various novel animal models including the fruit fly, Drosophila melanogaster, there have been significant advances in the understanding of sleep. Although the physiological functions of sleep have not been fully elucidated, accumulating evidence indicates that sleep is necessary to maintain the plasticity of neuronal circuits and, hence, is essential in learning and memory. Calcineurin (Cn) is a heterodimeric phosphatase composed of CnA and CnB subunits and known to function in memory consolidation in the mammalian brain, but its neurological functions in the fruit fly are largely unknown. Here, we show that Cn is an important regulator of sleep in Drosophila. A pan-neuronal RNA interference-mediated knockdown of Cn expression resulted in sleep loss, whereas misexpression of the constitutively active form of a CnA protein led to increased sleep. Furthermore, CnA knockdown also impaired the retention of aversive olfactory memory. These results indicate a role for Cn and calcium-dependent signal transduction in sleep and memory regulation and may bring insight into the relationship between them.
Introduction
Sleep has long been studied and described in mammalian species, but is now generally accepted as a universal neurological function experienced by a wide variety of species from humans, fish, lobsters, to insects (Mendoza-Angeles et al., 2007; Cirelli and Tononi, 2008; Zimmerman et al., 2008; Cirelli, 2009). Characteristics of sleep in the fruit fly, Drosophila melanogaster, are similar to those in mammalian species not only in terms of behavioral properties, such as circadian and homeostatic regulation and the changes in arousal threshold, but also in molecular and genetic properties (Hendricks et al., 2000; Shaw et al., 2000). One of the most striking findings was that dopamine is essential for maintaining wakefulness in Drosophila, like in mammals (Andretic et al., 2005; Kume et al., 2005; Kume, 2006). Thus, sleep and the circadian rhythm that regulates it are fundamental functions that appear to be evolutionarily conserved. Insects and mammals share similar molecular mechanisms generating circadian rhythm, the components of which are called clock genes. Clock genes such as the Drosophila period and mouse/human Per genes are highly homologous. Although several behavioral and neurological studies have pointed out that sleep functions in memory consolidation, elimination, and maintenance in humans and other mammals (Diekelmann and Born, 2010), the physiological basis of the involvement of sleep remains unclear. However, with the availability of novel animal models for sleep including remote species, there is increasing evidence indicating that sleep is required for plasticity of neuronal function and is essential in learning and memory in both vertebrates and invertebrates (Cirelli and Tononi, 2008; Seugnet et al., 2008; Donlea et al., 2009; Gilestro et al., 2009).
Calcineurin (Cn), also termed protein phosphatase 2B, is a heterodimeric Ca2+/calmodulin-dependent serine/threonine protein phosphatase. It is composed of an ∼60 kDa catalytic subunit, CnA, and a 19 kDa regulatory subunit, CnB, and is highly enriched in mammalian brains. A variety of proteins, including calcium channels, α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptors, N-methyl-d-aspartic acid (NMDA) receptors, synapsin I, dopamine- and cAMP-regulated phosphoprotein-32 (DARPP-32), and cAMP response element-binding protein (CREB), is dephosphorylated by Cn-mediated pathway and thus modulates neuronal plasticity (Greengard et al., 1999; Winder and Sweatt, 2001; Groth et al., 2003). Forebrain-specific CnB (CNB1) knock-out mice show impaired neuronal plasticity and working memory deficit (Zeng et al., 2001). Although no sleep-related phenotypes have been linked to Cn in mammals so far, Cn was uncovered as one of the sleep-induced genes in the cerebral cortex of rats (Cirelli et al., 2004).
In the present study, we investigated the role of Cn in the regulation of sleep and memory in Drosophila melanogaster using loss-of-function and gain-of-function approaches and examined the molecular link between memory and sleep.
Materials and Methods
Fly strains and conditions.
Flies were cultured at 25°C in 50–60% relative humidity on a conventional corn meal, yeast, and glucose agar medium. They were maintained under 12 h light (zeitgeber time, ZT 0–12) followed by 12 h dark (ZT 12–24) cycles defined as the LD condition. Transgenic lines expressing RNAi in the following genes were obtained from the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al., 2007): CanA-14F (Transformant ID: 30105), Pp2B-14D (46873), CanA1 (49986), CanB (21611), and CanB2 (28762). The UAS-Dicer-2 flies and the w1118, which is the genetic background of the RNAi lines, were also from VDRC. The tubulin-Gal80ts (tub-Gal80ts, Stock number: 7108) and elav-Gal4 (elavC155: 458) lines were from the Bloomington Drosophila Stock Center, Indiana University. NP6106 (Stock number: 105129) and UAS-mCD8::GFP (108068) lines were from the Drosophila Genetics Resource Center, Kyoto Institute of Technology, Japan. To express the full-length or the constitutively active forms of CanA-14F proteins with a C-terminal Myc tag, the corresponding cDNA fragments were generated by reverse transctiption-polymerase chain reaction (RT-PCR) as follows using total RNA from heads of adult w1118 flies as a template. A 1814 bp BglII-XbaI fragment containing the coding sequence of CanA-14F and a Myc-encoding sequence immediately upstream of the translational stop codon (CanA-14F-myc) was amplified by PCR with the following primers: 14F-F1, 5′-GCGCAGATCT ATCGGGCAAC CATGTCTTCG-3′; and 14F-R1, 5′-GCTCTAGATT ACAGATCCTC CTCGGAGATC AGCTTCTGCT CTCCATTGAT TGAATTCGCA TTTG-3′. Similarly, a 1454 bp BglII-XbaI fragment encoding the CanA-14F protein truncated at amino acid residue 465 followed by Myc-tag (CanA-14Fact-myc) was amplified by PCR with the following primers: 14F-F1 described above and 14F-R2, 5′-GCTCTAGATT ACAGATCCTC CTCGGAGATC AGCTTCTGCT CCTCGTCGTC GGAGAGTGGC T-3′. Each PCR fragment was digested with BglII and XbaI and cloned into the pUAST vector. Selected clones were validated by sequencing the insert DNA. UAS-CanA-14F-myc and UAS-CanA-14Fact-myc transgenic flies were generated through transformation of w1118 flies following a standard microinjection method (Genetic Services). RNAi-expressing flies and control flies were obtained by crossing the pan-neuronal Gal4 driver elav-Gal4 with the UAS-Dicer-2 on the second chromosome to each of the RNAi lines or w1118, respectively. The elav-Gal4 driver was used in combination with the tub-Gal80ts on the second chromosome to conditionally express the full-length and the constitutively active form of Myc-tagged CanA-14F proteins. Unless otherwise noted, 2- to 4-d-old posteclosion male flies were used in this study.
Measurements of transcript levels.
The efficiency and specificity of Cn RNAi were examined by RT-PCR and quantitative RT-PCR (qPCR). For RT-PCR analysis, flies expressing an RNAi or control flies were collected from each culture vial at ZT 6. Total RNA was extracted from 10 to 20 heads by using RNeasy Micro Kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized from the total RNA using oligo (dT)20 primer and ReverTra Ace reverse transcriptase (Toyobo) and used as PCR template. The primers were 5′-CCCCCAAAAT GGAAGCTAAT-3′ and 5′-TGGCAACTGC TTCGTGTCTA-3′ for CanA-14F, 5′-CGGAAAGAGC TAATTGAAAT CC-3′ and 5′-GCTGCTAAAT GTTTGGAACA GA-3′ for Pp2B-14D, 5′-AAGTTCCCAT AGGGGGTGAA G-3′ and 5′-CTGACCTTAC CCTAACGAGG TG-3′ for CanA1, 5′-TGCTGTCGAG CGAGTAGAGA-3′ and 5′-ATGCTGGCCT TTGGTTACTT-3′ for CanB, 5′-GGCGAGCCAG TTCAGTTAGT-3′ and 5′-GGCGCATTAT TCCACTTTTC-3′ for CanB2, 5′-GAAGAAGCGC ACCAAGCACT-3′ and 5′-TTGAATCCGG TGGGCAGCAT-3′ for RpL32 (rp49), and 5′-TGGTACGACA ACGAGTTTGG-3′ and 5′-TTTCAGGCCG TTTCTGAAGT-3′ for GAPDH2. Densitometric analysis was performed with the ImageJ 1.43 software (National Institutes of Health). The mRNA levels of each Cn and of RpL32 were first normalized by those of a housekeeping gene, GAPDH2, in all the RNAi and control flies. Then, the values of RNAi flies were divided by those of control flies and expressed as a percentage. For qPCR analysis, flies expressing an RNAi or control flies were collected from each of three culture vials at ZT 6 for biological replicates. Total RNA was extracted from 30 to 100 heads using TRIzol (Invitrogen) according to the manufacturer's instructions. RT reactions were performed as described above. The cDNA was used for qPCR using THUNDERBIRD SYBR qPCR Mix (Toyobo). The primers were 5′-GAATACCACG GACGAATCAG G-3′ and 5′-GACCTTCCCA CCCGTAAGAG-3′ for CanA-14F, 5′-GGCGAGGTCT GAATTGGTCT-3′ and 5′-TAAGGTTCCG TTCGGTTGGT-3′ for Pp2B-14D, 5′-ACGAGCAGGA CGACTGTGAA-3′ and 5′-AAATCAGTGT TGAGCAGGCA AG-3′ for CanA1, 5′-TCTGTGAGGG CGTTTGAGG-3′ and 5′-TCATTGCATT CTGTGGATGA TAGAG′ for CanB, and 5′-ACGTCGGCAT CCTTCGATT-3′ and 5′-GCTCGCCAAA GGCTCATAAC-3′ for CanB2. The primer sets for RpL32 (rp49) and GAPDH2 were same as above. Before qPCR, all the primer sets were confirmed to yield one major band corresponding to each targeted mRNA on the gel when used in conventional RT-PCR. Each expression level was first normalized to GAPDH2. Then, the values were normalized to the average of independent control samples, which was set at 100%. In conditional misexpression of CanA-14F-myc and CanA-14Fact-myc under control of the TARGET (temporal and regional gene expression targeting) system (McGuire et al., 2003), the induction of Cn transgene expression was assessed by RT-PCR. Total RNA was extracted from 10 to 20 heads of Cn-misexpressing male flies collected at the permissive temperature (19°C) or the restrictive temperature (30°C) by using RNeasy Micro Kit. cDNA was synthesized from the total RNA as described above and used as PCR template. RT-PCR was performed using the primers forward 5′-CGACCACCAA GTATCTGTTC C-3′ and reverse 5′-CCTCGGAGAT CAGCTTCTGC-3′ to amplify Cn transgene fragments containing the Myc-encoding sequence. GAPDH2 expression levels were used as an internal control.
