Abstract
The synapse-associated protein of 47 kDa (SAP47) is a member of a phylogenetically conserved gene family of hitherto unknown function. In Drosophila, SAP47 is encoded by a single gene (Sap47) and is expressed throughout all synaptic regions of the wild-type larval brain; specifically, electron microscopy reveals anti-SAP47 immunogold labeling within 30 nm of presynaptic vesicles. To analyze SAP47 function, we used the viable and fertile deletion mutant Sap47156, which suffers from a 1.7 kb deletion in the regulatory region and the first exon. SAP47 cannot be detected by either immunoblotting or immunohistochemistry in Sap47156 mutants. These mutants exhibit normal sensory detection of odorants and tastants as well as normal motor performance and basic neurotransmission at the neuromuscular junction. However, short-term plasticity at this synapse is distorted. Interestingly, Sap47156 mutant larvae also show a 50% reduction in odorant–tastant associative learning ability; a similar associative impairment is observed in a second deletion allele (Sap47201) and upon reduction of SAP47 levels using RNA interference. In turn, transgenically restoring SAP47 in Sap47156 mutant larvae rescues the defect in associative function. This report thus is the first to suggest a function for SAP47. It specifically argues that SAP47 is required for proper behavioral and synaptic plasticity in flies—and prompts the question whether its homologs are required for proper behavioral and synaptic plasticity in other species as well.
Introduction
The relationship between brain and behavior is the core topic of neuroscience. Given the multitude of events associated with any given behavior, it seems reasonable to address this issue in the form of its first time derivative, i.e., to ask which change in the brain is necessary and sufficient for a given change in behavior. In this sense, studying associative learning and memory, rather than being particularly difficult, may be tackling the relation between brain and behavior in a particularly accessible form. Within this context, we focus on the role of a specific synaptic protein [the synapse-associated protein of 47 kDa (SAP47) (Reichmuth et al., 1995; Funk et al., 2004; Hofbauer et al., 2009)] for associative function and synaptic physiology in larval Drosophila. This seems timely, given the importance of synaptic processes for learning in general [e.g., Lechner and Byrne (1998) and Pittenger and Kandel (2003)] and in larval Drosophila in particular (e.g., Michels et al., 2005).
The Sap47 gene of Drosophila (Fig. 1A) was identified by a monoclonal antibody from a hybridoma library raised against Drosophila brain (Hofbauer et al., 2009). It codes for a synaptic protein with a BSD domain and is highly conserved between nematode, fly, fish, and human (Reichmuth et al., 1995; Funk et al., 2004). The BSD domain is localized in a conserved central region and is found in BTF2-like transcription factors, SAP47 homolog proteins, and DOS2-like proteins. The domain is characterized by three predicted α helices and a conserved phenylalanine-tryptophan amino acid pair (Doerks et al., 2002). SAP47 is abundant in synaptic terminals (Reichmuth et al., 1995; Funk et al., 2004), but a role in synaptic function and/or behavior has not been determined, in any species.
Figure 1.
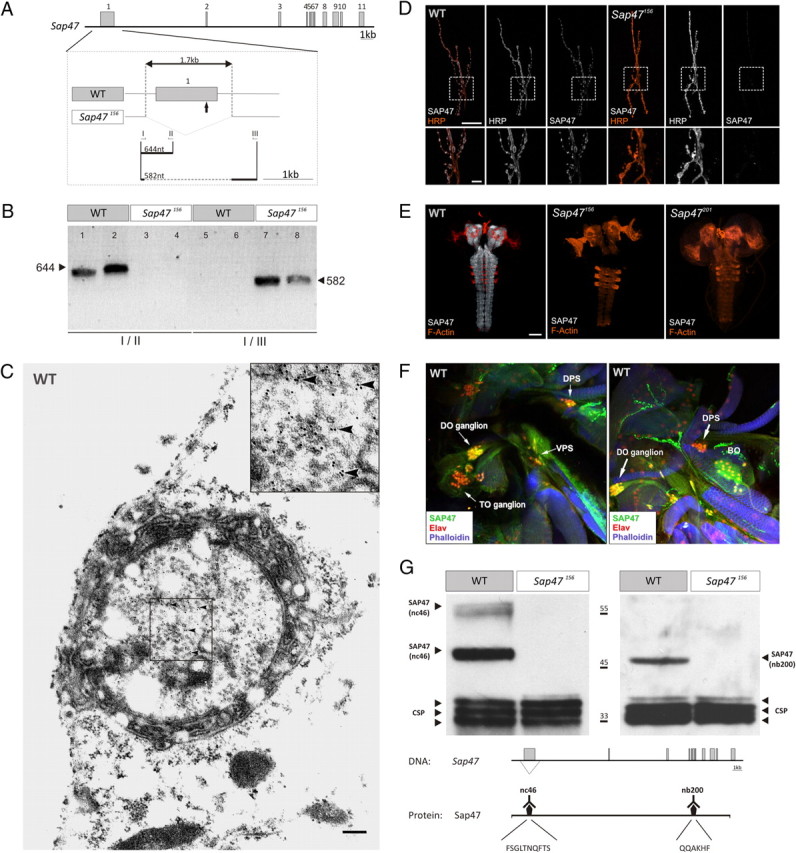
