Abstract
The membrane-associated palmitoylated protein 5 (MPP5 or PALS1) is thought to organize intracellular PALS1-CRB-MUPP1 protein scaffolds in the retina that are involved in maintenance of photoreceptor–Müller glia cell adhesion. In humans, the Crumbs homolog 1 (CRB1) gene is mutated in progressive types of autosomal recessive retinitis pigmentosa and Leber congenital amaurosis. However, there is no clear genotype–phenotype correlation for CRB1 mutations, which suggests that other components of the CRB complex may influence the severity of retinal disease. Therefore, to understand the physiological role of the Crumbs complex proteins, especially PALS1, we generated and analyzed conditional knockdown mice for Pals1. Small irregularly shaped spots were detected throughout the PALS1 deficient retina by confocal scanning laser ophthalmoscopy and spectral domain optical coherence tomography. The electroretinography a- and b-wave was severely attenuated in the aged mutant retinas, suggesting progressive degeneration of photoreceptors. The histological analysis showed abnormal retinal pigment epithelium structure, ectopic photoreceptor nuclei in the subretinal space, an irregular outer limiting membrane, half rosettes of photoreceptors in the outer plexiform layer, and a thinner photoreceptor synaptic layer suggesting improper photoreceptor cell layering during retinal development. The PALS1 deficient retinas showed reduced levels of Crumbs complex proteins adjacent to adherens junctions, upregulation of glial fibrillary acidic protein indicative of gliosis, and persisting programmed cell death after retinal maturation. The phenotype suggests important functions of PALS1 in the retinal pigment epithelium in addition to the neural retina.
Introduction
Polarization of cells depends on the asymmetric distribution of multiprotein complexes, and these complexes provide the basis for formation of cell-type-specific junctions that are crucial for their function (Bulgakova et al., 2008). Central components of many protein complexes are scaffolding proteins, and membrane-associated guanylate kinases (MAGUKs) form a family of scaffolding proteins involved in cell polarity at tight junctions (Funke et al., 2005).
PALS1 (Protein associated with Lin7) belongs to the MAGUK family of proteins, which are scaffolding factors containing one PDZ, one SH3, and two L27 domains (Kamberov et al., 2000). The PDZ domain of PALS1 links it to CRB1, 2, and 3, whereas the N-terminal L27 domain links it to PALS1-associated tight junction protein (PATJ) or multi-PDZ domain protein 1 (MUPP1). CRB-PALS1-PATJ and -MUPP1 are the core proteins in the Crumbs complex, and the complex is conserved in mammalian cells and localizes at tight junctions or subapical regions (SARs) immediately adjacent to adherens junctions in polarized epithelia (Kamberov et al., 2000; Roh et al., 2002; Makarova et al., 2003).
In humans, mutations have been identified in the CRB1 gene in individuals with Leber congenital amaurosis (LCA), and retinitis pigmentosa (RP) type 12 characterized by preservation of para-arteriolar retinal pigment epithelium (PPRPE), RP with Coats-like exudative vasculopathy, and early-onset RP without PPRPE (den Hollander et al., 1999; den Hollander et al., 2004; Gosens et al., 2008; Richard et al., 2006). Various animal models have been used to study the function of Crumbs. In Drosophila, loss of Crumbs results in the reduction of the stalk membrane, and the photoreceptor cells undergo light-dependent degeneration (Johnson et al., 2002). Loss of CRB1 function in the mouse impairs the integrity of the outer limiting membrane (OLM), resulting in the delamination of parts of the photoreceptor layer and neuronal cell death (Mehalow et al., 2003; van de Pavert et al., 2004). However, the severity of the phenotype is strongly dependent on the genetic background as different mutations in CRB1/Crb1 cause various retinal phenotypes in human and mice. The lack of a clear genotype–phenotype correlation for CRB1/Crb1 mutations may suggest that other components of the Crumbs complex have a function influencing the severity of the retinal disease. For instance, as orthologs to Pals1, Nagie oko zebrafish lacking PALS1 display severe defects in the organization of the retinal cell layers (Wei and Malicki, 2002; Wei et al., 2006), and Drosophila flies lacking the PALS1 ortholog Stardust develop an eye phenotype characterized by a shortened stalk membrane and altered rhabdomere morphogenesis (Hong et al., 2003; Nam and Choi, 2003; Berger et al., 2007). Previously, we demonstrated the function of PALS1 by in vitro study using Pals1 shRNA in cultured retinas, which resulted in the loss of localization of CRB1, CRB2, MUPP1, VELI3, and partially of CRB3 (van Rossum et al., 2006). Here, to address the involvement of PALS1 as a component of the Crumbs complex in retinal dystrophy, we generated conditional shPals1 knockdown mice and analyzed the consequence of lowered PALS1 levels in the retina.
Materials and Methods
Generation of conditional shPals1 knockdown mice.
Animal care and experiments were performed in accordance with guidelines established at the institutions in which the experiments were performed and approved by the Animal Care and Use Committee. The shPals1 oligonucleotides, TGTTCATTGAACATGGTGAATTCAAGAGATTCACCATGTTCAATGAACTTTTTTC and TCGAGAAAAAAGTTCATTGAACATGGTGAATCTCTTGAATTCACCATGTTCAATGAACA (van Rossum et al., 2006), were annealed and subcloned into the HpaI/XhoI site of pSico-pgk-puro (Ventura et al., 2004) behind the second loxP site. A 2.9 kb PstI/KpnI fragment containing the U6 promoter, the two TATA-loxP sites, the intervening pgk-puro sequence, the shPals1 sequence, and the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) sequences was isolated from pSico-U6-loxP-pgk-puro-loxP-shPals1 and electroporated into mouse E14 embryonic stem (ES) cells, and verified by long-template PCR of a construct-specific 2.3 kb fragment using specific primers in the U6 promoter (5′-AGACAAATGGCAGTATTCATCCAC-3′) and WPRE (5′-CCACATAGCGTAAAAGGAGCAACATAG-3′) of the targeting vector. After verifying correctly targeted shPals1 ES clones, 10 different Pals1 shRNA cell clones were selected and 3 clones were used for the blastocyst injection, of which 2 gave germ line transmission (clones 7 and 13). In the studies described here, we used transgenic shPals1 mice derived from ES clone 13. Chimeric conditional floxed Pals1 shRNA mice were generated and heterozygous shPals1 (shPals1flox/+, from here on referred to as shPals1) and control mice were maintained as a cross of C57BL/6 and 129/Ola (50%/50%). To obtain shPals1 conditional knockdown mice, RxCre (Swindell et al., 2006) and Chx10Cre (Rowan and Cepko, 2004) mice, maintained as a cross of C57BL/6 and 129/Ola (50%/50%), were used for Cre-mediated recombination. RxCre mice were obtained on FVB background, and thereafter backcrossed onto C57BL/6 and 129/Ola (50%/50%), with <1% FVB background, and had no mutations in the phosphodiesterase 6b (pde6b) gene. Genotyping of the mice was done by PCR on genomic DNA. For the RxCre genotyping, 5′-GTTGGGAGAATGCTCCGTAA-3′ and 5′-GTATCCCACAATTCCTTGCG-3′ primers were used, for the Chx10Cre genotyping, 5′-GGGCACCTGGGACCAACTTCACGA-3′ and 5′-CGGCGGCGGTCACGAACTCC-3′, and for the shPals1, 5′-TGTAAAACGACGGCCAGTTGCAAGAACTCTTCCTCACG-3′ and 5′-CAGGAAACAGCTATGACCAGGCCTTCCATCTGTTGCT-3′ were used. For all analyses, wild-type, shPals1, RxCre, Chx10Cre, shPals1flox/+;RxCre/+ (from here on referred to as shPals1-RxCre) or shPals1flox/+;Chx10Cre (as shPals1-Chx10Cre) mouse littermates of either sex were used. Cre-mediated recombination was verified by PCR using the following primer pair: 5′-GGGACAGCAGAGATCCAGTT-3′ and 5′-TTCTCGAGGTCGACGGTATC-3′. We have used shPals1 and RxCre as controls for shPals1-RxCre, and shPals1 and Chx10Cre as controls for shPals1-Chx10Cre mutant mice.
Western blot analysis.