Locomotor activity and sleep analysis.
Flies were placed individually in glass tubes (length, 65 mm; inside diameter, 3 mm) containing food at one end and were entrained for at least 3 d to LD conditions before transferring to constant dark (DD) conditions. Locomotor activity was monitored by recording infrared beam crossings by individual flies in 1 min bins using the Drosophila activity monitoring system (Trikinetics). Data were collected continuously for 3 d under either LD or DD conditions. Based on previous reports (Hendricks et al., 2000; Shaw et al., 2000; Huber et al., 2004; Kume et al., 2005), sleep was defined as periods of inactivity lasting 5 min or longer. Total activity, total sleep, sleep-bout length, and sleep-bout number in LD and DD conditions were analyzed by a Microsoft Excel-based software as described previously (Kume et al., 2005) and averaged over 3 d for each condition. This software was used to scan activity data over 3 d in 1 min bins (i.e., a total of 4320 bins). Five or more continuous bins with the value of zero (lack of activity) were regarded as one sleep bout, and the bins composing sleep bouts were assigned to sleep. To quantify sleep, the number of bins assigned to sleep were counted and expressed as a percentage of the total bins. The number and the length of each sleep bout were also recorded, and the mean bout length of sleep was calculated.
Mechanical stimulation during sleep.
To examine responsiveness to stimuli during sleep, an automatic mechanical stimulator was developed by coupling the Drosophila Activity Monitor 2 (Trikinetics) with a small vortex mixer, the mini MIXER N-20M (Nissin) controlled by a custom-built software on a LabVIEW (National Instruments) platform. Flies were individually placed in glass tubes in the mechanical stimulator and maintained in DD followed by 3 d LD cycle. Mechanical stimuli were applied to the animals by vortexing at regular intervals, at circadian time (CT) 14, 16, 18, 20, and 22, and repeated for 2 d. Strengths of the stimuli were adjusted by changing power supply voltage and duration of action of the mixer. Typically, 3.9 V for 200 or 600 ms were used for weak or strong stimuli, respectively. Activity was monitored every 15 s and a lack of activity for longer than 5 min was considered as sleep. Any locomotor activity during the 1 min interval between 15 s to 75 s after a stimulus was scored as a response to the stimulus. Otherwise, it was scored as no response. We did not use activity data within 15 s after stimulation because when a fly is resting near the infrared beam, which is around the center of the glass tube, mechanical stimulation sometimes induced a few activity counts even though the fly was not actually awakened. Data from the flies that were not sleeping before the stimulus were excluded from the analysis. Spontaneous arousal was estimated based on the activity data from flies used for mechanical stimulation experiments but during the intervals between stimulations. These recordings were taken 30 min before each stimulus, at CT 13.5, 15.5, 17.5, 19.5, and 21.5. Spontaneous arousal was quantified as the number of the sleeping flies that started to move within 1 min after the above indicated time points and was expressed as a percentage of the total number of sleeping flies at that time.
Sleep deprivation.
Flies were placed individually in glass tubes in the automatic mechanical stimulator described above and maintained in DD. Mechanical stimuli (5.2 V for 10 s) were delivered at 5 min intervals for 12 h during the subjective night to deprive them of sleep. Locomotor activity during sleep deprivation was recorded every 1 min to calculate sleep loss for each fly. Only flies that were active >80% during sleep deprivation were analyzed. Sleep, as defined by lack of mobility for periods lasting ≥5 min, was calculated for each 2 h period. Undisturbed groups of each genotype were used to obtain corresponding baseline values.
Aversive odor memory.
Olfactory conditioning with two odors (4-methylcyclohexanol and 3-octanol) was performed as described previously (Tully and Quinn, 1985; Aso et al., 2010). Flies were trained by providing the conditioning stimulus of one odor (CS+) for 1 min along with 12 pulses of electric shocks (90 V DC). Subsequently, they received another odor (CS−) for 1 min but without electroshock. After a given retention time, the conditioned response of the trained flies was measured with a choice between CS+ and CS− for 2 min in a T-maze. If no flies moved to either arms of the T-maze or if a small number of flies were trapped in the middle compartment, it indicated that the flies did not choose either odor. Approximately 100 male flies for each genotype were examined in one experiment. A performance index was calculated by taking the mean preference of two groups of flies that had been trained reciprocally in terms of the two odors used as CS+ and CS−.
In situ hybridization.
RNA probes for visualizing CanA-14F, CanB, and CanB2 were generated as follows. EcoRI/XbaI fragments containing a portion of cDNA of CanA-14F, CanB, and CanB2 were PCR amplified using total RNA from heads of adult w1118 flies with the following primers: 5′-CCGAATTCCT CTAGGGATCC GGTTGGTC-3′ and 5′-CCTCTAGACA ACTGGCAACT GCTTCGT-3′ for CanA-14F, 5′-CCGAATTCGT ATAGCCGATT TTCCGTAATG AG-3′ and 5′-CCTCTAGACG AGAGATGAAC TTAAGTGTTA GGC-3′ for CanB, and 5′-CCGAATTCGC AACACGGACA TTCACAAG-3′ and 5′-CCTCTAGAGT AGGAGGCGCA TTATTCCA-3′ for CanB2. The PCR products were digested with EcoRI and XbaI and cloned into the pCS2+ plasmid vector. Selected clones were validated by restriction mapping and sequencing of the plasmid DNA. The latter was used as a template for synthesizing DIG-labeled RNA probes using a DIG RNA labeling kit (Roche Diagnostics). For the antisense probe, EcoRI-linearized plasmid and T7 RNA polymerase were used. For the sense probe, XbaI-linearized plasmid and SP6 RNA polymerase were used. Drosophila male heads were fixed overnight in 4% PFA in PBS at 4°C. The proboscis was removed to allow complete penetration of the fixative. After dehydration, heads were embedded in paraffin and 6-μm-thick sections were obtained. In situ hybridization using DIG-labeled RNA probes was performed following the procedure of Thimgan et al. (2006) with minor modifications. After hybridization for 20 h and washes, sections were incubated with anti-DIG antibody conjugated with alkaline phosphatase (Roche Diagnostics) diluted 1:2000, and the signals were detected by using 4-nitro blue tetrazolium chloride (NBT; Roche Diagnostics) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP; Roche Diagnostics) as substrates. Nuclear Fast Red (Vector Laboratories) was used for nuclear counterstaining.
Immunohistochemistry.
Whole-mount immunofluorescence staining of adult brains was performed according to the published methods (Wu and Luo, 2006). We used mouse anti-Calcineurin B antibody (clone CN-B1, Sigma-Aldrich) at 1:100 dilution and goat anti-mouse Alexa 568 (Invitrogen) at 1:200 dilution as secondary antibodies. To test the specificity of the anti-Calcineurin B antibody, immunostained brain tissues of control and CnB#2 RNAi flies were processed and imaged in parallel using identical settings on a Leica TCS SP2 confocal microscope.