Characterizing the Sap47 gene and the Sap47156 mutant. A, Gene structure. Shown is the exon–intron structure of the Sap47 gene in wild type (WT: exons gray). The deletion in Sap47156 is shown in higher magnification. The arrow below the first exon indicates the translation start. Other arrows and roman numerals indicate binding sites of primers used for PCR in B. B, PCR. PCR with primer pair I/II generates a 644 bp fragment only in WT, because there is no binding site for primer II in the Sap47156 mutant; primer pair I/III generates a 582 bp fragment only in the Sap47156 mutant, because an elongation time was chosen that is too short for amplification of the long WT fragment. Two samples were run for each condition. C, Electron microscopy. Shown is a synaptic bouton with synaptic vesicles and immunolocalization of SAP47 in a WT larva using electron microscopy at the larval neuromuscular junction. Bound primary mouse anti-SAP47 antibodies were detected with gold-conjugated secondary anti-mouse antibodies. Gold particles (arrowheads) are localized close to synaptic vesicles. The inset represents the boxed area in higher magnification. Scale bar, 200 nm. D, Neuromuscular junction. At the neuromuscular junction (muscle pair 6/7), the Sap47156 strain does not show any anti-SAP47 immunoreactivity, whereas in WT, synaptic boutons are stained (white). Preparations are double labeled with a Texas Red-coupled anti-HRP antibody to label cell membranes (orange), and the nc46 antibody to label SAP47, and are viewed under a confocal microscope. Scale bar, 20 μm. The lower panels represent an enlarged view of the boxed area. Scale bar, 5 μm. E, Whole-mount larval brains. In both mutant strains (Sap47156 and Sap47201), no anti-SAP47 immunoreactivity is detectable in whole-mount preparations of the larval brain, whereas the neuropil regions in WT are strongly stained [antibody: nc46 (white)]. For orientation, F-actin is visualized with phalloidin (orange). Scale bar, 50 μm. F, Cephalic sensory systems. SAP47 immunoreactivity is detectable in single confocal slices of the cephalic sensory systems of third-instar WT larvae. DO, TO, the dorsal, posterior, and ventral pharyngeal sense organs [DPS, PPS (not shown), and VPS], and the Bolwig organ are at least partially stained [antibody: nc46 (green)]. For orientation, F-actin is visualized with phalloidin (blue) and an anti-Elav antibody is used to stain neuronal nuclei (red). Note that in the DO ganglion SAP47 is found in cell nuclei. G, Western blot. There is no SAP47 signal detectable on Western blot in the Sap47156 mutant, whereas WT shows the expected (Reichmuth et al., 1995; Funk et al., 2004) strong band at 47 kDa with both monoclonal antibodies used [left: nc46, with its epitope FSGLTNQFTS, which is within the Sap47156 deletion; right: nb200, with its epitope QQAKHF, which is downstream, C-terminal of the Sap47156 deletion (Hofbauer et al., 2009)]. As loading control, a monoclonal antibody against the synaptic protein CSP is used (antibody: ab49).
In this study, we tested for an association of SAP47 with synaptic vesicles using immunogold labeling in electron microscopy, and for the expression pattern of SAP47 in the brain of larval Drosophila by confocal immunohistochemistry. To analyze the function of SAP47, we used the fully viable and fertile deletion mutant Sap47156 (Funk et al., 2004). After extensive outcrossing of this mutant with the wild-type strain Canton-S (henceforth called WT), we tested for the genomic status of WT and Sap47156 by PCR, and for a possible residual expression of SAP47 in Sap47156 mutants. We then asked whether basic synaptic transmission at the neuromuscular junction as well as short-term synaptic plasticity at this synapse may be distorted, and whether sensory and motor abilities are intact in larvae lacking SAP47. We then moved on to test whether Sap47156 mutants were impaired in an odorant–reward associative conditioning task (see review by Gerber and Stocker, 2007), and whether knocking down SAP47 by RNAi mimicked this impairment (Brand and Perrimon, 1993; Smith et al., 2000; Kalidas and Smith, 2002). We finally tested whether the associative defect of Sap47156 mutant larvae could be rescued by restoring SAP47. We conclude that SAP47 functions in associative behavioral and short-term synaptic plasticity, without, however, contributing to basic synaptic transmission or to task-relevant sensory-motor function.
