Abstract
Several studies have linked the KIBRA rs17070145 T polymorphism to superior episodic memory in healthy humans. One study investigated the effect of KIBRA on brain activation patterns (Papassotiropoulos et al., 2006) and observed increased hippocampal activation in noncarriers of the T allele during retrieval. Noncarriers were interpreted to need more hippocampal activation to reach the same performance level as T carriers. Using large behavioral (N = 2230) and fMRI (N = 83) samples, we replicated the KIBRA effect on episodic memory performance, but found increased hippocampal activation in T carriers during episodic retrieval. There was no evidence of compensatory brain activation in noncarriers within the hippocampal region. In the main fMRI sample, T carriers performed better than noncarriers during scanning but, importantly, the difference in hippocampus activation remained after post hoc matching according to performance, sex, and age (N = 64). These findings link enhanced memory performance in KIBRA T allele carriers to elevated hippocampal functioning, rather than to neural compensation in noncarriers.
Introduction
The heritability of human episodic memory is ∼50% (McClearn et al., 1997), but only a limited number of genes have consistently been associated with memory performance (Koppel and Goldberg, 2009). The T allele of a common KIBRA (WW and C2 domain containing 1) C/T polymorphism (rs17070145) was first linked to increased episodic memory performance in a genome-wide association study and two independent replication samples by Papassotiropoulos and colleagues (2006). In subsequent studies, the T allele has been associated with increased episodic memory performance (Bates et al., 2009; Preuschhof et al., 2010; Vassos et al., 2010; but see Need et al., 2008).
Brain activation has been considered an important intermediate phenotype to extend and validate findings from behavioral genetic studies (Green et al., 2008; Rasch et al., 2010). In the original study by Papassotiropoulos et al. (2006), fMRI was used to investigate brain activation in 15 T carriers and 15 noncarriers matched for delayed recall. Noncarriers were found to over-recruit hippocampal/medial-temporal lobe (MTL) regions during episodic memory retrieval, interpreted as noncarriers needing relatively higher brain activation to reach the same performance level as T carriers.
A few other imaging genetics studies have shown increased hippocampal activation during an episodic memory task in carriers of a genotype related to poorer memory (Huentelman et al., 2007; Buchmann et al., 2008). However, in other studies, increased hippocampal/MTL activation has been reported for carriers of an allele associated with favorable memory (Hariri et al., 2003; Egan et al., 2004; Bertolino et al., 2006; Hashimoto et al., 2008), indicating elevated hippocampal functioning in carriers of the allele associated with enhanced memory.
Rasch et al. (2010) recently called for fMRI studies with large enough samples to study the impact of polymorphisms on hippocampal activation both in presence of performance differences and in performance-matched groups. Following this suggestion, we thoroughly examined the role of the KIBRA rs17070145 polymorphism on episodic memory performance and associated patterns of brain activity, with a primary focus on the hippocampal/MTL region. We predicted superior memory performance for T carriers. Of main interest was whether associated patterns of hippocampus fMRI BOLD activity would indicate elevated hippocampal functioning in T carriers or instead compensatory mechanisms in noncarriers.
Materials and Methods
Behavioral study
Subjects.
The behavioral data come from the Betula project, a longitudinal study on memory, health, and aging conducted in northern Sweden (Nilsson et al., 1997). To maximize sample size, we used cognitive data from a total of 2230 nondemented participants from the second wave of data collection that occurred 1993–1995 (age range: 35–85 years; for age, sex and genotype frequencies, see Table 1). Subjects were randomly recruited from the population registry, and participants gave their informed consent according to the guidelines of the Swedish Research Council. There were no significant differences between KIBRA genotypes and number of apolipoprotein E (APOE) ε4 alleles (χ2(4) = 3.44, p = 0.49) or genotypes of catechol-O-methyltransferase (COMT) val158met (χ2(4) = 6.7, p = 0.15) and BDNF val66met (χ2(4) = 2.90, p = 0.57).
Table 1.