Statistical analysis.
Data were analyzed as described in the figure legends using Microsoft Excel and the freely available statistical software package R (R Development Core Team, 2010, http://www.R-project.org/).
Results
Cn knockdown decreases sleep in Drosophila
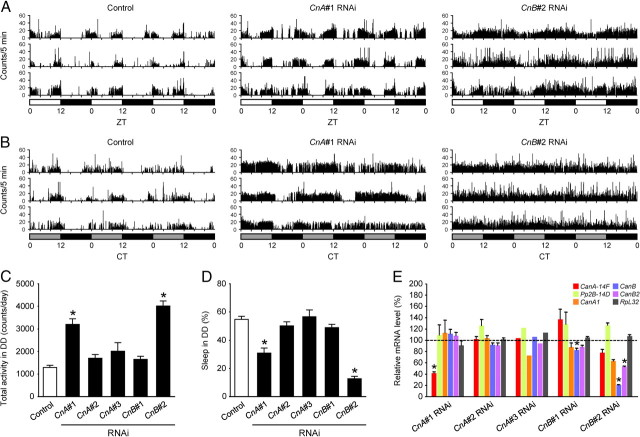
Drosophila melanogaster has five Cn genes encoding three isoforms of CnA subunits (CanA-14F, Pp2B-14D, and CanA1) and two isoforms of CnB subunits (CanB and CanB2) (Takeo et al., 2006). CanA-14F and Pp2B-14D are highly homologous to each other (82% protein sequence identity) and share 62–63% identity with CanA1. The protein sequences of CanB and CanB2 are 98% identical. To examine the potential roles of Cn in sleep regulation, we used RNAi knockdown of all three CnA and two CnB using the Gal4/upstream activating sequence system (UAS) with five independent UAS-RNAi transgenic lines (Dietzl et al., 2007) designated CnA#1, #2, #3, and CnB#1 and #2. elav-Gal4 driver was used to induce transgenic RNAi expression in all neurons, in combination with UAS-Dicer-2 to enhance transgenic RNAi effects (Dietzl et al., 2007). We entrained these RNAi-expressing flies for at least 3 d in LD conditions and recorded locomotor activity in LD or DD conditions for 3 d. Sleep was defined as no detectable activity for 5 min or longer, as described previously (Hendricks et al., 2000; Shaw et al., 2000; Huber et al., 2004; Kume et al., 2005). All the five RNAi-expressing fly lines were viable and showed no apparent abnormality, except that the wings of CnB#2 RNAi flies were occasionally crinkled. Therefore, only flies with normal wings were included in the behavioral analysis. Among the five RNAi-expressing fly lines tested, flies that expressed CnA#1 and CnB#2 RNAi displayed significant hyperactivity compared with control flies under both LD (Fig. 1A) and DD (Fig. 1B,C) conditions. On average, total daily activity increased significantly by two- to threefold in these two RNAi-expressing flies relative to control flies (Fig. 1C). In contrast, total daily sleep of CnA#1 and CnB#2 RNAi flies decreased to about half or one-fifth that of control flies, respectively (Fig. 1D). CnA#1 RNAi flies exhibited a substantial circadian rhythmicity of locomotor activity at an ∼24 h period in DD, whereas almost all CnB#2 RNAi flies showed very low-amplitude rhythms because of the high level of baseline activity at all times (Fig. 1B). The efficiency and targeting specificity of each RNAi transgene were assessed by both the quantification of RT-PCR products on conventional gel electrophoresis (Table 1) and qPCR (Fig. 1E) as described in Materials and Methods. The mRNA levels of the target genes were quantified and first normalized to those of GAPDH2. Then the values of each of the RNAi flies were divided by the values of control flies and expressed as the relative values to the control flies. As for the CnA subunit, CnA#1 specifically downregulated its intended target gene CanA-14F both by the gel (Table 1) and qPCR (Fig. 1E). CnA#2 failed to inhibit the expression of its target, Pp2B-14D. CnA#3 downregulated its target CanA1, to some degree, both by the gel and qPCR. As for the CnB subunit, CnB#1 downregulated CanB gene by the gel, and the effect was smaller but significant by qPCR. CnB#2 RNAi lines, targeting CanB2 gene, had overlapping effects on both CanB and CanB2 by the gel and qPCR. The subunit specificity of the CnA and CnB RNAi was evident from the fact that they did not significantly suppress the expression of CnB and CnA genes, respectively, and that of the internal control, ribosomal protein RpL32 gene. It was noted that CnB#2 RNAi line affected CanB more strongly than it did CanB2. Hence, we considered CnB#2 RNAi as a common CnB RNAi. Although the specificities of each subunit subtype were not very clear, because knocking down of either CnA or CnB subunit was effective without affecting other genes, these data indicated that sleep is affected by Cn knockdown and knocking down of CanA-14F alone has a significant effect.
Figure 1.
Sleep loss due to CnA or CnB knockdown. A, B, Locomotor activity profiles of three representative control (left panel), CnA#1 designed against CanA-14F (middle), and CnB#2 designed against CanB2 (right) RNAi-expressing flies by elav-Gal4;UAS-Dicer-2 driver for 3 d in LD (A) and DD conditions (B). White, gray, and black bars under the horizontal axis indicate day, subjective day, and night, respectively. Total daily activity (C) and total sleep (D) for control, three CnA (#2, #3, each designed against Pp2B-14D and CanA1) RNAi, and two CnB (#1 designed against CanB) RNAi-expressing fly lines in DD conditions. Asterisks indicate statistically significant differences compared with control as given by Tukey–Kramer HSD test for normally distributed data (p < 0.05). n = 32, 30, 30, 8, 32, and 40 (all males) for control, CnA#1, CnA#2, CnA#3, CnB#1 and CnB#2 RNAi flies, respectively. Data are presented as mean ± SEM. E, Efficiency and specificity of Cn gene knockdown. The expression levels of CanA-14F, Pp2B-14D, CanA1, CanB, CanB2, and RpL32 genes in the head of male flies expressing various Cn RNAi transgenes in all neurons are expressed as the relative values to the control flies. As described in Materials and Methods, each mRNA level was quantified by qPCR and first normalized to GAPDH2. Then, the values were normalized to the average of independent control samples, which were set at 100%. Statistical significance between control and RNAi flies: *p < 0.05, Student's t test, n = 3 for each group, except for CnA#3 RNAi (n = 1). Data are presented as mean ± SEM.
Table 1.
Efficiency and specificity of Cn gene knockdown
| Name | VDRC |
Relative expression levels of each gene (%)a |
||||||
|---|---|---|---|---|---|---|---|---|
| Transformant ID | Target gene | CanA-14F | Pp2B-14D | CanA1 | CanB | CanB2 | RpL32 | |
| CnA#1 RNAi | 30105 | CanA-14F | 36 | 94 | 83 | 78 | 97 | 75 |
| CnA#2 RNAi | 46873 | Pp2B-14D | 97 | 141 | 105 | 61 | 101 | 73 |
| CnA#3 RNAi | 49986 | CanA1 | 66 | 89 | 54 | 107 | 96 | 88 |
| CnB#1 RNAi | 21611 | CanB | 68 | 94 | 100 | 53 | 107 | 92 |
| CnB#2 RNAi | 28762 | CanB2 | 183 | 147 | 179 | 19 | 53 | 99 |
aThe expression levels of CanA-14F, Pp2B-14D, CanA1, CanB, CanB2, and RpL32 genes in the head of CnA#1, #2, #3, CnB#1, and #2 RNAi male flies with pan-neuronal expression of RNAi transgene indicated by the VDRC Transformant ID are expressed as the percentages to the control flies. As described in Materials and Methods, the expression levels of each gene were quantified by RT-PCR followed by densitometry and first normalized to GAPDH2. Then, the values were normalized to those of the control flies, which were set at 100%.
Cn knockdown decreases sleep-bout length
To examine in detail how Cn RNAi affected sleep, we analyzed sleep parameters such as total sleep, sleep-bout length, and sleep-bout number in CnA#1 and CnB#2 RNAi-expressing flies for subjective day and night under DD conditions. Similar to total daily sleep, total sleep for subjective day and subjective night in both RNAi flies decreased significantly compared with control flies (Fig. 2A). Nighttime sleep was more abundant than daytime sleep in control and CnA#1 RNAi flies. In contrast, CnB#2 RNAi flies showed a small diurnal variation in sleep amount. In control flies, sleep-bout length at nighttime was significantly longer than that at daytime, whereas this diurnal variation was totally abolished in flies expressing either of the RNAi transgenes (Fig. 2B). Compared with controls, CnA#1 RNAi flies had significantly shorter sleep-bout length only during nighttime. CnB#2 RNAi flies displayed a markedly shorter sleep-bout length compared with controls or CnA#1 RNAi flies regardless of night or day. In both RNAi-expressing fly lines, sleep-bout number was reduced significantly compared with control flies but only during the subjective day (Fig. 2C).
