Abstract
microRNA-9-2 and microRNA-9-3 double-mutant mice demonstrate that microRNA-9 (miR-9) controls neural progenitor proliferation and differentiation in the developing telencephalon by regulating the expression of multiple transcription factors. As suggested by our previous study, the Foxg1 expression was elevated, and the production of Cajal-Retzius cells and early-born neurons was suppressed in the miR-9-2/3 double-mutant pallium. At embryonic day 16.5 (E16.5), however, the Foxg1 expression was no longer elevated. The expression of an AU-rich RNA-binding protein Elavl2 increased at E16.5, Elav2 associated with Foxg1 3′ untranslated region (UTR), and it countered the Foxg1 suppression by miR-9. Later, progenitor proliferation was reduced in the miR-9-2/3 double-mutant pallium with the decrease in Nr2e1 and Pax6 expression and the increase in Meis2 expression. The analyses suggest that microRNA-9 indirectly inhibits Pax6 expression by suppressing Meis2 expression. In contrast, together with Elavl1 and Msi1, microRNA-9 targets Nr2e1 mRNA 3′ UTR to enhance the expression. Concomitantly, cortical layers were reduced, each cortical projection was malformed, and the tangential migration of interneurons into the pallium was impaired in the miR-9-2/3 double mutants. miR-9 also targets Gsh2 3′ UTR, and Gsh2, as well as Foxg1, expression was elevated in the miR-9-2/3 double-mutant subpallium. The subpallium progenitor proliferation was enhanced, and the development of basal ganglia including striatum and globus pallidus was suppressed. Pallial/subpallial boundary shifted dorsally, and the ventral pallium was lost. Corridor was malformed, and thalamocortical and corticofugal axons were misrouted in the miR-9-2/3 double mutants.
Introduction
Development of mammalian telencephalon depends on the highly ordered regulation of neural progenitor proliferation/differentiation and neuronal migration. In the pallium, younger projection neurons migrate radially and settle between the marginal zone and subplate to create the cortical plate in an inside-out manner. Cajal-Retzius cells in the marginal zone provide a critical instructive cue in the six-layer formation (Huang, 2009). The cortical projection neurons are generated from two major progenitor pools: radial glia in the ventricular zone (VZ) and intermediate progenitor cells (IPCs) in the subventricular zone (SVZ) (Guillemot et al., 2006). Several transcription factors control the proliferation/differentiation of these cells. Foxg1 regulates progenitor proliferation and Cajal-Retzius cell differentiation (Dou et al., 1999; Hanashima et al., 2002, 2004; Martynoga et al., 2005; Muzio and Mallamaci, 2005). Ngn1/2 and Tbr1 regulate the production of early-born neurons, Tbr2 directs IPC generation, and Pax6, Nr2e1/Tlx, and Id4 regulate the generation of later-born neurons (Yun et al., 2004; Bedford et al., 2005; Guillemot et al., 2006; Molyneaux et al., 2007). In the subpallium, lateral (LGE) and medial (MGE) ganglionic eminences generate distinct nuclei, including the striatum and the globus pallidus, respectively. The proliferation/differentiation of the subpallial progenitor cells are regulated by Mash1, Dlx1/2/5/6, Nkx2.1, and Gsh2 (Anderson et al., 1997; Horton et al., 1999; Sussel et al., 1999; Toresson et al., 2000; Yun et al., 2001; Flames et al., 2007; Molero et al., 2009).
The expression of these genes must be finely regulated for proper neurogenesis. microRNAs are small, noncoding RNAs that typically bind to specific sequences in the 3′ untranslated regions (3′ UTR) of target mRNAs to fine-tune protein expression (Bartel, 2009). Conditional mutants of the Dicer1 gene, which is essential to the biosynthesis of almost all microRNAs, have indicated that microRNAs are required for brain development (De Pietri Tonelli et al., 2008). However, the contribution of each microRNA has been essentially unexplored. microRNA-9 (miR-9) and complementary miR-9* are abundantly expressed in vertebrate brains (Lagos-Quintana et al., 2002; Griffiths-Jones, 2006; Kloosterman et al., 2006). In zebrafish, a knockdown study demonstrated that miR-9 modulates the organizing activity of the midbrain–hindbrain boundary by suppressing Fgf signaling pathway (Leucht et al., 2008). Several studies have suggested that miR-9/miR-9* targets Nr2e1, REST, CoREST, and BAF53a to suppress progenitor proliferation and promotes neural differentiation (Packer et al., 2008; Yoo et al., 2009; Zhao et al., 2009). miR-9 promotes proliferation of human embryonic stem cell-derived neural progenitors by targeting stathmin (Delaloy et al., 2010). We have proposed that miR-9 targets Foxg1 to regulate Cajal-Retzius cell production (Shibata et al., 2008). In this study, we generated mutant mice lacking miR-9-2 and miR-9-3 (referred to as miR-9-2/3 double mutants) to examine the miR-9/miR-9* functions in telencephalic development. miR-9-2/3 double mutants exhibited dysregulation of pallial and subpallial progenitor proliferation/differentiation. Concomitantly, the double mutants exhibited multiple defects in the telencephalic structures. These defects are considered to be brought about by the dysregulation of Foxg1, Nr2e1, Gsh2, and Meis2 expression. Furthermore, we propose that miR-9 functions are modulated by RNA-binding proteins.
Materials and Methods
Generation of miR-9-2 and miR-9-3 mutant mice.
microRNA-9-2 and microRNA-9-3 mutant mice (Center for Developmental Biology accession numbers CDB0072K and CDB0071K; http://www.cdb.riken.jp/arg/mutant%20mice%20list.html) were generated by gene targeting with TT2 embryonic stem cells as described previously (Yagi et al., 1993). Genomic DNA fragments were isolated by long-amplification PCR with C57BL/6 BAC clones to construct the targeting vector (Murata et al., 2004). The PrDT-A pA cassette is the diphtheria toxin A fragment gene with an MC1 promoter and β-globin poly(A) signal, and the PrNeo poly(A) cassette is the neomycin-resistant gene with a Pgk1 promoter and simian virus 40 (SV40) poly(A) signal; they were flanked by loxP sequences as described (http://www.cdb.riken.jp/arg/cassette.html). Chimeras were mated, and mutant mice were maintained by mating with C57BL/6 mice. Southern hybridization to identify the mutants was performed as described (Murata et al., 2004). Details of the targeting strategy will be provided on request. Genotyping of mice or embryos was routinely performed with specimens of tail, yolk sac, or a part of embryonic tissue by PCR using a mixture of two or three primers. Primer sets used are p1 (AAACTCCTTCAAGGTCACCGAG), p2 (CCCTTTTCCCTGCTAGCTCTAAG), p3 (ACCCGTGATATTGCTGAAGAGCTTGG), p4 (CCTTTGCCTTCACCCCATAA), and p5 (CGTCCGACCAGAACTATAGAGGTTTA), of which locations are indicated in supplemental Figure 2 (available at www.jneurosci.org as supplemental material). The PCR with p1–p3 primers yields 517 and 624 bp products for the wild-type (WT) and mutant (MT) miR-9-2 alleles and with p3–p5 606 and 856 bp products for the wild-type and mutant miR-9-3 alleles, respectively. Two mutant mouse lines were established from independent homologous recombinant embryonic stem cell clones of both miR-9-2 and miR-9-3 loci; no difference in phenotype was observed between the two mutant lines of each locus. Two independent lines were also established for miR-9-2/3 double mutants, giving no apparent difference in phenotype. miR-9-2 single homozygous pups died within 1 month of birth; miR-9-3 single homozygous pups grew to adulthood, but both male and female homozygous mutants were infertile. miR-9-3 homozygous and miR-9-2 heterozygous double mutants died within 1 month of birth, and miR-9-2/3 double homozygous mutants were obtained by crosses between double heterozygotes. Mice were housed in environmentally controlled rooms of the Animal Housing Facility of the Center for Developmental Biology, RIKEN Kobe under the institute guidelines for animal and recombinant DNA experiments.
Plasmid construction.
The procedure to generate luciferase reporter and expression vector was described previously (Shibata et al., 2008). To generate luciferase reporter constructs, wild-type and mutant DNA fragments harboring the putative miR-9 responsive elements from the Meis2, Islet1, Gsh2, and Pax6 3′ UTRs were synthesized as DNA oligonucleotides. Their sizes were 48–52 bp, and target sequences were placed in the middle of each fragment: top strand, 5′-ggCGTCTCTGGGCTGATTACTTAACCAAAGACAAAATAATGAGTCCTAA-3′ and bottom strand, 5′-ccTTAGGACTCATTATTTTGTCTTTGGTTAAGTAATCAGCCCAGAGACG-3′ for Meis2; top strand, 5′-ggCGTCTCTGGGCTGATTACTTAACGTTTGACAAAATAATGAGTCCTAA-3′ and bottom strand, 5′-ccTTAGGACTCATTATTTTGTCAAACGTTAAGTAATCAGCCCAGAGACG-3′ for mutant Meis2; top strand, 5′-ccTGTCTGTCCAAGAACTTTTCCCCCAAAGATGTGTATAGTTATTGGTTA-3′ and bottom strand, 5′-ggTAACCAATAACTATACACATCTTTGGGGGAAAAGTTCTTGGACAGACA-3′ for Islet1; top strand, 5′-ccTGTCTGTCCAAGAACTTTTCCCCGTTTGATGTGTATAGTTATTGGTTA-3′ and bottom strand, 5′-ggTAACCAATAACTATACACATCAAACGGGGAAAAGTTCTTGGACAGACA-3′ for mutant Islet1; top strand, 5′-ggCTCCTCCCCATCAGCACAGGGACCAAAGTTCTAGTCCATTGCCCTCAT-3′ and bottom strand, 5′-ccATGAGGGCAATGGACTAGAACTTTGGTCCCTGTGCTGATGGGGAGGAG-3′ for Gsh2; top strand, 5′-ggCTCCTCCCCATCAGCACAGGGACGTTTGTTCTAGTCCATTGCCCTCAT-3′ and bottom strand, 5′-ccATGAGGGCAATGGACTAGAACAAACGTCCCTGTGCTGATGGGGAGGAG-3′ for mutant Gsh2; and top strand, 5′-ggGGAGCCCGGGAATGGACTAGAACCAAGGACCTTTGCGTACAGAAGGC-3′ and bottom strand, 5′-ccGCCTTCTGTACGCAAAGGTCCTTGGTTCTAGTCCATTCCCGGGCTCC-3′ for Pax6. Underlining indicate sequences predicted to base pair with the seed sequence of miR-9; bold letters indicate mutated nucleotides. To construct reporters with mutant Foxg1 3′ UTR in Figure 4D (Foxg1 3′ UTR MT1, MT2, MT1 + 2, and MT3), T and A were substituted by G and C, respectively, as indicated. For the Nr2e1 reporter construct, a DNA fragment containing the full-length Nr2e1 3′ UTR (1398 bp) was amplified from clone 3425403H08, obtained from the Functional Annotation of Mouse-3 (FANTOM3) library (Carninci et al., 2005). Foxg1 reporter (luc–Foxg1), pEF1α–miR-9-2Wt and pEF1α–miR-9-2Mt were constructed previously (Shibata et al., 2008).
Figure 4.