Demographics and mean cognitive performance by T carrier status in the behavioral sample (N = 2230)
| Genotype | CC | CT/TT | Statistic | p |
|---|---|---|---|---|
| Females/males | 577/463 | 677/513 | χ2(1) = 0.45 | 0.5 |
| Age (SD) | 58.6 (14) | 58.2 (14) | t(2228) = −0.69 | 0.5 |
| Immediate free recall, composite score (SD) | 17.23 (4.7) | 17.82 (4.4) | F(1,2226) = 9.24 | 0.002 |
| SPT, immediate free recall | 8.51 (3.1) | 8.55 (3.2) | F(1,2226) = 0.02 | 0.9 |
| SPT, delayed cued recall | 9.87 (2.9) | 9.98 (2.8) | F(1,2225) = 1.01 | 0.3 |
| VT, immediate free recall | 4.82 (2.7) | 4.93 (2.9) | F(1,2226) = 0.84 | 0.4 |
| VT, delayed cued recall | 6.17 (3.3) | 6.26 (3.4) | F(1,2226) = 0.34 | 0.6 |
| Block design score (SD) | 26.4 (10.7) | 26.8 (10.6) | F(1,2226) = 0.43 | 0.5 |
SPT, Subject performed task; VT, verbal task.
Neuropsychological testing.
The main episodic memory variable used was a composite score of four different tests of immediate free recall of words. In each test, 12 words were read out loud to the subject (one word every 2 s), and immediately afterwards, subjects were instructed to recall the words verbally at the same pace, disregarding order. The four tests differed in the presence of a concurrent card sorting task during (1) encoding, (2) retrieval, (3) both encoding and retrieval, and (4) no distraction (Nyberg et al., 1997). Four additional episodic tasks were considered: immediate free recall and delayed category cued recall of subject performed tasks and of verbal tasks (Nilsson et al., 1997). Finally, a visuospatial block design task was included. In this task, white and red cubes were arranged to match a given two-dimensional figure outlined on paper (Wechsler, 1981).
Statistics.
ANCOVA using age and sex as covariates was used for analysis on the genotype level. All variables were normally distributed with equal variance (Levene's test of equality of error variances, p > 0.1).
Brain imaging study (functional MRI)
Subjects.
The participants belong to a large-scale fMRI sample containing a total of 376 individuals from the Betula project, for which longitudinal cognitive data and KIBRA rs17070145 genotyping were available for 293 individuals (age range: 55–80 years). The two youngest age cohorts, 55 and 60 years olds, were chosen for the main analysis. Sixteen participants were excluded in a blind data review due to poor performance on the scanner task (predefined as <50% retrieval responses and/or 10 of 24 correct responses and/or misunderstanding the instructions), missing or unreliable genotyping data, or technical problems. One subject showed a strongly deviating hippocampus beta value during retrieval (>2 SD below mean). As the subject was also found to suffer from systemic lupus erythematosus, the data was removed from the analysis as an outlier (CC genotype). Based on results from between-genotypes post hoc analysis of the behavioral data, we grouped individuals of CT and TT genotype for the fMRI analysis (see Results, below). Analyses were performed on the main sample of 83 55- to 60-year-olds as well as on a subsample of 64 participants, matched according to recall accuracy on the scanner task, sex, and age, which resulted in the removal of 19 females from the larger CT/TT group. For comparison, the same fMRI analysis was performed on participants in the age range of 65–75 years. Table 2 contains information on sex, age, and scanner task performance in the two allele groups for the fMRI subsamples. There were no significant differences in number of APOE ε4 alleles or in genotype frequencies of BDNF Val66Met and COMT Val158Met in any of the fMRI samples tested (Pearson χ2, p > 0.1).
Table 2.
Age, sex, and fMRI task performance by T carrier status in the fMRI samples
| Genotype | 55- to 60-year-olds (N = 83) |
55- to 60-year-olds, matched (N = 64) |
65- to 75-year-olds (N = 113) |
|||
|---|---|---|---|---|---|---|
| CC | CT/TT | CC | CT/TT | CC | CT/TT | |
| Females/males | 12/20 | 31/20 | 12/20 | 12/20 | 26/25 | 36/26 |
| Age | 57.50 | 57.65 | 57.50 | 57.50 | 69.02 | 69.68 |
| Names recalled | 15.44* | 16.59* | 15.44 | 15.53 | 14.41 | 14.37 |
| Response time (s) | 2.64 | 2.56 | 2.64 | 2.61 | 2.65 | 2.66 |
*p = 0.05, one-tailed.