Figure 2.
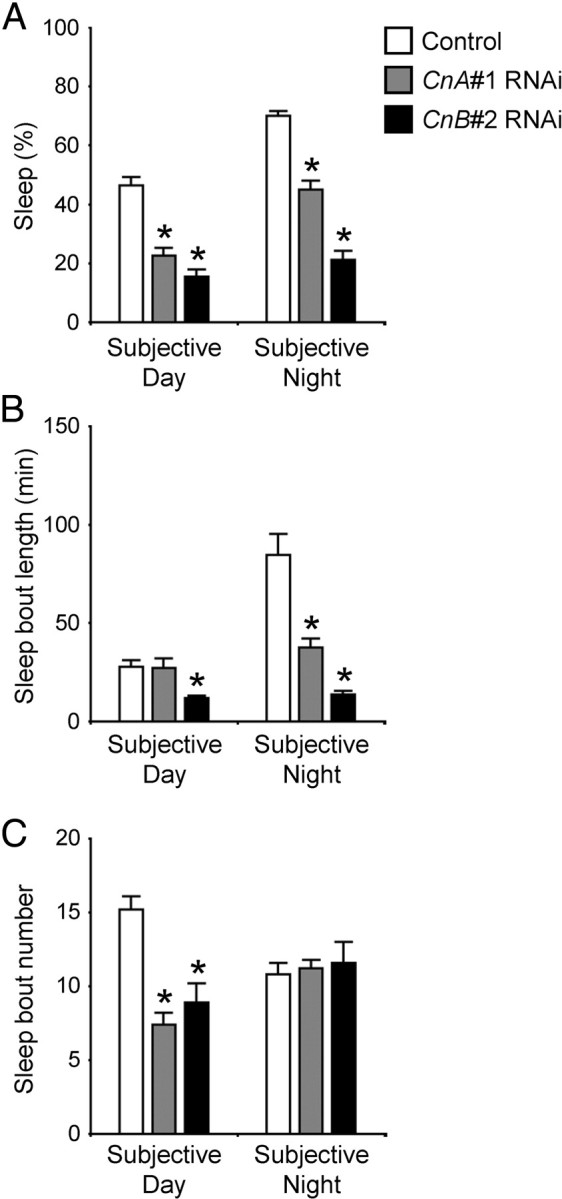
CnA and CnB knockdown flies exhibit a short sleep-bout length. Total sleep (A), sleep-bout length (B), and sleep-bout number (C) for control (white bars), CnA#1 RNAi (gray bars), and CnB#2 RNAi (black bars) during subjective day and subjective night in DD. The numbers of male flies tested are control = 57, CnA#1 RNAi = 57, CnB#2 RNAi = 27. Flies with no sleep during 12 h were excluded from the calculations of sleep-bout length in B. Asterisks indicate statistically significant differences from control determined by Tukey–Kramer HSD test for normally distributed data (p < 0.05). Data are presented as mean ± SEM.
Misexpression of constitutively active form of CanA-14F induces sleep
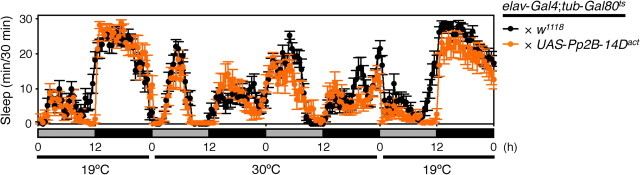
To further investigate the involvement of Cn in sleep regulation in Drosophila, we tested whether misexpression of Cn in all neurons affects sleep levels. We generated two transgenic lines: UAS-CanA-14F-myc and UAS-CanA-14Fact-myc. CanA-14F-myc encodes the full-length CanA-14F with a C-terminal Myc tag. CanA-14Fact-myc encodes a Myc-tagged constitutively active form of CanA-14F that lacks the C-terminal region containing the putative calmodulin-binding site and an autoinhibitory domain. This truncated CnA has been reported to function as a constitutively active form (Kincaid et al., 1988; Hashimoto et al., 1990; Brown et al., 1994). Pan-neuronal expression of CanA-14Fact-myc by elav-Gal4 throughout development caused lethality (data not shown). Therefore, we used the TARGET system to regulate the transgene expression temporally (McGuire et al., 2003). In this system, the transcriptional activator activity of Gal4 is suppressed by a temperature-sensitive variant of Gal80 protein (Gal80ts) under the control of tubulin promoter at 19°C. Temperature elevation to 30°C inactivates Gal80ts, thereby restoring Gal4 activity. Progeny from the control (elav-Gal4;tub-Gal80ts × w1118) and the experimental (elav-Gal4;tub-Gal80ts × UAS-CanA-14F-myc or elav-Gal4;tub-Gal80ts × UAS-CanA-14Fact-myc) crosses were raised at 19°C to adulthood. Adult progeny flies from the experimental group were viable and did not have visible developmental defects. Sleep was monitored in the two experimental lines and the control lines at 19°C for 2 d in LD and DD (Fig. 3A,B, respectively; only the second day is shown). At the beginning of Day 3, flies were transferred to 30°C for 2 d to induce UAS transgene expression and then returned to 19°C to determine whether the effects were reversible. The temperature shift to 30°C markedly decreased nighttime sleep in control flies and in those from the progeny of the elav-Gal4;tub-Gal80ts × UAS-CanA-14F-myc cross in both LD and DD, with no significant difference in their sleep levels before or after temperature shift (Fig. 3A–C). On the other hand, sleep levels of the elav-Gal4;tub-Gal80ts × UAS-CanA-14Fact-myc flies were significantly higher than those of the control groups throughout the day in both LD and DD conditions at 30°C. These effects on sleep were most pronounced during the nighttime, when the control flies and those expressing the full-length CanA-14F were highly active at 30°C. These sleep phenotypes of CanA-14F gain-of-function mutant flies were opposite those observed in RNAi-expressing flies. Sleep pattern of CanA-14Fact-misexpressing flies was largely unaffected after a return to 19°C, suggesting nondevelopmental effects on the neurons by Cn activity. We assessed the induction levels of the Cn constructs by RT-PCR on mRNA isolated from heads of flies at 30°C. At 19°C before temperature elevation, CanA-14F-myc and CanA-14Fact-myc expression levels were very low or undetectable, whereas an appreciable induction of both transgenes was detected 12 h after transfer to 30°C (Fig. 3D). To further confirm whether the decrease in sleep was due to a specific function of CanA-14F, we tested a constitutively active form of Pp2B-14D (Pp2B-14Dact) (Sullivan and Rubin, 2002; Takeo et al., 2006). Conditional misexpression of Pp2B-14Dact in all neurons by the TARGET system did not influence the sleep pattern (Fig. 4).
Figure 3.
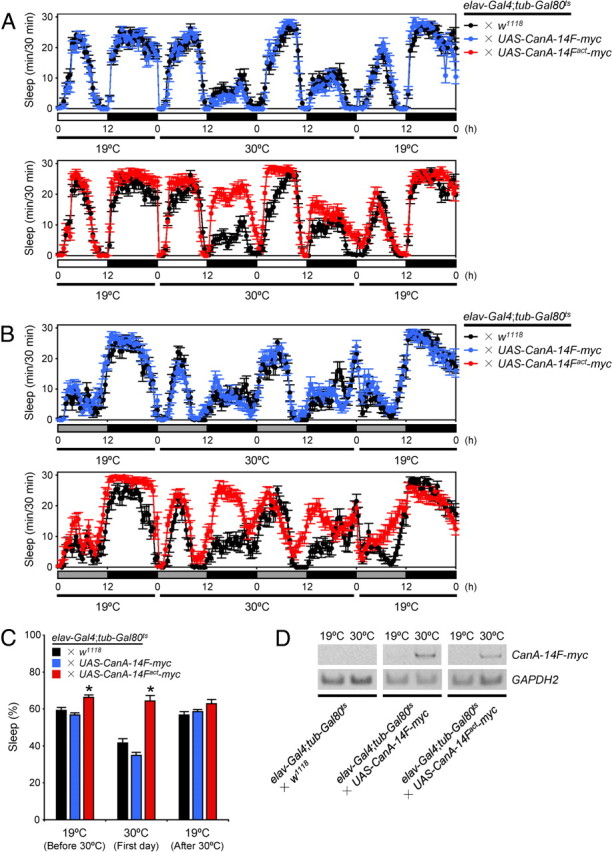
Misexpression of a constitutively active form of CanA-14F protein induces sleep. A, B, Sleep profiles in 30 min intervals for control (elav-Gal4;tub-Gal80ts × w1118, black circles, n = 17), male flies misexpressing CanA-14F-myc (elav-Gal4;tub-Gal80ts × UAS-CanA-14F-myc, blue circles, n = 23) or CanA-14Fact-myc (elav-Gal4;tub-Gal80ts × UAS-CanA-14Fact-myc, red circles, n = 24) at 30°C in 12 h light:12 h dark (LD) (A) and constant dark (DD) (B) conditions. Flies from all three groups were raised at 19°C until adulthood to suppress UAS transgene expression. Behavior was monitored over 2 d at 19°C (only the second day is shown) in LD and DD and shifted to 30°C for 2 d to allow UAS transgene expression, and then placed back to 19°C. C, Total daily sleep for controls (black), flies misexpressing CanA-14F-myc (blue), and CanA-14Fact-myc (red) at 19°C and 30°C in LD. Asterisks indicate statistically significant differences as determined by one-way ANOVA with Tukey–Kramer HSD post hoc test for normally distributed data (p < 0.05). Data are presented as mean ± SEM. D, Verification of UAS transgene expression in each misexpressed line at 30°C. Control male flies or those expressing CanA-14F-myc or CanA-14Fact-myc were collected at 12 h after the temperature shift from 19°C to 30°C at ZT 0. cDNA was prepared from total head RNA. The transgene fragments containing the Myc-encoding sequence were amplified by RT-PCR. GAPDH2 served as an internal control.
