Abstract
Pitx3 is a critical homeodomain transcription factor for the proper development and survival of mesodiencephalic dopaminergic (mdDA) neurons in mammals. Several variants of this gene have been associated with human Parkinson's disease (PD), and lack of Pitx3 in mice causes the preferential loss of substantia nigra pars compacta (SNc) mdDA neurons that are most affected in PD. It is currently unclear how Pitx3 activity promotes the survival of SNc mdDA neurons and which factors act upstream and downstream of Pitx3 in this context. Here we show that a transient expression of glial cell line-derived neurotrophic factor (GDNF) in the murine ventral midbrain (VM) induces transcription of Pitx3 via NF-κB-mediated signaling, and that Pitx3 is in turn required for activating the expression of brain-derived neurotrophic factor (BDNF) in a rostrolateral (SNc) mdDA neuron subpopulation during embryogenesis. The loss of BDNF expression correlates with the increased apoptotic cell death of this mdDA neuronal subpopulation in Pitx3−/− mice, whereas treatment of VM cell cultures with BDNF augments the survival of the Pitx3−/− mdDA neurons. Most importantly, only BDNF but not GDNF protects mdDA neurons against 6-hydroxydopamine-induced cell death in the absence of Pitx3. As the feedforward regulation of GDNF, Pitx3, and BDNF expression also persists in the adult rodent brain, our data suggest that the disruption of the regulatory interaction between these three factors contributes to the loss of mdDA neurons in Pitx3−/− mutant mice and perhaps also in human PD.
Introduction
The majority of the brain's dopamine (DA) is synthesized by neurons located in two nuclei of the mammalian ventral midbrain (VM): the substantia nigra pars compacta (SNc), innervating predominantly the dorsolateral striatum, and the ventral tegmental area (VTA) projecting mainly to limbic and cortical areas of the brain (Björklund and Dunnett, 2007). The SNc neurons control the execution of voluntary movements, and their selective degeneration in Parkinson's disease (PD) is responsible for the characteristic motor symptoms of this disease (Dauer and Przedborski, 2003).
During development, mesodiencephalic DA (mdDA) precursors arise from progenitors located in the midbrain floor plate (Smidt and Burbach, 2007). It is assumed that SNc neurons derive from a rostrolateral and VTA neurons from a caudomedial mdDA precursor subpopulation (Smits et al., 2006). Several secreted and transcription factors are involved in the specification of the mdDA fate in neuroepithelial progenitors and in their differentiation into mature mdDA neurons, as well as in their survival throughout development and adulthood (Prakash and Wurst, 2006; Smidt and Burbach, 2007). The transcription factor Pitx3 has gained particular interest, as Pitx3 expression in the brain is restricted to mdDA neurons and Pitx3 mutant mice display a progressive and preferential loss of the SNc mdDA neurons during embryonic and postnatal development (van den Munckhof et al., 2003; Smidt et al., 2004). Pitx3 cooperates with the nuclear receptor Nurr1 to activate transcription of several genes encoding DA biosynthetic enzymes, transporters, and receptors (Jacobs et al., 2009). The gene encoding the retinoic acid (RA)-synthesizing enzyme Aldh1a1 is a target of Pitx3, but maternal RA complementation is only able to partially rescue SNc neurodegeneration in Pitx3 mutant mice (Jacobs et al., 2007). Pitx3 might therefore activate additional target genes to promote the survival of SNc mdDA neurons.
Glial cell line-derived neurotrophic factor (GDNF) and brain-derived neurotrophic factor (BDNF) were the first potent survival factors identified for mdDA neurons (Airaksinen and Saarma, 2002; Zuccato and Cattaneo, 2009). Previous reports indicated that GDNF is highly transcribed in the striatum when nigrostriatal innervation takes place, suggesting that GDNF is a target-derived neurotrophic factor for mdDA neurons (Golden et al., 1999; Oo et al., 2005). The Ret tyrosine kinase receptor and its GDNF family coreceptors (GFR) α1 and α2, however, are expressed strongly in the VM throughout the mdDA neurogenic period (Golden et al., 1999). Furthermore, conditional deletion of the BDNF gene during midbrain development results in a selective reduction of these neurons at birth (Baquet et al., 2005).
We have previously shown that Pitx3 activates BDNF expression in primary cell cultures (Li et al., 2009). In the present study, we demonstrate that GDNF, Pitx3, and BDNF are engaged in a feedforward regulatory pathway in the rodent VM during development and in adulthood. This feedforward interaction promotes the survival of a rostrolateral mdDA neuronal subset during embryogenesis and possibly also of adult and aging SNc neurons and protects mdDA neurons against neurotoxic insults in vitro, suggesting that it might also be relevant for the pathogenesis of PD.
Materials and Methods
Animals.
C57BL/6 mice were purchased from Charles River. Adult male and pregnant female Sprague Dawley rats were purchased from the Experimental Animal Center (Shanghai, China). Generation and genotyping of Pitx3+/GFP knock-in mice is described by Zhao et al. (2004). Mice carrying the floxed Ret allele and a transgene encoding the Cre recombinase driven by the dopamine transporter (Dat) promoter (Kramer et al., 2007) often undergo Cre-mediated excision in the germline; their progeny thus carries one Ret-null allele in all cells (Ret+/− mice). Ret+/− mice were intercrossed and their offspring was genotyped by PCR. Newborn Ret−/− pups lacked kidneys and died shortly after birth, thus confirming that these mice were Ret-null mutants. Collection of embryonic stages was done from timed pregnant females; noon of the day of vaginal plug detection was designated as embryonic day 0.5 (E0.5). Animal treatment was conducted in accordance with the Laboratory Animal Care Guidelines approved by Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) or under federal guidelines as approved by the HMGU Institutional Animal Care and Use Committee (Munich, Germany).
Radioactive in situ hybridization. Paraffin sections (8 μm) were processed for radioactive ([α-35S]UTP, GE Healthcare) in situ hybridization as described by Fischer et al. (2007). Riboprobes used were Pitx3 (Brodski et al., 2003), an 813 base pair (bp) fragment of mouse GDNF (bp 29–841; GenBank accession no. NM_010275.2), and a 545 bp fragment of mouse BDNF (bp 548–1092, GenBank accession no. NM_001048139.1) that detects all murine BDNF isoforms. Specificity of the GDNF and BDNF antisense riboprobes was tested by using the corresponding sense riboprobes in some experiments. Images were taken using bright- and dark-field optics on an Axioplan2 microscope or StemiSV6 stereomicroscope, AxioCam MRc camera, and Axiovision 4.6 software (Zeiss) and processed with Adobe Photoshop 7.0 or CS software (Adobe Systems).
Immunostaining and cell counting.
Immunostaining of cultured cells, paraffin sections and free-floating cryosections (40 μm) was performed as described previously (Brodski et al., 2003; Peng et al., 2007). Polyclonal rabbit antisera were directed against NF-κB (1:100; Cell Signaling Technology), BDNF (1:500 (Sc-546); Santa Cruz Biotechnology), Pitx3 (1:300; Zymed), and cleaved (activated) Caspase-3 (cCasp3, 1:100; Cell Signaling Technology). The immunostaining for cCasp3 on cultured cells and tissue sections was amplified using the Tyramide Signal Amplification kit (PerkinElmer) according to the manufacturer's instructions. Monoclonal mouse antibodies were directed against tyrosine hydroxylase (TH) at 1:6000 (Sigma) and 1:600 (MAB318, Millipore Bioscience Research Reagents). Polyclonal chicken antisera were directed against GFP (1:2000; Aves Labs). Fluorescent images were taken with an Axiovert 200M inverted microscope (Zeiss), and Olympus BX51 microscope (Olympus), or an LSM510 confocal microscope (Zeiss), and processed with Adobe Photoshop 7.0 or CS software.
TH+, Pitx3+, and DAPI+ cells in E14 rat VM cultures were counted in 10 random fields per well for six wells, and data were collected from three independent experiments. Pitx3+ cells were counted on every fourth serial coronal hemisection through the midbrain of E12.5 and on every fourth serial sagittal section through the midbrain of E14.5 Ret+/+, Ret+/− and Ret−/− embryos. GFP+ and cCasp3+ cells were counted on every fourth serial coronal hemisection through the midbrain of E12.5 Pitx3+/GFP and Pitx3GFP/GFP embryos. GFP+, TH+, cCasp3+, and DAPI+ cells in E11.5 Pitx3+/GFP and Pitx3GFP/GFP VM cultures were counted in 12 random fields per well, and data were collected from at least three independent experiments.
Primary VM cultures and neurotrophin treatments.
Primary VM cultures were prepared from E14 rat embryos or E11.5 Pitx3+/GFP and Pitx3GFP/GFP mouse embryos as described by Du et al. (2005). For GDNF treatments, E14 rat primary VM cells were treated after 7 days in vitro (DIV) with 10 ng/ml recombinant rat GDNF (Sigma) in 0.9% saline or with 0.9% saline alone (control) for 3, 24 or 48 h. In some cases, 10 μg/ml NF-κB inhibitor (SN50; Calbiochem) were added for 1 h to the culture medium before the GDNF treatment. Cells were harvested for immunostaining and RT-PCR assays after 3 or 24 h and for Western blot analyses after 2 d of GDNF treatment. For BDNF treatments, dissociated E11.5 Pitx3+/GFP or Pitx3GFP/GFP mouse VM cells were plated at a density of 1.4 × 105 cells/well and cultured in DMEM/F12 with 2% B27 supplement (Invitrogen) containing 20 ng/ml recombinant human BDNF (R & D Systems) in 0.1% bovine serum albumin (BSA) or 0.1% BSA alone. BDNF or vehicle (0.1% BSA) was added with each medium change every second day. Cells were fixed for immunostaining after 4 d.
siRNA treatments.
Three different mouse Pitx3-specific short hairpin RNA (shRNA) oligonucleotide pairs were designed and synthesized by Shanghai GeneChem. Downregulation of Pitx3 mRNA by these shRNAs was tested in SH-SY5Y cells stably expressing mouse Pitx3 (Peng et al., 2007). Under these conditions, one Pitx3-specific shRNA oligonucleotide (antisense: 5′-GCACGCCUCU-UUCAGCUAUdTdT-3′; sense: 5′-AUAGCUGAAAGAGGCGUGCdTdT-3′; targeting sequences around position 956 to 974 in Pitx3 exon 4, GenBank accession no.. NM_008852.4) resulted in ∼80% knock-down of Pitx3 mRNA (data not shown). A nonsilencing shRNA oligonucleotide, as indicated by the manufacturer (Shanghai GeneChem) (antisense: 5′-CGUGACACGUUCGGAGAAdTdT-3′; sense: 5′-UUCUCCGAA-CGUGUCACGUdTdT-3′) was used as control. E14 rat primary VM cells were transfected with Pitx3 or control shRNA oligonucleotides (40 pmol) after 7 DIV using Lipofectamine 2000 (Invitrogen) right after the addition of GDNF to the culture medium. Cells were harvested after 24 h (for RT-PCR assays) or 2 d (for Western blot analyses).
6-Hydroxydopamine treatments. Dissociated E11.5 Pitx3+/GFP or Pitx3GFP/GFP mouse VM cells were plated at a density of 1.4 × 105 cells/well and cultured in DMEM/F12 with 2% B27 supplement for 3 DIV. Cells were treated after 3 DIV with 20 ng/ml BDNF or 20 ng/ml recombinant human GDNF (R & D Systems) or with vehicle (0.1% BSA) alone 2 h before the addition of 10 μm 6-hydroxydopamine (6-OHDA; catalog no. H116, Sigma) or vehicle alone (PBS) to the culture medium. Cells were fixed 24 h after the addition of 6-OHDA for immunostaining.
Luciferase reporter assays.