Retinas were dissected and homogenized by sonication in ice-cold RIPA buffer containing protease inhibitors. The protein concentrations of cell extracts were measured by Bradford's assay and retina lysate was loaded on a gradient SDS-PAGE gel (NuPAGE gels, Invitrogen) and subsequently transferred onto PVDF membranes. The membranes were then blocked, incubated with primary antibodies (anti-PALS1, 1/250, ProteinTech Group and anti-Actin, 1/500, Millipore Bioscience Research Reagents) and secondary antibodies (Streptavidin IRDye conjugated, Rockland Immunochemicals) in 0.3% BSA/TBS (50 mm Tris, pH 7.5, 150 mm NaCl), and washed in TBST (TBS and 0.05% Tween 20). Bands were visualized using the Odyssey Infrared Imaging system (LI-Cor Biosciences) and quantified by ImageJ (National Institutes of Health).
Confocal scanning laser ophthalmoscopy and spectral domain optical coherence tomography.
Confocal scanning laser ophthalmoscopy (cSLO) imaging was performed with HRA 1 and HRA 2 (Heidelberg Engineering) featuring up to two Argon wavelengths (488/514 nm) in the short wavelength range and two infrared diode lasers (HRA 1: 795/830 nm, HRA 2: 785/815 nm) in the long wavelength range as previously described (Seeliger et al., 2005). For spectral domain optical coherence tomography (SD-OCT) imaging, Spectralis HRA+OCT device (Heidelberg Engineering) was used as described previously (Fischer et al., 2009). Imaging was performed using the proprietary software package Eye Explorer version 3.2.1.0 (Heidelberg Engineering). Resulting data were exported and processed in Corel DrawX3.
Electroretinography.
Ganzfeld electroretinograms (ERGs) were recorded according to the procedures described previously (Seeliger et al., 2001; Tanimoto et al., 2009). Briefly, after overnight dark adaptation, mice were anesthetized with ketamine (66.7 mg/kg body weight) and xylazine (11.7 mg/kg body weight), and the pupils were dilated with tropicamide eye drops (Mydriaticum Stulln, Pharma Stulln). Single-flash responses were obtained under dark-adapted (scotopic) and light-adapted (photopic) conditions. Light adaptation was achieved with a background illumination of 30 cd/m2 starting 10 min before photopic recordings. Single white-flash stimuli ranged from −4 to 1.5 log cd · s/m2 under scotopic and from −2 to 1.5 log cd · s/m2 under photopic conditions, divided into 10 and 8 steps, respectively. Ten responses were averaged with interstimulus interval of 5 or 17 s (for 0–1.5 log cd · s/m2).
Electron microscopy analysis.
For immuno-electron microscopy (immuno-EM) analysis, eyes were processed according to the procedures described previously (Klooster et al., 2009). Briefly, eyes were fixed for 1 h at room temperature in 0.1 m phosphate-buffered (pH 6.5) 4% formaldehyde. After fixation, the eyes were cryoprotected and embedded in Tissue Tek (Sakura Finetek Europe). They were sectioned and incubated for 20–68 h with rabbit anti-PALS1 (1/200, Millipore Bioscience Research Reagents, ProteinTech). After rinsing, the sections were incubated with a PowerVisionPoly-HRP-Goat Anti-rabbit IgG (ImmunoVision Technologies). To visualize the peroxidase, the sections were incubated in a Tris–HCl diaminobenzidine (DAB) solution containing 0.03% H2O2. The DAB reaction product was then intensified by a gold-substituted silver peroxidase method. Sections were rinsed in sodium cacodylate buffer 0.1 m, pH 7.4, and postfixed for 20 min in 1% OsO4 supplemented with 1% potassium ferricyanide in sodium cacodylate buffer 0.1 m, pH 7.4. After washing in the sodium cacodylate buffer, the material was dehydrated and embedded in epoxy resin. Ultrathin sections were observed and photographed in FEI Technai 12 electron microscope. Electron micrographs were acquired as TIFF files with IMAGE II cameras.
Transmission electron microscopy analysis were performed according to the procedures described previously (van Rossum et al., 2006) with modification. Briefly, the eyes were fixed in 4% paraformaldehyde, 2.5% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.3, and postfixed in 1% osmium supplemented with sodium cacodylate buffer (0.1 m, pH 7.4). After fixation, eyes were dehydrated in a graded series of ethanol and embedded in epon 812 (Polysciences). Ultrathin sections (80 nm) were made with an Ultramicrotome UCT (Leica). Then, the sections were examined with a Zeiss EM 912 electron microscope (Zeiss), and images were captured using Gatan Micrograph software and a Bioscan camera.
Morphological and immunohistochemical analysis.
Mouse eyes (n = 3–6/age group) were harvested for morphological analysis as described previously (van de Pavert et al., 2007a; van Rossum et al., 2006). For morphological analysis, eyes were enucleated and fixed at room temperature with 4% paraformaldehyde (PFA) in 0.1 m phosphate buffer (PB, pH 7.4) for 30 min. After fixation, the eyes were dehydrated for 30 min in 30%, 50%, 70%, 90%, and 96% ethanol and embedded in Technovit 7100 (Kulzer) to make eye-embedded plastic blocks. The blocks were cut in 5 μm sections with a microtome and stained with 0.5% toluidine blue. For immunohistochemical analysis, eyes were fixed in 4% PFA in 0.1 m PB, dehydrated in 30% sucrose, and embedded in Tissue-Tek O.C.T. compound. Immunostainings were performed on cryosections using the following primary and secondary antibodies: rabbit anti-PALS1 (1/200, Millipore Bioscience Research Reagents, ProteinTech), mouse anti-β-catenin (1/300, BD), rabbit anti-CRB1 (1/100, AK2) (van de Pavert et al., 2004), rabbit anti-glial fibrillary acidic protein (GFAP) (1/200, DAKO), mouse anti-MUPP1 (1/300, BD), mouse anti-glutamine synthetase (1/300, BD), rabbit anti-MPP3 (1/100, ProteinTech), rabbit anti-Recoverin (1/200, Millipore Bioscience Research Reagents), mouse anti-PKC α (1/300, BD), and rabbit anti-Caspase-3 (cleaved form, 1/200, Cell Signaling). Cone outer segments and pedicles were stained with fluorescein-labeled peanut agglutinin lectin (PNA, 1/200, Vector Laboratories), Fluorescent-labeled secondary antibodies were donkey anti-chicken, goat anti-mouse, or goat anti-rabbit IgGs conjugated to Cy3, Alexa488, or FITC (Jackson ImmunoResearch Laboratories and Invitrogen). For retinal flat-mount analysis, eyes were fixed in 4% PFA in 0.1 m PB, and separated into the eyecup and the retina, and immunostainings were performed as mentioned above. For confocal microscopy analysis, the eyecup was flattened RPE side up on slides and coverslipped with an aqueous mounting medium and examined using a Zeiss microscope (Zeiss LSM510). Analysis and measurement of pictures were performed using Zeiss LSM browser v3.2 image software.
Quantification and measurement.
To quantify the apoptotic cells, retina sections (P6 and P18) were stained with cleaved Caspase-3 (c-CAS3) antibody and counterstained with TO-PRO-3 for nuclei. Total numbers of apoptotic cells were determined by manually counting c-CAS3 antibody-positive cells in the unit area (100 μm2) on digital images generated by Zeiss LSM browser v3.2 image software. For the measurement of cone outer segments, retina sections (P10, P18, and P30) were stained with fluorescein-labeled PNA lectin and the lengths of the stainings were measured by Zeiss LSM browser v3.2 image software. For ONL thickness, the thickness was measured at 0.5, 1, 1.5, 2, and 2.5 mm distance from the optic nerve head using Zeiss LSM browser v3.2 image software. For statistic analysis, Student's t test was applied for calculating significance and significance levels were set at <0.05.