Materials and Methods
We used third-instar feeding-stage larvae collected 5 d after egg laying. Flies were kept in mass culture and were maintained at 25°C, 60–70% relative humidity, and a 14/10 h light/dark cycle. Electrophysiology and behavioral experiments were performed blind with respect to genotype and treatment condition; these were decoded only after the experiments.
Single-larva PCR.
To confirm the deletion status of the Sap47156 mutant (Funk et al., 2004), single-larva PCR was performed according to Gloor et al. (1993) (Fig. 1A,B; supplemental Fig. 10A, available at www.jneurosci.org as supplemental material): the primer I binding site (5′-GAGAAGAGCTCGACTTTCCAG-3′) was upstream of the deletion, the binding site of primer II (5′-CTTCGCTCTCTTGGACTCG-3′) was within the deletion, and the binding site of primer III (5′-CCTATCCACTCAGTTTGAGGG-3′) was downstream of the deletion. The PCR product of primer pair I/II should generate a 644 nt fragment only in WT, whereas primer pair I/III should produce a 582 nt product only in Sap47156 mutants, because an elongation time was chosen that was too short to amplify the predicted 2309 nt WT fragment.
Probing for expression of long isoforms of SAP47.
Given that the predicted long isoforms of SAP47 cannot readily be detected on a Western blot, we probed for the expression of long isoforms of Sap47 on the cDNA level. Total RNA was isolated from WT larvae by homogenizing 200 larvae in 1 ml of TRIzol (Invitrogen) followed by 5 min incubation at room temperature. After adding 200 μl of chloroform, the samples were centrifuged (12,000 × g) and supernatant was selected for an isopropanol precipitation. Then, RNA was resuspended in 100 μl of DEPC-water. cDNA was produced using oligo-dT-primer with the RevertAid First Strand cDNA Synthesis Kit (Fermentas). PCR specific for long Sap47 isoforms was subsequently performed using the following primers: (A) CTC CGC AAG GGC GCA GGA (forward primer); (B) TTC AGT GAT GAT CTT GGG CAC CAG (reverse primer); and (C) CCC AGC TCT TTG CCG GC (reverse primer). PCR was performed using standard protocols, followed by electrophoresis in 0.8–1.0% agarose gels.
Fly strains.
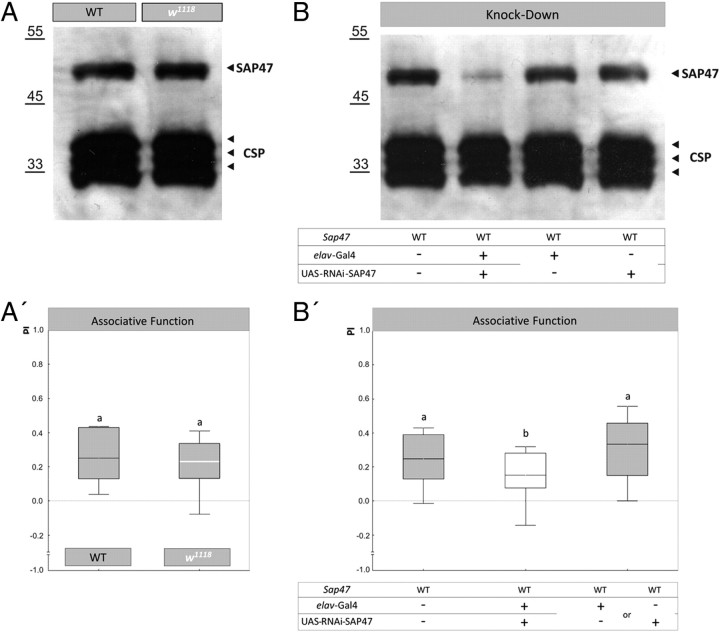
We compared WT to the deletion mutant Sap47156. The Sap47156 strain carries a deletion generated by imprecise P-element excision; it is characterized by a 1727 bp deletion, which removes part of the promoter region, the first exon, and a small part of the first intron of the Sap47 gene (Fig. 1A; supplemental Fig. 10A, available at www.jneurosci.org as supplemental material) (Funk et al., 2004). This strain was outcrossed to WT for nine generations to remove marker genes and to effectively adjust genetic background, which may otherwise distort results (de Belle and Heisenberg, 1996; Diegelmann et al., 2006). The second mutant Sap47 allele used was Sap47201, also generated by P-element jump-out and suffering a ∼5.8 kb deletion containing the promoter region, the first exon, and a larger part of the first intron; this strain still is in the w1118 mutant background. The status of the white gene is inconsequential for associative function in our paradigm (see Fig. 5A,A′).