Scanner task.
The episodic memory task used was a face–name paired-associate task (adopted from Persson et al., 2011). A blocked design consisting of three conditions was used: encoding, retrieval, and control/baseline. During encoding, face–name pairs were presented to the participants, who were instructed to memorize the name in association with each face and press a button. During retrieval, the same faces were presented together with three letters, of which one corresponded to the first letter in that face–name pair. Subjects were instructed to indicate the corresponding letter by pressing a button and respond by guessing if the name was forgotten. In the baseline condition, subjects fixated on a cross presented in the center of the visual field and pressed a button with their index finger as fast as possible whenever the cross was replaced with a circle. Six blocks of encoding and retrieval and eight blocks of baseline alternated throughout the experiment, each with a total length of 27.5 s. Instructions were shown for 4 s before each new block. For each encoding and retrieval block, four stimuli items were presented for 4 s, each with an interstimulus interval varying between 1500 and 4500 ms. At baseline, the circle was presented four times for 500 ms, with intervals varying between 3000 and 6000 ms. The whole experiment lasted for <10 min. All subjects performed a 1 min practice version of the task immediately before scanning until the instructions were properly understood. In addition, all subjects were offered a chance to familiarize themselves with the scanner environment in a 0T mock scanner before the fMRI investigation.
Image acquisition.
Functional magnetic resonance imaging was done on a 3T GE scanner with a 32 channel head coil. A gradient echoplanar imaging sequence was used with the following scanner parameters: repetition time, 2000 ms; echo time, 50 ms; flip angle, 80°; field of view, 25 cm. Thirty-seven transaxial slices with a thickness of 3.4 mm (0.5 mm gap) were acquired. Ten dummy scans were collected to allow for equilibration of the fMRI signal. Stimuli were presented on a computer screen seen by the participants through a tilted mirror attached to the head coil. Responses were made on a response pad using the right index, middle, and ring fingers. E-Prime (www.pstnet.com/eprime; Psychology Software Tools) was used for stimulus presentation and recording of responses. To reduce the scanner noise, headphones and earplugs were used, and cushions in the coil reduced head movement.
Preprocessing and data analysis.
Preprocessing of data files and data analysis was performed in SPM5 (Wellcome Department of Cognitive Neurology, London, UK) implemented in Matlab 7.8 (Mathworks). Software developed in-house (DataZ) was used for batching and visualization of statistical maps and BOLD signal changes. Before statistical analysis, all images were preprocessed in the following steps: correction for differences in slice acquisition timing within each volume, movement correction by unwarping and realignment to the first image of each volume, normalization to the MNI standard template brain, and spatial smoothing with an 8 mm FWHM Gaussian kernel. The potential problem of misalignment was considered using an alternative normalization approach using diffeomorphic anatomical registration using exponentiated lie algebra (DARTEL). Here, a sample-specific template was created from high-resolution T1-weighted images and each subject was nonlinearly normalized (using a high degree of deformation) to the template followed by affine registration to the MNI template (Nyberg et al., 2010).
The first-order analyses were set up by including the experimental conditions as regressors of interest in the general linear model, convolved with the hemodynamic response function. Six realignment parameters were included as covariates of no interest to account for movement artifacts. Group analyses were based on a random-effects model. Because of our a priori hypothesis of differential activation within the hippocampal region, we used a statistical threshold of p < 0.01 uncorrected for multiple comparisons as the level of significance within this area. At a power of 0.80 and an uncorrected alpha level of 0.05, 35 + 55 subject is sufficient for an effect size of the current main imaging finding (d = 0.62). A small volume correction would require 156 + 250 subjects (G*Power 3.1.2.). Outside the MTL region, a statistical threshold of p < 0.05, FWE corrected was used.