Results
Generation of shPals1 conditional knockdown mice
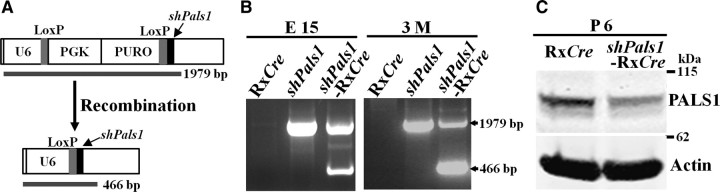
The effectiveness of the shPals1 sequences in cultured retinas and in utero brain ventricle electroporation has previously been shown (van Rossum et al., 2006; Kim et al., 2010). The conditional targeting construct (Fig. 1A) was electroporated into mouse E14 ES cells, and cells harboring the complete expression cassette were used for blastocyst injection and generation of shPals1 mice. Cre-mediated recombination was verified by PCR using retinal chromosomal DNA from wild-type, RxCre, shPals1, and shPals1-RxCre. PCR analysis confirmed the recombined DNA product (lower band, 466 bp) in shPals1-RxCre (Fig. 1B), but also indicated the presence of cells that escaped from recombination (upper band, 1979 bp). Next, we performed Western blot analysis to evaluate the reduction of PALS1 levels using total retina lysates (n = 6–8 eyes; Fig. 1C) at postnatal day 6 (P6). Upon quantification of the blots, PALS1 levels were decreased to 52 ± 3% (n = 10, ± SD) in shPals1-RxCre retinas compared to RxCre control retinas.
Figure 1.
Decreased levels of PALS1 in the retina after conditional gene silencing. A, Schematic representation of shPals1-RxCre retinas after Cre-mediated recombination. After Cre recombination, the U6 promoter drives the expression of shRNA against the Pals1 gene as illustrated. The small arrow indicates the insertion of the short hairpin Pals1 sequence behind the second loxP. PGK, Phosphoglycerate kinase promoter; PURO, puromycin gene; U6, U6 promoter. B, Genotyping of the mice (RxCre, shPals1, and shPals1-RxCre) was done by long-distance-PCR on genomic DNA isolated from E15 or 3-month-old retinas. The upper PCR bands (1979 bp) are from non-recombined and the lower bands (466 bp) are from recombined DNA products. 3M, 3 Months of age; E15, embryonic day 15. C, PALS1 protein levels were decreased to 52 ± 3% in P6 shPals1-RxCre retinas. Total retina cell lysates were used for SDS-PAGE and Western blot analysis. PALS1 antibody was used to detect PALS1 protein in the retinas and actin antibody was used for loading control. Expression levels were normalized to actin.
Reduced retinal PALS1 levels impairs vision in adult mice
To determine the phenotype, we analyzed the retinas of 1-, 3-, 6-, and 12-month-old shPals1-RxCre mice by cSLO, optical coherence tomography (OCT), and electroretinography (ERG). In cSLO and OCT analysis, all control littermate animals showed normal retinal morphology and retinal vasculature (Fig. 2A). However, the shPals1-RxCre mice showed changes in fundus appearance as well as vascular alterations especially in aged shPals1-RxCre mice. In 1- and 3-month-old shPals1-RxCre mice, an almost normal fundus appearance by cSLO examination was observed, but sites of cellular mislocalization as well as roundish structures in the outer plexiform layer (OPL) were found with OCT imaging (Fig. 2A) corresponding to the half rosettes observed in histological sections (Fig. 3A). In 6-month-old shPals1-RxCre mice, spotty and patchy areas as well as vascular leakage were observed with cSLO imaging (Fig. 2A, arrows). In 12-month-old shPals1-RxCre mice, the spotty and patchy areas and vascular leakage were more obvious and spread to the entire retina in cSLO imaging (Fig. 2A, arrows). OCT imaging revealed further sites of cellular mislocalization, as well as roundish structures in the OPL, and a reduction of the retinal thickness of the outer retina (red lines), as well as cellular mislocalization. Examinations of the retinal function in shPals1-RxCre mice by ERG showed reduced scotopic and photopic responses, with a- and b-wave amplitudes (reflecting activities of photoreceptors and ON-bipolar cells, respectively) lower than littermate controls (shPals1 and RxCre), indicating alterations of both rod and cone system components (Fig. 2B). Upon aging of shPals1-RxCre retinas, the ERG signals were severely attenuated, suggesting progressive degeneration of photoreceptors and reduced vision compared to controls (Fig. 2C).
Figure 2.
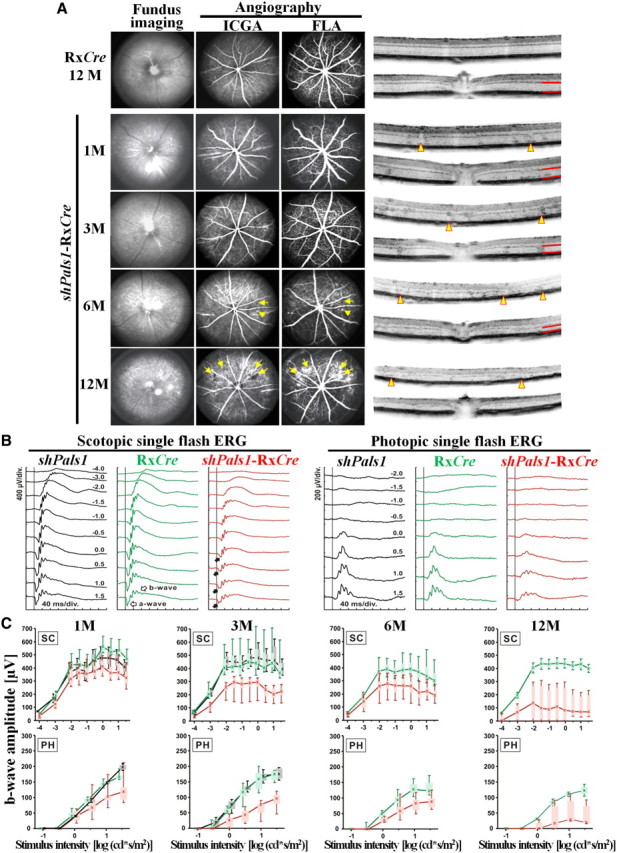
Decreased levels of PALS1 in shPals1-RxCre retinas caused early onset of loss of vision. A, cSLO and SD-OCT analysis from 1-, 3-, 6-, and 12-month-old shPals1-RxCre and control mice (RxCre). Spotty and patchy areas as well as vascular abnormalities (arrows in angiography) were found in shPals1-RxCre mouse retinas with cSLO fundus imaging. OCT analysis further revealed reduction of the retinal thickness in the outer retina as well as cellular mislocalization. Red lines indicate the thickness of the photoreceptor layer. Arrowheads indicate folded ONL regions and borders of photoreceptor degeneration. ICGA, Indocyanine green angiography; FLA, fluorescence angiography. B, Scotopic and photopic single-flash ERG responses from representative animals for a given genotype at the age of 3 months. The reduced scotopic ERG a-wave indicated photoreceptor degeneration (filled arrows in the right column). C, Single-flash ERG age series in shPals1-RxCre mutant (red), RxCre (green, control), or shPals1 (black, control) mice. Scotopic (SC, top) and photopic (PH, bottom) b-wave amplitudes were plotted as a function of the logarithm of the flash intensity. Data were obtained from 1-, 3-, 6-, and 12-month-old shPals1-RxCre, 1-, 3-, 6-, and 12-month-old RxCre, and 1- and 3-month-old control shPals1 animals. Boxes indicate the 25% and 75% quantile range, whiskers indicate the 5% and 95% quantiles, and solid lines connect the medians of the data. Note the decline of the b-wave amplitude in shPals1-RxCre mice with increasing age up to 12 months of age under both scotopic and photopic conditions, indicating degeneration of both rod and cone system components.
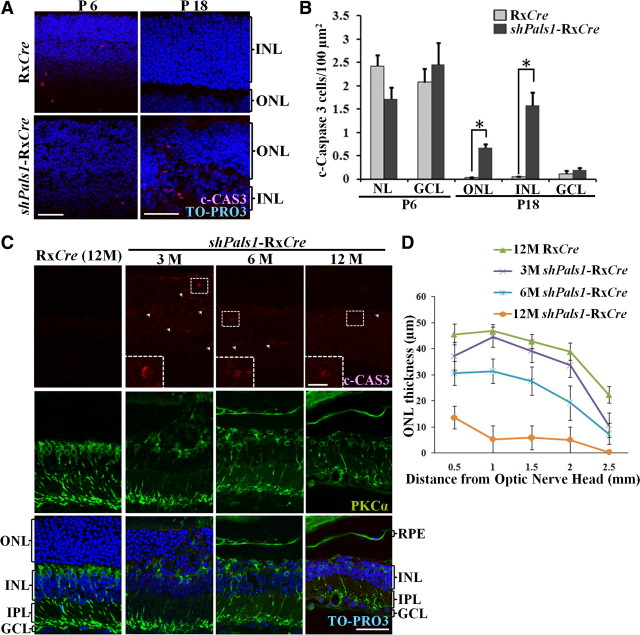
Figure 3.