A search for conserved NF-κB binding sites (BSs) within the Pitx3 promoter (−10kb to +1kb) and conserved Pitx3/Bicoid-like transcription factor BSs within the BDNF promoter region (−2.5kb to +0.5kb) was done using CONREAL (CONserved Regulatory Elements anchored ALignment; http://conreal.niob.knaw.nl/) and Gene2promoter (Genomatix) software, respectively. A 2514 bp Pitx3 promoter fragment (position −2425 to +89, GenBank accession no. NC_000085) and a 2616 bp BDNF promoter fragment (position −1930 to +686 relative to transcript variant 4, GenBank accession no. NM_001048142.1) were amplified from C57BL/6 mouse genomic DNA using the primer pairs Pitx3 forward, 5′-AAGCAAACCTTTTCTAAGGC-3′; Pitx3 reverse, 5′-TGTGGCGGCGGCGTTCT-3′; BDNF forward, 5′-TCTCGGATACCCATTTTTGC-3′; BDNF reverse, 5′-CCCAACAGCTGTCGCTCTAT-3′; and cloned into a pGL3 basic vector (Promega). Site-directed mutagenesis of two conserved Pitx3/Bicoid BSs within the BDNF promoter region was done using the primer 5′-CACCTGTCTGAGGTCTAGAAGAGAGGCTCCTTCTTC-3′ (BS2) and 5′-CCAAGGGAAGAGGACGACTTGCGCCATGGGAAAGCTTCATGCGTCCCGGGGATCC-3′ (BS9), and the QuickChange Multi Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions.
SH-SY5Y cells were transfected with 400 ng/well pGL3-Pitx3 promoter vector and 8 ng/well pRL-SV40 vector using Lipofectamine 2000 (Invitrogen). SN50 (10 μg/ml) was added 30 min before 10 ng/ml or 20 ng/ml GDNF was given to the medium. HEK293 cells were cotransfected with 200 ng/well pGL3-BDNF wild-type or mutant promoter vector, 400 ng/well pcDNA3.1-Pitx3 (Peng et al., 2007) or pcDNA3.1 alone, and 8 ng/well pRL-SV40 using Lipofectamine LTX and Plus Reagent (Invitrogen). Cells were lysed in Passive Lysis Buffer 24 h (BDNF promoter assays) or 36 h (Pitx3 promoter assays) post-transfection, and Firefly and Renilla Luciferase luminescence were measured in a Centro LB 960 luminometer (Berthold Technologies) using the Dual-Luciferase Reporter Assay system (Promega) according to the manufacturers' instructions. Firefly luminescence was normalized against Renilla luminescence for each well, and relative values (fold induction) were calculated by setting the normalized value of the control transfection as 1.
Semiquantitative RT-PCR and real-time PCR assays.
Semiquantitative RT-PCR was performed as reported in (Peng et al., 2007). PCR primers and conditions are provided in Table 1. For quantitative (q) PCR assays, 1 μl of a 1:3 diluted cDNA was amplified in a DNA Engine Opticon (MJ Research) together with 0.5× SYBR Green I (Roche Applied Science) and 5 pmol of each primer (Table 1). A standard curve was constructed using plasmid DNA containing the Pitx3 cDNA (from 102 to 107 molecules) (Peng et al., 2007). Signals from specific Pitx3 and BDNF PCR products were normalized against β-actin, and relative values were calculated by setting the normalized value of controls as 1. Samples from three independent experiments were analyzed in duplicate reactions for each cDNA, and all reactions were repeated more than twice.
Table 1.
Primers used for RT-PCR and qPCR experiments
| Gene | Forward primer/reverse primer | Length of product (bp) | Tm (°C)a | Cycles |
|---|---|---|---|---|
| Pitx3 RT-PCR (NM_019247) | 5′-GAGCACAGTGACTCGGAGAAGG-3′/5′-AAGGCGAACGGGAAGGTC-3′ | 413 | 60 | 28 |
| BDNF RT-PCR (NM_012513) | 5′-AGCGTGAATGGGCCCAGGGCA-3′/5′-TGTGACCGTCCCACCGGACA-3′ | 369 | 60 | 28 |
| GAPDH RT-PCR (NM_017008) | 5′-CCATGTTTGTGATGGGTGTGAACCA-3′/5′-GCCAGTGGATGCAGGGATGATGTTC-3′ | 251 | 58 | 22 |
| Pitx3 qPCR (NM_019247) | 5′-GAGTTTGGGCTGCTTGGTGAGGC-3′/5′-CCATGTTCTGGAAGCGGAGGGTGT-3′ | 92 | 60 | 35 |
| BDNF qPCR (NM_012513) | 5′-GATGCCGCAAACATGTCTATGA-3′/5′-TAATACTGTCACACACGCTCAGCTC-3 | 82 | 60 | 35 |
| Beta-actin qPCR (NM_031144) | 5′-CCCGAGGCTCTCTTCCAGCC-3′/5′-TAGAGGTCTTTACGGATGTCAACGT-3′ | 110 | 60 | 35 |
aTm, Melting temperature.
Western blot analyses.
Cells and brain tissues were processed as described by Peng et al. (2007). Antibodies used were rabbit anti-Pitx3 (1:500, Zymed), anti-BDNF (1:500, catalog no. sc-546, Santa Cruz Biotechnology), and anti-β-actin (1:2000, Sigma). Optical densities of the specific protein bands were quantified with an Image Analyzer (Quantity One-4.2.0; Bio-Rad).
Intrastriatal injections of GDNF.
Adult male rats (n = 5) were anesthetized with chloral hydrate (400 mg/kg, i.p.). Twenty microliters of a 15 ng/μl GDNF solution in 0.9% saline were injected into the right striatum (according to bregma: anterior-posterior, +0.7; medial-lateral, ±3.5; dorsal-ventral, −4.0) with a Hamilton syringe. Vehicle (0.9% saline) was injected into the contralateral striatum. The injection rate was 1 μl/min, and the cannula was left in place for another 10 min before it was retracted. After 48 h, animals were sacrificed by CO2 asphyxiation. For immunohistochemical analyses, animals were intracardially perfused with PBS followed by 4% paraformaldehyde (PFA), and brains were collected, postfixed in 4% PFA for 24 h, cryosectioned, and processed as described above. For Western blot analyses, brains were immediately removed and ventral midbrain tissue was dissected and processed as described before.
Statistics.
All values shown are mean ± SEM. Statistical significance between groups was assessed by paired t tests, independent samples t tests, or one-way ANOVA followed by post hoc S-N-K multiple comparisons using the SPSS 10.0 software (SPSS Inc.). A value of p < 0.05 was considered significant.
Results
Induction of Pitx3 expression by NF-κB-mediated GDNF/Ret signaling
To determine whether GDNF-mediated signaling is involved in the transcriptional activation of the Pitx3 gene during embryogenesis, we first compared the spatiotemporal expression patterns of GDNF and Pitx3 in the embryonic mouse VM. GDNF is expressed in the mantle zone (MZ) of the midbrain basal plate (ventrolateral midbrain) at E10.5, one day before the onset of Pitx3 transcription in this brain region (Fig. 1a,c,e). Pitx3 expression is detected at E11.5 in the MZ of the adjacent floor plate. At this stage, GDNF expression is restricted to a smaller domain in the midbrain basal plate (Fig. 1b,d,f), and becomes undetectable in the mouse VM at E12.5 (data not shown).
Figure 1.
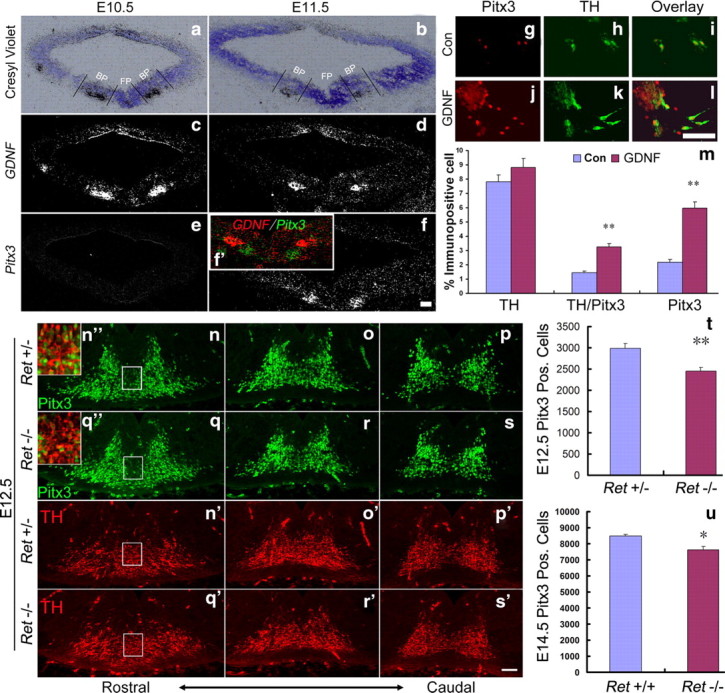
GDNF/Ret signaling is required for activation of Pitx3 expression in an mdDA neuronal subset. a–f, Representative midbrain coronal sections of wild-type (C57BL/6) mouse embryos at E10.5 (a, c, e) and E11.5 (b, d, f), hybridized with riboprobes for GDNF (c, d) and Pitx3 (e, f). a and b are Nissl-stained bright-field views of the dark-field pictures shown in c and d. Inset (f′) is a pseudo-colored overlay and magnification of the VM from the consecutive sections shown in d and f (red, GDNF; green, Pitx3). GDNF is expressed in the midbrain basal plate (BP) at E10.5 and E11.5 adjacent to the midbrain floor plate (FP), where Pitx3 starts to be transcribed at E11.5. g–l, Immunostaining of untreated [control (Con), g–i] and GDNF-treated (GDNF, j–l) E14 rat primary VM cultures with antibodies for Pitx3 (g, j) and TH (h, k) (merged images in i, l). m, Quantification of TH+, TH+/Pitx3+, and Pitx3+ cells relative to the total number of cells in untreated (Con, blue bars) and GDNF-treated (GDNF, red bars) primary rat VM cultures revealed a significant increase of Pitx3+ and Pitx3+/TH+ cells, but not of TH+ cells, in the GDNF-treated cultures as compared to untreated controls (TH+ cells: control, 7.82 ± 0.47%; GDNF-treated, 8.83 ± 0.61%, mean ± SEM, not significant, p = 0.26; TH+/Pitx3+ cells: control, 1.43 ± 0.13%; GDNF-treated, 3.25 ± 0.23%; mean ± SEM, **p < 0.01 in the independent t test; Pitx3+ cells: control, 2.19 ± 0.19%; GDNF-treated, 5.95 ± 0.46%, mean ± SEM; **p < 0.01 in the independent t test). Data were derived from three independent experiments. n–s′, Representative VM close-up views on coronal sections of Ret+/− (n–p, n′–p′) and Ret−/− (q–s, q′–s′) mouse embryos at E12.5, immunostained for Pitx3 (n–s) and TH (n′–s′). Insets (n″, q″) are merged images and higher magnifications of the boxed areas in n, n′, q, and q′. t, u, Quantification on coronal or sagittal midbrain sections revealed a significant decrease of Pitx3+ cells by 17.7% in E12.5 Ret−/− embryos (t) and 10.4% in E14.5 Ret−/− embryos (u) as compared to their wild-type (Ret+/− and Ret+/+) littermates (Pitx3 Pos. (Pitx3+) cells: E12.5 Ret+/−, 2981 ± 125, n = 3; E12.5 Ret−/−, 2455 ± 85, n = 3; E14.5 Ret+/+, 8491 ± 105, n = 4; E14.5 Ret−/−, 7609 ± 224, n = 3 mean ± SEM; **p < 0.01 in the paired t test; *p < 0.05 in the independent t test). Scale bars: (in f, l, s′) 50 μm.
Given the spatiotemporal correlation of GDNF expression and the initiation of Pitx3 transcription in the embryonic VM, we next asked whether GDNF can promote the expression of Pitx3 in primary VM cells in vitro. Treatment of E14 rat primary VM cell cultures (grown in the absence of neurotrophins for 7 DIV) with 10 ng/ml GDNF for 24 h significantly increased the number of Pitx3+ and Pitx3+/TH+ cells within the VM cell population by 1.7- and 1.3-fold, respectively (Fig. 1g–m). Under these experimental conditions, GDNF treatment did not significantly increase the number of TH+ neurons (Fig. 1m).