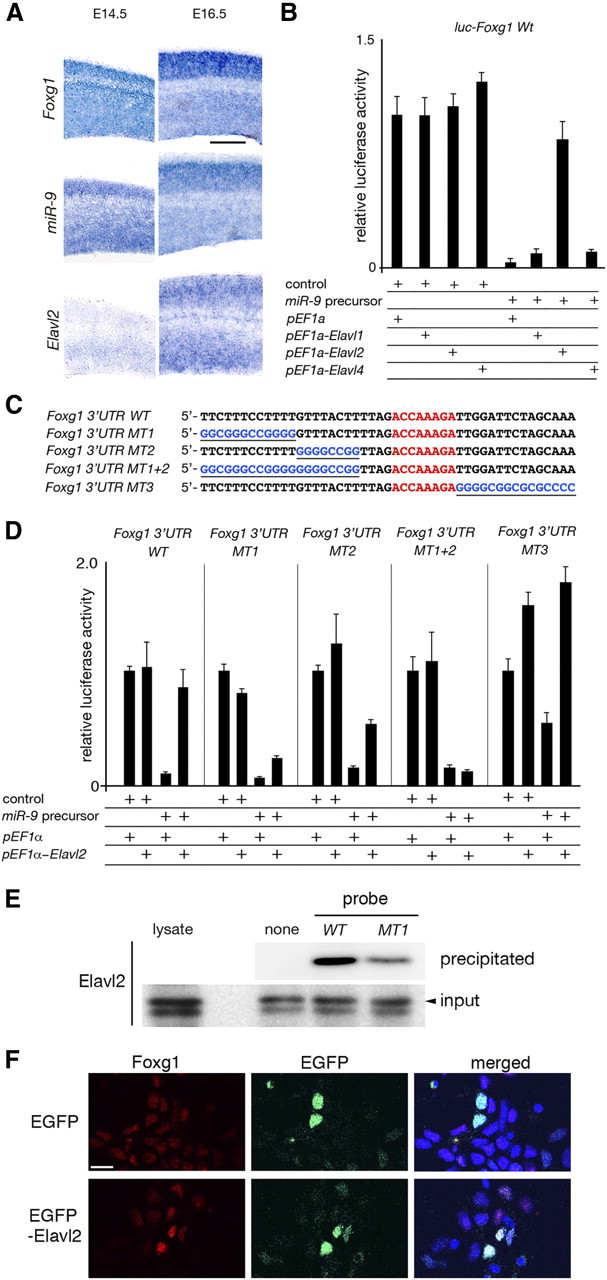
Elavl2 counteraction of the miR-9-2 suppression of Foxg1 expression. A, RNA in situ hybridization of Foxg1, miR-9, and Elavl2 expression in the wild-type neopallium at E14.5 and E16.5. B, Effects of Elavl paralogs on the miR-9-2 suppression of luciferase expression from a luciferase reporter conjugated to the Foxg1 3′ UTR in P19 cells. See Materials and Methods for relative luciferase activity. C, Sequences of the wild-type Foxg1 U-rich 3′ UTR and its mutant forms; miR-9 responsive elements are indicated by red letters, and substituted sequences are underlined and in blue letters. D, Luciferase expression from luciferase reporters conjugated to the wild-type and mutant Foxg1 3′ UTRs described in C. E, Elavl2 protein interaction with Foxg1 3′ UTR. Protein lysates from E17.5 cortices were incubated with biotinylated Foxg1 3′ UTR WT or Foxg1 3′ UTR MT1 RNA probe. Probes were precipitated with streptavidin-coated beads, and bound Elavl2 protein was detected by Western blotting using anti-Elavl2 antibody. F, Increase in Foxg1 expression by Elavl2 overexpression in E12.5 cortical cells. E12.5 cortical cells were dissociated, electroporated with control EGFP or EGFP--Elavl2 expression vector, and cultured for 48 h. Foxg1 expression was examined by immunostaining. Scale bars: A, 200 μm; F, 10 μm.
To express Elavl1, Elavl2, Elavl4, and Msi1 in the P19 cells, the protein-coding regions of Elavl1 (clone A630073F18; FANTOM3), Elavl2 (IMAGE identification number 6821133; Open Biosystems), Elavl4 (clone 3110037G03; FANTOM3), and Musashi1 (clone D130076N11; FANTOM3) cDNAs were amplified by PCR and inserted into the pEF-1α vector (Invitrogen). Primer sets used were as follows: forward, 5′-ataagaatgcggccgcATGTCTAATGGTTATGA-3′ and reverse, 5′-atagtttagcggccgcTTATTTGTGGGACTTGT-3′ for Elavl1; forward, 5′-ataagaatgcggccgcAACAAAGCAATATGAGGT-3′ and reverse, 5′-atagtttagcggccgcTTAGGCTTTGTGCGTTT-3′ for Elavl2; forward, 5′-ataagaatgcggccgc ATGGAGTGGAATGGCTT-3′ and reverse, 5′-atagtttagcggccgcTCAGGATTTGTGGGCTT-3′ for Elavl4; and forward, 5′-ataagaatgcggccgccaccATGTTCATCGGAGGACTCAG-3′ and reverse, 5′-atagtttagcggccgcTTACCTCCGTCTGCCTG-3′ for Msi1. Lowercase letters indicate linker sequences, and underlining indicate the NotI site. To express wild-type or mutant miR-9-2 in the P19 cells, ∼200 bp of wild-type or mutant DNA fragments obtained in the previous study (Shibata et al., 2008) were inserted into a modified pGK vector (Gene Bridge) at the NotI site to produce pGK–miR-9-2Wt or pGK–miR-9-2Mt. For electroporation into the telencephalon, the protein-coding region of a human Gsh2 cDNA clone (IMAGE identification number 30915389; Open Biosystems) and mouse Foxg1 cDNA (IMAGE identification number 6314329; Open Biosystems) were amplified by PCR and inserted into pCMS–EGFP vector (SV40–EGFP; Clontech). Primer sets used were as follows: forward, 5′-gctctagacaccATGTCGCGCTCCTTCTA-3′ (underlining indicates XbaI site) and reverse, 5′-gctctagaTCATAAGGGGGAAATCT-3′ for Gsh2; and forward, 5′-ggaattccaccATGCTGGACATGGGAGATAG-3′ (underlining indicates EcoRI site) and reverse, 5′-gctctagaCTAATGTATTAAAGGGTTGG-3′ (underlining indicates XbaI site) for Foxg1. Lowercase letters indicate linker sequences.
In situ hybridization.
Section in situ hybridization using antisense DIG-labeled RNA was performed as described previously (Shibata et al., 2008). All cDNAs used for the DIG–RNA probe synthesis were obtained from the FANTOM3 library, except for Wnt3a and Svet1. cDNAs for Wnt3a and Svet1 probes were gifts from Drs. A. McMahon (Harvard University, Cambridge, MA) and P. Gruss (Max Planck Institute of Biophysical Chemistry, Göttingen, Germany), respectively. To detect miR-9-1 and miR-9-3 precursors, 244 and 397 bp DNA fragments containing loop and miR-9* sequences were amplified from the mouse genome using T7-tagged reverse primers, and amplified PCR fragments were used as templates to generate probes. Primer sets used were as follows: forward, 5′-AGTGGTGTGGAGTCTTCATAAAGCT-3′ and reverse, 5′-ccggatcctaatacgactcactatagggCCAGAGCGGCTTCTTGCTT-3′ (lowercase letters indicate T7 promoter) for miR-9-1; and forward, 5′-AGTGCCACAGAGCCGTCATAA-3′ and reverse, 5′-ccggatcctaatacgactcactatagggGCGTGGATTGTTCTAGCCTGAT-3′ for miR-9-3. For the detection of miR-9-2, a cDNA clone (IMAGE identification number 6819393; Open Biosystems) was used as a template to generate probes. The in situ hybridization data were confirmed by repeating the assay more than twice with independent embryos from different litters.
Quantitative reverse transcription-PCR for RNAs.
cDNAs were prepared from more than three independent wild-type and miR-9-2/3 double-mutant cerebral hemispheres from different litters, and quantitative reverse transcription (RT)-PCR was performed as described previously (Shibata et al., 2008). At least three replicates per transcript were used for every reaction. Assays were repeated at least three times. Data are expressed as means ± SD. The copy number of transcripts was normalized against the TATA-binding protein transcript level, and the ratio is shown as relative transcript level in Figures 1A, 2C, 6D, 7B,F, and 8B,D and supplemental Figures 1B and 2G (available at www.jneurosci.org as supplemental material). For all tested primer sets, correlation (R2) was higher than 0.98, and the slope was −3.1 to −3.6 in each standard curve. Primers to detect Foxg1, the TATA binding protein, miR-9-1, miR-9-2, and miR-9-3 RNAs were described previously (Shibata et al., 2008). Primers to detect the expression of the following genes were designed in a single exon encoding the 3′ UTR: forward, 5′-CTAACCATGGCCAAGAGCTCA-3′ and reverse, 5′-CTGACCAAAGCATGTGGCTG-3′ for Foxp2; forward, 5′-ACAGGCTGAGCCAGGTTCTG-3′ and reverse, 5′-AATGATTCAGCGGGATCTTCC-3′ for Nr2e1; forward, 5′-GAAATGGCGGTTAGAAGCACTT-3′ and reverse, 5′-GGGCTCCAGTTCAGGACAGTT-3′ for Pax6; forward, 5′-CCAGGCCCTCCAGAGACAC-3′ and reverse, 5′-TCATTGGTGCCTGGGCTTAC-3′ for Elavl1; forward, 5′-CTGCCTCTCTCCCTGGTCG-3′ and reverse, 5′-GGGCAACTGGCTAATCCAAAA-3′ for Msi1; forward, 5′- GGGAGTGACAGCGAAAACCA-3′ and reverse, 5′-TGGCCCTTATCAGTTGGCTT-3′ for Sox5; and forward, 5′-GCTCTTTCACTTGACCCTCGA-3′ and reverse, 5′-GAGACCTGCACTTATCCCCG-3′ for Tbr1. Primers to detect the expression of transcripts surrounding each miR-9 precursor as shown in supplemental Figure 1B (available at www.jneurosci.org as supplemental material): forward, 5′-ACCCGGCAAGTGGTGACTAC-3′ and reverse, 5′-TTAGGTGACAGGCGTGTTCCT-3′ for Mef2C; forward, 5′-GGCTGGTTCTCTGTCCTGCT-3′ and reverse, 5′-CCCTTCTAGGGCACTGCATC-3′ for Polγ; forward, 5′-CCCATTAGCTGTGGCCAGAA-3′ and reverse, 5′-GGTGGCAGTCA CCTAGGCTG-3′ for Rhcg; forward, 5′-ATTCCCCTGCGTACCTTGC-3′ and reverse, 5′-CAGCTTCCGCACGACTTTTT-3′ for hs_Smarce1; forward, 5′-GCATAGGCAGGCAGATGGAA-3′ and reverse, 5′-GAGGCCCTGCTTTAAGCAATAG-3′ for the RT-PCR product “a” in AK043150; forward, 5′-GTTGGCTAACGATACTGGAGATTACTT-3′ and reverse, 5′-TTATTCCAAACTTTTCCCCTTCC-3′ for “b” in BC096523; forward, 5′-TTACTCTGCGCCTAGCATTGG-3′ and reverse, 5′-TTTTTTGTGGCGTAAATGTCACTT-3′ for “c” in BC096523; forward, 5′-CGCTGTACACGTGCCAAAGTT-3′ and reverse, 5′-GGGTCTTCTCGACTGCCTCTAC-3′ for “d” in AK048540; forward, 5′-AGCCTGGAGACTCGCCTGAT-3′ and reverse, 5′-GGTTGTGGTGATGGTGGTT-3′ for “e” in AK032343; forward, 5′-AACCACCATCACCACAACC-3′ and reverse, 5′-TCTTTTCTCCTGGCCCAC-3′ for “f” in AK032343; forward, 5′-TGATGACTAAGAAGATTGTCCTGTGAA-3′ and reverse, 5′-TCAGAGCCTGTGCCATCACTT-3′ for “g” in AK032343; and forward, 5′-TGTTCTGTGCCTGAGAACGG-3′ and reverse, 5′-CGAGTGTGCCCTTGAACCTC-3′ for “h” in AK032343.
Figure 1.
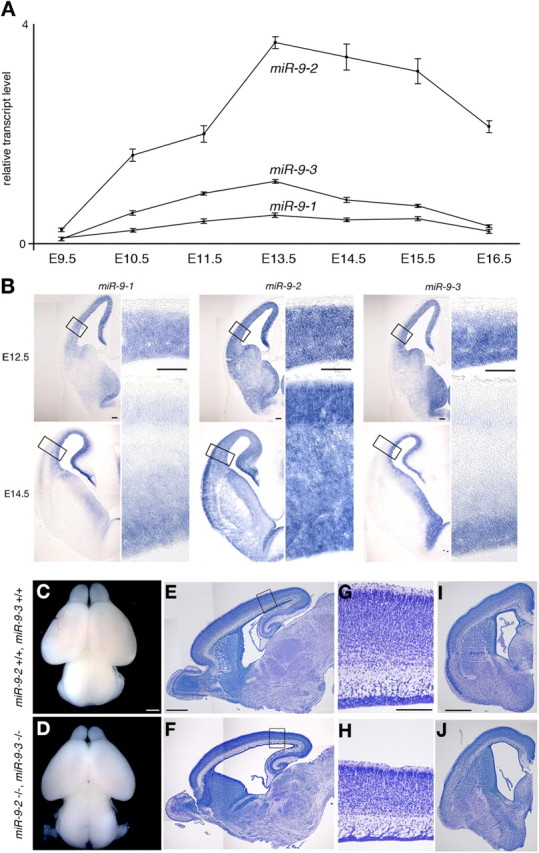
miR-9-2/3 double mutation. A, Quantitative RT-PCR analyses of expression levels of miR-9 precursors in developing cerebral hemispheres at the indicated stages. See Materials and Methods for relative transcript level. B, Expression pattern of each miR-9 precursor in E12.5 and E14.5 telencephalon by RNA in situ hybridization. In each miR-9 precursor expression, the right panels give enlarged views of the areas boxed in the left panels. C–J, Gross phenotypes of miR-9-2/3 double mutants at P1. Dorsal views (C, D), parasagittal sections (E, F), and frontal sections at the telencephalic level (I, J) of F2 wild-type and miR-9-2/3 double-mutant brains, respectively. G and H gives enlarged views of the cortices boxed in E and F. E–J, Nissl staining. Scale bars: B, 100 μm; C–F, I, J, 500 μm; G, H, 200 μm.