Genotyping.
Genomic DNA was isolated from samples of whole blood and diluted to 2.5–5.0 ng/μl. Genotyping was performed using the Sequenom iPLEX gold assay and MassARRAY MALDI-TOF mass spectrometry platform in accordance with the manufacturer's instructions. Primers for PCR amplification (5′-ACGTTGGATGAGGCTGTGGAATCTCTTGAC-3′ and 5′-ACGTTGGATGCCAGCTGCTCCTTGATCCT-3′), unextendend primer (5′-CTAAAGCTCAGGAACAGTT-3′) and extension primers (5′-CTAAAGCTCAGGAACAGTTA-3′ and 5′-CTAAAGCTCAGGAACAGTTG-3′) were designed using the Sequenom MassARRAY System Designer software.
A total of 51 additional single-nucleotide polymorphisms that were not analyzed within this study were genotyped on the same platform. Individuals with sample call rate <0.9 or indications of genotyping errors were excluded. The genotypic distribution of KIBRA rs17070145 did not deviate significantly from Hardy-Weinberg equilibrium in the behavioral sample (p > 0.001, exact test, PLINK toolbox). Genotype counts of all samples were as follows: behavioral sample: 1040 CC, 932 CT, and 258 TT genotype; fMRI sample, 55- to 60-year-olds, unmatched: 32 CC, 41 CT, and 10 TT genotype; fMRI sample, 55- to 60-year-olds, matched: 32 CC, 27 CT, and 5 TT genotype; fMRI sample, 65- to 75-year-olds: 51 CC, 50 TC, and 12 TT genotype.
Results
Behavioral results
After controlling for age and sex, there was a significant effect of KIBRA genotype on the composite measure of immediate free recall of words (F(2,2225) = 4.82, p = 0.008, two-tailed; age covariate: F(1,2225) = 827, p < 0.001; sex covariate: F(1,2225) = 55.5, p < 0.001). Post hoc testing revealed a significant difference between CC genotype and each of the other two genotypes (CT and TT, p < 0.05, Bonferroni corrected), whereas the difference between CT and TT genotype were not significant (p > 0.1). Based on these results and in agreement with previous studies (Papassotiropoulos et al., 2006), we assumed a recessive model of inheritance and grouped carriers of the T allele for subsequent analyses. Here, a main effect of T allele status was observed on immediate free recall (β = 0.502, F(1,2226) = 9.24, p = 0.002, two-tailed; Table 1). On immediate recall, there was a significant interaction of KIBRA with age (p < 0.05, larger effect with increased age) but not with sex. No significant effect of KIBRA genotype was seen on any of the other cognitive variables (p > 0.1; Table 1), although a trend in the same direction was seen for all cognitive tasks. Finally, in the younger main fMRI sample of 83 participants, the scanner task performance was significantly higher in T carriers, but no such effect was seen in the older group (Table 2). The Pearson correlation between performance on the immediate word recall composite and the scanner face–name task was 0.217 (p < 0.05) in the fMRI sample.
Brain imaging results
We examined the effect of KIBRA on hippocampal/MTL fMRI activation during episodic encoding and retrieval in the main sample (age range: 55–60 years; N = 83). First, we attempted to replicate the finding of higher retrieval activity for noncarriers of the T allele (Papassotiropoulos et al., 2006), but this contrast (CC > CT/TT) did not reveal any significant results in the hippocampal/MTL region even at a very liberal threshold (p < 0.05, uncorrected). Instead, when testing the opposite contrast (CT/TT > CC), higher hippocampal/MTL activity was observed for T carriers compared with noncarriers, with local maximum in the right hippocampus (36, −28, −10; t = 2.80, k = 26, p = 0.003). The fMRI signal change in this cluster correlated with response time (RT) (Pearson correlation = −0.242, p = 0.027; i.e., faster response time correlated positively with increased fMRI signal change) but not with recall accuracy (p > 0.1). There was also a negative relation between recall accuracy and RT (r = −0.428, p < 0.001; i.e., higher accuracy with faster response).