To explore whether GDNF signaling is required for induction of Pitx3 expression in vivo, we counted the number of Pitx3+ cells in GDNF receptor Ret knock-out embryos. We found that the number of Pitx3+ cells in Ret−/− embryos was reduced by 17.7% at E12.5 and by 10.4% at E14.5 compared to their heterozygous or wild-type littermates (Fig. 1n–u). Pitx3 expression was particularly lost in ventromedial (Fig. 1n″,q″, insets) and lateral (data not shown) TH+ cells in the Ret−/− embryos, suggesting that GDNF signaling might specifically induce Pitx3 expression in a TH+ mdDA neuron subpopulation. The partial recovery of Pitx3+ cell numbers in the E14.5 Ret−/− embryos might be due to compensation by other GDNF receptors, such as GFRα1 and NCAM, and could explain the lack of an mdDA phenotype in postnatal DAT-Retflx//flx mice (Kramer et al., 2007; Paratcha and Ledda, 2008).
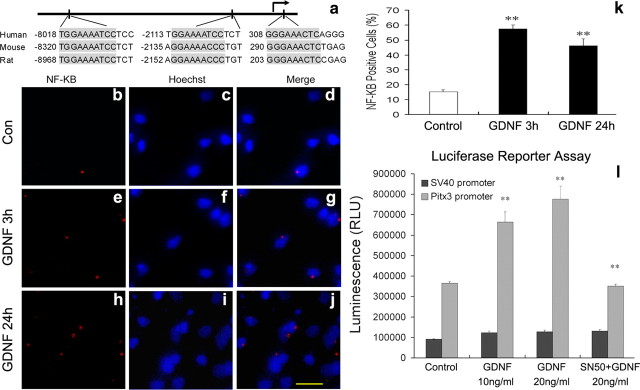
We next tested whether GDNF signaling directly activates the Pitx3 promoter in rodent VM cells. GDNF-mediated signaling enhances the nuclear translocation of NF-κB protein complexes and subsequent activation of NF-κB target genes in mdDA neurons (Cao et al., 2008; Wang et al., 2008). Analysis of the human, mouse, and rat Pitx3 promoter regions using bioinformatics prediction tools revealed the existence of conserved NF-κB BSs in these regions (Fig. 2a), and treatment of E14 rat primary VM cultures after 7 DIV with GDNF for 3 or 24 h increased the nuclear translocation of NF-κB, as expected (Fig. 2b–k). To determine whether this treatment resulted in increased activation of the Pitx3 promoter, we made use of a Pitx3 promoter/reporter construct containing a conserved NF-κB BS. GDNF treatment of SH-SY5Y cells transfected with this Pitx3 reporter construct resulted in a dose-dependent activation (80–110% increase) of the Pitx3 promoter (Fig. 2l). This activation was prevented when we treated the cells with SN50, an inhibitor of NF-κB nuclear translocation (Lin et al., 1995). We thus concluded that GDNF/Ret signaling is necessary and sufficient for the activation of the Pitx3 promoter in the embryonic rodent VM, and we identified NF-κB as a mediator of this process.
Figure 2.
NF-κB-mediated GDNF-signaling activates murine Pitx3 transcription. a, The human, mouse, and rat Pitx3 promoter region contains three highly conserved NF-κB BS (consensus sequence: 5′-GGGRNTYYCC-3′, where R = A or G, Y = C or T, and N = any nucleotide). Position of the first nucleotide of each BS is relative to the transcription start site (arrow). b–j, Immunostaining for NF-κB (b, e, h) and nuclear staining (Hoechst; c, f, i) (merged images in d, g, j) of untreated (b–d) E14 rat primary VM cultures or after treatment with GDNF for 3 h (e–g) or 24 h (h–j). k, Quantification of NF-κB+ cells relative to the total number of cells in these cell cultures with or without GDNF-treatment (control: 15.3 ± 1.3%; 3 h GDNF: 57.6 ± 2.5%; 24 h GDNF: 46.1 ± 4.8%, mean ± SEM; **p < 0.005 in the independent t test; n = 3). l, Transfection of SH-SY5Y cells with pGL3-Pitx3 promoter vector (light gray bars) and subsequent treatment of these cells with 10 or 20 ng/ml GDNF resulted in a dose-dependent activation of the Pitx3 promoter as compared to untreated controls. Dark gray bars represent values from pRL-SV40 vector (internal control). GDNF-mediated activation of the Pitx3 promoter was blocked by the simultaneous application of SN50, an inhibitor of NF-κB nuclear translocation (control: SV40 promoter, 96954 ± 2863; Pitx3 promoter, 365722 ± 7460; GDNF at 10 ng/ml: SV40 promoter, 123,900 ± 6980; Pitx3 promoter, 663,831 ± 51,615; GDNF at 20 ng/ml: SV40 promoter, 128,212 ± 7174; Pitx3 promoter, 776,831 ± 62,941; SN50 + GDNF at 20 ng/ml: SV40 promoter, 131,242 ± 7724; Pitx3 promoter, 350,674 ± 9528, mean ± SEM; **p < 0.005 in one-way ANOVA for repeated measurements). Data were derived from three independent experiments. RLU, Relative luciferase unit. Scale bar: (in j), 20 μm.
GDNF-mediated activation of Pitx3 expression induces BDNF transcription in vitro
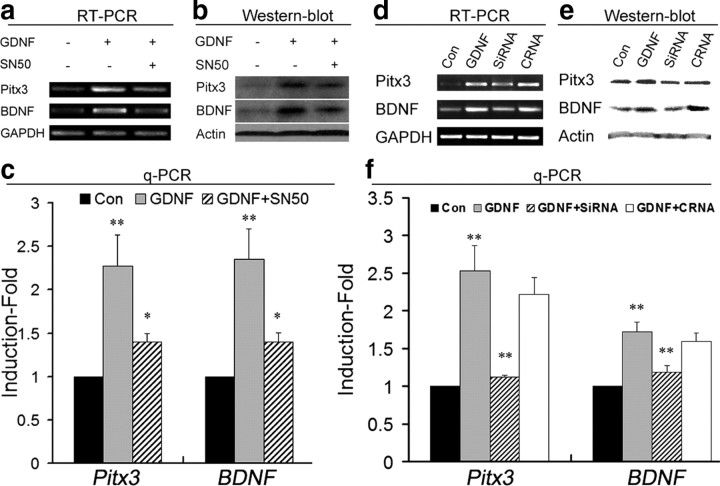
We have previously shown that overexpression of Pitx3 upregulates the transcription and secretion of BDNF protein from neuroblastoma cells and primary VM as well as astrocyte cultures (Peng et al., 2007; Yang et al., 2008). We therefore hypothesized that GDNF-mediated activation of Pitx3 expression might subsequently induce the transcription of BDNF in these cells. To test this hypothesis, we treated E14 rat primary VM cultures after 7 DIV with GDNF and evaluated the transcription of Pitx3 and BDNF under these conditions. As expected, GDNF treatment increased the transcription of Pitx3 and BDNF in these cultures by 1.3- and 1.4-fold, respectively, and this effect of GDNF on Pitx3 and BDNF transcription was blocked by the application of the NF-κB inhibitor SN50 (Fig. 3a,c). These results were also confirmed at protein levels (Fig. 3b). To further establish whether the GDNF-induced activation of BDNF transcription was mediated by Pitx3 and not by other Pitx3-independent pathways, we depleted Pitx3 in these cultures by siRNA-mediated knock-down. Quantitative RT-PCR indicated that transfection of a Pitx3 siRNA after 1 h of GDNF treatment-reduced Pitx3 mRNA levels by 56%, concomitant with a 32% reduction of BDNF mRNA levels in these cultures (Fig. 3d,f). A slight but not significant decrease in Pitx3 and BDNF mRNA levels was also observed after transfection of a control (nonsilencing) siRNA oligonucleotide (Fig. 3d,f). This reduction was most likely unspecific and due to the lipofection of the oligonucleotides, as a similar decrease in mRNA levels was observed in sham-lipofected GDNF-treated cultures as compared to untransfected controls (data not shown). We also used Western blotting to confirm that Pitx3 knock-down leads to a reduction of Pitx3 and BDNF protein levels in GDNF-treated primary VM cultures (Fig. 3e). These results therefore suggest that Pitx3 mediates the transcriptional activation of the BDNF gene following treatment of primary VM cells with GDNF.
Figure 3.
GDNF-mediated induction of Pitx3 expression activates BDNF transcription. a, b, RT-PCR (a) and Western blot (b) analyses of untreated (first lane), GDNF-treated (second lane), and SN50 + GDNF-treated (third lane) E14 rat primary VM cultures showed increased Pitx3 and BDNF expression after GDNF-treatment, which was blocked by simultaneous treatment with NF-κB inhibitor SN50. c, qPCR analysis indicated an 1.3-fold increase of Pitx3 and 1.4-fold increase of BDNF mRNA levels after GDNF-treatment and 70% reduction in both cases after SN50 application (Pitx3: GDNF, 2.27 ± 0.36; GDNF + SN50, 1.40 ± 0.09, mean ± SEM.; BDNF: GDNF, 2.35 ± 0.35; GDNF + SN50, 1.40 ± 0.11, mean ± SEM; *p < 0.05; **p < 0.005 in the one-way ANOVA for repeated measurements; n = 4). Con, Control. d, e, RT-PCR (d) and Western blot (e) analyses of untreated (first lane), GDNF-treated (second lane), Pitx3 siRNA + GDNF-treated (third lane), and control siRNA + GDNF-treated (fourth lane) E14 rat primary VM cultures revealed that siRNA-mediated knockdown of Pitx3 (siRNA), but not a control siRNA (CRNA) abolished the increased expression of BDNF in these cultures after GDNF-treatment. f, qPCR analysis indicated that Pitx3 siRNA + GDNF treatment resulted in a 56 and 32% reduction of Pitx3 and BDNF mRNA levels, respectively, as compared to GDNF only-treated cultures and a control siRNA (Pitx3: GDNF, 2.53 ± 0.33; GDNF + Pitx3 siRNA, 1.12 ± 0.02; GDNF + control siRNA, 2.21 ± 0.22, mean ± SEM; BDNF: GDNF, 1.73 ± 0.12; GDNF + Pitx3 siRNA, 1.18 ± 0.09; GDNF + control siRNA, 1.59 ± 0.11, mean ± SEM; **p < 0.005 in the one-way ANOVA for repeated measurements). Data were derived from at least three independent experiments in each case.
Pitx3 is required for BDNF transcription in an mdDA neuronal subset in vivo
Our results raised the possibility that Pitx3 is an essential regulator of BDNF gene expression in the rodent VM. To address this possibility, we analyzed the time course of BDNF expression in wild-type (Pitx3+/+ and Pitx3+/GFP mice) and Pitx3-null mutant (Pitx3GFG/GFP) mice, which allow the fate mapping of Pitx3-expressing (GFP+) cells (Maxwell et al., 2005). In wild-type mouse embryos, Pitx3 expression in the VM was first detected around E11.0–E11.5 (Fig. 1e,f) (Smidt et al., 1997); at this or earlier stages, BDNF expression was not detected in the VM (Fig. 4a–c and data not shown). We found that BDNF was widely transcribed in the wild-type and Pitx3+/GFP VM at E12.5 and exhibited a particularly prominent expression domain in the rostrolateral and medial VM overlapping with GFP+ (Pitx3+) mdDA neurons (Fig. 4e–g″ and data not shown). Remarkably, BDNF expression in this rostrolateral and medial domain was entirely lost in the Pitx3GFP/GFP embryos at E12.5, although the corresponding GFP+ neurons were still present in this domain and despite the persistent expression of BDNF in the adjacent Pitx3-negative (GFP−) VM tissue of the null mutants (Fig. 4 h–j″).
Figure 4.