Figure 2.
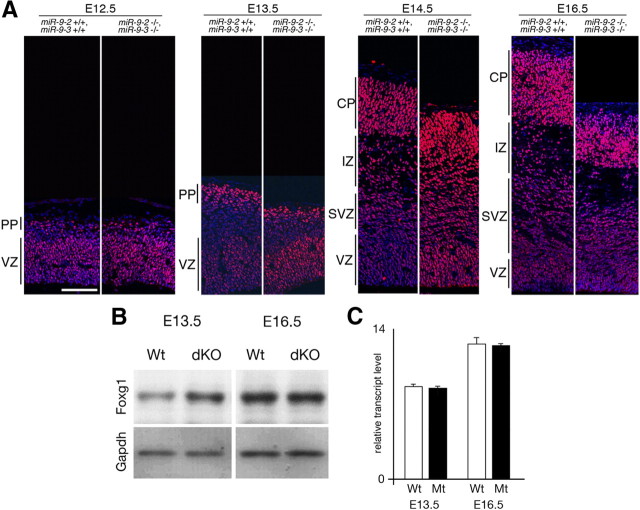
Foxg1 expression in miR-9-2/3 double-mutant pallium. A, Immunostaining with an anti-Foxg1 antibody of cortices at indicated stages. Layers are indicated as VZ, preplate (PP), SVZ, intermediate zone (IZ), and cortical plate (CP). B, Western blotting for Foxg1 expression in cerebral hemispheres dissected at E13.5 and E16.5. Relative intensities of Foxg1 bands in the double mutant are 2.0-fold and 1.0-fold of the wild type at E13.5 and E16.5, respectively, as estimated by normalizing the intensities against those of Gapdh bands. C, Quantitative RT-PCR for Foxg1 transcripts. Wt, Wild type; Mt, miR-9-2/-3 double mutant; dKO, double knock-out. Scale bar, 100 μm.
Figure 6.
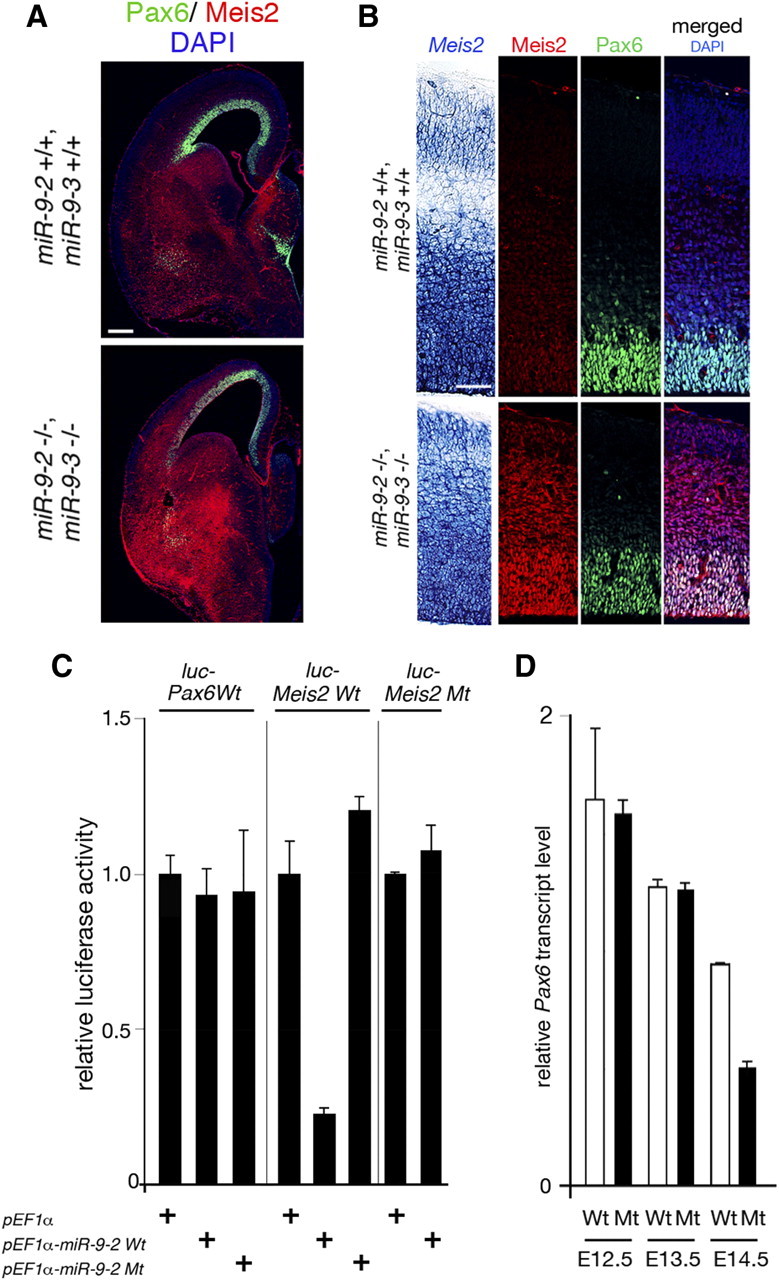
miR-9 regulation of Pax6 expression. A, Immunohistology of Pax6 (green) and Meis2 (red) protein expression in the E15.5 telencephalon. B, RNA in situ hybridization of Meis2 expression, anti-Meis2 or anti-Pax6 staining, and merged views of Meis2 and Pax6 protein expression with DAPI staining in the E15.5 pallium. C, miR-9-2 suppression of luciferase expression from a reporter conjugated to the wild-type and mutant Pax6 and Meis2 3′ UTRs; in mutant miR-9-2 (miR-9-2Mt), the mutation was introduced in the miR-9 seed sequences. D, Quantitative RT-PCR analyses of Pax6 mRNA expression at indicated stages. Scale bars: A, 200 μm; B, 50 μm.
Figure 7.
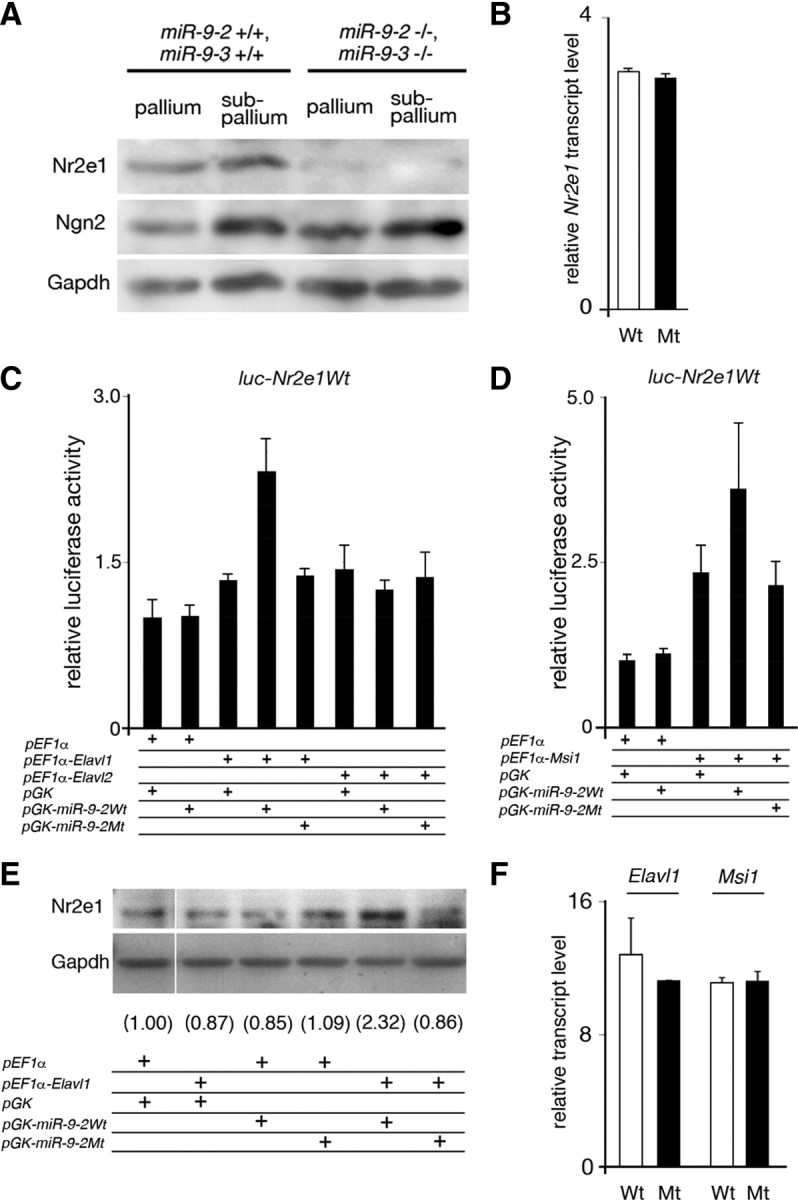
miR-9 regulation of Nr2e1 expression. A, Western blotting for Nr2e1 expression in E13.5 pallium and subpallium, with Ngn2 as a negative control and Gapdh as a loading control. B, Quantitative RT-PCR for Nr2e1 mRNA expression in cerebral hemispheres dissected at E16.5. C, D, Effects of Elavl1 and Elavl2 (C) and Msi1 (D) on luciferase expression from a luciferase reporter conjugated to the Nr2e1 3′ UTR in P19 cells. No miR-9 suppression of luciferase expression was observed from luciferase reporters conjugated to the 1.4 kb full-length Nr2e1 3′ UTR (C, D) as used by Zhao et al. (2009) or with four tandem repeats of its 50 nt fragment containing miR-9 responsive element (data not shown). E, Increase in the endogenous Nr2e1 expression in P19 cells by Elavl1 and miR-9. P19 cells were transfected with Elavl1 and/or miR-9 expression vectors as indicated, and Nr2e1 expression was detected by Western blotting. Numerals give the relative intensity of each Nr2e1 band to that in control in which pEF1α and pGK vectors were transfected; each band intensity was normalized by the intensity of Gapdh band. F, Quantitative RT-PCR analysis of Elavl1 and Msi1 mRNA expression in wild and miR-9-2/3 double-mutant cortices at E16.5.
Figure 8.
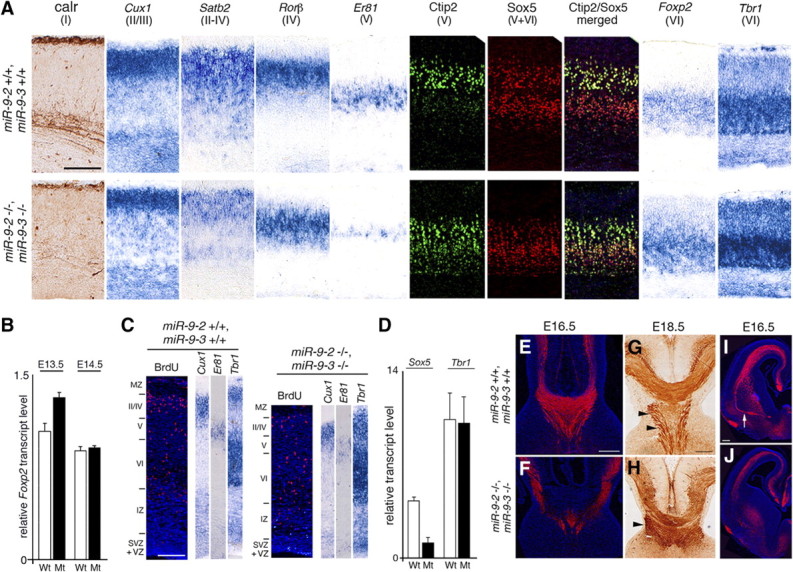
Layer formation in miR-9-2/3 double-mutant neopallium. A, Marker analysis for layer formation at E18.5. B, Quantification of Foxp2 mRNA expression at indicated stages. C, Birthdating analysis of the E18.5 cortex by BrdU labeling at E14.5. D, Quantitative RT-PCR analysis of Sox5 and Tbr1 mRNA expression in the pallium at E14.5. E–J, Anti-L1 staining for corpus callosum, fornix/hippocampal commissure (arrowheads), and anterior commissure (arrow). Sections in E, F, I, and J were immunostained as described in Materials and Methods, whereas the section in G and H was stained with 3,3′-diaminobenzidine tetrahydrochloride after immunoreaction with the secondary antibody conjugated to horseradish peroxidase. Scale bars: A, E–J, 200 μm; C, 100 μm.
miR-9 and miR-9* quantification.