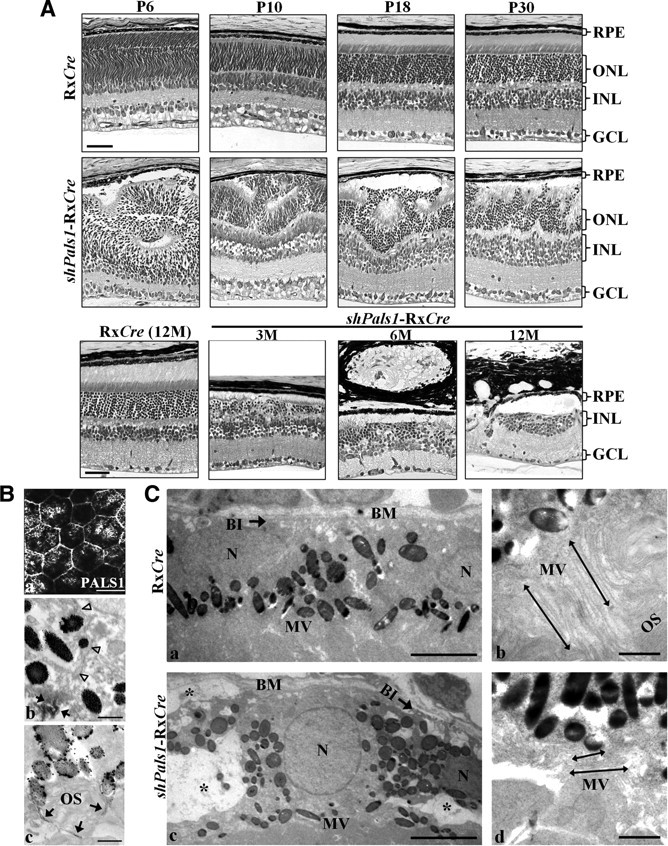
Decreased levels of PALS1 in shPals1-RxCre retinas caused retinal degeneration and abnormal RPE structure. A, Technovit retinal sections of control RxCre mice at postnatal day 6 (P6), P10, P18, and P30 and 12 months of age (12M), and shPals1-RxCre mice at P6, P10, P18, and P30 and 3, 6, and 12 months of age (3M, 6M, 12M). The phenotype in shPals1-RxCre retinas is detectable initially at the peripheral part of the retina at P6, and it spreads to the entire retina following retinal maturation. Even though the retinal layers (ONL, INL, GCL) are separated, retinal folds and half rosettes of the ONL are detected, and photoreceptor nuclei protruded into the subretinal space and ingressed into the OPL from P10 onwards. In aged mutant retinas (3–12M), the ONL and OPL become thinner due to gradual retinal degeneration. The ONL is also severely damaged and abnormal large vacuoles are detected in the RPE layer. Scale bar, 50 μm. B, Representative PALS1 staining images from immuno-EM and IHC analysis are shown from wild-type at P10. a, IHC flat mount of RPE cells stained with anti-PALS1. At immuno-EM level, the RPE monolayer has apical microvilli toward the photoreceptor outer segments and the cells are interconnected by tight junctions (b, c). Arrowheads indicate the basolateral membrane. b, c, Arrows indicate PALS1 at the tight junction (b) and in apical microvilli surrounding a photoreceptor outer segment (c). Scale bars: 20 μm (a), 0.5 μm (b, c). C, EM analysis of RxCre and shPals1-RxCre mice at P10. RPE cells appeared to be disorganized with short microvilli (double-headed arrows in d), reduced/disorganized basal infoldings (arrow in c), and in addition, numerous vacuoles were present within the cells (c, asterisk). N, RPE cell nucleus; OS, photoreceptor outer segments; Mv, microvilli; BM, Bruch's membrane; BI, basal infoldings. Scale bars: 5 μm (a, c), 1 μm (b, d).
Reduced PALS1 levels cause retinal disorganization and degeneration
To investigate the retinal shPals1-RxCre phenotype, we collected eyes from P6 to 1 year and analyzed the morphology of the retinas (Fig. 3A). In control retina at P10, the three nuclear layers, outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell nuclear layer (GCL) are well laminated, but photoreceptor segments are still short and the OPL and inner plexiform layer (IPL) synapses are not yet fully maturated (Cepko et al., 1996; Reese, 2011). At P18, the retina is almost matured and the retina morphology is similar to adult retinas. However, the shPals1-RxCre retinas showed abnormal retina structure. At P6, the GCL and IPL are well laminated, but regions of the developing INL, ONL, and OPL are disorganized. From P10 to P30, the three layers were relatively well separated, but showed retinal folds and half rosettes of photoreceptors in the ONL and OPL, and ectopic photoreceptor nuclei in the subretinal space. In addition, the OLM showed gaps and irregularities. In aged shPals1-RxCre retinas, the ONL and OPL became progressively thinner and the retina showed gradual degeneration of the ONL as well as INL. Overall, the phenotype in shPals1-RxCre retinas started with sporadically detectable disorganizations at the peripheral part of the retina at the early postnatal days (P6 till P30) and progressed throughout the entire retina following retinal maturation.
Tangential imaging of wild-type retina (P10) shows the localization of PALS1 in the apical cell membrane of retinal pigment epithelium (RPE) cells and in the apical microvilli (Fig. 3Ba). To get more detail, we examined the retina at EM level. Immuno-EM images of wild-type retinas (P10) display the localization of PALS1 at the tight junctions of RPE cells (arrows in Fig. 3Bb) without detectable staining at the basolateral membrane (Fig. 3Bb, arrowheads). PALS1 staining was also detected in RPE apical microvilli (Fig. 3Bc, arrows). In EM examination of shPals1-RxCre retinas at P10, basal infoldings (Fig. 3Cc, arrows) were disorganized and vacuoles (asterisk) were increased in number and size. The apical microvilli of the mutant RPE were either absent or shorter (Fig. 3Cd, double-headed arrows) and did not interdigitate with the outer segments of photoreceptor cells (Fig. 3Cb, double-headed arrows). From the EM results, we suggest that PALS1 is essential for the RPE structure, and especially their apical microvilli that have important roles in phagocytosis of shed photoreceptor outer segments and removal of waste products from photoreceptor cells (Bok, 1993; Bonilha et al., 2004).
The retinal phenotype depends on dosage and location of the decrease of PALS1
In RxCre mice, Cre starts to be expressed as early as embryonic day 9 in the forebrain in the eye determining field (Swindell et al., 2006). The region expressing Cre encompasses the future neural retina and the RPE. In Chx10Cre mice, Cre starts to be expressed a little later at E10.5 in the neural retina but not in the RPE (Rowan and Cepko, 2004). To examine the function of PALS1 in the early neural retina, we examined shPals1-Chx10Cre mice. In shPals1-Chx10Cre retinas, conditional recombination occurred as in shPals1-RxCre retinas (Fig. 4A), and PALS1 levels were decreased to 66 ± 4% in shPals1-Chx10Cre retinas compared to Chx10Cre control retinas (n = 6–8 eyes; n = 8 blots, ±SD; Fig. 4B). The shPals1-Chx10Cre retinas showed a significant milder phenotype than shPals1-RxCre retinas. Retinas of shPals1-Chx10Cre at 12 months of age showed normal vascular pattern in cSLO fundus imaging, but OCT imaging revealed lamination defects at the OPL (Fig. 4C). In ERG analysis, the mutant mice showed only slightly reduced scotopic and photopic responses compared to control littermates (Fig. 4D). Sections of plastic embedded retinas confirmed the morphological abnormalities observed by OCT imaging (Fig. 4E). At 3 months of age, we detected a morphological phenotype in shPals1-Chx10Cre retinas much milder than in shPals1-RxCre retinas. In shPals1-Chx10Cre retinas, we only sporadically detected gaps at the OLM, ectopic photoreceptor nuclei in the subretinal space and OPL, and upregulated GFAP (Fig. 4E and data not shown). The shPals1-Chx10Cre retinas of 12 months of age showed a thinner photoreceptor layer and OPL with shortened photoreceptor segments, and at foci disorganization of the OLM, ONL, and INL. EM analysis at P10 did not show differences between shPals1 and shPals1-Chx10Cre retinas (Fig. 4F). The milder phenotype in shPals1-Chx10Cre retinas may be the result of less efficient gene silencing of Pals1, and an intact RPE required for maintenance of the outer retina (Longbottom et al., 2009). In summary, the data from shPals1-RxCre and shPals1-Chx10Cre retinas suggests an important function for PALS1 in the RPE in addition to the neural retina.