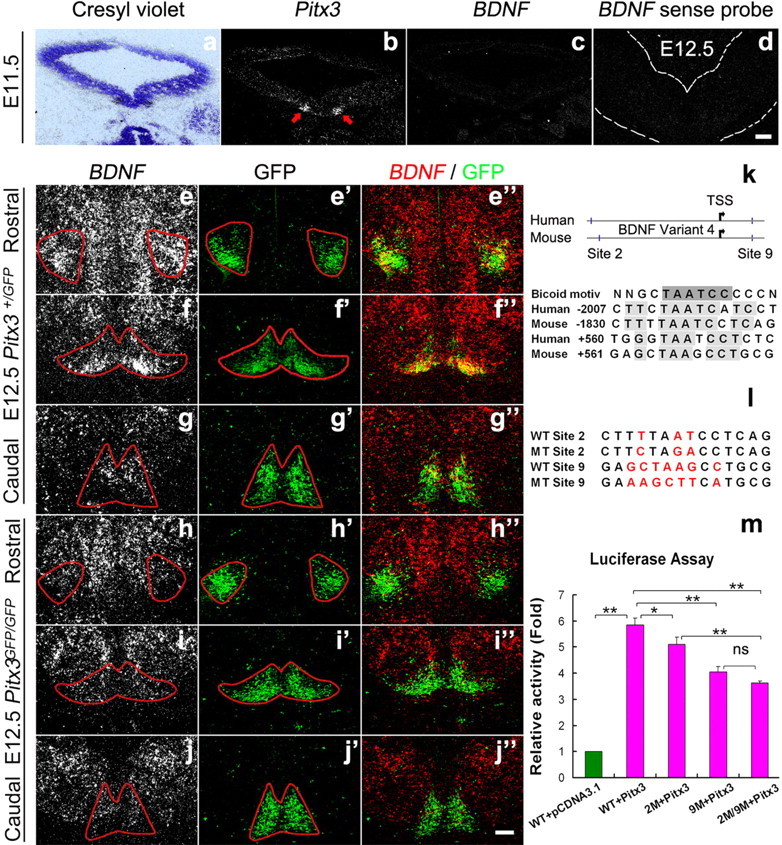
Pitx3 is required for the onset of BDNF transcription in an mdDA neuronal subpopulation at E12.5. a–d, Representative midbrain coronal sections of wild-type mouse embryos at E11.5 (a–c, consecutive sections) and E12.5 (d), hybridized with Pitx3 (b), BDNF antisense (c) and BDNF sense (d) riboprobes. a, Nissl-stained bright-field view of the dark-field picture shown in b. BDNF is not expressed in the murine VM at E11.5, although Pitx3 starts to be expressed in a bilateral VM domain at this stage (red arrows in b). e–j″, Representative coronal sections at different rostrocaudal levels of the midbrain from E12.5 Pitx3+/GFP (e–g″) and Pitx3GFP/GFP (h–j″) mice, hybridized with BDNF antisense riboprobe (e–j), or immunostained for GFP (e′–j′). Merged images are shown in (e″–j″). BDNF expression is not induced in a rostrolateral and medial GFP+ mdDA domain of the Pitx3GFP/GFP embryos at E12.5 (h–j″). Note that the BDNF sense probe does not give a signal at this stage in wild-type embryos (section shown in d), demonstrating the specificity of the BDNF antisense riboprobe. k–m, Two highly conserved Pitx3/Bicoid-like transcription factor BSs in the human and mouse BDNF promoter regions [Site 2 and Site 9, positions relative to the transcription start site (TSS, arrow)] were tested for BDNF promoter activation by Pitx3 (k). Cotransfection of the pGL3-BDNF promoter vector containing these two conserved Pitx3 BSs (WT, wild type) and pcDNA3.1-Pitx3 vector (red bars) into HEK293 cells resulted in a 4.9-fold activation of the BDNF promoter relative to the control (pcDNA3.1 only, green bar) (Pitx3: 5.86 ± 0.24, mean ±S EM; **p < 0.005 in the one-way ANOVA for repeated measurements) (m). Site-directed mutagenesis (MT) of Pitx3 BS 2 (MT Site 2) and/or 9 (MT Site 9) (l) significantly attenuated BDNF promoter activation by Pitx3 (BDNF WT promoter: 5.86 ± 0.24; BDNF promoter with MT Site 2 (2M): 5.11 ± 0.27; BDNF promoter with MT Site 9 (9M): 4.04 ± 0.22; BDNF promoter with MT Site 2 and MT Site 9 (2M/9M): 3.6 ± 0.08, mean ± SEM; *p < 0.05; **p < 0.005 in the one-way ANOVA for repeated measurements) (m). Data were derived from three independent experiments. Scale bars: (in d and j″), 100 μm.
To determine whether Pitx3 can directly activate the transcription of the BDNF gene, we searched for conserved Pitx3/Bicoid-like BSS within the mouse and human BDNF promoters. We found two highly conserved Pitx3/Bicoid-like BSs in these two species, one located ∼1.8–2.0 kb upstream (referred to as BS2) and the other one located ∼560 bp downstream (referred to as BS9) of the putative transcription start site for the mouse BDNF variant 4 (Fig. 4k). Cotransfection of a luciferase reporter construct containing 2.6 kb of mouse BDNF promoter sequences (including these two highly conserved Pitx3 BSs and seven other putative Pitx3 BSs) and a Pitx3 expression vector resulted in a 4.9-fold increase in luciferase (promoter) activity relative to the control (Fig. 4m). Moreover, site-directed mutagenesis of either one of these two conserved Pitx3 BSs decreased the Pitx3-induced activation of the BDNF promoter by 15% (BS2) and 37% (BS9) (Fig. 4l,m), whereas mutation of both BS2 and BS9 resulted in a 47% decrease of BDNF promoter activity after Pitx3 cotransfection (Fig. 4m).
To establish whether the loss of BDNF expression in the rostrolateral and medial VM of the Pitx3GFP/GFP embryos is rapidly followed by the death of GFP+ (Pitx3+) mdDA neurons in these embryos, we assessed the proportion of apoptotic (cCasp3+) and GFP+ cells relative to the total number of GFP+ mdDA neurons in wild-type (Pitx3+/GFP) and mutant (Pitx3GFP/GFP) embryos at E12.5 (Fig. 5a–e). This proportion was significantly increased by ∼3-fold in the rostral but not caudal VM of the Pitx3GFP/GFP embryos as compared to their wild-type littermates (Fig. 5e), indicating that the lack of BDNF expression indeed correlates with a reduced survival of mdDA neurons in the rostral VM of the Pitx3GFP/GFP embryos.
Figure 5.
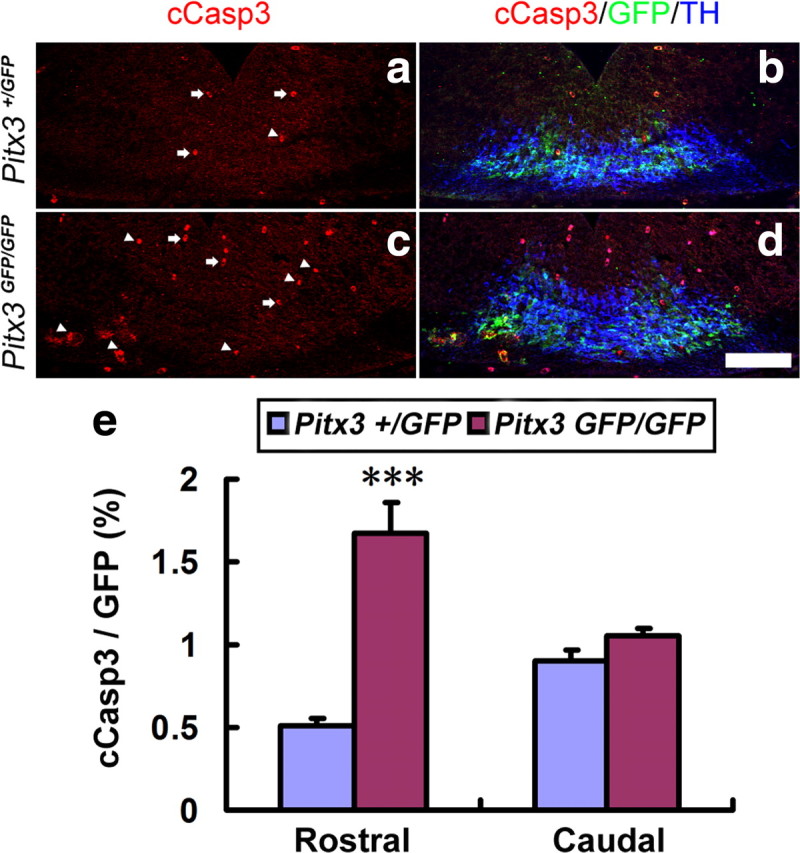
The loss of BDNF expression correlates with an increased apoptotic cell death of mdDA neurons in the E12.5 Pitx3GFP/GFP rostral VM. a–d, Triple immunostaining for cCasp3, GFP, and TH on representative midbrain coronal sections of Pitx3+/GFP (a, b) and Pitx3GFP/GFP (c, d) embryos at E12.5 revealed an increase of apoptotic Pitx3+ mdDA neurons [cCasp3+ (red) and GFP+ (green) double-labeled cells, white arrowheads) in the mutant VM. White arrows point at erythrocytes that were stained unspecifically for cCasp3. e, Quantification of cCasp3+/GFP+ double-labeled cells relative to the total amount of GFP+ (Pitx3+) cells in these sections showed a significant increase of apoptotic mdDA neurons in the rostral (cCasp3+/GFP+ cells: E12.5 Pitx3+/GFP, 0.506 ± 0.047%; E12.5 Pitx3GFP/GFP, 1.676 ± 0.185%; n = 3, mean ± SEM; ***p < 0.001 in the independent samples t test), but not caudal (cCasp3+/GFP+ cells: E12.5 Pitx3+/GFP, 0.902 ± 0.068%; Pitx3GFP/GFP, 1.056 ± 0.042%, n = 3, mean ± SEM), VM of the Pitx3GFP/GFP embryos. Scale bar: (in d), 100 μm.
We next investigated the effect of Pitx3 inactivation on BDNF expression in late gestational and postnatal mdDA neurons. BDNF expression was still not detected in the rostrolateral and medial VM domains of Pitx3GFP/GFP embryos at E14.5 (Fig. 6a–f″) and E16.5 (data not shown). Loss of Pitx3 resulted in a notable decrease of TH+ SNc mdDA neurons in the null mutant VM at postnatal day 30 (P30), concomitant with a strong reduction of BDNF-expressing cells in the same region (Fig. 6 g–l″). Notably, BDNF transcription appeared to be very low or almost undetectable in the caudomedial mdDA domain throughout embryonic development (Figs. 4e–g″, 6a–c″), and this mdDA neuronal subset was less affected in the Pitx3GFP/GFP-null mutants at embryonic and postnatal stages (Figs. 4h–j″, 5e, 6d–f″,j–l″; Maxwell et al., 2005). Collectively, our results indicate that Pitx3 directly activates the BDNF promoter in an mdDA neuron subpopulation located in the rostrolateral and medial VM of the mouse embryo and suggest that the Pitx3-mediated activation of BDNF expression is necessary for the survival of this mdDA neuronal subset at embryonic and postnatal stages.
Figure 6.