Total RNAs were isolated from wild-type and miR-9-2/3 double-mutant telencephalon using a mirVana miRNA Isolation kit (Ambion). Quantitative RT-PCR was performed using a Taqman microRNA Reverse Transcription kit (Ambion) and Taqman Universal Master MixII, noUNG (Ambion) according to the instructions of the manufacturer. The snoRNA202 was used as an endogenous internal control for normalization; after the normalization, each transcript level was shown as the relative ratio to the level in E16.5 wild-type cerebral hemispheres (supplemental Fig. 2H,I, available at www.jneurosci.org as supplemental material). Assays were repeated at least three times with two independent embryos from different litters. Data are presented as means ± SD.
Immunoblot, immunohistochemistry, and histology.
Immunoblot and immunohistochemistry were performed as described (Shibata et al., 2008). Each immunoblotting was repeated more than two times with independent embryos from different litters, and representative results are presented in Figures 2B, 7A,E, 9R, and 10L. Quantification of band intensity was performed with NIH ImageJ software (Abramoff et al., 2004). Immunohistology was also confirmed by repeating the assay more than twice with independent embryos from different litters. For immunohistochemistry, the brain was dissected from each embryo and fixed with 4% paraformaldehyde overnight at 4°C, followed by embedding in OCT and sectioning at 14 μm. For antigen retrieval, sections were treated with HistoVT One (Nacalai Tesque) for 20 min at 72°C. For the detection of bromodeoxyuridine (BrdU) distribution in the pallium, sections were treated in 1N HCl for 10 min at room temperature and then in 2N HCl for 40 min at 37°C to denature DNA, followed by a rinse with PBS and the antigen retrieval. Nissl staining was performed using standard procedure. terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) assay was performed with the In Situ Cell Death Detection kit (Roche) according to the instructions of the manufacturer. The sources of antibodies used are as follows: primary antibodies used for immunohistochemistry were anti-Foxg1 (1:500) (Watanabe et al., 2005), anti-active caspase3 (1:500; Promega), anti-Gsh2 (1:5000) (Toresson et al., 2000), anti-Tuj1 (1:500; Covance), anti-calretinin (1:1000; Millipore Bioscience Research Reagents), anti-Id4 (1:500; Santa Cruz Biotechnology), anti-Nestin (1:100; BD Pharmingen), anti-BrdU (1:100; BD Pharmingen), anti-Ctip2 (1:500; Abcam), anti-Sox5 (1:200; Abcam), anti-Ki67 (1:200; Thermo Fisher Scientific), anti-L1 (1:200; Millipore Bioscience Research Reagents), anti-TAG1 (whole serum; Developmental Studies Hybridoma Bank), anti-GAP43 (1:2000; Zymed), anti-chondroitin sulfate proteoglycans (CSPGs) (1:200; Sigma), anti-Meis2 (1:500; Santa Cruz Biotechnology), and anti-Pax6 (1:500; Covance). Secondary antibodies were conjugated with Alexa Fluor 488 or 594 (1:200; Invitrogen). The anti-TAG1 antibody (4D7) developed by M. Yamamoto was obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biology (Iowa City, IA). Primary antibodies used for immunoblot were anti-Nr2E1 (1:500; Perseus Proteomics), anti-Ngn2 (1:200; US Biological), anti-Foxg1 (1:1000) (Watanabe et al., 2005), anti-Gsh2 (1:5000) (Toresson et al., 2000), anti-Islet1 (1:200; Abcam), anti-Elavl2 (1:500; Abcam), and anti-glyceraldehyde-3-phosphate dehydrogenase (Gapdh) (1:5000; Abcam). Secondary antibodies were conjugated to horseradish peroxidase (1: 2000; Millipore or Cell Signaling Technology).
Figure 9.
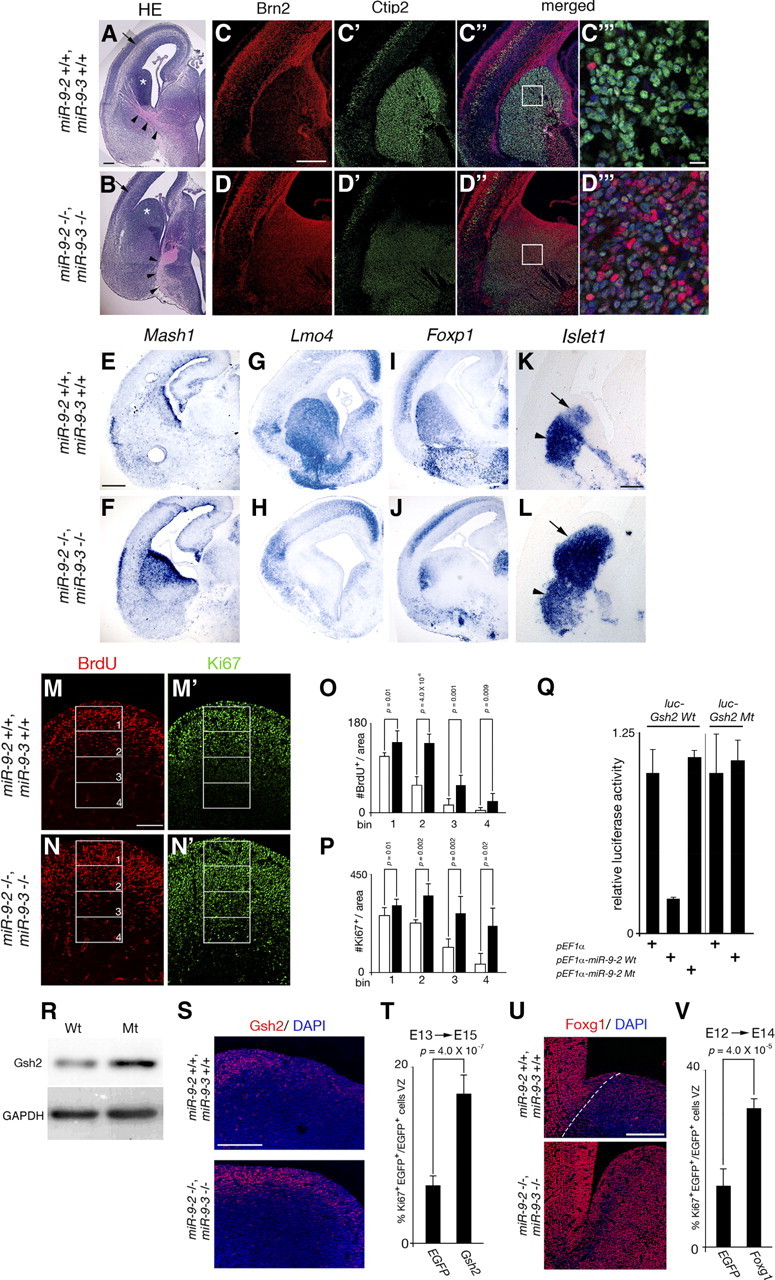
miR-9 regulation of cell proliferation and neural differentiation in the subpallium. A, B, Coronal sections of the E15.5 telencephalon stained with hematoxylin/eosin. Arrowheads indicate TCA trajectories, and arrows the intermediate zones. C, D, Anti-Brn2 staining of the E18.5 subpallium for the proliferative zone. C′, D′, Anti-Ctip2 staining for the striatal neurons in the adjacent sections. C″, D″, Their merged views. C‴, D‴, The enlarged views of the regions boxed in C″, D″. E–L, RNA in situ hybridization of Mash1 (E, F) expression for the proliferative zone at E17.5, of Lmo4 (G, H) and Foxp1 (I, J) expression for the striatal neurons at E17.5, and of Islet1 (K, L) expression for striatal neurons (arrowheads) and SVZ (arrows) at E15.5. M–N′, Typical examples of 30 min BrdU labeling for S-phase cells (M, N) and Ki67 staining for cycling cells (M′, N′) in the E15.5 LGE. O, P, The number of BrdU-positive (O) and Ki67-positive (P) cells in the four bins along the ventricular–pial axis of the LGE shown in M–N′. Quantification was performed as described in Materials and Methods. Q, Luciferase expression from a luciferase reporter conjugated to the wild-type and mutant Gsh2 3′ UTR: the mutation was introduced in the miR-9 responsive element. R, Western blotting for Gsh2 expression in cerebral hemispheres at E15.5. Quantification of the Gsh2 protein level by normalizing with Gapdh levels (loading control) indicated 1.7-fold increase in miR-9-2/3 double-mutant telencephalon. S, Anti-Gsh2 staining in the wild-type and double-mutant E15.5 LGE region. T, V, The effects of Gsh2 (T) and Foxg1 (V) overexpression by exo utero electroporation at E13 or E12 on cell proliferation in the subpallium at E15 or E14, respectively. The percentage of Ki67 and EGFP double-positive cells among total EGFP-positive cells is indicated. Quantification was performed as described in Materials and Methods. U, Anti-Foxg1 staining in the wild-type and miR-9-2/3 double-mutant E15.5 LGE region. A–L, M–N′, S, and U are all coronal views. Nr2e1 is known to promote cell proliferation in subpallium (Stenman et al., 2003c), and thus the reduction in Nr2e1 expression in E15.5 miR-9-2/3 double-mutant subpallium (Fig. 7A) cannot account for the hyperproliferation of progenitors in the miR-9-2/3 double-mutant subpallium. The areas of each bin in (M–N′) are 180 × 100 μm. Scale bars: A, B, M–N′, 100 μm; C–D″, E–L, 300 μm; C‴, D‴, 10 μm; S, U, 200 μm.
Figure 10.
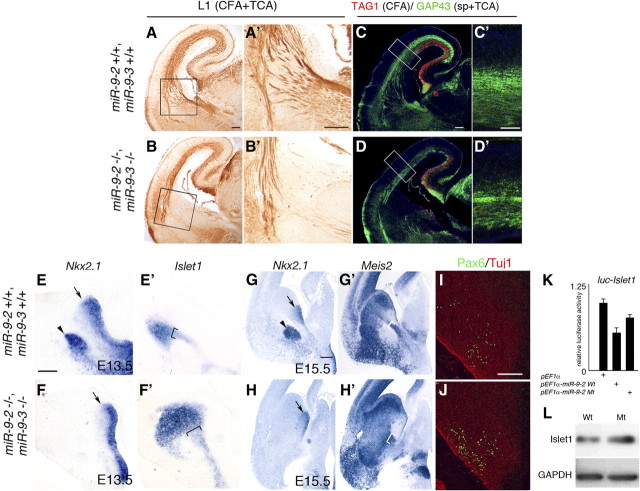
Corticofugal and thalamocortical projections with corridor malformation in miR-9-2/3 double-mutant subpallium. A, B, Anti-L1 staining for CFAs and TCAs at E18.5. A′, B′, Enlarged views of the internal capsule areas boxed in A and B. C, D, Anti-TAG1 (red) and anti-GAP43 (green) staining for CFAs and TCAs/subplate at E18.5. C′, D′, Enlarged views of the areas boxed in C and D. Scale bars: A–D, 100 μm; C′, D′, 50 μm. RNA in situ hybridization of Nkx2.1 for the proliferating field of the MGE and globus pallidus at E13.5 (E, F), of Islet1 for corridor in sections adjacent to E and F (E′,F′), and of Nkx2.1 (G, H) and Meis2 (G′, H′) for the ventral telencephalon in adjacent sections at E15.5. I, J, Anti-Pax6 (green) and Tuj1 (red) staining in the E15.5 subpallium. Arrowheads indicate the globus pallidus in E and G; arrows indicate the MGE VZ in E–H, and brackets indicate the corridor in E′–H′. Scale bars: E–F′, 50 μm; G–H′, 200 μm; I, J, 100 μm. K, miR-9-2 suppression of luciferase expression from a reporter conjugated to the Islet1 3′ UTR. L, Moderate increase of Islet1 expression in E13.5 miR-9-2/3 double-mutant cerebral hemispheres.