Figure 5.
SAP47 knockdown. A, Western blot of WT and white mutants. Given that all strains are in the w1118 mutant background (see Materials and Methods), a test of SAP47 expression in w1118 is warranted, which shows no difference in SAP47 expression to WT. Antibodies used are the monoclonal nc46 for SAP47 detection and, as loading control, the monoclonal ab49, the latter labeling CSP, another presynaptic protein. A′, white mutants are not impaired in associative function. WT and w1118 perform equally well in the associative learning paradigm. The PREF scores underlying PI values are documented in supplemental Figure 3 (available at www.jneurosci.org as supplemental material). N = 16, 16. Shared lettering above plots signifies p > 0.05 in a Mann–Whitney U test. B, Western blot of SAP47 knockdown. The elav-Gal4 driver-control and the UAS-RNAi-SAP47 effector-control strains show no difference to WT in terms of SAP47 expression level, but the knockdown larvae show an obvious reduction. B′, SAP47 knockdown larvae are impaired in associative function. Associative function is reduced to ∼50% of control levels upon expression of an RNA-interference mRNA, using elav-Gal4 as driver- and UAS-RNAi-SAP47 as effector-strain. Both controls perform equally well (see supplemental Fig. 8A, available at www.jneurosci.org as supplemental material) and are therefore pooled. Notably, this RNAi-induced reduction of associative function is as severe as the one seen upon a total lack of SAP47 in the Sap47156 mutant (Fig. 2). The PREF scores underlying the PI values are documented in supplemental Figure 4 (available at www.jneurosci.org as supplemental material). N = 32, 19, 36. Different lettering above plots signifies p < 0.05/2 in Mann–Whitney U tests.
For a knockdown of the SAP47 protein, we used the widely expressed elav-Gal4 driver strain [elav-Gal4 [X] (Robinow and White, 1988); called c155 by Lin and Goodman (1994)] from the Bloomington stock center. These elav-Gal4 flies were crossed to UAS-RNAi-SAP47-effector flies [[VII10]; the construct consists of an ∼1.1 kb fragment of Sap47 cDNA (exons 1, 3–7) inserted in sense and antisense orientation; for more detail, see Funk et al. (2004)] to yield animals that carry both transgenes heterozygously; therefore, in these animals SAP47 expression should be reduced pan-neuronally. Genetic controls were heterozygous for either the driver or the effector transgene, respectively.
For pan-neural rescue expression of SAP47, we combined the driver strain elav-Gal4 [X] (c155) into the Sap47156 mutant background by classical genetics. As effector strains for the rescue experiments, we used either UAS-Sap47-RF; Sap47156 [III] or UAS-Sap47-RA; Sap47156 [II; III].
The UAS-Sap47-RF; Sap47156 [III] strain was generated on the basis of Wegener (2008) [her supplemental Fig. S2, UAS-Sap47-1; Sap47156; the coding region corresponds to Flybase (http://flybase.bio.indiana.edu): Transcript Dmel\Sap47-RF: FlyBase ID: FBtr0301655; for the predicted amino acid sequence, see supplemental Fig. 10B, available at www.jneurosci.org as supplemental material]. To generate the experimental genotype for rescue experiments, we crossed the driver strain (elav-Gal4; Sap47156) to the UAS-Sap47-RF; Sap47156 effector strain, yielding double heterozygous larvae in which expression of the 47 kDa isoform of SAP47 is restored. Genetic controls were in the Sap47156 mutant background and were heterozygous for either the driver or the effector transgene, respectively.
For expressing the full-length isoform of SAP47, the UAS-Sap47-RA; Sap47156 [II; III] effector strain was generated. The full-length cDNA clone LD36546 [(http://flybase.bio.indiana.edu): Transcript Dmel\Sap47-RA; FlyBase ID: FBcI0175830; for the predicted amino acid sequence, see supplemental Fig. 10C, available at www.jneurosci.org as supplemental material] from the Drosophila Genomics Resource Center (The Center for Genomics and Bioinformatics, Indiana University, Bloomington) was used to amplify the full-length cDNA by PCR with primers containing a NotI and KpnI restriction site. The used primer pair was as follows: primer 1: 5′-ATA AGA ATG CGG CCG CCG CAG TTG TTG TTT CC-3′ and primer 2: 5′-GAG CGG TAC CGG TTT CGA ATA GTT TTG TAT TTT GTT TGG-3′. The resulting PCR fragment was excised as a 2822 bp NotI/KpnI fragment, ligated into NotI/KpnI-cut pUAST (Brand and Perrimon, 1993), and transformed into recombination-deficient SURE2 supercompetent cells (Stratagene). Germ-line transformation then was performed into a w1118 strain (Bestgene). The resulting effector strain was combined into Sap47156 mutant background by classical genetics. To generate the experimental genotype for the rescue experiments, we crossed the driver strain (elav-Gal4; Sap47156) to the UAS-Sap47-RA; Sap47156 effector strain, yielding double heterozygous larvae in which expression of the full-length isoform of SAP47 of ∼70 kDa was restored. Genetic controls were also in the Sap47156 mutant background, and were heterozygous for either the driver or the effector transgene, respectively.