Figure 4.
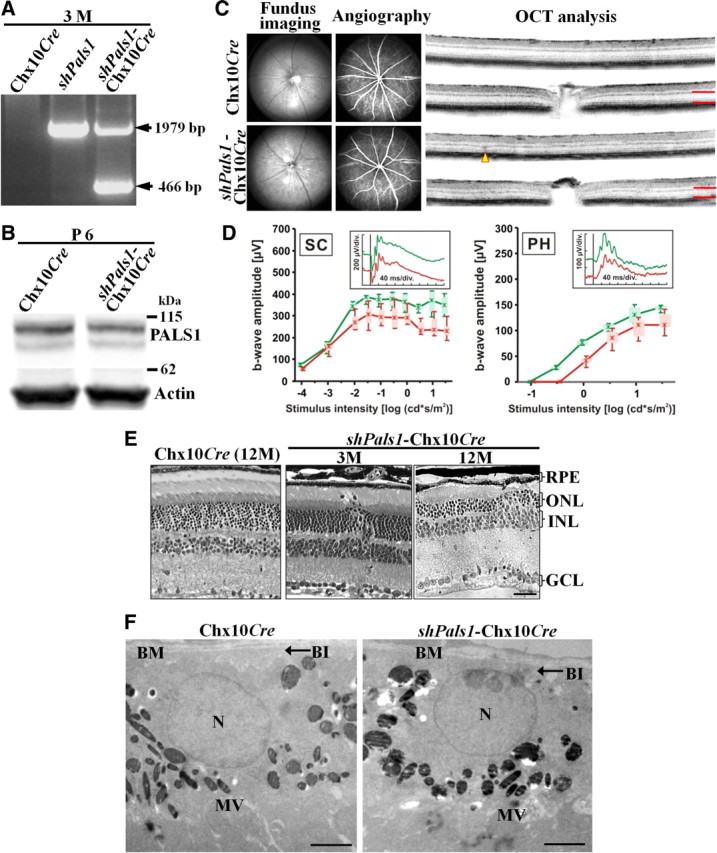
Decreased levels of PALS1 in shPals1-Chx10Cre retinas caused late onset of loss of vision. A, Genotyping of the mice (Chx10Cre, shPals1, and shPals1-Chx10Cre) was done on genomic DNA isolated from 3-month-old retinas. The lower bands (466 bp) are from shPals1-Chx10Cre recombined DNA products. B, PALS1 protein levels were decreased to 66 ± 4% in P6 shPals1-Chx10Cre retinas. PALS1 antibody was used to detect PALS1 protein in the retinas and actin antibody was used for loading control. Expression levels were normalized to actin. C, cSLO and OCT analysis of 12-month-old shPals1-Chx10Cre retinas. shPals1-Chx10Cre retinas showed almost normal fundus appearance and regular vascular pattern in cSLO fundus imaging but some mislocalized structures in the OPL are found with OCT imaging. Red lines indicate the thickness of the photoreceptor layer and arrowhead indicates folded ONL regions. D, Electroretinographic data from 12-month-old shPals1-Chx10Cre mutant mice and control Chx10Cre mice showing scotopic (SC, left) and photopic (PH, right) b-wave amplitudes from controls (green) and shPals1-Chx10Cre mice (red) as a function of the logarithm of the flash intensity. Boxes indicate the 25% and 75% quantile range, whiskers indicate the 5% and 95% quantiles, and solid lines connect the medians of the data. Insets, Scotopic (left) and photopic (right) single-flash ERGs with 1.5 log cd · s/m2 intensity of a Chx10Cre control mouse (green traces) and a shPals1-Chx10Cre mouse (red traces). The 12-month-old shPals1-Chx10Cre mice showed only slightly reduced scotopic and photopic responses compared to the Chx10Cre littermate controls. E, Morphological analysis of control Chx10Cre and shPals1-Chx10Cre retinas. At 3 months of age (3M), the shPals1-Chx10Cre retinas showed mild and sporadic phenotype. At 12 months of age (12M), the mutant retinas showed disturbed ONL and INL layers and also reduced width of the OPL. Scale bar, 50 μm. F, EM analysis of shPals1-Chx10Cre at P10. The RPE of shPals1-Chx10Cre showed normal cell structure as Chx10Cre controls. N, RPE cell nucleus; Mv, microvilli; BM, Bruch's membrane; BI, basal infoldings. Scale bar, 5 μm.
Reduced PALS1 levels disturbs the Crumbs complex and cause gliosis
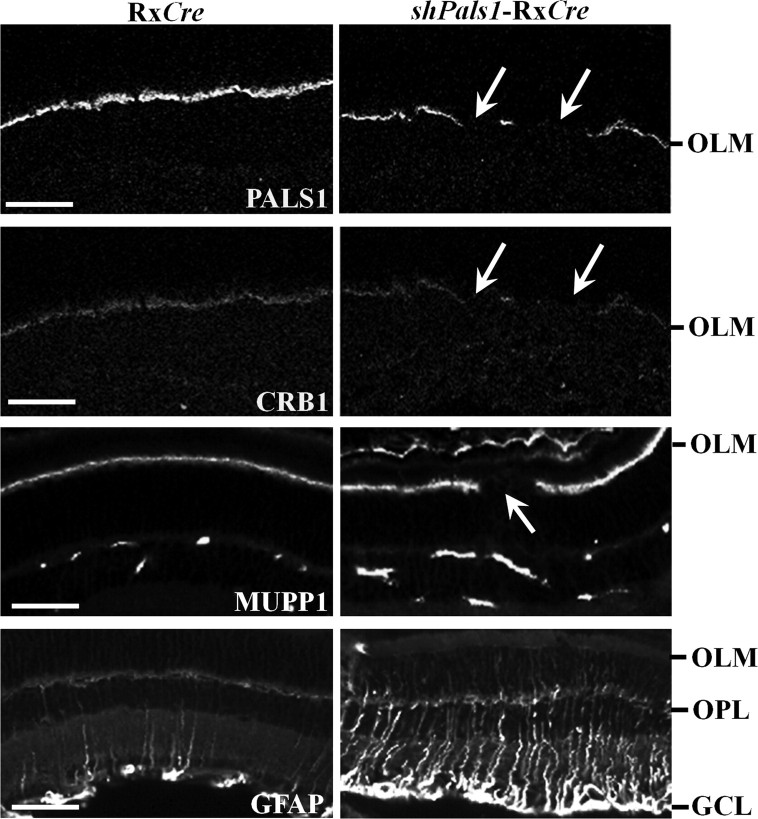
The phenotype in shPals1-RxCre and shPals1-Chx10Cre mice resembles in part the phenotype observed in Crb1 mutant mice (Mehalow et al., 2003; van de Pavert et al., 2004, 2007a,b). A striking difference is that in Crb1 mutant mice the phenotype is limited to one quadrant of the inferior retina, whereas in shPals1 conditional knockdown retinas, the phenotype is not restricted to one quadrant, resulting in significant loss of retinal activity. We therefore examined by immunohistochemistry the localization of Crumbs complex members at the SAR immediately adjacent to adherens junctions at the OLM. Whereas, at 3 month of age, in control retinas PALS1, CRB1, and MUPP1 stained the SAR adjacent to the OLM, in shPals1-RxCre retinas the staining for SAR proteins is reduced or lost especially at regions of protrusions of photoreceptor nuclei into the subretinal space and ingression of photoreceptor nuclei into the OPL (Fig. 5). Staining for markers of the adherens junctions at the OLM (cadherins, β-catenin, p120) gave similar results (data not shown). In Crb1 mutant mice, upregulation of markers for intermediate filaments in Müller glia cells such as GFAP was detected at foci of retinal disorganization (van de Pavert et al., 2004, 2007a,b). In shPals1-RxCre retinas, we detect upregulation of GFAP throughout the retina suggestive of gliosis (Fig. 5). Previous coimmunoprecipitation studies showed a complex of retinal PALS1 with CRB1 and MUPP1, immunohistochemistry showed their colocalization, and shPals1 gene silencing studies in cultured retinas showed that PALS1 is required for the localization of these proteins (van de Pavert et al., 2004; van Rossum et al., 2006). From these, we suggest that reduced levels of PALS1 in the retina result in disruptions at the OLM and apical protein complex.