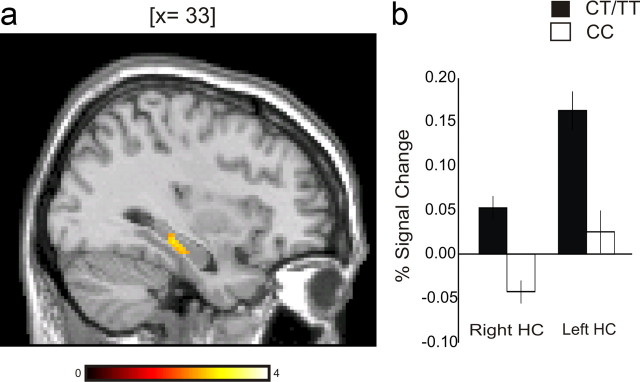
Second, to control for performance differences between genotypes, we performed the same analyses on the matched subgroup (N = 64). Vastly similar hippocampal group differences were seen as for the main sample [local maxima in the right (x, y, z = 36, −28, −10), t = 3.14, p = 0.0013 (Fig. 1) and left (x, y, z = −16, −34, −2), t = 2.61, p = 0.0056 (Fig. 1b) hippocampus]. Moreover, analyses done with the complementary normalization method (DARTEL; see Materials and Methods, above) revealed the same basic fMRI results.
Figure 1.
a, Increased right hippocampal activation in KIBRA rs17070145 T allele carriers (CT/TT genotype) relative to noncarriers (CC) during retrieval. b, Percentage signal change in local maximum of right (x, y, z = 36, −28, −10) and left (x, y, z = −16, −34, −2) hippocampus (HC). a, b, Matched groups (N = 64).
No significant group differences were observed in the hippocampal/MTL region during encoding, and there were no group differences in brain activation outside the hippocampal region during encoding or retrieval. Finally, in the older subsample (65–75 years old), we performed similar analyses and observed no group differences in the hippocampal region during encoding or retrieval.
Discussion
We used large-scale behavioral and functional MRI to study memory performance and hippocampal activation in relation to the KIBRA C/T polymorphism (rs17070145). Behaviorally, we confirmed previous findings of increased episodic memory performance in KIBRA T carriers (Papassotiropoulos et al., 2006; Bates et al., 2009; Preuschhof et al., 2010; Vassos et al., 2010). For the brain imaging data, Papassotiropoulos et al. (2006) reported enhanced hippocampal activation in noncarriers of the T allele, but here, a genotype effect on hippocampal activation was seen in the opposite direction. Critically, the same basic result was evident both when genotypes differed in memory performance and in subgroups matched for scanner task performance.
The reason for the discrepancy in hippocampus activity between studies is not clear, but it should be noted that the present study included almost three times as many subjects and is both older and geographically distant from the previous population (Papassotiropoulos et al., 2006). Furthermore, both studies involved scanner tasks taxing associative episodic memory, but the present face–name cued recall task was likely more demanding than the previous one, where items were repeated.
Numerous previous studies link hippocampus activation with successful encoding and episodic remembering in healthy subjects (Eichenbaum et al., 2007). Specifically, higher hippocampal activity during acquisition reflects more efficient memory encoding, and the degree of retrieval success relates to the magnitude of hippocampus activity (Nyberg et al., 1996; Eldridge et al., 2000). Here, increased hippocampal activation was related to faster response times during retrieval. Thus, the present observation of higher hippocampus activity in T carriers suggests that the superiority of KIBRA T carriers in episodic memory is mediated through improved hippocampal functioning.
It should be stressed that elevated hippocampal activity in T carriers remained after controlling for performance differences. Rasch et al. (2010) argued that this type of result might indicate that the genetic effect on brain activation is not related to its effect on memory functions, or alternatively that differences in hippocampus brain activity despite no performance differences could be interpreted as reflecting deeper encoding/retrieval processing that does not immediately translate into performance differences. In support of the latter view, there is much evidence from imaging studies that increased hippocampal activation can reflect more qualitative aspects of memory processing. Here, group differences were seen at retrieval and, using the remember/know paradigm, Eldridge et al. (2000) showed that hippocampus activation is stronger when remembering is built on conscious recollection of the encoded material compared with when subjects only experience a sense of familiarity. Further, Wais (2008) found a positive relation between retrieval activation in hippocampus and memory strength, as measured with confidence ratings. Thus, matching for performance does not eliminate all differences between groups, but serves to verify that the results are not driven by a difference in memory performance per se. The stronger retrieval-related hippocampus activation in T carriers, which was also seen when there were no performance differences, possibly reflecting richer and/or stronger memories, may well translate into a memory advantage over time.