Image processing.
Several photographs of a section taken at the same time were assembled into single images shown in Figures 1E,F, 2A, 5A,E, 6A,B, and 9A–D″,M–N′,S,U.
Figure 5.
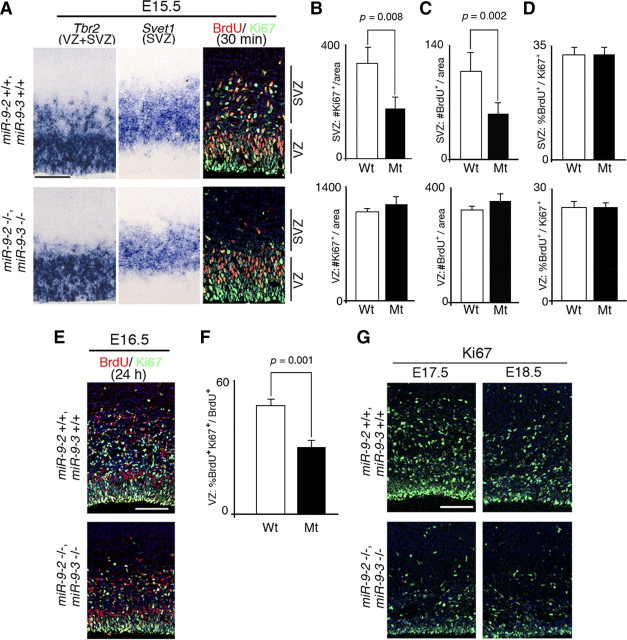
Progenitor proliferation in later miR-9-2/3 double-mutant pallium. A, Typical examples of RNA in situ hybridization of Tbr2 and Svet1 expression for the VZ and SVZ, and 30 min BrdU labeling and Ki67 staining for S phase and cycling cells in the pallium at E15.5, respectively. B, C, Quantification of Ki67-positive (B) and BrdU-positive (C) cells in the SVZ and VZ. D, Cell cycle length as indicated by the number of BrdU and Ki67 double-positive cells among Ki67-positive cycling cells in the SVZ and VZ. E, Typical examples of 24 h BrdU labeling and Ki67 staining for cells reentering the cell cycle in the VZ at E16.5. F, Quantification of the cells reentering the cell cycle as the ratio of Ki67 and BrdU double-positive cells among BrdU-positive cells. G, Typical examples of Ki-67 staining of E17.5 and E18.5 cortices. Quantification was performed as described in Materials and Methods. Scale bars: A, E, G, 100 μm.
RNA affinity precipitation assay.
The RNA affinity precipitation assay was performed as described previously (Wakiyama et al., 2007) with minor modifications. Mouse cortices at E17.5 were homogenized in radioimmunoprecipitation assay buffer, and homogenate was adjusted to 5 mg/ml. Ten microliters of lysates were diluted in 500 μl of binding buffer [20 mm HEPES-KOH at pH 7.4, 150 mm NaCl, 1.5 mm MgCl2, 5% glycerol, 0.1% Triton X-100, 1.5 mm dithiothreitol, 0.2 mg/ml heparin, 0.2 mg/ml tRNA (Ambion), 0.25% BSA (Ambion), protease inhibitor cocktail (Roche Diagnostics), and ribonuclease inhibitor (Invitrogen)]. The mixtures were incubated with or without biotinylated RNA (2 μg) for 10 min at 16°C. Adding Dynabeads Myone Streptavidin T1 (Invitrogen), preequilibrated in the binding buffer, the mixtures were further incubated for 15 min at 4°C. The beads were then captured with a magnet, washed four times with 500 μl of the wash buffer (20 mm HEPES-KOH at pH 7.4, 150 mm NaCl, 1.5 mm MgCl2, 5% glycerol, 0.1% Triton X-100, 1.5 mm dithiothreitol, and 0.2 mg/ml heparin), and boiled with 20 μl of SDS sample buffer. The supernatant was resolved on 10% polyacrylamide gels and subjected to the Elavl2 immunoblot. Biotinylated Foxg1 3′ UTR (both wild-type Foxg1 3′ UTR WT and mutated Foxg1 3′ UTR MT1) fragments were purchased from Gene Design.
Cell culture and transfection.
Cell culture and transfection for luciferase assays were performed with P19 cells as described (Shibata et al., 2008); P19 cells were treated with 500 nm retinoic acid for 2 d after transfection. Dissociation of and electroporation into cortical cells in culture were conducted as described previously (Shibata et al., 2008).
Luciferase assay.
Luciferase assay was performed with P19 cells using a Dual-Glo Luciferase Assay system (Promega) as described previously (Shibata et al., 2008); Renilla reniformis luciferase expression vector was cotransfected for normalization. For each sample, four replicates were measured in a single assay. Assays were repeated at least three times, and representative results are shown. Data are expressed as means ± SD of four replicates. In Figure 4, B and D, miR-9-2 precursor RNA and its control oligo RNA were purchased from Ambion and transfected into P19 cells. In Figures 6C, 7C,D, 9Q, and 10K, miR-9 was expressed by transfecting miR-9-2 precursor in pEF1α or pGK expression vector, and the vector served as control. The relative luciferase activity is given as the relative ratio to the activity observed by the control (oligo RNA, pEF1α, or pGK) transfection.
BrdU labeling.
For the pulse labeling of cells in S phase, timed pregnant female mice at E12.5, E13.5, and E15.5 were injected intraperitoneally with a single pulse of BrdU (Roche) (50 mg/kg body weight) and killed after 30 min or after 24 h for the dissection of embryonic brains. For birthdating analyses, BrdU was injected at E14.5, and the distribution of BrdU-positive cells in the embryonic pallium was determined at E18.5 as described previously (Schuurmans et al., 2004).
Quantification of BrdU-positive, Ki67-positive, and pH3-positive cells.
In Figure 3, V and W, three anatomically matched cortical sections 84 μm apart were selected from wild-type and miR-9-2/3 double-mutant cortices. They were immunostained with the anti-BrdU antibody, two anatomically matched 270 × 200 μm areas, including the entire ventricular–pial axis, were selected, and the BrdU-positive cells in the areas were counted. For E13.5 cortex, areas were divided into the VZ and SVZ according to Tbr2 expression, and the BrdU-positive cells in VZ and SVZ were counted. In Figure 3X, three anatomically matched cortical sections 84 μm apart from three cortices of both genotypes were stained with the anti-phospho-histone H3 antibody, and two anatomically matched 200 × 200 μm areas were selected and divided into the VZ and SVZ according to Tbr2 expression for counting. In Figure 5B–D, three anatomically matched cortical sections 168 μm apart from wild-type and miR-9-2/3 double-mutant cortices were immunostained with anti-BrdU and anti-Ki67 antibodies, and five anatomically matched 200 × 100 μm areas in the VZ and 200 × 200 μm (wild-type) or 200 × 150 μm (miR-9-2/3 double mutant) areas in the SVZ were selected for counting. In Figure 5F, two anatomically matched 400 × 100 μm areas of both genotypes were selected for the counting. The VZ and SVZ in Figure 5B–F were defined by Tbr2 or Svet1 expression, respectively, detected by in situ hybridization of adjacent sections. In Figure 9M–P and supplemental Figure 7A–F (available at www.jneurosci.org as supplemental material), BrdU- and Ki67-positive cells were counted in four and three, respectively, 180 × 100 μm bins along the ventricular–pial axis of the LGE. Three anatomically matched cortical sections 168 μm (see Fig. 9M–P) or 84 μm (supplemental Fig. 7A–F, available at www.jneurosci.org as supplemental material) apart were chosen for the counting. All these assays were repeated more than twice with three independent embryos from different litters. Data are presented as means ± SD. Unpaired two-tailed Student's t tests were performed, and p values are given in each figure.
Figure 3.
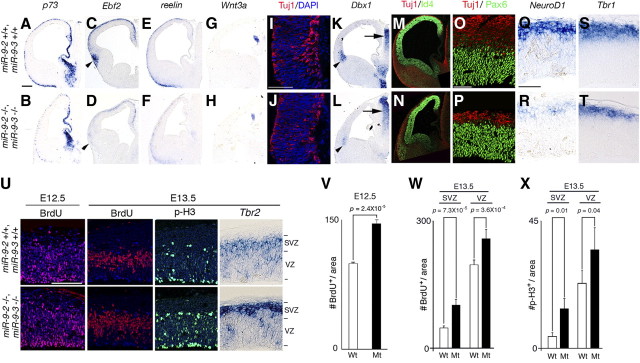
Early neurogenesis in miR-9-2/3 double-mutant pallium. A–R, Marker analyses of early-born neurons at E12.5. A–H, K, L, and Q–T give RNA in situ hybridization for genes indicated; I, J, M–P, immunostaining with an anti-Tuj1 antibody (red) and 4′,6′-diamidino-2-phenylindole (DAPI) (blue) at the cortical hem region (I, J), with anti-Tuj1 antibody and an anti-Id4 antibody (green) in telencephalon (M, N) or with anti-Tuj1 antibody and anti-Pax6 antibody (green) in the pallium (O, P). Arrowheads in C, D, K, and L indicate the ventral pallium, and arrows in K and L indicate the normal Dbx1 expression in the thalamus. Scale bars: A–H, K–N, 200 μm; I, J, 100 μm; O–T, 50 μm. U, Typical examples of distribution of BrdU-incorporated cells after 30 min labeling, of distribution of phospho-histone H3-positive cells, and of RNA in situ hybridization for SVZ marker, Tbr2 in the neopallium at either E12.5 or E13.5. V, Quantification of BrdU-positive cells in the entire E12.5 pallium. W, X, Quantification of BrdU-positive cells (W) and phospho-histone H3-positive cells (X) in the VZ and SVZ at E13.5. Quantification in V–X was performed as described in Materials and Methods. Scale bar (in U), 100 μm.
Exo utero electroporation.
Exo utero electroporation of DNA constructs into the cortex was performed as described previously (Shibata et al., 2008). Gsh2 (see Fig. 9T) or Foxg1 (see Fig. 9V) transgene under the cytomegalovirus promoter was conjugated to enhanced green fluorescent protein (EGFP) under the SV40 promoter and electroporated into the subpallium. In an assay, two anatomically matched sections 84 μm apart were selected from electroporated hemispheres. They were immunostained with the Ki67 and GFP antibody, anatomically matched 400 × 200 μm LGE areas were selected, and Ki67- and GFP-positive cells were counted. The assays were repeated more than two times with four independent embryos. Data are presented as means ± SD. Unpaired two-tailed Student's t tests were performed, and p values are given in the figures.
Results
Targeted deletion of miR-9-2 and miR-9-3
miR-9/mR-9* has three precursors, miR-9-1, miR-9-2, and miR-9-3 encoded in chromosomes 3, 13, and 7, respectively (Griffiths-Jones, 2006) (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). Quantitative RT-PCR revealed that miR-9-2 is expressed most abundantly, miR-9-3 much less so, and miR-9-1 the least during telencephalon development (Shibata et al., 2008) (Fig. 1A). In situ hybridization demonstrated that, at E12.5 and E14.5, the miR-9-2 is expressed throughout the telencephalon, whereas miR-9-1 and miR-9-3 are mostly localized in the proliferative zone (Fig. 1B). To assess the functions of miR-9/miR-9* in telencephalon development, we mutated the miR-9-2 and miR-9-3 loci by replacing 72 and 90 nt, respectively, covering the mature miR-9/miR-9* with a neomycin resistance cassette (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). The double mutation abolished the miR-9-2 and miR-9-3 expression; no compensatory increase in miR-9-1 expression was apparent (supplemental Fig. 2G, available at www.jneurosci.org as supplemental material). Consequently, mature miR-9 expression was reduced to 24 and 23% of wild type in the miR-9-2/3 double mutants at E12.5 and E16.5, respectively (supplemental Fig. 2H, available at www.jneurosci.org as supplemental material). The miR-9* expression was decreased to 29 and 38% at E12.5 and E16.5, respectively (supplemental Fig. 2I, available at www.jneurosci.org as supplemental material).