Please note that all transgenic fly strains used were in the white mutant background (w1118) to keep track of the transgenes. Therefore, genetic controls were established by crossing the respective transgenic strains to a w1118 strain (in the case of the RNAi experiment) or a w1118; Sap47156 strain (in the case of the rescue experiments). To see whether the w1118 mutation may have an effect, either on associative learning (Diegelmann et al., 2006; Yarali et al., 2009) or on SAP47 expression, we compared w1118 with WT larvae in odorant–reward associative ability as well as by Western blot analysis (see Fig. 5A,A′).
Immunocytochemistry: electron microscopy.
For immunogold localization of SAP47, preparations of the nerve–muscle synapse were made in ice-cold HL3 Ringer's solution (Stewart et al., 1994), fixed in 4% paraformaldehyde in 0.1 cacodylate buffer, pH 7.2, for 90 min on ice, washed three times for 30 min in this buffer, incubated in 10 mm ammonium chloride for 15 min, and washed two times for 15 min in H2O. After dehydration in ascending ethanol series, 15 min at −20°C for each step, the tissue was incubated for 1 h in a 1 + 1 mixture of LR-White (Polysciences Europe) and ethanol at −20°C, followed by two 1 h incubation periods in pure LR-White at −20°C, before the preparations were stored at 4°C for 3 d to achieve complete penetration of the resin. Polymerization was allowed to proceed for 5 d at 41°C. After verification of section plane from 2 μm sections in the light microscope, ultrathin (70 nm) sections were cut and transferred to copper grids; grids were then washed 5 min on a drop of PBS at pH 7.2, blocked for 5 min on a drop of 1% BSA in PBS, and incubated for 24 h at 4°C on a drop of the mouse monoclonal antibody nc46 (Hofbauer et al., 2009) (see also Funk et al., 2004), diluted 1:25 in PBS. After storage of the grids for 1 h at 37°C in a moist chamber to increase antibody affinity, they were washed thoroughly in PBS and incubated for 1 h on a drop of 1:10 diluted anti-mouse IgG conjugated to 12 nm gold particles (Dianova) and subjected to a final wash.
Immunocytochemistry: neuromuscular junction.
Third-instar larvae were dissected in calcium-free saline (Stewart et al., 1996) and fixed in 4% paraformaldehyde/phosphate buffer for 30 min at room temperature. After three 10 min washes in PBST, fillets were blocked with 1.5% normal horse serum diluted in PBST for 30 min at room temperature. The used primary monoclonal anti-SAP47 mouse antibody (nc46, 1:10 in PBST) is added and all fillets were kept overnight at 4°C. As secondary antibody, a rabbit anti-mouse Ig conjugated with Alexa Fluor 488 (1:500 in PBST) (Molecular Probes, Invitrogen Detection Technologies) was used; to label cell membranes, a Texas Red-coupled rabbit anti-HRP antibody (1:250) (Jackson ImmunoResearch Laboratories) was added to the incubation solution. Confocal data were acquired as image stacks of separate channels with a Leica TCS SP 1 laser scanning confocal microscope, combined, and visualized with Leica TCS NT software. Final pictures were obtained as maximum intensity projections.
Immunocytochemistry: whole-brain and cephalic sensory system preparation.