Figure 5.
PALS1 localizes with CRB1 and MUPP1 to the subapical region adjacent to adherens junctions at the outer limiting membrane. Immunohistochemistry on 3-month-old retinas showing confocal images of staining for proteins associated with the Crumbs complex, PALS1, CRB1, and MUPP1, and for GFAP, a Müller glial cell marker. Staining for the Crumbs complex shows fragmented staining (arrows) of PALS1, CRB1, and MUPP1 in the shPals1-RxCre retinas, which indicate the dislocalization of the Crumbs complex proteins at the SAR adjacent to adherens junctions at the OLM. GFAP staining shows increased reactive gliosis in the mutant retinas. Scale bar, 50 μm.
Retinal cell specification is not affected, whereas retina lamination is aberrant
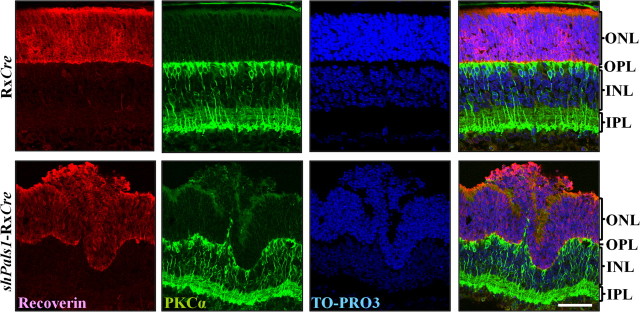
Retinal progenitor cells initiate a cell-type-specific differentiation program, controlling their migratory behavior and their morphogenesis. In this way, the different cell types achieve their laminar positioning, adult morphologies, and synaptic connections (Cepko et al., 1996; Reese, 2011). Lack of functional PALS1 in zebrafish nagie oko mutant retinas results in retinas that do not show the normal nuclear layering, the cells occupy highly abnormal positions, but cell-specification mechanisms are not affected (Wei and Malicki, 2002). To verify the cell specification and patterning in shPals1-RxCre retinas, we stained for markers of photoreceptors and bipolar and Müller glia cells. Control retinas at P10 show lamination of the retina, and normal location of bipolar cells and photoreceptors (Fig. 6). In shPals1-RxCre retinas, all cell types were generated and most but not all retinal cells were properly located in the retina. The mutant retina showed folds, some photoreceptor cells ectopically localized in the subretinal space or OPL, and some bipolar cells ectopically localized in the ONL, suggesting incorrect patterning of retinal cells. We suggest that reduced levels of PALS1 do not affect retinal cell-specification mechanisms, but affect correct patterning of newly born retinal cells, especially photoreceptors, and that may have negative consequences for establishing and maintenance of stable neuronal connections at the OPL.
Figure 6.
Retinal cell specification is normal, but cell patterning is aberrant in shPals1-RxCre retinas. At P10, in RxCre control retina, the staining of PKCα (green), a bipolar cell marker, and recoverin (red), a photoreceptor cell and cone bipolar cell marker, show well separated, properly layered cells. At P10, in shPals1-RxCre retinas, even though the retina layers are folded and some photoreceptor cells are dislocated into the subretinal space, most retina cells are separated and properly located in the retina. TO-PRO-3 was used for nuclear DNA staining (blue). Scale bar, 50 μm.
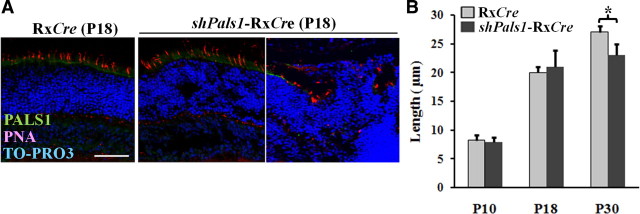
Lack of the PALS1 ortholog Stardust in the fruit fly Drosophila leads to loss of Crumbs and Patj from the stalk membrane, and to shortened stalk membranes, and increased sensitivity to light, although different Stardust mutant alleles give different phenotypes (Hong et al., 2003; Nam and Choi, 2003; Berger et al., 2007). Lack of Crumbs in Drosophila photoreceptors leads to loss of Stardust and Patj from the stalk membrane, and to shortened stalk membranes, and increased sensitivity to light (Johnson et al., 2002). Similarly, lack of CRB ortholog Crb2b in zebrafish Danio rerio leads to reduced photoreceptor segments length (Omori and Malicki, 2006). We therefore studied the length of cone outer segments in shPals1-RxCre retinas by measuring peanut agglutinin (PNA) stains of P10, P18, and P30 retinas, but detected no significant effect on outer segment length upon decreased levels of PALS1 except for foci of severely disorganized retina (Fig. 7A,B). The reduced length of cone outer segments at P30 is likely to be the consequence of the increased retinal degeneration throughout the retina (Fig. 7B). The data suggest that moderately reduced levels of PALS1 do not directly affect cone photoreceptor outer segment length.
Figure 7.
Reduced levels of PALS1 do not affect the size of cone photoreceptor outer segments. A, PNA staining (red) for cone outer segment on representative sections of RxCre control retina and two different regions (less disorganized, left panel, and more disorganized, right panel) from the same shPals1-RxCre mutant retina at P18. The subapical region adjacent to the OLM is stained by anti-PALS1 (green). Scale bar, 50 μm. B, Histogram shows the length of cone outer segments in the mutant retinas and control retinas at P10, P18, and P30. The PNA stained cone outer segments at less disturbed regions were measured to verify the involvement of PALS1 in the growth of cone outer segment during development, but there was no significant difference in P10 and P18 except for in P30 (B). Asterisk (*) indicates significant difference compared to the control (Student's t test, p < 0.0007). Error bars indicate +SEM.
Retinas with reduced levels of PALS1 undergo persistent programmed cell death
shPals1-RxCre retinas showed disorganization during late retinal development followed by progressive degeneration. Cell death is a normal process during retinal development (Young, 1984). It takes place in two waves concomitant with the processes of neurogenesis, cell migration, and cell differentiation at an early phase and when connections are established and synapses are formed, resulting in selective elimination of inappropriate connections at a later phase (Vecino et al., 2004). To study the incidence of apoptosis in shPals1-RxCre retinas, we used cleaved-Caspase-3 (c-CAS3) antibody as a marker for apoptosis. The late phase of apoptosis normally occurs around P6, and apoptosis rates are similar as in the control retinas (Fig. 8A,B). The late phase of apoptosis normally ends after P10, but shPals1-RxCre retinas showed persistent apoptosis, suggesting continuous clearance of inappropriate connections. Untimely apoptosis is detected in the photoreceptor layer as well as INL, whereas the ganglion layer is spared. In aged retinas of 3, 6, and 12 months of age, we detected loss of photoreceptors and bipolar cells (Fig. 8C,D). Our data on shPals1-RxCre retinas suggest that reduced levels of PALS1 cause inappropriate localization of photoreceptor and INL neurons that subsequently undergo programmed cell death.
Figure 8.
Retinas lacking PALS1 undergo persistent programmed cell death at maturation of the retina. A, Immunohistochemistry for c-CAS3 on representative sections of shPals1-RxCre and RxCre retinas at P6 and P18. Apoptotic cells were stained with c-CAS3 antibodies (red). Nuclear DNA was counterstained with TO-PRO-3 (blue). Scale bar, 50 μm. B, Histogram depicting the number of c-CAS3-positive cells in shPals1-RxCre and control retinas at P6 and P18. Asterisk (*) indicates significant difference compared to the control (Student's t test, p < 0.0017 for P18 OLM, p < 0.0029 for P18 INL). Error bars indicate +SEM. C, Immunohistochemistry for apoptotic cells and bipolar cells on representative sections of shPals1-RxCre and control RxCre retinas at 12 months of age. Apoptotic cells were stained with c-CAS3 antibodies (red, arrowheads), and bipolar cells with anti-PKCα (green). Nuclear DNA was counterstained with TO-PRO-3 (blue). Dashed quadrants in the upper panels indicate examples of a c-CAS3-positive cell, and are shown enlarged in the panel down on the left (scale bar, 10 μm). The number of photoreceptor and bipolar cells decreased over time. Scale bar, 50 μm. D, Quantification of the outer nuclear layer thickness. ONL thickness measured every 500 μm from the optic nerve to an area near the peripheral edge of retinal sections in control (12M) and shPals1-RxCre (3, 6, 12M). Error bars indicate ±SEM.