The miR-9-2/3 double mutants obtained by crosses between double heterozygotes were live born but exhibited severe growth retardation with small cerebral hemispheres and olfactory bulbs (Fig. 1C,D), and they died within 1 week. miR-9-2/3 double mutants exhibited defects in other parts of the brain, especially in the inferior colliculus, but this study focused on the functions of miR-9 in telencephalon development. Histological analyses at postnatal day 1 (P1) (Fig. 1E–J) showed marked reduction of the cortical layers and VZ; concomitantly, the lateral ventricles were expanded. In the subpallium, the proliferating zone was hyperplastic, whereas differentiated structures were reduced. Neither miR-9-2 nor miR-9-3 single homozygous mutants exhibited any apparent defects in neurogenesis of pallium or subpallium (supplemental Fig. 4, available at www.jneurosci.org as supplemental material) (data not shown), and the miR-9-2/3 double-mutant defects are apparently attributable to miR-9/miR-9* reduction (supplemental Figs. 1, 2, available at www.jneurosci.org as supplemental material).
miR-9 suppresses Foxg1 protein expression for the proper differentiation of Cajal-Retzius cells and early-born neurons
Foxg1 mRNA has miR-9 responsive element in its 3′ UTR (supplemental Fig. 3, available at www.jneurosci.org as supplemental material); it does not have miR-9* responsive element. The presence or absence of miR-9 and miR-9* responsive elements in a mRNA was assessed as described previously (Shibata et al., 2008) (also see Discussion). As suggested by our previous study (Shibata et al., 2008), the Foxg1 protein expression increased in the miR-9-2/3 double-mutant VZ at E12.5–E14.5 (Fig. 2A,B; Table 1); Foxg1 mRNA level was essentially unaffected (Fig. 2C). Concomitantly, cortical hem and the pallium/subpallium boundary (PSB)-derived Cajal-Retzius cells, marked by p73 and Ebf2, respectively, were substantially reduced in the marginal zone at E12.5 and E13.5 (Fig. 3A–D) (data not shown). Consequently, reelin-positive Cajal-Retzius cells of multiple origins were greatly reduced in the marginal zone (Fig. 3E,F). The Wnt3a-positive domain and the differentiation of early neurons marked by Tuj1 or calretinin were also reduced in the cortical hem region (Fig. 3G–J) (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material). Dbx1-positive ventral pallium from which Ebf2-positive Cajal-Retzius cells originate (Hanashima et al., 2007) was greatly reduced (Fig. 3K,L). The septum-derived Cajal-Retzius cells also appeared reduced in miR-9-2/3 double mutants (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material), but more detailed analyses are required with specific markers. Cxcl12-positive meninges that are proposed to be the substrate for the tangential migration of Cajal-Retzius cells from the cortical hem (Huang, 2009) and BMP7- and Msx1-positive choroidal roof and choroid plexus developed normally (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material). There were also no changes in the number of TUNEL-positive or active caspase3-positive cells (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material).
Table 1.
miR-9/miR-9* target candidates
| Gene | Responsive elementa |
Reporter assayb |
Expression in double mutantsc |
||||
|---|---|---|---|---|---|---|---|
| miR-9 | miR-9* | I | II | Reference | Protein level | Reference | |
| Foxg1 | + | − | ↓ | DS (Elavl2) | Shibata et al., 2008; Fig. 4 | ↑ (E13.5)þNSC(E16.5) | Fig. 2 |
| CoREST | + | + | ↓ (miR-9*) | NE | Packer et al., 2008 | NSC (E13.5) | Data not shown |
| REST | + | + | ↓ (miR-9) | NE | Packer et al., 2008 | NSC (E13.5) | Data not shown |
| BAF53a | + | + | ↓ (miR-9*) | NE | Yoo et al., 2009 | NSC (E13.5, 15.5) | Data not shown |
| Nr2e1 | + | − | NSC | ↑ (Elavl1, Msi1) | Fig. 7C,D | ↓ (E13.5) | Fig. 7A |
| Nr2e1 | + | − | ↓ | NE | Zhao et al., 2009 | ↓ (E13.5) | Fig. 7A |
| Id4 | + | − | NSC | NE | Shibata et al., 2008 | NSC (E12.5) | Fig. 3M,N |
| Pax6 | + | − | NSC | NE | Fig. 6C | ↓ (E15.5) | Fig. 6A,B |
| Meis2 | + | − | ↓ | NE | Fig. 6C | ↑ (E15.5) | Fig. 6A,B |
| Gsh2 | + | − | ↓ | NE | Fig. 9Q | ↑ (E15.5) | Fig. 9R,S |
| Islet1 | + | − | m↓ | NE | Fig. 10K | m↑ (E13.5) | Fig. 10L |
a+ indicates the presence and − the absence of the miR-9 or miR-9* responsive element in the mRNA of the gene indicated.
b Reporter assay with the cells given in the reference without (I) or with (II) the overexpression of an RNA-binding protein indicated in the parentheses. ↓ indicates the decrease and ↑ the increase in the expression by miR-9 or miR-9*. NSC, No significant change; DS, desupression; NE, not examined; m, moderate.
cChanges of the protein expression in miR-9–2/3 double mutants at the stage indicated in parentheses.
Foxg1 has also been suggested to promote the proliferation of cortical progenitor cells in the VZ (Hanashima et al., 2002, 2004; Martynoga et al., 2005). In the E12.5 pallium, Id4- or Pax6-positive neuroepithelial cells were not significantly affected (Fig. 3M–P), and Nestin-positive radial glia cells were almost normal in the miR-9-2/3 double mutants (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material). However, early-differentiated Tuj1-, NeuroD1-, or Tbr1-positive neurons were significantly reduced in the preplate at E12.5 (Fig. 3M–T). Coincidentally, the calretinin-positive marginal zone and subplate and the CSPG- or GAP43-positive subplate were reduced in the miR-9-2/3 double mutants at E18.5 (supplemental Fig. 5B,C, available at www.jneurosci.org as supplemental material) (Fig. 10C–D′). BrdU 30 min pulse labeling at E12.5 demonstrated that cell proliferation was increased by 56% in the miR-9-2/3 double-mutant cortices compared with control cortices (Fig. 3V). At E13.5 BrdU incorporation was increased by 112 and 31% in the SVZ and VZ of the double-mutant cortices, respectively (Fig. 3W). Phospho-histone H3-positive mitotic cells were also increased by 230 and 47% in the SVZ and VZ, respectively (Fig. 3X). Concomitant with the increase in progenitor proliferation in the SVZ, Tbr2 expression was enhanced in the SVZ (Fig. 3U). This is consistent with the report that IPCs are reduced by Foxg1 haploinsufficiency (Siegenthaler et al., 2008). Among genes suggested to regulate the production of early-born neurons (see Introduction), Ngn1, Ngn2, or Tbr1 mRNAs do not have miR-9/miR-9* responsive element. Among miR-9/miR-9* targets demonstrated in vitro for progenitor proliferation (see Introduction), the protein expression of CoREST, REST, or BAF53a was not significantly changed in the E13.5 miR-9-2/3 double-mutant telencephalon (Table 1).
Elavl2 attenuates the miR-9 suppression of Foxg1 expression
The increase in Foxg1 expression in miR-9-2/3 double-mutant VZ was apparent at E14.5 but not from E16.5 onward (Fig. 2A,B). Even at these stages, miR-9 and Foxg1 expression overlapped (Fig. 4A), implying that miR-9 suppression of Foxg1 expression was attenuated as development proceeded. The miR-430 suppression of Nanos1 and Tdrd7 protein expression has been reported to be cancelled by an AU-rich RNA binding protein Dnd1 in germ cells (Kedde et al., 2007). The 3′ UTR of Foxg1 has U-rich sequences upstream of the miR-9 responsive element (Fig. 4C). Elavl1/HuR, Elavl2/HuB, and Elavl4/HuD are the AU-rich RNA-binding proteins reportedly expressed in the pallium (Deschênes-Furry et al., 2006). Among them, Elavl2 was found to counter the miR-9 suppression of luciferase expression from a reporter conjugated to the Foxg1 3′ UTR (Fig. 4B). This desuppression of miR-9 activity by Elavl2 required the U-rich sequence located 5′ upstream of the miR-9 responsive element in the Foxg1 3′ UTR (Fig. 4C,D); Foxg1 3′ UTRs bearing mutations in the U-rich sequences (Foxg1 3′ UTR MT1, MT2, and MT1 + 2) were suppressed by miR-9, but these suppressions were not released by Elavl2. Foxg1 3′ UTR MT1 + 2 completely lost the ability to be desuppressed by Elavl2. In addition, RNA affinity precipitation assay demonstrated that endogenous Elavl2 associates with Foxg1 3′ UTR WT RNA probe; the association was inefficient with the Foxg1 3′ UTR MT1 probe (Fig. 4E). Furthermore, the Elavl2 overexpression in the dissociated cortical cells at E12.5 upregulated Foxg1 expression (Fig. 4F). Elavl2 was expressed in the E16.5 VZ as well as the cortical plate and intermediate zone, but the expression was faint in the E14.5 VZ (Fig. 4A). These results suggest that Foxg1 expression is suppressed by miR-9 at earlier stages, but the suppression is countered by Elavl2, whose expression increases at later stages. Concomitantly, no significant increase in the cycling Ki67-positive or BrdU-incorporated cells after 30 min of labeling was apparent in the miR-9-2/3 double-mutant VZ at E15.5 as described below (Fig. 5A–C).
miR-9 enhances Nr2e1 and Pax6 expression, and progenitor proliferation was suppressed in later ventricular and subventricular zones
In P1 miR-9-2/3 double mutant, cortical layers were hypoplastic (Fig. 1E–J). In E15.5 miR-9-2/3 double-mutant pallium, the thickness of Tbr2-positive VZ (Tarabykin et al., 2001; Englund et al., 2005) was almost unaffected, whereas Tbr2- and Svet1-positive SVZ (Tarabykin et al., 2001) was reduced (Fig. 5A). The number of cycling Ki67-positive cells or of BrdU-positive S-phase cells was significantly reduced in the double-mutant SVZ (Fig. 5B,C). There was no significant difference in the ratio of BrdU-positive cells among Ki67-positive cells between wild-type and mutant SVZ (Fig. 5D), suggesting that IPCs in the mutant pallium had a cell cycle length similar to that of the wild type. We also examined cell cycle reentry in the VZ at E16.5 by labeling with BrdU for 24 h; the frequency of the cells reentering the cell cycle is given as the ratio of BrdU and Ki67 double-positive cells over the total number of BrdU-positive cells. This ratio was significantly reduced in the miR-9-2/3 double-mutant VZ (Fig. 5E,F). Consistently, the number of Ki67-positive proliferating cells in the VZ significantly decreased at E17.5 and E18.5 (Fig. 5G), suggesting the depletion of the progenitor pool in the VZ at later stages.
Among genes suggested to regulate cortical progenitor proliferation and differentiation at later phases (see Introduction), Id4, Pax6, and Nr2e1 mRNAs have miR-9 target sequences (supplemental Fig. 3, available at www.jneurosci.org as supplemental material); none has miR-9* responsive element. miR-9-2 did not affect the luciferase expression from a reporter conjugated to the Id4 3′ UTR (Shibata et al., 2008); moreover, Id4 protein expression was almost unchanged in the miR-9-2/3 double-mutant pallium (Fig. 3M,N). In contrast, Pax6 and Nr2e1 protein expressions were reduced in the E15.5 miR-9-2/3 double mutants (Figs. 6A,B, 7A; Table 1). However, we were unable to demonstrate significant change of luciferase expression from a reporter conjugated to the Pax6 3′ UTR by miR-9 (Fig. 6C). The Pax6 mRNA expression reduces with telencephalon development (Fig. 6D). The double mutation did not affect the Pax6 mRNA expression initially, but the reduction was more pronounced in the miR-9-2/3 double-mutant telencephalon later than E14.5 (Fig. 6D). Meis2 has been known to suppress the Pax6 expression (Agoston and Schulte, 2009), and Meis2 mRNA has miR-9 responsive element (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Indeed, wild-type miR-9-2 suppressed luciferase expression from a reporter conjugated to the wild-type Meis2 3′ UTR, whereas mutant miR-9-2 did not (Fig. 6C). Furthermore, miR-9-2 did not suppress luciferase expression from a reporter conjugated to the Meis2 3′ UTR with a mutation in the miR-9 responsive element (Fig. 6C). Meis2 protein expression was indeed enhanced in miR-9-2/3 double-mutant pallium, although its expression was not significantly changed at the mRNA level (Fig. 6A,B; Table 1).