Brains were dissected in Ringer's solution on ice and incubated in fixative (2.5% paraformaldehyde and 0.3% Triton X-100 in 1×PBS) for 2 h. After washing them three times in PBST (0.3% Triton X-100 in 1×PBS), blocking was performed with 3% normal goat serum (NGS) in 1×PBST for 1.5 h. The primary monoclonal anti-SAP47 mouse antibody nc46 (1:10 in 3% NGS-PBST) was added and all brains were incubated overnight at 4°C. Samples were washed six times in PBST for 10 min and incubated overnight with Alexa Fluor 568 Phalloidin (1:200) (Invitrogen; Lot 41A1–4) for counterstaining of F-actin, and the secondary antibody (goat anti-mouse Ig, conjugated with Alexa Fluor 488 1:200 in PBST; Molecular Probes, Invitrogen Detection Technologies) at 4°C. Preparations of the cephalic sensory system were additionally stained with a rat anti-Elav antibody (Jackson ImmunoResearch Laboratories) as neuronal nucleus marker. As secondary antibody, goat anti-rat Ig conjugated with Alexa Fluor 647 1:200 (Molecular Probes, Invitrogen Detection Technologies) was used. Preparations were examined under a Leica TCS SP 1 laser scanning confocal microscope, combined, and visualized with Leica TCS NT software.
Western blotting.
After homogenization of 10 larval brains in Laemmli buffer and electrophoretic separation (SDS-PAGE), proteins were transferred onto a nitrocellulose membrane (45 μm, Schleicher and Schuell) by semidry blotting (Kyhse-Andersen, 1984) with a semidry electroblotter (PEQLAB). After blotting, the membrane was transferred in 5% powdered milk in TBST and washed for three times in TBST. Blots were probed with either the nc46 antibody or the mouse monoclonal antibody nb200; these antibodies recognize different epitopes of the protein within (nc46) and more C-terminal of (nb200) the deletion (Fig. 1G) (Hofbauer et al., 2009) (see also Funk et al., 2004), as well as with the mouse monoclonal ab49 antibody against the cysteine string protein (CSP) [corresponding to the DCSP1 antibody in Hofbauer et al. (2009)] (see also Arnold et al., 2004), marking a band at 32 kDa as loading control in all cases. Antibodies were used at the following dilutions in 1× TBST: nc46 (1:100), nb200 (1:50), and ab49 (1:400). To detect antibody-labeled proteins, IgG-HRP-conjugated goat anti-mouse (1:3750 in TBST) and ECL-Western blotting detection reagents (GE Healthcare) were applied.
Electrophysiology.
Two-electrode voltage-clamp recordings were performed on ventral longitudinal muscle 6 of male third-instar larvae in extracellular hemolymph-like solution HL3 containing 70 mm NaCl, 5 mm KCl, 20 mm MgCl2, 10 mm NaHCO3, 5 mm trehalose, 115 mm sucrose, 5 mm HEPES, and 1 mm CaCl2. Recordings were made from cells with input resistances of at least 4 MΩ and initial membrane potentials between −50 and −70 mV. Intracellular electrodes with resistances of 10–35 MΩ filled with 3 m KCl were used. The holding potential was −60 mV for evoked EPSCs (eEPSCs) and −80 mV for miniature EPSCs (mEPSCs). EPSCs were recorded at a stimulation frequency of 0.1 Hz. For the analysis of short-term plasticity, trains of 100 stimuli at 60 Hz were applied. Care was taken to ensure the recruitment of both motoneurons innervating muscle 6. Recordings were analyzed with pClamp10 (Molecular Devices). Experiments were performed blind with respect to genotype.
Associative function.
Larval learning experiments represent a modified version of the mass assay described by Neuser et al. (2005) (for sketches, see Fig. 2A,B). Notably, unless mentioned otherwise, we here used only one odorant, n-amyl acetate (AM, CAS: 628–63-7, purity: 99%, Merck) to simplify the paradigm (Selcho et al., 2009; Saumweber et al., 2011). That is, we train groups of 30 larvae each and compare olfactory choice performance after either of two reciprocal training regimens: one group was exposed to the odorant AM in the presence of a positive reinforcer and to a no-odor situation without the reinforcer (AM+/noAM); the second group was trained reciprocally, i.e., by unpaired presentations of odorant and reinforcer (AM/noAM+). Then, animals were tested for their choice between AM and noAM. Associative learning was indicated by systematic differences in test performance between the reciprocal treatment conditions. The reciprocally trained groups were run alternately, which allows stringent pairing of data for the calculation of a performing index (PI) [see below and discussion in Hendel et al. (2005)]. For a differential, two-odor version of our paradigm (Fig. 2B), we used 1-OCT (OCT, CAS: 111-87-5; purity: 99%, Sigma-Aldrich) as second odor.
Figure 2.