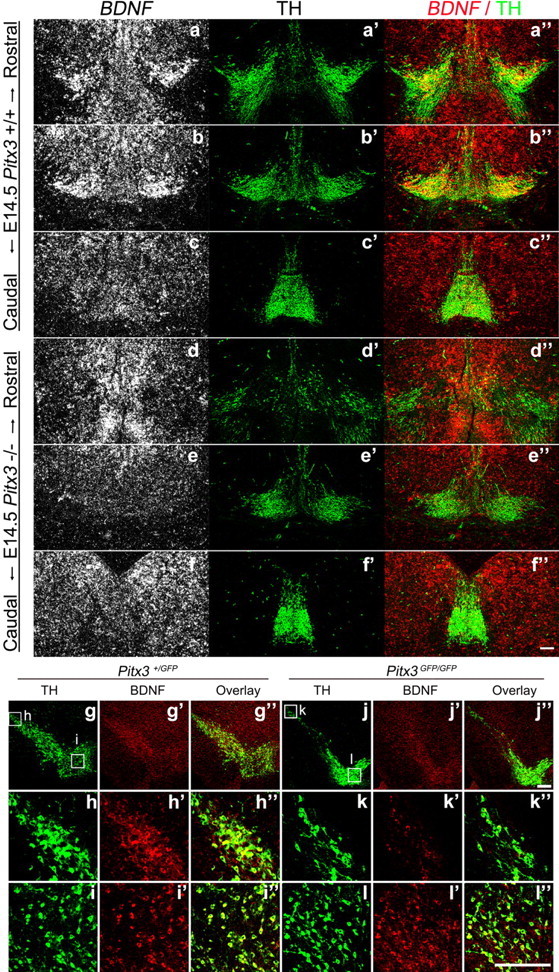
Lack of BDNF expression in an mdDA neuronal subpopulation that disappears at later embryonic/postnatal stages in the absence of Pitx3. (a–l″) Representative coronal midbrain sections from E14.5 (a–f″, different rostrocaudal levels) and P30 (g–l″) Pitx3+/GFP (a–c″; g–i″) and Pitx3GFP/GFP (d–f″; j–l″) mice hybridized with a BDNF riboprobe (a–f) or immunostained for TH (a′–f′, g–l) and BDNF (g′–l′). Merged images are shown in a″–f″ and g″–l″. BDNF is expressed strongly in a rostrolateral TH+ mdDA domain in the wild-type (Pitx3+/GFP) embryos (a–c″; g–i″), but this BDNF+ domain was completely lost in the VM of Pitx3GFP/GFP embryos at E14.5 (d, d″, e, e″,f, f″), concomitant with a strong reduction at E14.5 (d′–f′) and followed by the almost complete loss at P30 (j–l″) of the rostrolateral (SNc) TH+ mdDA neurons in these mutants. Scale bars: (in f″, J″, and l ″),100 μm.
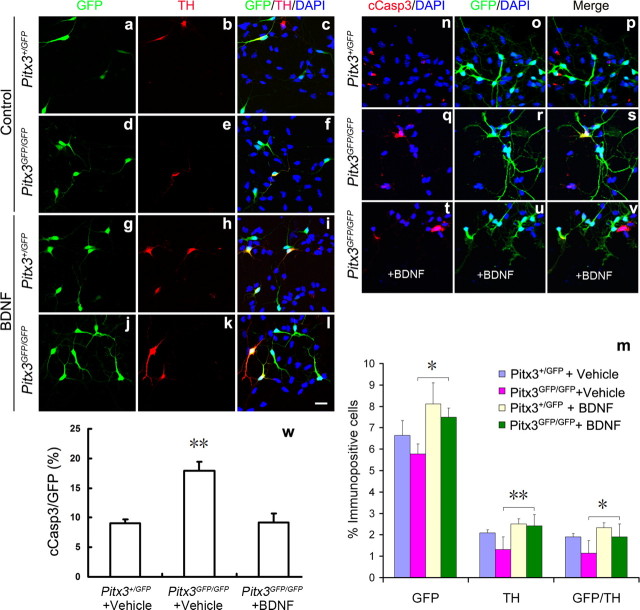
BDNF augments the survival of mdDA neurons in Pitx3-null mutant primary VM cultures
To further investigate whether the lack of BDNF expression contributes to the loss of mdDA neurons in Pitx3GFP/GFP mice (Fig. 5 and Maxwell et al., 2005), we tested whether BDNF treatment is sufficient to rescue the numbers of mdDA neurons in primary VM cultures derived from E11.5 Pitx3GFP/GFP embryos. We found a reduction of GFP+ cell numbers (cells that would express Pitx3 in the wild type) by 13%, TH+ cells by 37.5%, and GFP/TH double-positive cells by 40% in the untreated Pitx3GFP/GFP cultures as compared to untreated wild-type (Pitx3+/GFP) cultures (Fig. 7a–f,m). This result is consistent with in vivo data from E12.5 Pitx3GFP/GFP mice showing a reduction of GFP+ cells by 21%, TH+ cells by 54%, and GFP/TH double-positive cells by 48% relative to the Pitx3+/GFP controls (Maxwell et al., 2005), and it confirms a selective loss of GFP+ and TH+ cells in the absence of Pitx3 also in primary VM cultures. Notably, BDNF treatment of Pitx3GFP/GFP VM cultures increased the numbers of GFP+ cells by 29.6%, TH+ cells by 84.7%, and TH/GFP double-positive cells by 66.7% relative to the untreated Pitx3GFP/GFP cultures, thereby reaching similar numbers as in the BDNF-treated wild-type (Pitx3+/GFP) cultures (Fig. 7g–m). To determine whether this was due to a survival-promoting effect of the BDNF treatment on the cultured mdDA neurons, we assessed the proportion of apoptotic (cCasp3+) mdDA (GFP+) neurons in the untreated and BDNF-treated VM cultures from wild-type (Pitx3+/GFP) and mutant (Pitx3GFP/GFP) embryos. As expected, this proportion was significantly increased in the untreated Pitx3GFP/GFP VM cultures as compared to the untreated controls (Fig. 7n–s,w). Notably, the increased death of GFP+ cells in the Pitx3GFP/GFP VM cultures was rescued back to control levels by the addition of BDNF to these cultures (Fig. 7t–w), indicating that in the absence of Pitx3, exogenous BDNF application prevents the cell death of cultured Pitx3-null (Pitx3GFP/GFP) mdDA neurons.
Figure 7.
BDNF augments the survival of mdDA neurons in Pitx3GFP/GFP primary VM cultures. a–v, Primary VM cultures derived from E11.5 Pitx3+/GFP (a–c, g–i, n–p) and Pitx3GFP/GFP (d–f, j–l, q–v) embryos were treated with vehicle (0.1% BSA) (a–f, n–s) or 20 ng/ml BDNF (g–l, t–v) for 4 d and immunostained for GFP (green in a, d, g, j, o, r, u), TH (red in b, e, h, k), and cCasp3 (red in n, q, t); merged images with DAPI stain in c, f, i, l, p, s, and v. m, Quantification of GFP and TH immunopositive cells in these four experimental groups revealed an increased number of GFP+ and TH+ mdDA neurons in the BDNF-treated relative to the untreated Pitx3GFP/GFP VM cultures (GFP+ cells: Pitx3+/GFP untreated, 6.64 ± 0.69%; Pitx3GFP/GFP untreated, 5.78 ± 0.46%; Pitx3+/GFP BDNF-treated, 8.11 ± 0.99%; Pitx3GFP/GFP BDNF-treated, 7.49 ± 0.44%; TH+ cells: Pitx3+/GFP untreated, 2.09 ± 0.14%; Pitx3GFP/GFP untreated, 1.31 ± 0.60%; Pitx3+/GFP BDNF-treated, 2.51 ± 0.24%; Pitx3GFP/GFP BDNF-treated, 2.42 ± 0.53%; TH+/GFP+ cells: Pitx3+/GFP untreated, 1.91 ± 0.15%; Pitx3GFP/GFP untreated, 1.14 ± 0.59%; Pitx3+/GFP BDNF-treated, 2.34 ± 0.22%; Pitx3GFP/GFP BDNF-treated, 1.90 ± 0.60%, mean ± SEM). w, Quantification of the relative amount of apoptotic (cCasp3+) mdDA neurons (GFP+) in these cultures showed an increase of apoptotic cells in the untreated Pitx3GFP/GFP VM cultures as compared to the untreated controls that was rescued by the addition of BDNF protein to the mutant cultures (cCasp3+/GFP+ cells: Pitx3+/GFP untreated, 9.11 ± 0.55%; Pitx3GFP/GFP untreated, 17.92 ± 1.46%; Pitx3GFP/GFP BDNF-treated, 9.12 ± 1.63%). Paired t test was used for statistical analysis of treatment effects within the same genotype, and one-way ANOVA was used for statistical analysis of genotype differences for the same treatment. *p < 0.05; **p < 0.005. Data were derived from three independent experiments. Scale bar: (in l), 20 μm.
Intrastriatal injection of GDNF upregulates Pitx3 and BDNF expression in the adult SNc
Our previous results strongly suggested the existence of a feedforward mechanism for the initiation and/or maintenance of GDNF, Pitx3, and BDNF expression in the murine VM during embryonic development. We therefore hypothesized that this feedforward mechanism might also persist in the adult brain, as it is known that GDNF released from striatal target cells is taken up and retrogradely transported to the soma of mdDA neurons, where it promotes their survival in the early postnatal and adult rodent brain (Tomac et al., 1995; Oo et al., 2003; Kholodilov et al., 2004). To test this hypothesis, we injected GDNF protein unilaterally into the striatum of adult rats, and 48 h later we analyzed the endogenous expression levels of Pitx3 and BDNF proteins in the ipsilateral (GDNF-treated) SNc relative to the contralateral (vehicle-treated) control side. We found that Pitx3 and BDNF protein levels were increased by 2.8- and 6.1-fold, respectively, in the ipsilateral (GDNF-treated) SNc, as determined by immunohistochemistry (Fig. 8a–l) and on Western blots (Fig. 8n–p). These data provide direct evidence that retrograde GDNF signaling stimulates Pitx3 and BDNF expression in mature SNc mdDA neurons.
Figure 8.
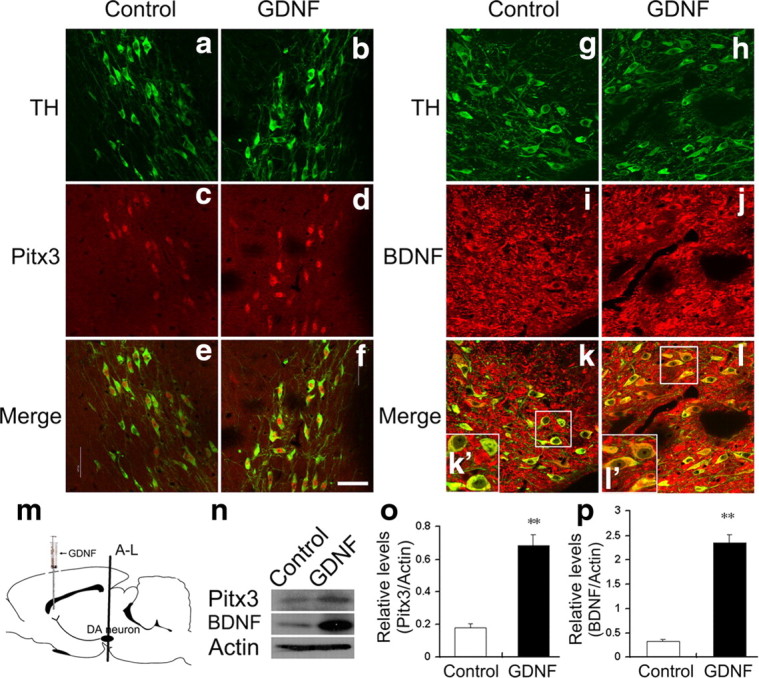
Intrastriatal injection of GDNF upregulates Pitx3 and BDNF expression in the adult SNc. a–l, Representative coronal sections from the SNc of adult male rats who had received an unilateral injection of 0.9% saline (control, a, c, e, g, i, k) and 15 ng/μl GDNF into the contralateral side of the striatum (b, d, f, h, j, l) 48 h earlier, immunostained for TH (a, b, g, h), Pitx3 (c, d) and BDNF (i, j). Merged images are shown in e, f, k, and l. Insets in k′ and l′ are higher magnifications of the boxed areas in k and l, respectively. m, Schematic drawing of the striatal injection site and position of the VM sections shown in a–l. n, Western blots using protein extracts from vehicle-treated control and GDNF-treated contralateral side of the VM showed that unilateral intrastriatal injection of GDNF leads to a 2.8-fold increase of Pitx3 expression (quantification in o) and 6.1-fold increase of BDNF expression (quantification in p) in the ipsilateral mdDA neurons (control: Pitx3, 0.18 ± 0.02; BDNF, 0.33 ± 0.04, mean ± SEM; GDNF-treated (GDNF): Pitx3, 0.68 ± 0.07; BDNF, 2.35 ± 0.18; **p < 0.01 in the independent samples t test for repeated measurements; n = 3). Scale bar: (in f), 50 μm.