Nr2e1 expression was not significantly changed at the mRNA level (Fig. 7B), and miR-9-2 did not suppress luciferase expression from a reporter conjugated to the Nr2e1 3′ UTR in P19 cells (Fig. 7C,D). It is reported that RNA-binding protein FXR1 activates protein expression from miR369-3-targeted tumor necrosis factor α mRNAs (Vasudevan et al., 2007). Among RNA-binding proteins expressed in the pallium (Deschênes-Furry et al., 2006), we found that Elavl1 (Ma et al., 1996) and Msi1 (Sakakibara et al., 1996) activated miR-9-2-directed luciferase expression from the reporter conjugated to the Nr2e1 3′ UTR. Neither Elavl1 nor Msi1 activated luciferase expression directed by mutant miR-9-2 (Fig. 7C,D). Moreover, wild-type miR-9-2, but not mutant miR-9-2, upregulated the expression of endogenous Nr2e1 in P19 cells, synergistically with Elavl1 (Fig. 7E). Elavl1 did not relieve the miR-9-directed suppression of luciferase expression from the reporter conjugated to the Foxg1 3′ UTR (Fig. 4B), suggesting that the Elavl1 activity is specific to Nr2e1. Elavl1 is expressed intensively in the VZ and moderately in the SVZ and cortical plate (data not shown); Msi1 is expressed in the VZ and SVZ (Sakakibara et al., 1996). Msi1 enhanced the luciferase expression by itself (Fig. 7D). However, neither Elavl1 nor Msi1 mRNA has miR-9 responsive elements, and their expressions were unchanged in the miR-9-2/3 double-mutant telencephalon (Fig. 7F).
Neocortical lamination is malformed in miR-9-2/3 double mutants
Cajal-Retzius cells were significantly decreased but present (Fig. 3A–F), Nestin-positive radial glial fibers that function as substrates for radial neural migration were oriented normally (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material), and inside-out lamination took place in the miR-9-2/3 double-mutant neopallium. Concomitantly with the defects in progenitor proliferation and differentiation, however, cortical layers were hypoplastic in the P1 miR-9-2/3 double mutants (Fig. 1G,H). At E18.5, Foxp2- or Tbr1-positive layer VI was almost normally generated, whereas calretinin-positive layer I, Cux1-positive layer II/III, Satb2-positive layer II-IV, Rorβ-positive layer IV, and calretinin-positive subplate were reduced (Fig. 8A). Normal generation of the layer VI was unexpected, because layer VI neurons are produced at approximately E12.5 (Langevin et al., 2007) when progenitor proliferation is enhanced in the double mutants. Tbr1-positive layer VI neurons as well as Tuj1- or NeuroD1-positive neurons were significantly reduced at E12.5 as described above (Fig. 3M–T). However, the layer VI neurons assessed by Foxp2 transcript level were not significantly changed at E13.5 or E14.5 (Fig. 8B). Conversely, birthdating analysis also indicated that the number of BrdU-positive cells labeled at E14.5 was significantly decreased in mutant layer II–III, defined by Cux1 expression at E18.5 (Fig. 8C). Notably, the Er81-positive layer V was also reduced, whereas Ctip2 expression was expanded into deeper layers in miR-9-2/3 double mutants (Fig. 8A); Er81 marks all layer V neurons, whereas Ctip2 marks corticospinal motor neurons (Molnár and Cheung, 2006). Moreover, the Sox5 expression in V and VI layers was also decreased in the double mutants to approximately one-third of the wild-type level (Fig. 8A,D). Neither Er81 nor Ctip2 mRNAs have miR-9 responsive elements. Er81 is known to be a downstream target of Pax6 in cortical progenitors (Tuoc and Stoykova, 2008), and we speculate that the reduction of Er81-positive layer V is attributable to the Pax6 reduction. Sox5 is also known to suppress Ctip2 expression in layer VI and the subplate (Kwan et al., 2008; Lai et al., 2008), and we assume that the expansion of Ctip2 expression is attributable to the Sox5 reduction. However, Sox5 mRNA does not have miR-9 responsive element; thus, miR-9 must enhance Sox5 expression indirectly.
As expected from the malformation of cortical layers, each cortical projection was greatly reduced in the miR-9-2/3 double mutants. Axons of commissural neurons labeled by L1 (corpus callosum, anterior commissure, and fornix/hippocampal commissure) were poorly formed (Fig. 8E–J), and corticofugal axons (CFAs) labeled by TAG-1 and L1 were also thin in the miR-9-2/3 double-mutant pallium (see Fig. 10A–D′). Furthermore, the cortical interneurons that are born in the subpallium and tangentially migrate into the pallium were scarcely present in the miR-9-2/3 double-mutant pallium (supplemental Fig. 6A–J, available at www.jneurosci.org as supplemental material); this is coincident with the malformation of the intermediate and marginal zones that provide interneuron passages (Métin et al., 2006) or the reduction of TAG-1- or reelin-positive cells that guide the migration (Denaxa et al., 2001; Morante-Oria et al., 2003). However, interneurons were apparently normally specified in MGE (supplemental Fig. 6K,L, available at www.jneurosci.org as supplemental material). Moreover, interneurons in the rostral migratory stream to the olfactory bulb were also generated apparently normally in the miR-9-2/3 double-mutant LGE (supplemental Fig. 6M,N, available at www.jneurosci.org as supplemental material).
miR-9 suppresses Gsh2 and Foxg1 expression and progenitor proliferation is enhanced in the miR-9-2/3 double-mutant subpallium
Morphologically, the miR-9-2/3 double-mutant subpallium was almost normal at E12.5 (Fig. 3A–H,K–N) (data not shown) but hyperplastic at E15.5; the cell-dense proliferating field was markedly expanded into the deep differentiating field (Figs. 1E,F, 9A,B). Indeed, the Brn2- and Mash1-positive fields expanded deeply in the miR-9-2/3 double-mutant subpallium (Fig. 9C,D, C″,D″,E,F). Concomitantly, the Ctip2-, Lmo4-, and Foxp1-positive differentiating field (Hermanson et al., 1999; Tamura et al., 2004; Arlotta et al., 2008) was greatly reduced (Fig. 9C′–D″,G–J). The Islet1-positive LGE SVZ (Stenman et al., 2003a) was also markedly expanded, whereas that in the differentiating area was reduced (Fig. 9K,L). At E15.5, the number of BrdU-positive and Ki67-positive cells in the lateral ganglionic eminences was increased in the miR-9-2/3 double-mutant LGE (Fig. 9M–P), particularly in the deeper area. However, such hyperproliferation was not apparent at E12.5 (supplemental Fig. 7A–F, available at www.jneurosci.org as supplemental material).
Among genes reported to be essential to progenitor cell proliferation in the VZ and SVZ of ganglionic eminences (see Introduction), we found none has miR-9* responsive element, and neither Mash1, Dlx1/2/5/6, nor Nkx2.1 has miR-9 responsive element. Gsh2 mRNA, however, has miR-9 responsive element in its 3′ UTRs (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Wild-type miR-9-2 effectively suppressed the expression of luciferase from a reporter conjugated to the wild-type Gsh2 3′ UTR but not the 3′ UTR, of which the miR-9 responsive element was transversely mutated (Fig. 9Q). Gsh2 protein level was indeed enhanced in the miR-9-2/3 double-mutant compared with the wild-type subpallial region (Fig. 9R,S; Table 1). In addition, Gsh2 overexpression in the LGE region by electroporation markedly enhanced the number of Ki67-positive cells (Fig. 9T). Foxg1 is also expressed in subpallium, but its role in subpallial progenitor proliferation and differentiation has been unclear in Foxg1 mutants because of the earlier defect in the subpallial specification (Martynoga et al., 2005; Manuel et al., 2010). Foxg1 protein expression was modest in the E15.5 wild-type subpallium but enhanced in the miR-9-2/3 double-mutant subpallium (Fig. 9U); this increase was not apparent in the E12.5 miR-9-2/3 double-mutant subpallium (supplemental Fig. 7G,H, available at www.jneurosci.org as supplemental material). In addition, Foxg1 overexpression enhanced the proportion of cycling cells in the subpallium (Fig. 9V).
Dorsal shift of pallial–subpallial boundary
The PSB is proposed to be determined by the counteraction of Pax6 and Gsh2 (Toresson et al., 2000; Yun et al., 2001). The PSB also shifts dorsally in Nr2e1 mutants (Stenman et al., 2003b). Consistent with the reduction of Pax6 and Nr2e1 expression and the increase of Gsh2 expression in the miR-9-2/3 double mutants, the Tbr2/Gsh2 boundary that marks the PSB shifted dorsally and coincided with the morphological flexure between the pallium and subpallium (supplemental Fig. 8A–D, available at www.jneurosci.org as supplemental material). The PSB is also marked morphologically by a fasciculated palisade of radial glial fibers (Stenman et al., 2003b). In miR-9-2/3 double mutants, the Nestin-positive glial palisade was aberrant and less fasciculated (supplemental Fig. 8E,F, available at www.jneurosci.org as supplemental material). With the dorsal shift of the PSB, the Dbx1-positive ventral pallium that juxtaposes to the PSB was greatly reduced as stated above (Fig. 3K,L), and Er81-positive basolateral amygdala that derives from the pallium (Stenman et al., 2003b) was lost in the miR-9-2/3 double mutants (supplemental Fig. 8G,H, available at www.jneurosci.org as supplemental material).
Thalamocortical axon and CFA projections are misrouted with corridor malformation in MGE
In the miR-9-2/3 double-mutant ventral telencephalon, CFAs poorly innervated the internal capsule (Fig. 10A–D′). Thalamocortical axons (TCAs) labeled by L1 and GAP43 were nearly lost before reaching the internal capsule, and a majority of fibers abnormally descended into the hypothalamus (Figs. 9A,B, 10A–D′). Islet1- and Meis2-positive corridor neurons, which originate from the LGE and locate between the Nkx2.1-positive proliferating zone and globus pallidus (López-Bendito et al., 2006) (Fig. 10E,E′,G,G′), are known to function as a gateway for TCA tracts to the internal capsule. Nkx2.1-positive globus pallidus was nearly lost (Fig. 10E–H), and Islet1- and Meis2-positive corridor neurons were expanded or dispersed in the miR-9-2/3 double mutants (Fig. 10E′,F′,G′,H′). In a mutant that has no Pax6 expression in the ventral telencephalon, the corridor cells are dispersed and TCA is not trajected into the telencephalon (Simpson et al., 2009). This Pax6 expression was, however, not significantly altered in the miR-9-2/3 double mutant at E15.5 (Fig. 10I,J). Not only Meis2, as described above, but also Islet1 mRNA has miR-9 responsive elements (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Meis2 protein level was increased in the miR-9-2/3 double-mutant subpallium (Fig. 6A,B). Wild-type miR-9-2 moderately suppressed luciferase expression from a reporter conjugated to the Islet1 3′ UTR (Fig. 10K). The increase in Islet1 protein expression was also moderate in the miR-9-2/3 double-mutant subpallium (Fig. 10L; Table 1).
Discussion
The miR-9-2/3 double mutants demonstrate that miR-9 orchestrates neurogenesis in the pallium and subpallium by regulating the expression of multiple genes that function in progenitor proliferation and differentiation. Furthermore, we propose that these miR-9 functions are modulated by the RNA-binding proteins. Increase in Foxg1 expression in miR-9-2/3 double mutant at early neurogenic stages is consistent with the reduction in Cajal-Retzius cells and early-born neurons (Dou et al., 1999; Hanashima et al., 2002, 2004; Martynoga et al., 2005; Muzio and Mallamaci, 2005). Nr2e1 and Pax6 reduction is also consistent with the decrease in later progenitor proliferation in pallium (Stenman et al., 2003b; Roy et al., 2004; Schuurmans et al., 2004) and the increase in Gsh2 and Foxg1 expression with the increase in progenitor proliferation in subpallium (Toresson et al., 2000; Yun et al., 2001) in this study. However, the Foxg1 increase was at a twofold level, and it is a question whether this level of Foxg1 increase can fully explain the reduction of Cajal-Retzius cells and early-born neurons; the question remains for future studies such as Foxg1 overexpression or the miR-9-2/3 compound mutation with heterozygous Foxg1 mutation. It should also be examined whether and how Nr2e1 or Pax6 overexpression can restore the later pallium phenotype in the miR-9-2/3 double mutants. Gsh2 increase in the miR-9-2/3 double mutants was also ∼1.7-fold and Islet1 increase was ∼1.4-fold. It is plausible that miR-9 targets many more genes in each event. The miR-9 regulation of each gene is moderate, and the regulation of a single gene by miR-9 might not be responsible in each phenotype; rather, miR-9 mutant phenotype might be explained by the cumulation of the moderate reduction of multiple genes that participate in an event.