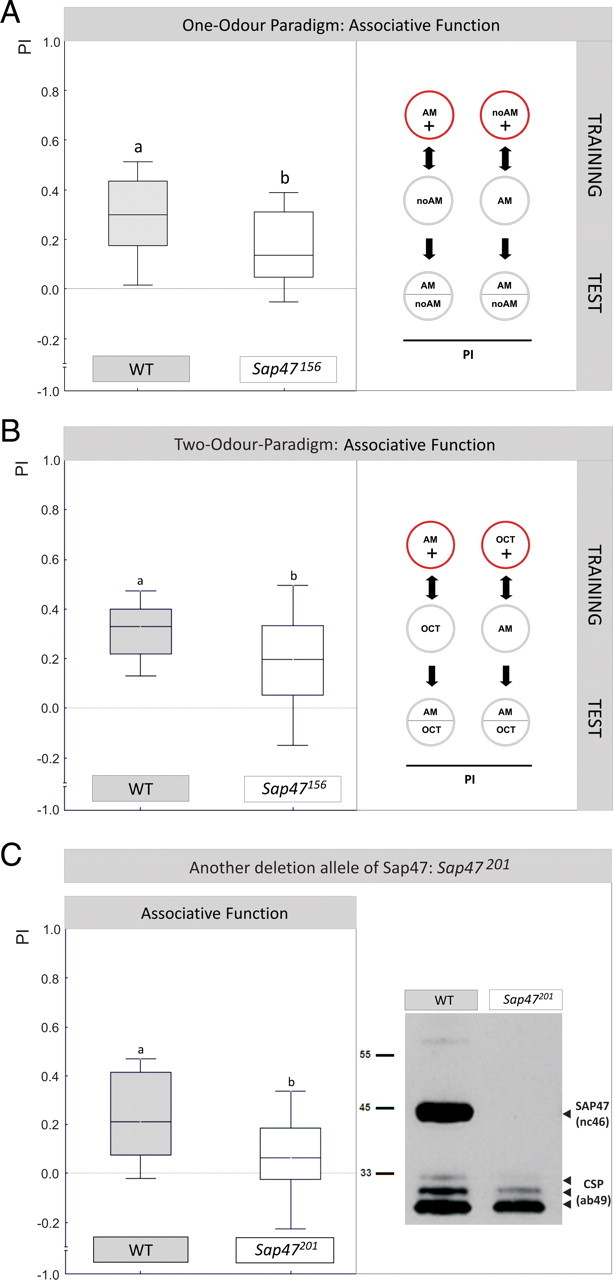
Associative function. A, Sap47156 mutants are impaired in associative function. WT and Sap47156 can learn, but performance indices in Sap47156 mutants are reduced by ∼50%. The inset illustrates the training regimen: For each genotype, one group is trained such that in the presence of amyl acetate animals are rewarded, whereas in the absence of amyl acetate they are not rewarded (AM+/noAM); the second group is trained reciprocally (AM/noAM+). Note that in half of the cases the sequence of trials is as indicated (i.e., the respective AM-containing trials first), but in the other half of the cases it is inverted; the sequence of training trials is without effect on test performance (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). The PI measures the extent to which both reciprocally trained groups differ in their AM preference during the test, and thus provides a measure of associative learning; the PREF scores underlying all PI values are documented in supplemental Figure 2A (available at www.jneurosci.org as supplemental material). N = 35, 35. Different lettering above plots signifies p < 0.05 in a Mann–Whitney U test. Box plots represent the median as the middle line, 25 and 75% quantiles as box boundaries, and 10 and 90% quantiles as whiskers. B, Also in a differential, two-odor paradigm Sap47156 mutants are impaired in associative function. Performance indices of Sap47156 mutants are also reduced by ∼50% compared to WT in a differential, two-odor version of the learning paradigm. The inset illustrates the training regime. N = 35 in both cases (one-sample sign tests: p < 0.05/2 for both genotypes). Different lettering above plots signifies p < 0.05 (U = 414.5; sample sizes as above) in a Mann–Whitney U test. The PREF scores underlying all PI values are documented in supplemental Figure 2B (available at www.jneurosci.org as supplemental material). C, Another deletion allele of Sap47 is also impaired in associative function. Left, Performance indices in the one-odor paradigm in Sap47201 mutants are reduced by ∼50% compared to WT. N = 16, 14, respectively (one-sample sign test: p = 0.21 for Sap47201 and p < 0.05/2 for WT). Different lettering above plots signifies p < 0.05 (U = 61; sample sizes as above) in a Mann–Whitney U test. The PREF scores underlying all PI values are documented in supplemental Figure 2C (available at www.jneurosci.org as supplemental material). Right, No signal is detectable on a Western blot of the Sap47201 mutant, whereas WT shows the expected band at 47 kDa. Anti-SAP47: nc46; anti-CSP: ab49 (loading control).