BDNF, but not GDNF, protects Pitx3−/− mdDA neurons against 6-OHDA neurotoxicity
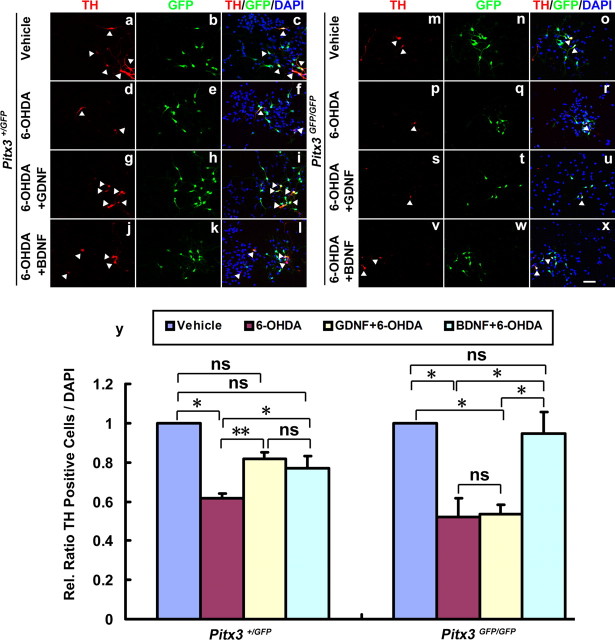
Given the previous result, we wanted to know whether the neuroprotective effect of GDNF against 6-OHDA toxicity on mdDA neurons might be due to Pitx3-mediated activation of BDNF expression in these neurons. Therefore, we treated wild-type (Pitx3+/GFP) and Pitx3-null mutant (Pitx3GFP/GFP) VM cultures after 3 DIV with 6-OHDA and assessed the effect of a previous 2 h incubation with GDNF or BDNF on these 6-OHDA-treated cultures. Treatment with 6-OHDA significantly reduced the numbers of TH+ cells in both wild-type and mutant cultures (Fig. 9a–f,m–r,y) as expected. Prior incubation with GDNF or BDNF significantly rescued the numbers of TH+ mdDA neurons in the wild-type (Pitx3+/GFP) VM cultures, reaching a similar level as that under normal (untreated) conditions (Fig. 9 g–l,y). Remarkably, the numbers of TH+ mdDA neurons in the 6-OHDA-treated Pitx3GFP/GFP VM cultures were only rescued back to normal (untreated) levels after a previous incubation of these cells with BDNF, but not with GDNF (Fig. 9s–y), indicating that GDNF cannot protect TH+ mdDA neurons against 6-OHDA-induced neurotoxicity in the absence of Pitx3.
Figure 9.
BDNF, but not GDNF, protects Pitx3-null mutant mdDA neurons against 6-OHDA neurotoxicity. a–x, Primary VM cultures derived from E11.5 Pitx3+/GFP (a–l) and Pitx3GFP/GFP (m–x) embryos were treated after 3 DIV with vehicle (0.1% BSA) (a–f, m–r), 20 ng/ml GDNF (g–i, s–u), or 20 ng/ml BDNF (j–l, v–x); after 2 h, cells were incubated with 10 μm 6-OHDA (d–l, p–x) or vehicle (PBS; a–c, m–o) for 24 h and immunostained for TH (red in a, d, g, j, m, p, s, and v) and GFP (green in b, e, h, k, n, q, t, and w); merged images with DAPI stain are shown in c, f, i, l, o, r, u, and x. y, Quantification of TH-immunopositive cells in these eight experimental groups revealed that in Pitx3+/GFP VM cultures both GDNF and BDNF protect TH+ mdDA neurons against 6-OHDA toxicity, but in Pitx3GFP/GFP VM cultures only BDNF (and not GDNF) protects these neurons against 6-OHDA toxicity [TH+/DAPI+ cells (white arrowheads): Pitx3+/GFP vehicle-treated, normalized to 1; Pitx3+/GFP 6-OHDA-treated, 0.615 ± 0.025; Pitx3+/GFP 6-OHDA + GDNF-treated, 0.816 ± 0.037; Pitx3+/GFP 6-OHDA + BDNF-treated, 0.768 ± 0.066, mean ± SEM, n = 3; Pitx3GFP/GFP vehicle-treated, normalized to 1; Pitx3GFP/GFP 6-OHDA-treated, 0.521 ± 0.096; Pitx3GFP/GFP 6-OHDA + GDNF-treated, 0.533 ± 0.051; Pitx3GFP/GFP 6-OHDA + BDNF-treated, 0.946 ± 0.111, mean ± SEM, n = 4]. Paired t test was used for statistical analysis of treatment effects. *p < 0.05; **p < 0.01; ns, not significant. Data were derived from at least three independent experiments. Scale bar: (in x), 50 μm.
To determine whether these two neurotrophic factors exerted their protective effect on the 6-OHDA-treated wild-type (Pitx3+/GFP) and mutant (Pitx3GFP/GFP) VM cultures by a reduction of neurotoxin-induced apoptotic cell death, we assessed the relative proportion of apoptotic (cCasp3+) and GFP+ mdDA neurons in the 6-OHDA-treated VM cultures alone and after previous incubation with GDNF or BDNF. As expected, treatment with 6-OHDA alone significantly increased the numbers of apoptotic mdDA neurons in these cultures, regardless of their genotype (Fig. 10a–f,m–r,y). In line with our previous findings, BDNF significantly decreased the numbers of apoptotic mdDA neurons down to normal (untreated) levels in both Pitx3+/GFP and Pitx3GFP/GFP 6-OHDA-treated cultures (Fig. 10j–l,v–x,y), whereas GDNF only decreased these numbers significantly in Pitx3+/GFP but not in Pitx3GFP/GFP 6-OHDA-treated cultures (Fig. 10 g–i,s–u,y). Altogether, these results strongly suggest that GDNF exerts its neuroprotective and survival-promoting effect on at least a subpopulation of mdDA neurons through the activation of Pitx3-mediated BDNF expression in these cells. In the absence of Pitx3, GDNF cannot protect the mutant mdDA neurons against 6-OHDA-induced neurotoxicity and apoptotic cell death, and these cells are only rescued by the exogenous application of BDNF acting downstream of GDNF and Pitx3, as suggested by our previous results.
Figure 10.
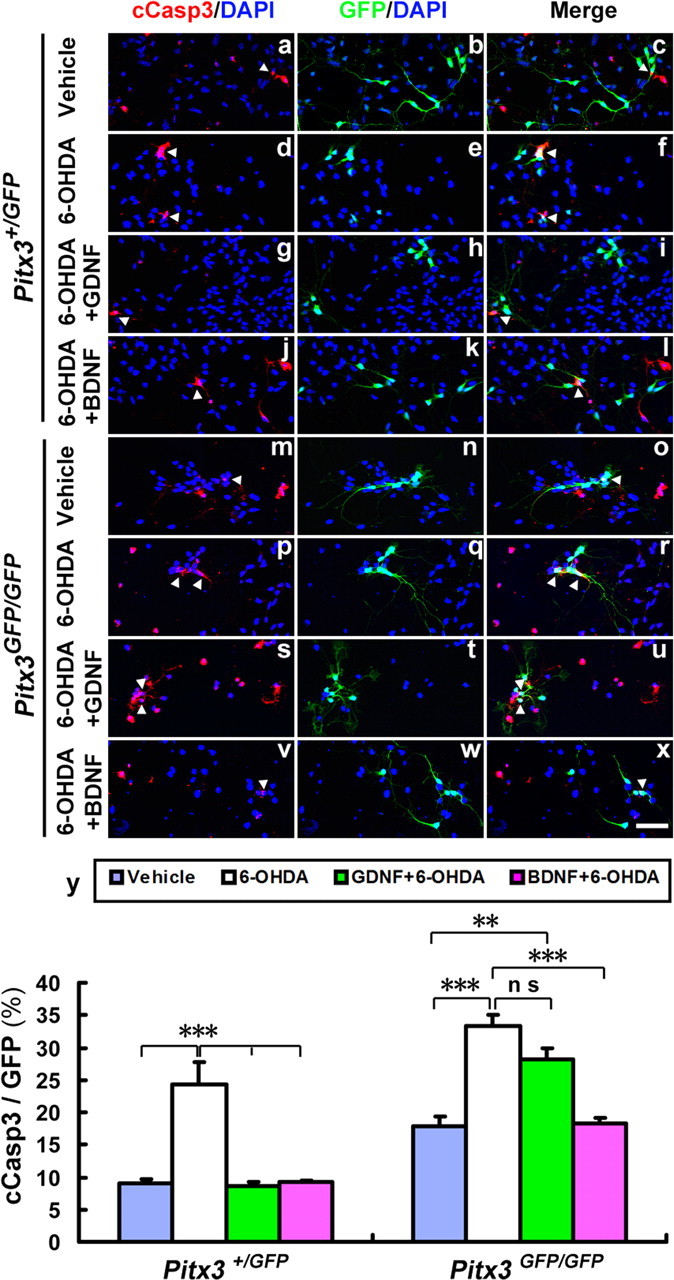
GDNF does not protect against 6-OHDA-induced apoptotic cell death of Pitx3-null mutant mdDA neurons. a–x, Primary VM cultures derived from E11.5 Pitx3+/GFP (a–l) and Pitx3GFP/GFP (m–x) embryos were treated after 3 DIV with vehicle (0.1% BSA) (a–f, m–r), 20 ng/ml GDNF (g–i, s–u), or 20 ng/ml BDNF (j–l, v–x); after 2 h, cells were incubated with 10 μm 6-OHDA (d–l, p–x) or vehicle (PBS; a–c, m–o) for 24 h and immunostained for cCasp3 (red in a, d, g, j, m, p, s, and v) and GFP (green in b, e, h, k, n, q, t, and w); blue, DAPI stain; merged images in c, f, i, l, o, r, u, and x. y, Quantification of the relative amount of apoptotic (cCasp3+) mdDA neurons (GFP+) in these eight experimental groups revealed that both GDNF and BDNF prevent the 6-OHDA-induced cell death of Pitx3+/GFP mdDA neurons, but only BDNF (and not GDNF) prevents the 6-OHDA-induced cell death of Pitx3GFP/GFP mdDA neurons [cCasp3+/GFP+ cells (white arrowheads): Pitx3+/GFP vehicle-treated, 9.11 ± 0.55%; Pitx3+/GFP 6-OHDA-treated, 24.29 ± 3.49%; Pitx3+/GFP 6-OHDA + GDNF-treated, 8.70 ± 0.55%; Pitx3+/GFP 6-OHDA + BDNF-treated, 9.25 ± 0.11%; mean ± SEM, n = 3; Pitx3GFP/GFP vehicle-treated, 17.92 ± 1.46%; Pitx3GFP/GFP 6-OHDA-treated, 33.36 ± 1.74%; Pitx3GFP/GFP 6-OHDA + GDNF-treated, 28.11 ± 1.86%; Pitx3GFP/GFP 6-OHDA + BDNF-treated, 18.35 ± 0.78%, mean ± SEM; n = 3]. One-way ANOVA was used for statistical analysis of treatment effects. **p < 0.005; ***p < 0.001. Data were derived from three independent experiments. Scale bar: (in x), 50 μm.
Discussion
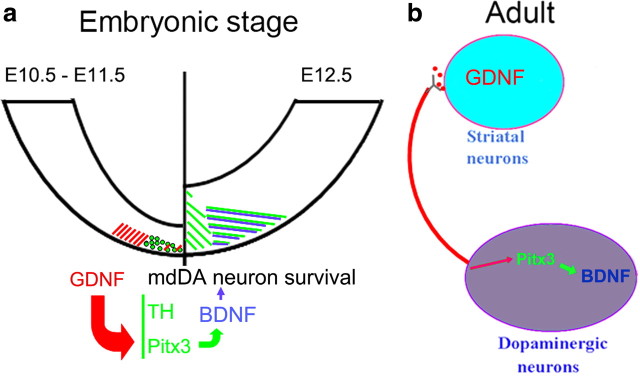
The secreted factors GDNF and BDNF and the transcription factor Pitx3 are individually required for the proper development of mdDA neurons or for their survival during postnatal and adult stages (Baquet et al., 2005; Smidt and Burbach, 2007; Pascual et al., 2008; Oo et al., 2009), but it remained unclear whether a regulatory interaction exists between these three factors during embryonic development or in the adult brain. Here we show that NF-κB-mediated GDNF/Ret signaling is, at least in part, sufficient for the activation of Pitx3 expression, which is in turn required for the transcription of BDNF in a rostrolateral and medial mdDA neuronal subpopulation, thereby promoting the survival of these neurons during mouse embryonic development (Fig. 11). We also show that this feedforward regulation of GDNF, Pitx3, and BDNF expression is still active in the adult rodent brain and that, in the absence of Pitx3, BDNF but not GDNF protects mdDA neurons against 6-OHDA toxicity in vitro. Altogether, our data indicate that the regulatory interaction between GDNF, Pitx3, and BDNF is necessary for the survival and protection of mdDA neurons against neurotoxic insults during embryogenesis, and that this regulatory interaction might also be relevant for the survival and neuroprotection of adult mdDA neurons (Fig. 11).