Discussion
In the current study, we have generated and analyzed shPals1-RxCre and shPals1-Chx10Cre mice. The phenotype in shPals1-RxCre is more severe than in shPals1-Chx10Cre retinas, suggesting a role for PALS1 in the RPE and neural retina during retinal development.
We have crossed the shPals1 mice with two different Cre lines, RxCre and Chx10Cre, both expressing Cre in retinal progenitor cells; but shPals1-RxCre and shPals1-Chx10Cre retinas show a difference in onset and severity of the retinal phenotype. We have also generated shPals1-NestinCre mice that gave as severe a retinal phenotype, in ERG, SLO, OCT, histology, and IHC analysis, as shPals1-RxCre (data not shown). The difference in severity of the phenotype might be explained by (1) difference in cell type-specific expression of Cre (expression in neural retina, or neural retina and RPE), (2) difference in time of expression of Cre and subsequently of Pals1 shRNA (from E9 in RxCre, or from E10.5 in Chx10Cre), or (3) difference in mosaic expression of Cre and silencing of PALS1 expression. The Chx10Cre mouse line used is known to express Cre in most but not all retinal progenitor cells, resulting in a mosaic retina of wild-type and mutant cells (Rowan and Cepko, 2004).
The RPE performs highly specialized, unique functions essential for homeostasis of the retina, and loss/malfunction of RPE causes loss of adhesion between the retina and RPE (Ashburn et al., 1980), and photoreceptor degeneration and atrophy of the choriocapillaris (Korte et al., 1984). Using EM analysis, we detected disturbances in mutant RPE cells such as shortened microvilli, disorganized basal infoldings, and increased number and size of vacuoles. Therefore, our data suggest that reduced levels of PALS1 in the RPE and neural retina cause the phenotype observed in shPals1-RxCre retinas. In zebrafish, PALS1 is expressed in the early RPE, and nagie oko zebrafish that completely lack PALS1 showed disturbances in RPE cells, and the phenotype could be rescued in part by expression of a nagie oko transgene in the RPE showing proper patterning of ganglion cells and cells of the INL (Zolessi et al., 2006; Zou et al., 2008; Randlett et al., 2011). Transgenic mouse lines expressing Cre in the RPE are available, but are unfortunately not useful to address the hypothesis because they either express Cre recombinase in the RPE as well as in the neural retina, or in a small subset of RPE cells, or they express postnatally (Mori et al., 2002; Delmas et al., 2003; Longbottom et al., 2009; Zhao et al., 2011).
In mouse neural retina, PALS1 localizes to the SAR adjacent to adherens junctions between photoreceptors and Müller glia cells and forming complexes with CRB1 and CRB2 in Müller glia cells, and with CRB2 in rod and cone photoreceptors (van Rossum et al., 2006). The phenotype observed in Figure 3 (e.g., gaps in the OLM, retinal folds, half-rosettes, ectopic photoreceptors in the subretinal space and OPL, progressive photoreceptor death) is in part similar to the phenotype observed in Crb1 knock-out mice. As shown in Figure 5, the Crumbs complex proteins were reduced or lost at the SAR. Therefore, PALS1 may be required for the recruitment of the Crumbs complex components, and reduced levels of PALS1 are likely to give a complex phenotype due to the reduced levels of PALS1-CRB1 and PALS1-CRB2 complexes in Müller glia cells and photoreceptors (van Rossum et al., 2006).
During retinal development, newborn neurons, which make up six of the seven major classes of cells in the retina, must be produced in proper ratios, migrate to the proper layer, and form synaptic connections with each other (Cepko et al., 1996; Wei and Malicki, 2002). Previous studies indicated that all retinal cell classes were present in nagie oko mutant retinas; the mutant showed, however, a severe disruption of retinal architecture, which indicated that zebrafish PALS1 did not have a role in cell-class specification (Wei et al., 2006). To verify the involvement of mouse PALS1 in cell specification, we used specific markers for retinal cells as shown in Figure 6. However, unlike the nagie oko mutant, most cells in the mutant retinas were well separated and migrated properly, even though the retina layers are folded and some photoreceptor cells are dislocated into the subretinal space. The data suggest that PALS1 levels are not critical for cell class specification, but are critical for proper stratification of the retina.
Similar PALS1/CRB complexes reside adjacent to adherens junctions in the developing cortex and developing and adult retina. Loss of PALS1 from the developing cortex and hippocampus resulted in premature withdrawal from the cell cycle, inducing excessive generation of early-born postmitotic neurons followed by massive programmed cell death in the mouse cortex and hippocampus (Kim et al., 2010). Even heterozygote mice with reduced levels of PALS1 in the cortex showed a significant phenotype, suggesting dosage sensitivity of PALS1. We analyzed the number of mitotic cells between E12 and E16 to examine the involvement of PALS1 in progenitor cell proliferation in the retina, but could not detect significant differences (data not shown). In shPals1-RxCre retinas, a reduction of up to 52% of PALS1 may not be sufficient to affect cell proliferation, but we cannot exclude the possibility that cell proliferation is inappropriately timed. The time course of neurogenesis in the retina is longer than in the cortex and hippocampus (Götz and Huttner, 2005), and levels of PALS1 might not be as critical in retinal progenitor cell proliferation as in cortical progenitor cell proliferation.
Footnotes
This work was supported by a grant (HEALTH-F2-2008-200234) from the European Commission, the Deutsche Forschungsgemeinschaft (DFG, Grants Se837/5-2, Se837/6-1, and Se837/7-1), and the German Ministry of Education and Research (BMBF, Grant 0314106). We thank Lucie Pellissier and Jeroen Dudok for comments that contributed to the final version of this manuscript, Marian Verhage for blastocyst injections, Penny Rashbass for anti-CRB2, and Inge Versteeg and Nienke Koot for technical assistance. Chx10Cre were provided by the The Jackson Laboratory. The transmission electron microscopy was done at the CNRS UMR 6216 and Université de la Méditérannée UMR 6216 IBDML imaging center called PICSL (Gis-Ibiza Plate-form), and the immuno-EM microscopy was done at The Netherlands Institute for Neuroscience.