Figure 4.
Sleep is unaffected by misexpression of a constitutively active form of Pp2B–14D protein. Sleep profiles were recorded under the same condition described in Figure 3 for control flies (elav-Gal4;tub-Gal80ts × w1118, black circles, n = 17) or male flies misexpressing Pp2B–14Dact (elav-Gal4;tub-Gal80ts × UAS-Pp2B–14Dact, orange circles, n = 14). Behavior was assayed for 2 d at 19°C (only the second day is shown) in DD and shifted to 30°C to allow UAS transgene expression for 2 d, and then placed back at 19°C. Data are presented as mean ± SEM.
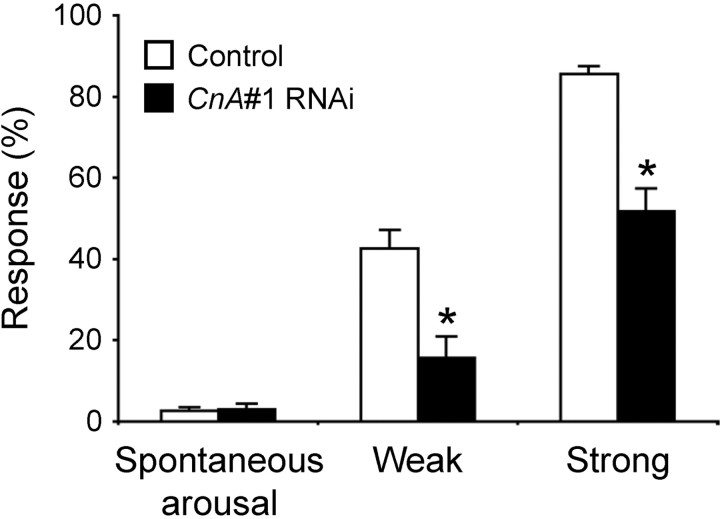
CnA#1 RNAi flies exhibit a decreased response to stimuli during sleep
Because previous studies showed that flies with short sleep phenotypes, including a dopamine transporter mutant, fumin, are hyperresponsive to stimuli, we tested arousal threshold of Cn knockdown flies by examining their response to mechanical stimuli of weak or strong intensities during the subjective night. The arousal threshold of the CnB#2 RNAi flies could not be determined quantitatively because they hardly slept (Fig. 1A,B). As shown in Figure 5, 16% of CnA#1 RNAi flies were aroused by a weak stimulus compared with 43% of control flies. In response to a strong stimulus, 52% of CnA#1 RNAi flies and 86% of controls were aroused. Thus, despite a short sleep phenotype, sleeping CnA#1 RNAi flies were hyporesponsive to mechanical stimuli compared with control flies. In contrast, spontaneous arousal during the subjective night was not significantly different between CnA#1 RNAi and control flies.
Figure 5.
Hyporesponsiveness to mechanical stimuli of sleeping CnA#1 RNAi flies. Mechanical stimuli were applied to the male flies and response rate (the ratio of the number of aroused flies to the number of the sleeping flies expressed as a percentage) was calculated. Compared with control flies (elav-Gal4;UAS-Dicer-2 × w1118, open bars), the CnA#1 RNAi flies (elav-Gal4;UAS-Dicer-2 × CnA#1, solid bars) showed a smaller response rate to both weak and strong stimuli (n = 19 or 20 for strongly stimulated CnA#1 RNAi flies and other stimulated groups, respectively). In contrast, there was no significant difference in the occurrence of spontaneous arousals (arousals without mechanical stimulations; see Materials and Methods for details) between control and CnA#1 RNAi flies (n = 40 for both genotypes) as calculated from behavioral data. Asterisks indicate statistically significant differences between control and CnA#1 RNAi flies (Student's t test; p < 0.05). Data are presented as mean ± SEM.
Homeostatic regulation of sleep is maintained in CnA#1 and CnB#2 RNAi flies
As in mammals, a clear rebound after sleep deprivation to compensate for sleep loss is observed in flies (Hendricks et al., 2000; Shaw et al., 2000). Because expression of CnA#1 and CnB#2 RNAi caused sleep loss and may have reduced sleep need (Figs. 1A,B, 2), we examined whether sleep rebound might be affected. To assess sleep homeostasis in CnA#1 and CnB#2 RNAi flies, we performed mechanical sleep deprivation of the flies for 12 h during subjective night and measured sleep rebound for 4 h following subjective day onset. Both RNAi flies exhibited a homeostatic response after sleep deprivation (Figs. 6, 7), indicating that they are able to compensate for acute sleep loss.
Figure 6.
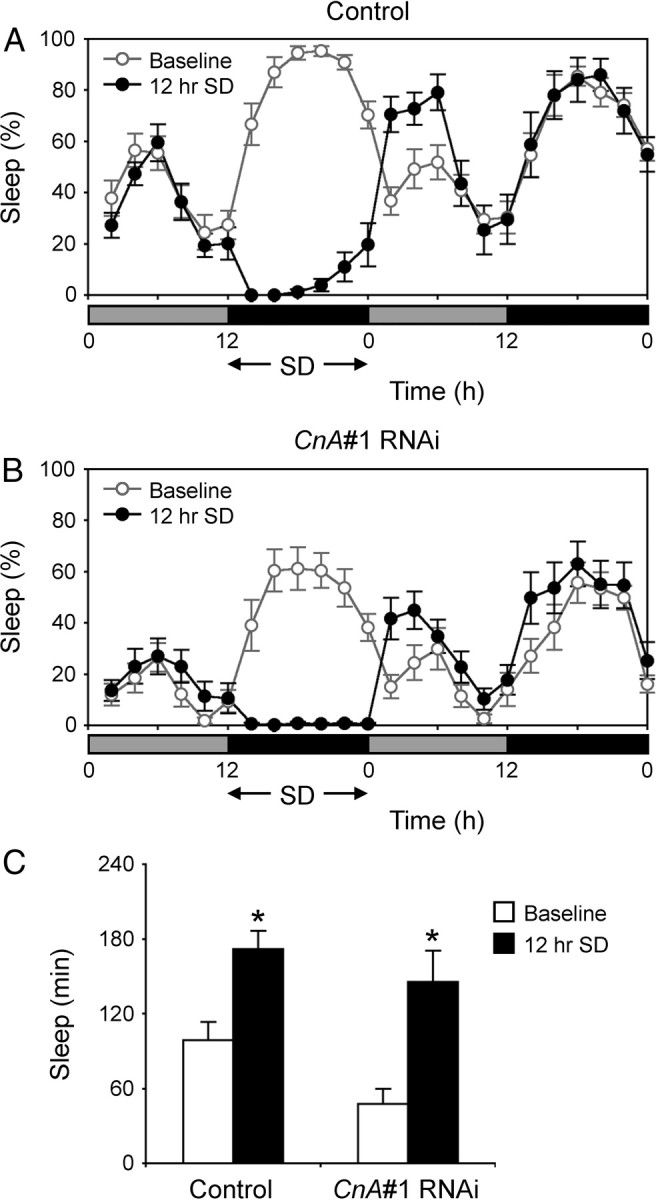
Homeostatic response to sleep deprivation in CnA#1 RNAi flies. A, B, Sleep profiles plotted in 2 h bins over 2 d under DD conditions for control flies (elav-Gal4;UAS-Dicer-2 × w1118) (A) or CnA#1 RNAi (elav-Gal4;UAS-Dicer-2 × CnA#1) male flies (B). Gray and black bars under the horizontal axis indicate subjective day and night, respectively. Twelve hours of sleep deprivation (SD) was started at CT 12. Only flies that were active >80% of the time during SD were analyzed. The values for baseline (open circles) and sleep-deprived siblings (closed circles) are the group average for 2 h (n = 11–16). C, The amount of sleep during the first 4 h of baseline (open bars) and recovery (solid bars). Significant sleep rebound was observed in both strains (Student's t test; *p < 0.01). Data are presented as mean ± SEM.