We also investigated an older fMRI sample (age range: 65–75 years), but found no differences in hippocampal activation in favor of any genotype, even though an effect of KIBRA on memory performance has been previously reported in older populations (Almeida et al., 2008; Schaper et al., 2008; Bates et al., 2009) and was also seen in the present behavioral sample. As increased hippocampal activation has been related to low performance in healthy elderly subjects (Miller et al., 2008a) and to subsequent cognitive decline in elderly subjects with mild cognitive impairment (Miller et al., 2008b), imaging results from this age group might be affected by a relation of KIBRA with pathological aging processes (Rodríguez-Rodríguez et al., 2009; Corneveaux, et al., 2010).
At a cellular level, KIBRA is expressed in various brain areas, including the hippocampus, where the protein is enriched in the postsynaptic density (Johannsen et al., 2008). The KIBRA protein has been linked to memory-related processes through interaction with several postsynaptic proteins involved in synaptic plasticity and induction of long-term potentiation, including dendrin, synaptopodin, and PKCζ molecules (Büther et al., 2004; Schneider et al., 2010). Yoshihama et al. (2009) recently showed, in the mouse hippocampus, that KIBRA colocalizes with PKMζ, the only known molecule both necessary and sufficient for long-term potentiation maintenance (Sacktor, 2008), and this interaction was suggested as the functional role of KIBRA in memory.
In conclusion, together with previous molecular findings, the present results suggest that enhanced episodic memory function in KIBRA T allele carriers is mediated through elevated hippocampal processing.
Footnotes
This research was supported by a grant from the Swedish Research Council and a Wallenberg Scholar Grant from the Knut and Alice Wallenberg Foundation (to L.N.). The Betula project is supported by a grant from the Swedish Science Council (to L.G.N. and L.N.). E.E. was supported by The Swedish Research Council, Torsten and Ragnar Söderberg's Foundation, Bertil Hållstens Foundation/Swedish Brain Foundation, and the Swedish Brain Power Initiative. We thank Alireza Salami and the staff at Betula and Umeå Center for Functional Brain Imaging.
The authors declare no financial conflict of interest.
References
- Almeida OP, Schwab SG, Lautenschlager NT, Morar B, Greenop KR, Flicker L, Wildenauer D. KIBRA genetic polymorphism influences episodic memory in later life, but does not increase the risk of mild cognitive impairment. J Cell Mol Med. 2008;12:1672–1676. doi: 10.1111/j.1582-4934.2008.00229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TC, Price JF, Harris SE, Marioni RE, Fowkes FG, Stewart MC, Murray GD, Whalley LJ, Starr JM, Deary IJ. Association of KIBRA and memory. Neurosci Lett. 2009;458:140–143. doi: 10.1016/j.neulet.2009.04.050. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Blasi G, Latorre V, Rubino V, Rampino A, Sinibaldi L, Caforio G, Petruzzella V, Pizzuti A, Scarabino T, Nardini M, Weinberger DR, Dallapiccola B. Additive effects of genetic variation in dopamine regulating genes on working memory cortical activity in human brain. J Neurosci. 2006;26:3918–3922. doi: 10.1523/JNEUROSCI.4975-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann A, Mondadori CR, Hänggi J, Aerni A, Vrticka P, Luechinger R, Boesiger P, Hock C, Nitsch RM, de Quervain DJ, Papassotiropoulos A, Henke K. Prion protein M129V polymorphism affects retrieval-related brain activity. Neuropsychologia. 2008;46:2389–2402. doi: 10.1016/j.neuropsychologia.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Büther K, Plaas C, Barnekow A, Kremerskothen J. KIBRA is a novel substrate for protein kinase Cζ. Biochem Biophys Res Commun. 2004;317:703–707. doi: 10.1016/j.bbrc.2004.03.107. [DOI] [PubMed] [Google Scholar]
- Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Craig DW, Coon KD, Dunckley T, Bandy D, Lee W, Chen K, Beach TG, Mastroeni D, Grover A, Ravid R, Sando SB, et al. Evidence for an association between KIBRA and late-onset Alzheimer′s disease. Neurobiol Aging. 2010;31:901–909. doi: 10.1016/j.neurobiolaging.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, Mattay VS, Bertolino A, Hyde TM, Shannon-Weickert C, Akil M, Crook J, Vakkalanka RK, Balkissoon R, Gibbs RA, Kleinman JE, Weinberger DR. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci U S A. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H, Yonelinas AP, Ranganath C. The medial temporal lobe and recognition memory. Annu Rev Neurosci. 2007;30:123–152. doi: 10.1146/annurev.neuro.30.051606.094328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge LL, Knowlton BJ, Furmanski CS, Bookheimer SY, Engel SA. Remembering episodes: a selective role for the hippocampus during retrieval. Nat Neurosci. 2000;3:1149–1152. doi: 10.1038/80671. [DOI] [PubMed] [Google Scholar]
- Green AE, Munafò MR, DeYoung CG, Fossella JA, Fan J, Gray JR. Using genetic data in cognitive neuroscience: from growing pains to genuine insights. Nat Rev Neurosci. 2008;9:710–720. doi: 10.1038/nrn2461. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Moriguchi Y, Yamashita F, Mori T, Nemoto K, Okada T, Hori H, Noguchi H, Kunugi H, Ohnishi T. Dose-dependent effect of the Val66Met polymorphism of the brain-derived neurotrophic factor gene on memory-related hippocampal activity. Neurosci Res. 2008;61:360–367. doi: 10.1016/j.neures.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Huentelman MJ, Papassotiropoulos A, Craig DW, Hoerndli FJ, Pearson JV, Huynh KD, Corneveaux J, Hänggi J, Mondadori CR, Buchmann A, Reiman EM, Henke K, de Quervain DJ, Stephan DA. Calmodulin-binding transcription activator 1 (CAMTA1) alleles predispose human episodic memory performance. Hum Mol Genet. 2007;16:1469–1477. doi: 10.1093/hmg/ddm097. [DOI] [PubMed] [Google Scholar]
- Johannsen S, Duning K, Pavenstädt H, Kremerskothen J, Boeckers TM. Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience. 2008;155:1165–1173. doi: 10.1016/j.neuroscience.2008.06.054. [DOI] [PubMed] [Google Scholar]
- Koppel J, Goldberg T. The genetics of episodic memory. Cogn Neuropsychiatry. 2009;14:356–376. doi: 10.1080/13546800902990438. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Johansson B, Berg S, Pedersen NL, Ahern F, Petrill SA, Plomin R. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science. 1997;276:1560–1563. doi: 10.1126/science.276.5318.1560. [DOI] [PubMed] [Google Scholar]
- Miller SL, Celone K, DePeau K, Diamond E, Dickerson BC, Rentz D, Pihlajamäki M, Sperling RA. Age-related memory impairment associated with loss of parietal deactivation but preserved hippocampal activation. Proc Natl Acad Sci U S A. 2008a;105:2181–2186. doi: 10.1073/pnas.0706818105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Fenstermacher E, Bates J, Blacker D, Sperling RA, Dickerson BC. Hippocampal activation in adults with mild cognitive impairment predicts subsequent cognitive decline. J Neurol Neurosurg Psychiatry. 2008b;79:630–635. doi: 10.1136/jnnp.2007.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KN, Wagoner AP, Gumbs CE, Giegling I, Möller HJ, Francks C, Muglia P, Roses A, Gibson G, Weale ME, Rujescu D, Goldstein DB. Failure to replicate effect of KIBRA on human memory in two large cohorts of European origin. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:667–668. doi: 10.1002/ajmg.b.30658. [DOI] [PubMed] [Google Scholar]