References
- Ashburn FS, Jr, Pilkerton AR, Rao NA, Marak GE. The effects of iodate and iodoacetate on the retinal adhesion. Invest Ophthalmol Vis Sci. 1980;19:1427–1432. [PubMed] [Google Scholar]
- Berger S, Bulgakova NA, Grawe F, Johnson K, Knust E. Unraveling the genetic complexity of Drosophila stardust during photoreceptor morphogenesis and prevention of light-induced degeneration. Genetics. 2007;176:2189–2200. doi: 10.1534/genetics.107.071449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D. The retinal pigment epithelium: a versatile partner in vision. J Cell Sci Suppl. 1993;17:189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- Bonilha VL, Bhattacharya SK, West KA, Crabb JS, Sun J, Rayborn ME, Nawrot M, Saari JC, Crabb JW. Support for a proposed retinoid-processing protein complex in apical retinal pigment epithelium. Exp Eye Res. 2004;79:419–422. doi: 10.1016/j.exer.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Bulgakova NA, Kempkens O, Knust E. Multiple domains of Stardust differentially mediate localisation of the Crumbs-Stardust complex during photoreceptor development in Drosophila. J Cell Sci. 2008;121:2018–2026. doi: 10.1242/jcs.031088. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Martinozzi S, Bourgeois Y, Holzenberger M, Larue L. Cre-mediated recombination in the skin melanocyte lineage. Genesis. 2003;36:73–80. doi: 10.1002/gene.10197. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, van de Pol DJ, Payne AM, Bhattacharya SS, Kellner U, Hoyng CB, Westerveld A, Brunner HG, Bleeker-Wagemakers EM, Deutman AF, Heckenlively JR, Cremers FP, Bergen AA. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12) Nat Genet. 1999;23:217–221. doi: 10.1038/13848. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Davis J, van der Velde-Visser SD, Zonneveld MN, Pierrottet CO, Koenekoop RK, Kellner U, van den Born LI, Heckenlively JR, Hoyng CB, Handford PA, Roepman R, Cremers FP. CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat. 2004;24:355–369. doi: 10.1002/humu.20093. [DOI] [PubMed] [Google Scholar]
- Fischer MD, Huber G, Beck SC, Tanimoto N, Muehlfriedel R, Fahl E, Grimm C, Wenzel A, Remé CE, van de Pavert SA, Wijnholds J, Pacal M, Bremner R, Seeliger MW. Noninvasive, in vivo assessment of mouse retinal structure using optical coherence tomography. PLoS One. 2009;4:e7507. doi: 10.1371/journal.pone.0007507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- Gosens I, den Hollander AI, Cremers FP, Roepman R. Composition and function of the Crumbs protein complex in the mammalian retina. Exp Eye Res. 2008;86:713–726. doi: 10.1016/j.exer.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Hong Y, Ackerman L, Jan LY, Jan YN. Distinct roles of Bazooka and Stardust in the specification of Drosophila photoreceptor membrane architecture. Proc Natl Acad Sci U S A. 2003;100:12712–12717. doi: 10.1073/pnas.2135347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Grawe F, Grzeschik N, Knust E. Drosophila crumbs is required to inhibit light-induced photoreceptor degeneration. Curr Biol. 2002;12:1675–1680. doi: 10.1016/s0960-9822(02)01180-6. [DOI] [PubMed] [Google Scholar]
- Kamberov E, Makarova O, Roh M, Liu A, Karnak D, Straight S, Margolis B. Molecular cloning and characterization of Pals, proteins associated with mLin-7. J Biol Chem. 2000;275:11425–11431. doi: 10.1074/jbc.275.15.11425. [DOI] [PubMed] [Google Scholar]
- Kim S, Lehtinen MK, Sessa A, Zappaterra MW, Cho SH, Gonzalez D, Boggan B, Austin CA, Wijnholds J, Gambello MJ, Malicki J, LaMantia AS, Broccoli V, Walsh CA. The apical complex couples cell fate and cell survival to cerebral cortical development. Neuron. 2010;66:69–84. doi: 10.1016/j.neuron.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klooster J, Yazulla S, Kamermans M. Ultrastructural analysis of the glutamatergic system in the outer plexiform layer of zebrafish retina. J Chem Neuroanat. 2009;37:254–265. doi: 10.1016/j.jchemneu.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Korte GE, Reppucci V, Henkind P. RPE destruction causes choriocapillary atrophy. Invest Ophthalmol Vis Sci. 1984;25:1135–1145. [PubMed] [Google Scholar]
- Longbottom R, Fruttiger M, Douglas RH, Martinez-Barbera JP, Greenwood J, Moss SE. Genetic ablation of retinal pigment epithelial cells reveals the adaptive response of the epithelium and impact on photoreceptors. Proc Natl Acad Sci U S A. 2009;106:18728–18733. doi: 10.1073/pnas.0902593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B. Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1) Gene. 2003;302:21–29. doi: 10.1016/s0378111902010843. [DOI] [PubMed] [Google Scholar]
- Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, Bechtold L, Haider NB, Tepass U, Heckenlively JR, Chang B, Naggert JK, Nishina PM. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet. 2003;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- Mori M, Metzger D, Garnier JM, Chambon P, Mark M. Site-specific somatic mutagenesis in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2002;43:1384–1388. [PubMed] [Google Scholar]
- Nam SC, Choi KW. Interaction of Par-6 and Crumbs complexes is essential for photoreceptor morphogenesis in Drosophila. Development. 2003;130:4363–4372. doi: 10.1242/dev.00648. [DOI] [PubMed] [Google Scholar]
- Omori Y, Malicki J. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol. 2006;16:945–957. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- Randlett O, Norden C, Harris WA. The vertebrate retina: a model for neuronal polarization in vivo. Dev Neurobiol. 2011;71:567–583. doi: 10.1002/dneu.20841. [DOI] [PubMed] [Google Scholar]
- Reese BE. Development of the retina and optic pathway. Vision Res. 2011;51:613–632. doi: 10.1016/j.visres.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Roepman R, Aartsen WM, van Rossum AG, den Hollander AI, Knust E, Wijnholds J, Cremers FP. Towards understanding CRUMBS function in retinal dystrophies. Hum Mol Genet. 2006;15(Spec No 2):R235–R243. doi: 10.1093/hmg/ddl195. [DOI] [PubMed] [Google Scholar]
- Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J Cell Biol. 2002;157:161–172. doi: 10.1083/jcb.200109010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Seeliger MW, Zrenner E, Apfelstedt-Sylla E, Jaissle GB. Identification of Usher syndrome subtypes by ERG implicit time. Invest Ophthalmol Vis Sci. 2001;42:3066–3071. [PubMed] [Google Scholar]
- Seeliger MW, Beck SC, Pereyra-Muñoz N, Dangel S, Tsai JY, Luhmann UF, van de Pavert SA, Wijnholds J, Samardzija M, Wenzel A, Zrenner E, Narfström K, Fahl E, Tanimoto N, Acar N, Tonagel F. In vivo confocal imaging of the retina in animal models using scanning laser ophthalmoscopy. Vision Res. 2005;45:3512–3519. doi: 10.1016/j.visres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Swindell EC, Bailey TJ, Loosli F, Liu C, Amaya-Manzanares F, Mahon KA, Wittbrodt J, Jamrich M. Rx-Cre, a tool for inactivation of gene expression in the developing retina. Genesis. 2006;44:361–363. doi: 10.1002/dvg.20225. [DOI] [PubMed] [Google Scholar]
- Tanimoto N, Muehlfriedel RL, Fischer MD, Fahl E, Humphries P, Biel M, Seeliger MW. Vision tests in the mouse: Functional phenotyping with electroretinography. Front Biosci. 2009;14:2730–2737. doi: 10.2741/3409. [DOI] [PubMed] [Google Scholar]
- van de Pavert SA, Kantardzhieva A, Malysheva A, Meuleman J, Versteeg I, Levelt C, Klooster J, Geiger S, Seeliger MW, Rashbass P, Le Bivic A, Wijnholds J. Crumbs homologue 1 is required for maintenance of photoreceptor cell polarization and adhesion during light exposure. J Cell Sci. 2004;117:4169–4177. doi: 10.1242/jcs.01301. [DOI] [PubMed] [Google Scholar]
- van de Pavert SA, Meuleman J, Malysheva A, Aartsen WM, Versteeg I, Tonagel F, Kamphuis W, McCabe CJ, Seeliger MW, Wijnholds J. A single amino acid substitution (Cys249Trp) in Crb1 causes retinal degeneration and deregulates expression of pituitary tumor transforming gene Pttg1. J Neurosci. 2007a;27:564–573. doi: 10.1523/JNEUROSCI.3496-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert SA, Sanz AS, Aartsen WM, Vos RM, Versteeg I, Beck SC, Klooster J, Seeliger MW, Wijnholds J. Crb1 is a determinant of retinal apical Muller glia cell features. Glia. 2007b;55:1486–1497. doi: 10.1002/glia.20561. [DOI] [PubMed] [Google Scholar]
- van Rossum AG, Aartsen WM, Meuleman J, Klooster J, Malysheva A, Versteeg I, Arsanto JP, Le Bivic A, Wijnholds J. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Muller glia cells. Hum Mol Genet. 2006;15:2659–2672. doi: 10.1093/hmg/ddl194. [DOI] [PubMed] [Google Scholar]
- Vecino E, Hernández M, García M. Cell death in the developing vertebrate retina. Int J Dev Biol. 2004;48:965–974. doi: 10.1387/ijdb.041891ev. [DOI] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31:150–157. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- Wei X, Zou J, Takechi M, Kawamura S, Li L. Nok plays an essential role in maintaining the integrity of the outer nuclear layer in the zebrafish retina. Exp Eye Res. 2006;83:31–44. doi: 10.1016/j.exer.2005.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell death during differentiation of the retina in the mouse. J Comp Neurol. 1984;229:362–373. doi: 10.1002/cne.902290307. [DOI] [PubMed] [Google Scholar]
- Zhao C, Yasumura D, Li X, Matthes M, Lloyd M, Nielsen G, Ahern K, Snyder M, Bok D, Dunaief JL, LaVail MM, Vollrath D. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolessi FR, Poggi L, Wilkinson CJ, Chien CB, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Lathrop KL, Sun M, Wei X. Intact retinal pigment epithelium maintained by Nok is essential for retinal epithelial polarity and cellular patterning in zebrafish. J Neurosci. 2008;28:13684–13695. doi: 10.1523/JNEUROSCI.4333-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]