Figure 7.
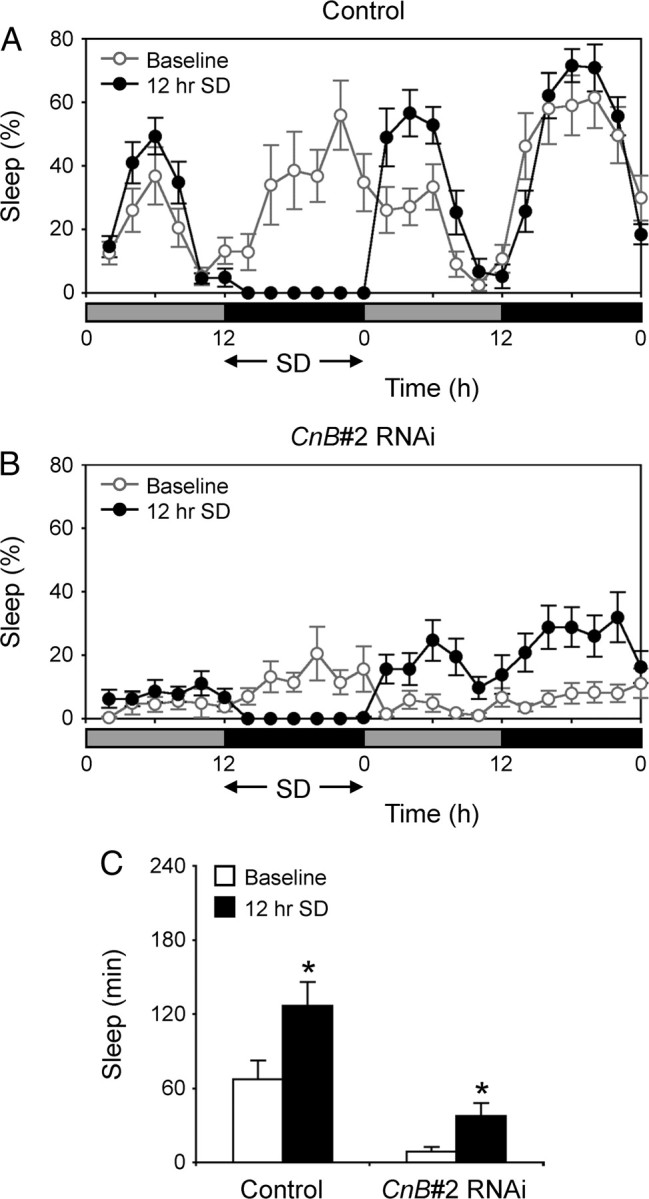
Homeostatic response to sleep deprivation in CnB#2 RNAi flies. A, B, Sleep profiles plotted in 2 h bins over 2 d under DD conditions for control flies (elav-Gal4;UAS-Dicer-2 × w1118) (A) or CnB#2 RNAi (elav-Gal4;UAS-Dicer-2 × CnB#2) male flies (B). The conditions are same as Figure 6 (n = 11–12). C, The amount of sleep during the first 4 h of baseline (open bars) and recovery (solid bars). Significant sleep rebound was observed in both strains (Student's t test; *p < 0.01). Data are presented as mean ± SEM.
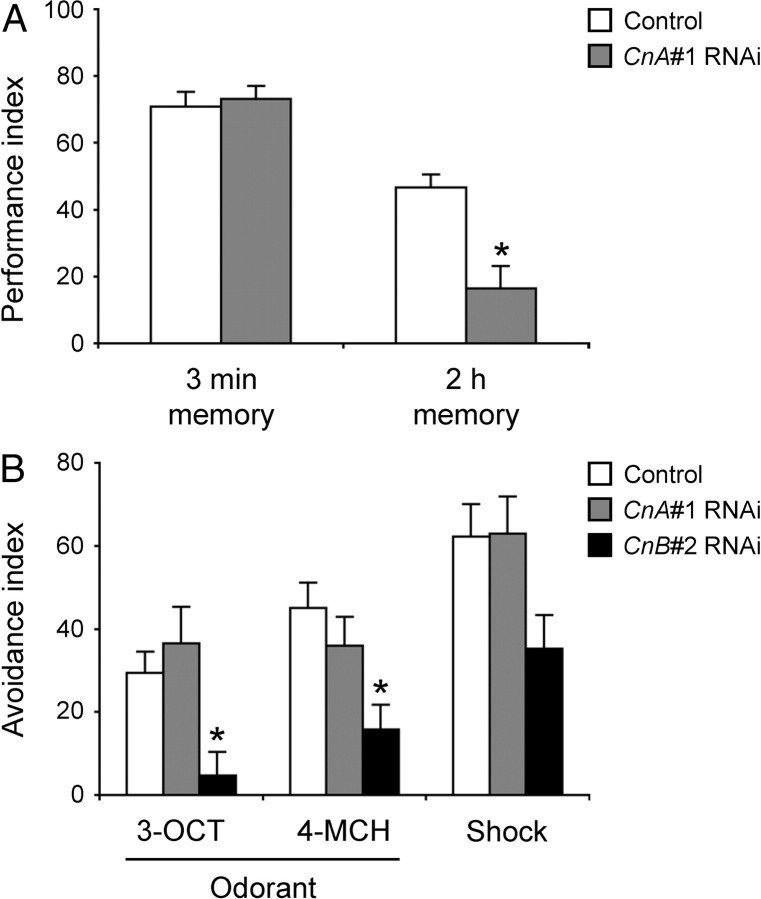
CnA#1 RNAi impairs memory retention
Cn has important functions in synaptic plasticity, learning, and memory in mammals. Therefore, we assessed the learning and memory ability of CnA#1 RNAi flies by using an olfactory aversive learning task. The immediate memory (3 min memory) of CnA#1 RNAi flies was indistinguishable from that of control flies (Fig. 8A). By contrast, the 2 h memory of the RNAi-expressing flies decreased significantly compared with control flies. Thus, a knockdown of CnA expression caused a defect in the maintenance of memory rather than the acquisition of memory. This defective memory retention is not attributable to abnormal sensorimotor responses because CnA#1 RNAi flies responded normally to both odors (3-octanol and 4-methylcyclohexanol) and to electrical shock (Fig. 8B). Because CnB#2 RNAi flies showed an impairment in both olfactory sensing and electrical shock response (Fig. 8B), we were unable to determine a specific role of CnB in learning and memory.
Figure 8.
CnA#1 RNAi flies show defective memory retention. A, The 3 min memory (immediate memory) and 2 h memory of control (elav-Gal4;UAS-Dicer-2 × w1118, white bars) and CnA#1 RNAi (elav-Gal4;UAS-Dicer-2 × CnA#1, gray bars) male flies were determined in an aversive olfactory conditioning test after training. B, Avoidance responses of control (white bars), CnA#1 RNAi (gray bars), and CnB#2 RNAi (elav-Gal4;UAS-Dicer-2 × CnB#2, black bars) male flies to 3-octanol (3-OCT), 4-methylcyclohexanol (4-MCH), and electric shock (Shock). Asterisks indicate statistically significant differences from control determined by Tukey–Kramer HSD test for normally distributed data (p < 0.05). Data are presented as mean ± SEM (n = 8–10 for each group).
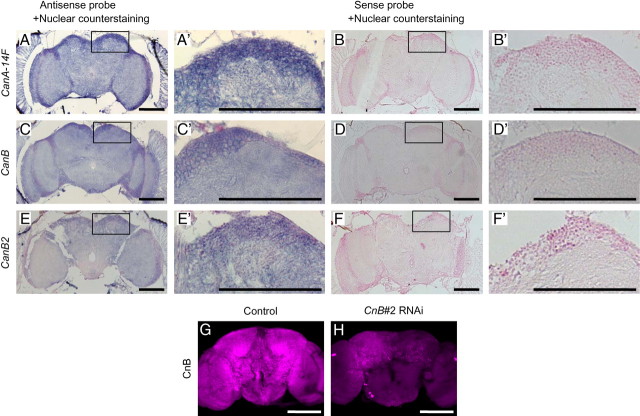
CanA-14F, CanB, and CanB2 are broadly expressed in the adult Drosophila brain
Based on their involvement in sleep regulation, we confirmed the expression of CanA-14F, CanB, and CanB2 genes in the adult Drosophila brain. In situ hybridization of specific antisense RNA probes to transcripts from adult head sections indicated widespread expression of all three genes in the brain, optic lobes, and retina (Fig. 9A,C,E). Furthermore, we used an enhancer-trap line, NP6106, that has a Gal4 (P{GawB} element) inserted at 103 bp upstream of the putative transcription start site of CanA-14F gene. NP6106 was crossed with a UAS-mCD8::GFP transgenic line, which expresses membrane-targeted GFP by Gal4. Progeny of this cross, expected to express GFP in CanA-14F-expressing cells, showed extensive expression of GFP in the adult brain (data not shown). As for the CnB genes, we used an antibody raised against mammalian CnB subunit, of which the target peptide sequence was sufficiently homologous to both Drosophila CanB and CanB2. CnB immunoreactivity was detected in the entire brain (Fig. 9G), which is consistent with in situ hybridization patterns of CanB and CanB2 genes (Fig. 9C,E). A dramatic reduction of CnB immunoreactivity in CnB#2 RNAi flies confirmed the specificity of the antibody to Drosophila CnB proteins (Fig. 9H). These results support the idea that Cn functions as a sleep regulator in the Drosophila brain.