Previous studies (Bhattacharyya et al., 2006; Kedde et al., 2007; Vasudevan and Steitz, 2007; Vasudevan et al., 2007) and this study strongly suggest that it depends on the context of RNA-binding proteins expressed whether a microRNA suppresses or enhances the protein expression or has no effect. The Foxg1 3′ UTR has U-rich sequences, and this study proposes that the miR-9 suppression of Foxg1 protein expression is countered by an AU-rich RNA binding protein, Elavl2, later than E15.5. We also propose that miR-9 targets Nr2e1 mRNA to enhance its protein expression in cooperation with Elavl1 and/or Msi1. Notably, miR-9 suppresses the expression of luciferase conjugated to the 1.4 kb Nr2e1 3′ UTR in HEK 293 cells and adult neural stem cells (Zhao et al., 2009). We speculate that the discrepancy is explained by differences in cell types or RNA binding protein repertoires. Extensive analysis of factors that bind to Foxg1 and Nr2e1 3′ UTRs site and stage specifically are required to clarify the precise mechanisms of the miR-9 regulation of Foxg1 and Nr2e1 expression. miR-9 was ineffective on Id4 and Pax6 3′ UTRs and only marginally effective on Islet1 3′ UTR in the luciferase assay with the P19 cells. However, miR-9 might also target these mRNAs efficiently in vivo in the presence of RNA binding proteins that are missing or insufficient in the P19 cells. At the same time, the possibility that miR-9 enhances Nr2e1 expression indirectly by suppressing a gene that inhibits Nr2e1 expression cannot be ruled out. This possibility will be tested in future studies by deleting the miR-9 responsive element in endogenous Nr2e1; Nr2e1 expression from such allele should not be affected by miR-9-2/3 deficiency if miR-9 directly targets Nr2e1.
Cajal-Retzius cells are reduced in miR-9-2/3 double mutants as expected from our previous study (Shibata et al., 2008). However, a significant number of Cajal-Retzius cells remain in the miR-9-2/3 double mutants, explaining the normal lamination in the inside-out pattern (Yoshida et al., 2006). The presence of miR-9-1 in the miR-9-2/3 double mutants might explain the incomplete loss of Cajal-Retzius cells (Fig. 3A–F). Alternatively, microRNAs fine-tune protein expression levels rather than completely switching gene expression on and off, and a significant number of Cajal-Retzius cells might be generated even if miR-9 were completely lost. This must await the analyses of miR-9-1/2/3 triple mutants, and the issue must be kept in mind in interpreting all phenotypes of miR-9-2/3 double mutants.
We propose that miR-9 targets Meis2 in both pallium and subpallium, but the roles of Meis2 in telencephalon development remain to be demonstrated. The mechanisms of the Meis2- and Islet-1-positive corridor expansion, including whether it is a real expansion or dispersion brought about by the loss of Nkx2–1-positive globus pallidus, and the significance of the corridor abnormality in TCA projection require additional studies. No reduction of corridor was reported in Islet1 knock-out mutants (Elshatory and Gan, 2008), but the effects of Islet1 overexpression in the corridor formation remain to be determined. The reduction of differentiating field in subpallium but normal differentiation of interneurons as well as details of the lack of tangential migration of interneurons in miR-9-2/3 double mutants also require additional study; reduction in striatum differentiation but normal interneuron generation was observed in Nr2e1 mutant LGE (Stenman et al., 2003c). The reason why the layer VI was apparently normal in the double mutants, against the increase in progenitor proliferation and decrease in neuronal differentiation at E12.5, remains uncertain; layer VI neuron production was caught up at E13.5 in miR-9-2/3 double mutants. To explain this, the origin of layer VI neurons or the birthdate analysis of double-mutant progenitor cells must first be examined in detail during E12.5–E13.5. miR-9* expression was decreased to 38% in miR-9-2/3 double-mutant telencephalon at E16.5, but this study could not demonstrate miR-9* functions in neurogenesis in telencephalon. More detailed studies are required to assess miR-9* regulation of REST, CoREST, and BAF53a expression; it might be stage- and/or site-dependent in terms of RNA binding partners. In addition, we could not find other miR-9* target candidates with the criteria described in the previous paper (Shibata et al., 2008). The criteria include the conservation of miR-9* responsive element among mouse, human, and chick orthologs, and the analyses of genes that conserve miR-9/miR-9* responsive elements between mouse and human orthologs may be desired. A lot of questions remain to future studies. Nevertheless, the study fully demonstrates that miR-9 is a crucial regulator of progenitor proliferation in both the pallium and subpallium through the regulation of multiple genes.
Finally, defects in subpallial development in the miR-9-2/3 double mutant are reminiscent of mutant mice in which exon 1 of huntingtin (Htt) was replaced with a human mutant Htt exon 1; progenitor proliferation is enhanced, and differentiation suppressed, in the striatum (Molero et al., 2009). Huntington's disease is a neurogenic disorder associated with dysfunction and degeneration of mainly medial spiny neurons of the striatum, having a midlife onset. However, the nexus of Huntington's disease might occur at an earlier time in the specification of the neurons (Molero et al., 2009). Nuclear localization of REST by Htt mutation with an expanded polyglutamate stretch is one putative molecular mechanism underlying Huntington's disease pathogenesis (Zuccato et al., 2003). REST suppresses the transcription of neuron restrictive silencer element-controlled neural genes. miR-9/miR-9* is reported to suppress REST and CoREST expression in vitro (Packer et al., 2008) and REST in turn to suppress miR-9 expression for progenitor proliferation (Conaco et al., 2006). Furthermore, miR-9/9* expression was significantly decreased in Huntington's disease patient cortices (Packer et al., 2008). In vivo functions of REST in neurogenesis in mouse telencephalon have been not demonstrated, and REST expression was not significantly altered in miR-9-2/3 double-mutant E13.5 telencephalon. However, more detailed analysis, especially on miR-9-1/2/3 triple mutants, is worthwhile to elucidate the significance of miR-9/miR-9* functions in the neurogenesis as well as in the pathological mechanisms of Huntington's disease.
Footnotes
This work was supported by a Grant-in-Aid for Creative Scientific Research from the Japan Society for the Promotion of Science. We are indebted to the Laboratory for Animal Resources and Genetic Engineering, RIKEN Center for Developmental Biology (CDB), for their help in the generation and housing of the mutant mice. We acknowledge Dr. Y. Sasai of RIKEN CDB for the Foxg1 antibody, Dr. K. Campbell of the University of Cincinnati College of Medicine for the Gsh2 antibody, and Dr. Y. Hayashizaki of RIKEN Omics Science Center and the Genome Resource and Analysis unit of RIKEN CDB for providing us FANTOM3 clones. We also thank those who provided us with probes for RNA in situ hybridization.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Agoston Z, Schulte D. Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development. 2009;136:3311–3322. doi: 10.1242/dev.037770. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Walker R, Kondo T, van Crüchten I, King ER, Sablitzky F. Id4 is required for the correct timing of neural differentiation. Dev Biol. 2005;280:386–395. doi: 10.1016/j.ydbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denaxa M, Chan CH, Schachner M, Parnavelas JG, Karagogeos D. The adhesion molecule TAG-1 mediates the migration of cortical interneurons from the ganglionic eminence along the corticofugal fiber system. Development. 2001;128:4635–4644. doi: 10.1242/dev.128.22.4635. [DOI] [PubMed] [Google Scholar]
- Deschênes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. Bioessays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- Dou CL, Li S, Lai E. Dual role of brain factor-1 in regulating growth and patterning of the cerebral hemispheres. Cereb Cortex. 1999;9:543–550. doi: 10.1093/cercor/9.6.543. [DOI] [PubMed] [Google Scholar]
- Elshatory Y, Gan L. The LIM-homeobox gene Islet-1 is required for the development of restricted forebrain cholinergic neurons. J Neurosci. 2008;28:3291–3297. doi: 10.1523/JNEUROSCI.5730-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marín O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Molnár Z, Tarabykin V, Stoykova A. Molecular mechanisms of cortical differentiation. Eur J Neurosci. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- Hanashima C, Shen L, Li SC, Lai E. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J Neurosci. 2002;22:6526–6536. doi: 10.1523/JNEUROSCI.22-15-06526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hanashima C, Fernandes M, Hebert JM, Fishell G. The role of Foxg1 and dorsal midline signaling in the generation of Cajal-Retzius subtypes. J Neurosci. 2007;27:11103–11111. doi: 10.1523/JNEUROSCI.1066-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O, Sugihara TM, Andersen B. Expression of LMO-4 in the central nervous system of the embryonic and adult mouse. Cell Mol Biol. 1999;45:677–686. [PubMed] [Google Scholar]
- Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- Huang Z. Molecular regulation of neuronal migration during neocortical development. Mol Cell Neurosci. 2009;42:11–22. doi: 10.1016/j.mcn.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, Lund AH, Perrakis A, Raz E, Agami R. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Langevin LM, Mattar P, Scardigli R, Roussigné M, Logan C, Blader P, Schuurmans C. Validating in utero electroporation for the rapid analysis of gene regulatory elements in the murine telencephalon. Dev Dyn. 2007;236:1273–1286. doi: 10.1002/dvdy.21126. [DOI] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- López-Bendito G, Cautinat A, Sánchez JA, Bielle F, Flames N, Garratt AN, Talmage DA, Role LW, Charnay P, Marín O, Garel S. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125:127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Manuel M, Martynoga B, Yu T, West JD, Mason JO, Price DJ. The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development. 2010;137:487–497. doi: 10.1242/dev.039800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Métin C, Baudoin JP, Rakić S, Parnavelas JG. Cell and molecular mechanisms involved in the migration of cortical interneurons. Eur J Neurosci. 2006;23:894–900. doi: 10.1111/j.1460-9568.2006.04630.x. [DOI] [PubMed] [Google Scholar]
- Molero AE, Gokhan S, Gonzalez S, Feig JL, Alexandre LC, Mehler MF. Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington's disease. Proc Natl Acad Sci U S A. 2009;106:21900–21905. doi: 10.1073/pnas.0912171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár Z, Cheung AF. Towards the classification of subpopulations of layer V pyramidal projection neurons. Neurosci Res. 2006;55:105–115. doi: 10.1016/j.neures.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Morante-Oria J, Carleton A, Ortino B, Kremer EJ, Fairén A, Lledo PM. Subpallial origin of a population of projecting pioneer neurons during corticogenesis. Proc Natl Acad Sci U S A. 2003;100:12468–12473. doi: 10.1073/pnas.1633692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Furushima K, Hirano M, Kiyonari H, Nakamura M, Suda Y, Aizawa S. ang is a novel gene expressed in early neuroectoderm, but its null mutant exhibits no obvious phenotype. Gene Expr Patterns. 2004;5:171–178. doi: 10.1016/j.modgep.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Muzio L, Mallamaci A. Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J Neurosci. 2005;25:4435–4441. doi: 10.1523/JNEUROSCI.4804-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Kuznicki K, Wu Q, Sun Z, Bock D, Schutz G, Vranich N, Monaghan AP. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci. 2004;24:8333–8345. doi: 10.1523/JNEUROSCI.1148-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesugi S, Miyata T, Ogawa M, Mikoshiba K, Okano H. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex. 2008;18:1865–1875. doi: 10.1093/cercor/bhm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TI, Pratt T, Mason JO, Price DJ. Normal ventral telencephalic expression of Pax6 is required for normal development of thalamocortical axons in embryonic mice. Neural Dev. 2009;4:19. doi: 10.1186/1749-8104-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003a;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J, Yu RT, Evans RM, Campbell K. Tlx and Pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development. 2003b;130:1113–1122. doi: 10.1242/dev.00328. [DOI] [PubMed] [Google Scholar]
- Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003c;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tamura S, Morikawa Y, Iwanishi H, Hisaoka T, Senba E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124:261–267. doi: 10.1016/j.neuroscience.2003.11.036. [DOI] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Stoykova A. Er81 is a downstream target of Pax6 in cortical progenitors. BMC Dev Biol. 2008;8:23. doi: 10.1186/1471-213X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Assimacopoulos S, Jones KR, Grove EA. Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development. 2006;133:537–545. doi: 10.1242/dev.02209. [DOI] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]