- Nilsson LG, Bäckman L, Erngrund K, Nyberg L, Adolfsson R, Bucht Gös, Karlsson S, Widing M, Winblad B. The Betula prospective cohort study: memory, health, and aging. Aging Neuropsychol Cogn. 1997;4:1–32. [Google Scholar]
- Nyberg L, McIntosh AR, Houle S, Nilsson LG, Tulving E. Activation of medial temporal structures during episodic memory retrieval. Nature. 1996;380:715–717. doi: 10.1038/380715a0. [DOI] [PubMed] [Google Scholar]
- Nyberg L, Nilsson LG, Olofsson U, Bäckman L. Effects of division of attention during encoding and retrieval on age differences in episodic memory. Exp Aging Res. 1997;23:137–143. doi: 10.1080/03610739708254029. [DOI] [PubMed] [Google Scholar]
- Nyberg L, Salami A, Andersson M, Eriksson J, Kalpouzos G, Kauppi K, Lind J, Pudas S, Persson J, Nilsson LG. Longitudinal evidence for diminished frontal cortex function in aging. Proc Natl Acad Sci U S A. 2010;107:22682–22686. doi: 10.1073/pnas.1012651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hänggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ. Common KIBRA alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- Persson J, Kalpouzos G, Nilsson LG, Ryberg M, Nyberg L. Preserved hippocampal activation in normal aging as revealed by fMRI. Hippocampus. 2011;21:753–766. doi: 10.1002/hipo.20794. [DOI] [PubMed] [Google Scholar]
- Preuschhof C, Heekeren HR, Li SC, Sander T, Lindenberger U, Bäckman L. KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory. Neuropsychologia. 2010;48:402–408. doi: 10.1016/j.neuropsychologia.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Rasch B, Papassotiropoulos A, de Quervain DF. Imaging genetics of cognitive functions: focus on episodic memory. Neuroimage. 2010;53:870–877. doi: 10.1016/j.neuroimage.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rodríguez E, Infante J, Llorca J, Mateo I, Sánchez-Quintana C, García-Gorostiaga I, Sánchez-Juan P, Berciano J, Combarros O. Age-dependent association of KIBRA genetic variation and Alzheimer′s disease risk. Neurobiol Aging. 2009;30:322–324. doi: 10.1016/j.neurobiolaging.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Sacktor TC. PKMz, LTP maintenance, and the dynamic molecular biology of memory storage. In: Sossin WS, LAcaille JC, Castellucci VF, Belleville S, editors. Progress in brain research 169. Amsterdam: Elsevier; 2008. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- Schaper K, Kolsch H, Popp J, Wagner M, Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging. 2008;29:1123–1125. doi: 10.1016/j.neurobiolaging.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Schneider A, Huentelman MJ, Kremerskothen J, Duning K, Spoelgen R, Nikolich K. KIBRA: a new gateway to learning and memory? Front Aging Neurosci. 2010;2:4. doi: 10.3389/neuro.24.004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassos E, Bramon E, Picchioni M, Walshe M, Filbey FM, Kravariti E, McDonald C, Murray RM, Collier DA, Toulopoulou T. Evidence of association of KIBRA genotype with episodic memory in families of psychotic patients and controls. J Psychiatr Res. 2010;44:795–798. doi: 10.1016/j.jpsychires.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Wais PE. FMRI signals associated with memory strength in the medial temporal lobes: a meta-analysis. Neuropsychologia. 2008;46:3185–3196. doi: 10.1016/j.neuropsychologia.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Manual for the Wechsler Adult Intelligence Scale-Revised. New York: The Psychological Corporation; 1981. [Google Scholar]
- Yoshihama Y, Hirai T, Ohtsuka T, Chida K. KIBRA co-localizes with protein kinase Mζ (PKMζ) in the mouse hippocampus. Biosci Biotechnol Biochem. 2009;73:147–151. doi: 10.1271/bbb.80564. [DOI] [PubMed] [Google Scholar]