Figure 9.
CanA-14F, CanB, and CanB2 genes are expressed ubiquitously in the adult brain. In situ hybridization and immunohistochemistry were performed as described in Materials and Methods. A, C, E, Representative frontal sections of the male fly head showing the brain, optic lobes, and retina labeled with DIG-labeled antisense RNA probes for CanA-14F (A), CanB (C), and CanB2 (E). B, D, F, Sense RNA probes were used as control. All tissue sections were counterstained with Nuclear Fast Red following in situ hybridization. The box areas are shown in higher magnification in A′–F′. Whole-mount adult male brains of control (elav-Gal4;UAS-Dicer-2 × w1118) (G) and CnB#2 RNAi (elav-Gal4;UAS-Dicer-2 × CnB#2) (H) were stained with an anti-Calcineurin B antibody. Representative maximum projection images of 1.5 μm confocal sections of the entire brain are shown. Scale bars, 100 μm.
Discussion
This study provides evidence that the Ca2+/calmodulin-dependent protein phosphatase Cn is involved in the regulation of sleep in flies. We showed that a pan-neuronal knockdown of either CnA or CnB, which constitutes a heterodimeric Cn functional protein, resulted in locomotor hyperactivity and sleep reduction (Fig. 1A–C). We also demonstrated that the conditional misexpression of a constitutively active form of CnA caused increased sleep (Fig. 3). We confirmed the expression of CanA-14F, CanB, and CanB2 genes in the brain. Together, our data indicate that Cn regulates the amount of sleep in Drosophila.
We examined three RNAi lines that were designed to knockdown the different CnA subtypes and found that only CnA#1 RNAi was efficient in specifically inhibiting transcription of its target gene, CanA-14F (Table 1 and Fig. 1E) and also caused a decrease in sleep when driven by pan-neuronal driver (Fig. 1). In the gain-of-function experiments, the constitutively active form, CanA-14Fact, affected sleep significantly, but Pp2B-14Dact did not (Figs. 3, 4). These data suggest that among the CnA subtypes, CanA-14F is the major contributor to sleep regulation in Drosophila. As for CnB subtype, only CnB#2 RNAi, which inhibited both CanB and CanB2 expression, significantly decreased sleep. Although RNAi data alone do not guarantee subtype specificities, these data clearly showed that Cn plays an important role in regulating the amount of sleep. We have developed targeted null mutants of all five Cn genes, and found that CanA-14F and CanB mutants exhibited strong alterations in sleep, although the other three also affected sleep to some extent (Nakai et al., in press). Those data extend and complement the conclusions drawn from the RNAi results presented here. In addition to causing an increase in sleep, expression of a constitutively active form of CanA-14F altered the circadian distribution profile of sleep (Fig. 3B), suggesting that CanA-14F is also involved in the association of sleep with the circadian clock. Although Cn has multiple targets in various tissues and its diverse roles have been extensively investigated, the specific functions and properties of the different CnA isoforms are largely unknown. In mammals too, the three isoforms of CnA (α, β, and γ) are highly homologous to each other. However, recent biochemical studies have revealed that differences in substrate specificities of the CnA isoforms may contribute to their specific physiological functions (Kilka et al., 2009). Thus, in the Drosophila brain substrates specific to CanA-14F may regulate sleep.
Interestingly, the involvement of Cn in locomotor regulation has been demonstrated in other animals. For example, in the nematode Caenorhabditis elegans, null mutants in CnB (cnb-1) display lethargic movement (Bandyopadhyay et al., 2002), whereas forebrain-specific Cn knock-out mice show increased locomotor activity (Miyakawa et al., 2003). Our findings have added another analogous function of Cn in locomotor activity in yet another species.
Several short sleep mutants have been described so far and they differ in their sleep and arousal properties. For example, we reported earlier that the dopamine transport-deficient fumin mutant showed hyperactivity and a lower arousal threshold (Kume et al., 2005). Other short sleep mutants, namely Shaker and Hyperkinetic, were also reported to have a lower arousal threshold (Cirelli et al., 2005; Bushey et al., 2007). These mutations result in hypersensitivity to weak stimuli while sleeping, which likely leads to a short and frequently interrupted sleep. Thus, it is suggested that their sleep may be shallower than that of the control flies. The sleep-deficient phenotype of CnA#1 RNAi flies described here is unique in that it is not associated with a lower arousal threshold (Fig. 5). It might be interpreted that the sleep of CnA#1 RNAi flies is short but deep. Loss of Drosophila homolog of fragile X mental retardation gene dFmr1 resulted in prolonged sleep (Inoue et al., 2002; Morales et al., 2002), whereas Bushey et al. (2009) reported that their hypermorph resulted in decreased sleep. Thus, they concluded that the amount of dFmr1 gene expression was inversely correlated with the amount of sleep and that dFmr1 protein levels affected the number but not the bout length of sleep episodes. Because the short sleep phenotype of CnA#1 and CnB#2 RNAi flies is largely due to the reduction in the duration of sleep bouts (Fig. 2), the basis of their short sleep may be due to another mechanism distinct from dFmr1 overexpression. In addition, CnA and CnB knockdown affects the amount of daily sleep without impairing the response to sleep deprivation and circadian rhythm (Figs. 6, 7). This suggests that these flies have a relatively normal regulatory mechanism of sleep. In addition, the sleep-reducing effect of Cn knockdown is more prominent during dark period than during light period (Fig. 1A). This was also true to the dopamine transport-deficient fumin mutant (Kume et al., 2005). The difference between daytime and nighttime sleep may bring more insight in sleep and arousal regulations.
The association between sleep and memory in Drosophila has recently been described in various reports. The dopamine transporter mutant fumin is a short sleeper and was also shown to have poor memory retention (Zhang et al., 2008). Likewise, sleep deprivation caused impairment in both aversive olfactory learning and memory (Li et al., 2009). Another short sleep mutant, Hyperkinetic, also had an impaired memory (Bushey et al., 2007). Seugnet et al. (2008) showed that sleep deprivation impaired aversive phototaxic suppression learning, which could be rescued by D1 dopamine receptor activation. Conversely, learning experiences increased sleep in normal flies, which was not observed in learning-deficient mutants (Donlea et al., 2009). Moreover, in the Drosophila brain, levels of synaptic component proteins decreased during sleep and increased during waking, suggesting that sleep is required for the maintenance of synapses (Gilestro et al., 2009). Our data of Cn provide not only another example of association between sleep and memory, but also a molecular clue to uncover a possible physiological relationship between them.
In Drosophila it is not clear which region(s) of the brain regulates sleep. The results of in situ hybridization and immunohistochemistry demonstrate the widespread expression of both CnA and CnB genes in the brain (Fig. 9). In situ hybridization patterns of CanB and CanB2 were consistent with those in the brains of Day 3 pupa (Gajewski et al., 2003). These results suggest several possibilities. The involvement of mushroom body (MB) in sleep regulation has been established by several lines of evidence. The ablation of MB results in short sleep, and the inhibition of MB function also alters sleep (Joiner et al., 2006; Pitman et al., 2006). The dopamine receptors DopR and DopR2 have been reported to be expressed in MB (Han et al., 1996; Kim et al., 2003), which may also play a role. Pars intercerebralis (PI) is another region, which was suggested by localization of epidermal growth factor (EGF) signaling (Foltenyi et al., 2007). Recently, signaling induced by the biogenic amine octopamine has been found to suppress sleep through insulin-secreting neurons of the PI (Crocker et al., 2010). Because Cn negatively regulates EGF receptor/Ras signaling pathway (Sullivan and Rubin, 2002), it is possible that Cn activity in PI is involved in regulating sleep. Although we have no data on what kind of molecules link Cn and sleep, one possibility lies in their relationship with dopamine. In the mammalian brain, Cn dephosphorylates dopamine- and cAMP-regulated phosphoprotein (DARPP-32) and inhibitor-1, the endogenous inhibitors of protein phosphatase 1 (PP1), which subsequently results in the activation of PP1 (Greengard et al., 1999; Winder and Sweatt, 2001; Groth et al., 2003).