Figure 11.
Feed forward regulation of GDNF, Pitx3, and BDNF expression in the embryonic and adult rodent brain. a, During development, GDNF (red) is transiently expressed in the midbrain BP of the E10.5/E11.5 mouse embryo, where it is required for the NF-κB-mediated induction of Pitx3 expression in an mdDA neuronal subset within the adjacent midbrain floor plate. Pitx3 (green) is in turn necessary for the activation of BDNF expression in a rostrolateral/medial mdDA neuronal subpopulation at E12.5. Neurotrophic support of this mdDA neuronal subset by BDNF (blue) might be required for their survival throughout development and their protection against neurotoxic insults. b, In the adult brain, striatal uptake and retrograde axonal transport of GDNF (red) to the cell soma maintains the proper levels of Pitx3 (green) and BDNF (blue) expression in SNc mdDA neurons and might thereby contribute to the sustained survival and neuroprotection of these neurons throughout adulthood.
GDNF/Ret signaling activates Pitx3 expression in an mdDA neuronal subpopulation
The relevance of GDNF signaling for the normal development of mdDA neurons in vivo has remained controversial until now. Inactivation of the murine GDNF, Ret, and Gfra1 genes apparently does not interfere with prenatal mdDA neuron differentiation or survival (Airaksinen and Saarma, 2002; Kramer et al., 2007; Paratcha and Ledda, 2008). Moreover, persistent overexpression of GDNF driven by the TH promoter leads to a reduction of SNc DA neurons (Chun et al., 2002), whereas intrastriatal or intranigral injections of GDNF protein during early postnatal stages, or the constitutive activation of the Ret receptor, result in increased numbers of TH+ SNc neurons and an enhanced DA metabolism (Beck et al., 1996; Mijatovic et al., 2007). The transient but detectable expression of GDNF in the midgestational VM shown here might have been missed by previous in situ hybridization studies due to lower sensitivity or slight differences in the staging of the embryos (Hellmich et al., 1996; Golden et al., 1999), but GDNF expression in the embryonic rat VM was also shown by Choi-Lundberg and Bohn (1995) using a more sensitive RT-PCR-based detection method. Our and previous findings therefore suggest that the transient expression of GDNF in the rodent VM might have two different functions during development: on the one hand, GDNF signaling might activate Pitx3 transcription in an mdDA precursor subpopulation; on the other hand, GDNF might provide transient trophic support to developing progenitors and postmitotic neurons in this region of the brain. The requirement of GDNF signaling for these two processes, however, might be compensated by other factors during embryonic development, leading to the absence of an mdDA phenotype in the GDNF, Ret, and Gfra1-null mutants at birth (Airaksinen and Saarma, 2002; Paratcha and Ledda, 2008). Our and previous results also suggest that the tight regulation of GDNF protein levels is critical for proper mdDA neuron development. The notion that a transient (rather than persistent) expression of GDNF during embryonic development might have an important permissive role is supported by the observation that sustained GDNF expression in differentiating TH+ SNc neurons results in a marked reduction of these neurons shortly after birth (Chun et al., 2002).
Pitx3 is necessary for the activation of BDNF transcription in an mdDA neuronal subset
Pitx3 is required for the initiation of TH expression in an mdDA neuronal subset during development and for the survival of these neurons during subsequent prenatal and postnatal stages (Maxwell et al., 2005; Smidt and Burbach, 2007). Aldh1a1 was identified as a target gene that could mediate the prosurvival activity of Pitx3, as maternal RA complementation partially rescues the loss of mdDA neurons in Pitx3 mutants (Jacobs et al., 2007). Here we identify BDNF as another target gene of Pitx3 in the rostrolateral and medial mdDA neuronal subpopulation that is most affected in Pitx3−/− mice (Smidt et al., 2004; Maxwell et al., 2005). Because BDNF treatment augmented the survival of mdDA neurons in primary VM cultures prepared from E11.5 Pitx3GFP/GFP-null mutant embryos, we suggest that the failure to induce BDNF expression in rostrolateral and medial (SNc) mdDA precursors during development contributes to the preferential degeneration of these neurons in the Pitx3GFP/GFP mice. Interestingly, retinoid signaling via retinoic acid receptors was recently shown to induce BDNF transcription and to prevent the inflammatory degeneration of mdDA neurons in vitro and in vivo (Katsuki et al., 2009). While our data strongly suggest a direct activation of the BDNF gene by Pitx3, we cannot exclude that Pitx3 might indirectly maintain BDNF expression through the induction of Aldh1a1 and subsequent local production of RA in an mdDA neuronal subset.
GDNF protects mdDA neurons against 6-OHDA toxicity only in the presence of Pitx3
Treatment of Pitx3GFP/GFP-null mutant VM cultures with 6-OHDA resulted in an increased survival of mdDA neurons in these cultures only after preincubation with BDNF, but not with GDNF, suggesting that GDNF acts upstream of Pitx3 and BDNF to protect mdDA neurons against cytotoxic insults and to increase their survival under these conditions. BDNF is in fact more potent than GDNF in promoting the survival of mdDA neurons after unilateral 6-OHDA lesion of the SNc in organotypic cultures of the rat VM (Stahl et al., 2011). Moreover, GDNF appears to promote primarily the survival of calbindin-expressing VTA neurons under basal conditions (Meyer et al., 1999) and of VTA and rostromedial SNc neurons after 6-OHDA lesion (Barroso-Chinea et al., 2005), which are the cell groups that are not affected by the loss of BDNF expression in our Pitx3GFP/GFP mice. Together, our and previous findings therefore suggest that GDNF acts as a potent survival factor for those medial mdDA neuronal subpopulations projecting to the GDNF-rich ventral striatum (Barroso-Chinea et al., 2005). However, the rostrolateral mdDA neurons projecting to the dorsolateral striatum (where lower levels of GDNF are expressed; Barroso-Chinea et al., 2005) depend on Pitx3-mediated local BDNF synthesis for their survival and neuroprotection.
The feedforward regulation of GDNF, Pitx3, and BDNF expression in the adult SNc might be relevant for the pathogenesis of PD
We also found that the intrastriatal delivery of GDNF protein in the adult rat brain increased the expression of Pitx3 and BDNF in the ipsilateral SNc. This finding raises the possibility that retrograde transport of GDNF or local signaling at axon terminals in the striatum controls the levels of Pitx3 and BDNF proteins in adult mdDA neurons (Fig. 11) and might have implications for the pathogenesis and treatment of PD. Among all neurotrophic factors, GDNF exhibits the most severe decrease in the SNc of PD patients (Chauhan et al., 2001). Reduced intranigral GDNF levels might lead to reduced Pitx3 expression and thus to decreased expression of BDNF in SNc mdDA neurons. This might render these neurons more susceptible to death in the diseased brain. A reduction of BDNF mRNA and protein levels in the SNc of PD patients has been consistently reported by several groups (Mogi et al., 1999; Parain et al., 1999; Howells et al., 2000; Chauhan et al., 2001). Interestingly, some of these studies also noted in the patients the preferential loss of a subset of SNc neurons that normally express high levels of BDNF (Parain et al., 1999; Howells et al., 2000). Moreover, several polymorphisms in noncoding regions of the PITX3 gene (which might alter PITX3 expression) are associated with idiopathic or early onset PD (Fuchs et al., 2009; Bergman et al., 2010; Haubenberger et al., 2011; Le et al., 2011), and one polymorphism in the promoter region of the human BDNF gene was associated with familial PD (Parsian et al., 2004). These findings, together with our results and the observation that conditional ablation of the BDNF gene during mouse development leads to the selective loss of a subset of TH+ neurons in the SNc, but not VTA (Baquet et al., 2005), raise the intriguing possibility that the rostrolateral (SNc) mdDA neuron subpopulation expressing high levels of BDNF is most affected by the loss of PITX3 in human PD patients and cannot be protected by the exogenous application of GDNF. We suggest that a developmental failure to induce normal expression of Pitx3 coupled to age-related decreases in GDNF levels (Miyazaki et al., 2003) might lead to a depletion of BDNF in SNc neurons and predispose them to neurodegeneration (Porritt et al., 2005). According to this model, alterations in this neurotrophic factor reinforcement loop might lead to reduced neurotrophic support of SNc mdDA neurons, thus contributing to PD. Further dissection of the molecular underpinnings of this regulatory network might therefore facilitate the development of novel therapeutic approaches for the treatment of PD.
Footnotes
This work was supported by research grants from National Natural Science Foundation of China (NSFC30730096 and NSFC39070925) and National Basic Research Program of China (2011CB945200 and 2010CB510003) and research grants from Bayerischer Forschungsverbund ‘ForNeuroCell II’ (F2-F2412.18/10 086) to W.W. and N.P., by the Initiative and Networking Fund in the framework of the Helmholtz Alliance of Systems Biology (CoReNe), Federal Ministry of Education and Research (BMBF) NGFNPlus DiGtoP (FKZ 01GS0858), and European Union (mdDANEURODEV FP7-Health-2007-B-222999, EuTRACC LSHG-CT-2006-037445) to W.W. We thank X. Li for helpful discussions and T. Wen and S. Laass for excellent technical assistance.
The authors declare no conflicts of interest or relevant financial interests.
References
- Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- Baquet ZC, Bickford PC, Jones KR. Brain-derived neurotrophic factor is required for the establishment of the proper number of dopaminergic neurons in the substantia nigra pars compacta. J Neurosci. 2005;25:6251–6259. doi: 10.1523/JNEUROSCI.4601-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Chinea P, Cruz-Muros I, Aymerich MS, Rodríguez-Díaz M, Afonso-Oramas D, Lanciego JL, González-Hernández T. Striatal expression of GDNF and differential vulnerability of midbrain dopaminergic cells. Eur J Neurosci. 2005;21:1815–1827. doi: 10.1111/j.1460-9568.2005.04024.x. [DOI] [PubMed] [Google Scholar]
- Beck KD, Irwin I, Valverde J, Brennan TJ, Langston JW, Hefti F. GDNF induces a dystonia-like state in neonatal rats and stimulates dopamine and serotonin synthesis. Neuron. 1996;16:665–673. doi: 10.1016/s0896-6273(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Bergman O, Håkansson A, Westberg L, Nordenström K, Carmine Belin A, Sydow O, Olson L, Holmberg B, Eriksson E, Nissbrandt H. PITX3 polymorphism is associated with early onset Parkinson's disease. Neurobiol Aging. 2010;31:114–117. doi: 10.1016/j.neurobiolaging.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Björklund A, Dunnett SB. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Brodski C, Weisenhorn DM, Signore M, Sillaber I, Oesterheld M, Broccoli V, Acampora D, Simeone A, Wurst W. Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain-hindbrain organizer. J Neurosci. 2003;23:4199–4207. doi: 10.1523/JNEUROSCI.23-10-04199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao JP, Wang HJ, Yu JK, Liu HM, Gao DS. The involvement of NF-kappaB p65/p52 in the effects of GDNF on DA neurons in early PD rats. Brain Res Bull. 2008;76:505–511. doi: 10.1016/j.brainresbull.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Chauhan NB, Siegel GJ, Lee JM. Depletion of glial cell line-derived neurotrophic factor in substantia nigra neurons of Parkinson's disease brain. J Chem Neuroanat. 2001;21:277–288. doi: 10.1016/s0891-0618(01)00115-6. [DOI] [PubMed] [Google Scholar]
- Choi-Lundberg DL, Bohn MC. Ontogeny and distribution of glial cell line-derived neurotrophic factor (GDNF) mRNA in rat. Brain Res Dev Brain Res. 1995;85:80–88. doi: 10.1016/0165-3806(94)00197-8. [DOI] [PubMed] [Google Scholar]
- Chun HS, Yoo MS, DeGiorgio LA, Volpe BT, Peng D, Baker H, Peng C, Son JH. Marked dopaminergic cell loss subsequent to developmental, intranigral expression of glial cell line-derived neurotrophic factor. Exp Neurol. 2002;173:235–244. doi: 10.1006/exnr.2001.7842. [DOI] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- Du F, Li R, Huang Y, Li X, Le W. Dopamine D3 receptor-preferring agonists induce neurotrophic effects on mesencephalic dopamine neurons. Eur J Neurosci. 2005;22:2422–2430. doi: 10.1111/j.1460-9568.2005.04438.x. [DOI] [PubMed] [Google Scholar]
- Fischer T, Guimera J, Wurst W, Prakash N. Distinct but redundant expression of the Frizzled Wnt receptor genes at signaling centers of the developing mouse brain. Neuroscience. 2007;147:693–711. doi: 10.1016/j.neuroscience.2007.04.060. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Mueller JC, Lichtner P, Schulte C, Munz M, Berg D, Wüllner U, Illig T, Sharma M, Gasser T. The transcription factor PITX3 is associated with sporadic Parkinson's disease. Neurobiol Aging. 2009;30:731–738. doi: 10.1016/j.neurobiolaging.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Golden JP, DeMaro JA, Osborne PA, Milbrandt J, Johnson EM., Jr Expression of neurturin, GDNF, and GDNF family-receptor mRNA in the developing and mature mouse. Exp Neurol. 1999;158:504–528. doi: 10.1006/exnr.1999.7127. [DOI] [PubMed] [Google Scholar]
- Haubenberger D, Reinthaler E, Mueller JC, Pirker W, Katzenschlager R, Froehlich R, Bruecke T, Daniel G, Auff E, Zimprich A. Association of transcription factor polymorphisms PITX3 and EN1 with Parkinson's disease. Neurobiol Aging. 2011;32:302–307. doi: 10.1016/j.neurobiolaging.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Hellmich HL, Kos L, Cho ES, Mahon KA, Zimmer A. Embryonic expression of glial cell-line derived neurotrophic factor (GDNF) suggests multiple developmental roles in neural differentiation and epithelial-mesenchymal interactions. Mech Dev. 1996;54:95–105. doi: 10.1016/0925-4773(95)00464-5. [DOI] [PubMed] [Google Scholar]
- Howells DW, Porritt MJ, Wong JY, Batchelor PE, Kalnins R, Hughes AJ, Donnan GA. Reduced BDNF mRNA expression in the Parkinson's disease substantia nigra. Exp Neurol. 2000;166:127–135. doi: 10.1006/exnr.2000.7483. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Smits SM, Noorlander CW, von Oerthel L, van der Linden AJ, Burbach JP, Smidt MP. Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development. 2007;134:2673–2684. doi: 10.1242/dev.02865. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, van Erp S, van der Linden AJ, von Oerthel L, Burbach JP, Smidt MP. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136:531–540. doi: 10.1242/dev.029769. [DOI] [PubMed] [Google Scholar]
- Katsuki H, Kurimoto E, Takemori S, Kurauchi Y, Hisatsune A, Isohama Y, Izumi Y, Kume T, Shudo K, Akaike A. Retinoic acid receptor stimulation protects midbrain dopaminergic neurons from inflammatory degeneration via BDNF-mediated signaling. J Neurochem. 2009;110:707–718. doi: 10.1111/j.1471-4159.2009.06171.x. [DOI] [PubMed] [Google Scholar]
- Kholodilov N, Yarygina O, Oo TF, Zhang H, Sulzer D, Dauer W, Burke RE. Regulation of the development of mesencephalic dopaminergic systems by the selective expression of glial cell line-derived neurotrophic factor in their targets. J Neurosci. 2004;24:3136–3146. doi: 10.1523/JNEUROSCI.4506-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Aron L, Ramakers GM, Seitz S, Zhuang X, Beyer K, Smidt MP, Klein R. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol. 2007;5:e39. doi: 10.1371/journal.pbio.0050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le W, Nguyen D, Lin XW, Rawal P, Huang M, Ding Y, Xie W, Deng H, Jankovic J. Transcription factor PITX3 gene in Parkinson's disease. Neurobiol Aging. 2011;32:750–753. doi: 10.1016/j.neurobiolaging.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Li J, Dani JA, Le W. The role of transcription factor Pitx3 in dopamine neuron development and Parkinson's disease. Curr Top Med Chem. 2009;9:855–859. [PMC free article] [PubMed] [Google Scholar]
- Lin YZ, Yao SY, Veach RA, Torgerson TR, Hawiger J. Inhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J Biol Chem. 1995;270:14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- Maxwell SL, Ho HY, Kuehner E, Zhao S, Li M. Pitx3 regulates tyrosine hydroxylase expression in the substantia nigra and identifies a subgroup of mesencephalic dopaminergic progenitor neurons during mouse development. Dev Biol. 2005;282:467–479. doi: 10.1016/j.ydbio.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Meyer M, Zimmer J, Seiler RW, Widmer HR. GDNF increases the density of cells containing calbindin but not of cells containing calretinin in cultured rat and human fetal nigral tissue. Cell Transplant. 1999;8:25–36. doi: 10.1177/096368979900800112. [DOI] [PubMed] [Google Scholar]
- Mijatovic J, Airavaara M, Planken A, Auvinen P, Raasmaja A, Piepponen TP, Costantini F, Ahtee L, Saarma M. Constitutive Ret activity in knock-in multiple endocrine neoplasia type B mice induces profound elevation of brain dopamine concentration via enhanced synthesis and increases the number of TH-positive cells in the substantia nigra. J Neurosci. 2007;27:4799–4809. doi: 10.1523/JNEUROSCI.5647-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H, Okuma Y, Nomura J, Nagashima K, Nomura Y. Age-related alterations in the expression of glial cell line-derived neurotrophic factor in the senescence-accelerated mouse brain. J Pharmacol Sci. 2003;92:28–34. doi: 10.1254/jphs.92.28. [DOI] [PubMed] [Google Scholar]
- Mogi M, Togari A, Kondo T, Mizuno Y, Komure O, Kuno S, Ichinose H, Nagatsu T. Brain-derived growth factor and nerve growth factor concentrations are decreased in the substantia nigra in Parkinson's disease. Neurosci Lett. 1999;270:45–48. doi: 10.1016/s0304-3940(99)00463-2. [DOI] [PubMed] [Google Scholar]
- Oo TF, Kholodilov N, Burke RE. Regulation of natural cell death in dopaminergic neurons of the substantia nigra by striatal glial cell line-derived neurotrophic factor in vivo. J Neurosci. 2003;23:5141–5148. doi: 10.1523/JNEUROSCI.23-12-05141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oo TF, Ries V, Cho J, Kholodilov N, Burke RE. Anatomical basis of glial cell line-derived neurotrophic factor expression in the striatum and related basal ganglia during postnatal development of the rat. J Comp Neurol. 2005;484:57–67. doi: 10.1002/cne.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oo TF, Marchionini DM, Yarygina O, O'Leary PD, Hughes RA, Kholodilov N, Burke RE. Brain-derived neurotrophic factor regulates early postnatal developmental cell death of dopamine neurons of the substantia nigra in vivo. Mol Cell Neurosci. 2009;41:440–447. doi: 10.1016/j.mcn.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parain K, Murer MG, Yan Q, Faucheux B, Agid Y, Hirsch E, Raisman-Vozari R. Reduced expression of brain-derived neurotrophic factor protein in Parkinson's disease substantia nigra. Neuroreport. 1999;10:557–561. doi: 10.1097/00001756-199902250-00021. [DOI] [PubMed] [Google Scholar]
- Paratcha G, Ledda F. GDNF and GFRalpha: a versatile molecular complex for developing neurons. Trends Neurosci. 2008;31:384–391. doi: 10.1016/j.tins.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Parsian A, Sinha R, Racette B, Zhao JH, Perlmutter JS. Association of a variation in the promoter region of the brain-derived neurotrophic factor gene with familial Parkinson's disease. Parkinsonism Relat Disord. 2004;10:213–219. doi: 10.1016/j.parkreldis.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Pascual A, Hidalgo-Figueroa M, Piruat JI, Pintado CO, Gómez-Díaz R, López-Barneo J. Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat Neurosci. 2008;11:755–761. doi: 10.1038/nn.2136. [DOI] [PubMed] [Google Scholar]
- Peng C, Fan S, Li X, Fan X, Ming M, Sun Z, Le W. Overexpression of Pitx3 upregulates expression of BDNF and GDNF in SH-SY5Y cells and primary ventral mesencephalic cultures. FEBS Lett. 2007;581:1357–1361. doi: 10.1016/j.febslet.2007.02.054. [DOI] [PubMed] [Google Scholar]
- Porritt MJ, Batchelor PE, Howells DW. Inhibiting BDNF expression by antisense oligonucleotide infusion causes loss of nigral dopaminergic neurons. Exp Neurol. 2005;192:226–234. doi: 10.1016/j.expneurol.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Prakash N, Wurst W. Development of dopaminergic neurons in the mammalian brain. Cell Mol Life Sci. 2006;63:187–206. doi: 10.1007/s00018-005-5387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Smidt MP, van Schaick HS, Lanctôt C, Tremblay JJ, Cox JJ, van der Kleij AA, Wolterink G, Drouin J, Burbach JP. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci U S A. 1997;94:13305–13310. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, Smits SM, Bouwmeester H, Hamers FP, van der Linden AJ, Hellemons AJ, Graw J, Burbach JP. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- Smits SM, Burbach JP, Smidt MP. Developmental origin and fate of meso-diencephalic dopamine neurons. Prog Neurobiol. 2006;78:1–16. doi: 10.1016/j.pneurobio.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stahl K, Mylonakou MN, Skare Ø, Amiry-Moghaddam M, Torp R. Cytoprotective effects of growth factors: BDNF more potent than GDNF in an organotypic culture model of Parkinson's disease. Brain Res. 2011;1378:105–118. doi: 10.1016/j.brainres.2010.12.090. [DOI] [PubMed] [Google Scholar]
- Tomac A, Widenfalk J, Lin LF, Kohno T, Ebendal T, Hoffer BJ, Olson L. Retrograde axonal transport of glial cell line-derived neurotrophic factor in the adult nigrostriatal system suggests a trophic role in the adult. Proc Natl Acad Sci U S A. 1995;92:8274–8278. doi: 10.1073/pnas.92.18.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Munckhof P, Luk KC, Ste-Marie L, Montgomery J, Blanchet PJ, Sadikot AF, Drouin J. Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development. 2003;130:2535–2542. doi: 10.1242/dev.00464. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Cao JP, Yu JK, Zhang LC, Jiang ZJ, Gao DS. Calbindin-D28K expression induced by glial cell line-derived neurotrophic factor in substantia nigra neurons dependent on PI3K/Akt/NF-kappaB signaling pathway. Eur J Pharmacol. 2008;595:7–12. doi: 10.1016/j.ejphar.2008.07.044. [DOI] [PubMed] [Google Scholar]
- Yang D, Peng C, Li X, Fan X, Li L, Ming M, Chen S, Le W. Pitx3-transfected astrocytes secrete brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor and protect dopamine neurons in mesencephalon cultures. J Neurosci Res. 2008;86:3393–3400. doi: 10.1002/jnr.21774. [DOI] [PubMed] [Google Scholar]
- Zhao S, Maxwell S, Jimenez-Beristain A, Vives J, Kuehner E, Zhao J, O'Brien C, de Felipe C, Semina E, Li M. Generation of embryonic stem cells and transgenic mice expressing green fluorescence protein in midbrain dopaminergic neurons. Eur J Neurosci. 2004;19:1133–1140. doi: 10.1111/j.1460-9568.2004.03206.x. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]