Another possible mechanism is an association of Cn with the NMDA receptor. Cn is activated by Ca2+ influx through the NMDA receptor. The Drosophila NMDA receptor subunits dNR1 and dNR2 are widely expressed in the adult brain and play a role in memory formation (Xia et al., 2005; Wu et al., 2007). Based on our preliminary data, which indicate that dNR1 plays a possible role in sleep regulation (J. Tomita, unpublished results), it might be informative to examine the function of Cn in NMDA receptor signaling.
Together, our results showed that Cn is one of the key molecules regulating sleep and that it is also involved in memory function. Studying Cn signaling in Drosophila may bring a better insight into the physiological networks between them.
Note added in proof.
In Figure 8B, a reactivity of naive flies to the electric shock or odors was measured in the T-maze by giving a 1 min choice between tow tubes: one with 1.5 s pulses at every 5 s and another without shock, or one with odor and another without odor.
Footnotes
This research was supported by grants-in-aid from the Ministry of Education, Culture, Science, Sports and Technology, Japan (15300108, 17300105, K.K.), Global Center of Excellence (J.T., T.U., S.K.), Deutsche Forschungsgemeinschaft (Emmy–Noether Program and Germany–Japan Collaboration Initiative, H.T.), and the Max-Planck-Gesellschaft, Deutscher Akademischer Austausch Dienst (Y.A.). J.T. and T.U. were supported by Japanese Society for Promotion of Science Fellowships. We thank the Vienna Drosophila RNAi Center, the Bloomington Drosophila Stock Center, and the Drosophila Genetics Resource Center for fly stocks; the Gene Technology Center in Kumamoto University and the members of Kume lab for fruitful discussions and advice, especially M. Yamazaki, K. Takahama, and E. Sakamoto for technical assistance and K. Katsumoto for in situ hybridization.
References
- Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Aso Y, Siwanowicz I, Bräcker L, Ito K, Kitamoto T, Tanimoto H. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20:1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay J, Lee J, Lee J, Lee JI, Yu JR, Jee C, Cho JH, Jung S, Lee MH, Zannoni S, Singson A, Kim DH, Koo HS, Ahnn J. Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol Biol Cell. 2002;13:3281–3293. doi: 10.1091/mbc.E02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Chen MX, Cohen PT. Identification of a cDNA encoding a Drosophila calcium/calmodulin regulated protein phosphatase, which has its most abundant expression in the early embryo. FEBS Lett. 1994;339:124–128. doi: 10.1016/0014-5793(94)80398-6. [DOI] [PubMed] [Google Scholar]
- Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci. 2007;27:5384–5393. doi: 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci. 2009;29:1948–1961. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci. 2009;10:549–560. doi: 10.1038/nrn2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Is sleep essential? PLoS Biol. 2008;6:e216. doi: 10.1371/journal.pbio.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekelmann S, Born J. The memory function of sleep. Nat Rev Neurosci. 2010;11:114–126. doi: 10.1038/nrn2762. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–108. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- Gajewski K, Wang J, Molkentin JD, Chen EH, Olson EN, Schulz RA. Requirement of the calcineurin subunit gene canB2 for indirect flight muscle formation in Drosophila. Proc Natl Acad Sci U S A. 2003;100:1040–1045. doi: 10.1073/pnas.0337662100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro GF, Tononi G, Cirelli C. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science. 2009;324:109–112. doi: 10.1126/science.1166673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–447. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Groth RD, Dunbar RL, Mermelstein PG. Calcineurin regulation of neuronal plasticity. Biochem Biophys Res Commun. 2003;311:1159–1171. doi: 10.1016/j.bbrc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Han KA, Millar NS, Grotewiel MS, Davis RL. DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron. 1996;16:1127–1135. doi: 10.1016/s0896-6273(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Perrino BA, Soderling TR. Identification of an autoinhibitory domain in calcineurin. J Biol Chem. 1990;265:1924–1927. [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- Inoue S, Shimoda M, Nishinokubi I, Siomi MC, Okamura M, Nakamura A, Kobayashi S, Ishida N, Siomi H. A role for the Drosophila fragile X-related gene in circadian output. Curr Biol. 2002;12:1331–1335. doi: 10.1016/s0960-9822(02)01036-9. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kilka S, Erdmann F, Migdoll A, Fischer G, Weiwad M. The proline-rich N-terminal sequence of calcineurin Abeta determines substrate binding. Biochemistry. 2009;48:1900–1910. doi: 10.1021/bi8019355. [DOI] [PubMed] [Google Scholar]
- Kim YC, Lee HG, Seong CS, Han KA. Expression of a D1 dopamine receptor dDA1/DmDOP1 in the central nervous system of Drosophila melanogaster. Gene Expr Patterns. 2003;3:237–245. doi: 10.1016/s1567-133x(02)00098-4. [DOI] [PubMed] [Google Scholar]
- Kincaid RL, Nightingale MS, Martin BM. Characterization of a cDNA clone encoding the calmodulin-binding domain of mouse brain calcineurin. Proc Natl Acad Sci U S A. 1988;85:8983–8987. doi: 10.1073/pnas.85.23.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K. A Drosophila dopamine transporter mutant, fumin (fmn), is defective in arousal regulation. Sleep Biol Rhythms. 2006;4:263–273. [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yu F, Guo A. Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep. 2009;32:1417–1424. doi: 10.1093/sleep/32.11.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mendoza-Angeles K, Cabrera A, Hernández-Falcón J, Ramón F. Slow waves during sleep in crayfish: a time-frequency analysis. J Neurosci Methods. 2007;162:264–271. doi: 10.1016/j.jneumeth.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Miyakawa T, Leiter LM, Gerber DJ, Gainetdinov RR, Sotnikova TD, Zeng H, Caron MG, Tonegawa S. Conditional calcineurin knockout mice exhibit multiple abnormal behaviors related to schizophrenia. Proc Natl Acad Sci U S A. 2003;100:8987–8992. doi: 10.1073/pnas.1432926100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Horiuchi J, Tsuda M, Takeo S, Akahori S, Matsuo T, Kume K, Aigaki T. Calcineurin and its regulator Sra/DSCR1 are essential for sleep in Drosophila. J Neurosci. 2011 doi: 10.1523/JNEUROSCI.1337-11.2011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol. 2008;18:1110–1117. doi: 10.1016/j.cub.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Sullivan KM, Rubin GM. The Ca2+-calmodulin-activated protein phosphatase calcineurin negatively regulates EGF receptor signaling in Drosophila development. Genetics. 2002;161:183–193. doi: 10.1093/genetics/161.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S, Tsuda M, Akahori S, Matsuo T, Aigaki T. The calcineurin regulator Sra plays an essential role in female meiosis in Drosophila. Curr Biol. 2006;16:1435–1440. doi: 10.1016/j.cub.2006.05.058. [DOI] [PubMed] [Google Scholar]
- Thimgan MS, Berg JS, Stuart AE. Comparative sequence analysis and tissue localization of members of the SLC6 family of transporters in adult Drosophila melanogaster. J Exp Biol. 2006;209:3383–3404. doi: 10.1242/jeb.02328. [DOI] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Winder DG, Sweatt JD. Roles of serine/threonine phosphatases in hippocampal synaptic plasticity. Nat Rev Neurosci. 2001;2:461–474. doi: 10.1038/35081514. [DOI] [PubMed] [Google Scholar]
- Wu CL, Xia S, Fu TF, Wang H, Chen YH, Leong D, Chiang AS, Tully T. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci. 2007;10:1578–1586. doi: 10.1038/nn2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, Lin IR, Saitoe M, Tully T, Chiang AS. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15:603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Chattarji S, Barbarosie M, Rondi-Reig L, Philpot BD, Miyakawa T, Bear MF, Tonegawa S. Forebrain-specific calcineurin knockout selectively impairs bidirectional synaptic plasticity and working/episodic-like memory. Cell. 2001;107:617–629. doi: 10.1016/s0092-8674(01)00585-2. [DOI] [PubMed] [Google Scholar]
- Zhang S, Yin Y, Lu H, Guo A. Increased dopaminergic signaling impairs aversive olfactory memory retention in Drosophila. Biochem Biophys Res Commun. 2008;370:82–86. doi: 10.1016/j.bbrc.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Zimmerman JE, Naidoo N, Raizen DM, Pack AI. Conservation of sleep: insights from nonmammalian model systems. Trends Neurosci. 2008;31:371–376. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]