Petri dishes (Sarstedt) with 85 mm inner diameter were filled with 1% agarose (electrophoresis grade; Roth), allowed to solidify, covered with their lids, and left untreated until the following day. As positive reinforcer, we used 2 mol of fructose (FRU, purity: 99%) added to 1 l of agarose 10 min after boiling.
As an odorant, we used amyl acetate diluted 1:1600 in paraffin oil, which by itself is not behaviorally active (Saumweber et al., 2011); OCT was used undiluted. Experiments were performed in red light under a fume hood at 21–24°C. Just before the experiments, we replaced the regular lids of the Petri dishes with lids perforated in the center by roughly sixty 1 mm holes to improve aeration.
A spoonful of food medium containing larvae was taken from the food bottle and transferred to a glass vial. Thirty animals were collected, briefly washed in tap water, and as a group transferred to the assay plates for the start of training. Each training trial lasted 5 min. Immediately before a trial, two custom-made Teflon containers for possible loading with odorant (5 mm inner diameter, lid with seven 0.5 mm holes) were placed onto the assay plate on opposite sides of the plate, 7 mm from the edges. Within each reciprocal training condition, for half of the cases we started with AM in the odorant containers and for the other half with noAM. Consequently, for half of the cases we started with an agarose plate that had FRU added to the substrate and for the other half with a plate without FRU. Then, the lid was closed and the larvae were allowed to move for 5 min. The larvae were then transferred to a plate with the alternative odorant condition and the respective other substrate for 5 min. This cycle was repeated three times. Fresh assay plates were used for each trial.
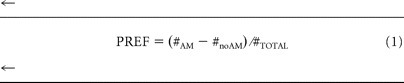
After this training, animals were tested for their odor choice. The larvae were placed in the middle of a fresh, pure agarose assay plate with a container of odorant on one side and an empty container on the other side (AM vs noAM); for half of the cases, AM was to the left, and for the other half of the cases, it was to the right. After 3 min, the number of animals on the “AM” or “noAM” side was counted. For both reciprocally trained groups, we then calculate an odor preference ranging from −1 to 1 as the number of animals observed on the AM side minus the number of animals observed on the noAM side, divided by the total number of larvae:
 |
For all learning experiments, these PREF scores are documented in the supplemental material. To determine whether these preferences are different depending on training regime, we took the paired data from alternately run, reciprocally trained groups and calculate a performing index ranging from −1 to 1 as follows:
After the data for one such PI value had been collected for one of the genotypes, the corresponding data for the other genotype were gathered; i.e., data from both genotypes were obtained alternately.
Detectability of odorant and reward.
To test whether larvae were able to detect the to-be-associated stimuli, animals were tested for their preference between AM and noAM as well as between FRU and noFRU. The assay for the detectability of the odorant was the same as described above for the test in Equation 1, except that experimentally naive animals were used.
To test the ability to detect FRU, split Petri dishes were prepared with one side pure agarose and the other with fructose added to the agarose (Hendel et al., 2005). Larvae were positioned in the middle of the Petri dish; after 3 min, the number of animals on either side was determined for calculation of the FRU preference in a way corresponding to Equation 1.
Sham training and tests for sequence effects.
Associative training by necessity involves exposure to the odorant, the reward, and handling. One may therefore argue that these aspects of training may induce motivational changes, fatigue, habituation, and/or adaptation, which may feign a learning deficit in the mutants if they were more- or less-susceptible to these kinds of effect than the wild type. Therefore, the response to the odorant needs to be tested after so-called “sham training” (Michels et al., 2005; Knapek et al., 2010). Two types of sham training controls were run: both consisted of the same treatments as in the learning experiment, except that either the reinforcer was omitted (SHAMOdor-noReward) or the odorant was omitted (SHAMReward-noOdor). After either of these sham training regimens, the ability of the larvae to detect the odorant was tested as detailed above.
The learning experiment allowed for a post hoc analysis of possible effects of the sequence of stimulus presentation on test performance: we compared the AM preferences of those groups that shared the same odorant–reward contingency, but differed in terms of the temporal pattern of the two types of trials [AM+/noAM vs noAM/AM+ (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material); noAM+/AM vs AM/noAM+ (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material)]. We did not find any effect of the timing of trial types, neither in WT (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) nor in Sap47156 (data not shown).
The same kind of analysis is possible for sham training experiments: we compared the AM preferences of the group of larvae exposed to AM during the first, third, and fifth trials and to noAM during the other trials (AM/noAM) to the AM preferences of the group that was exposed to AM during the second, fourth, and last trials (noAM/AM) (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). Similarly, we compared the AM preferences between those groups that did or did not receive the reward during the first, third, and fifth trials before test (FRU/noFRU vs noFRU/FRU) (supplemental Fig. 1D, available at www.jneurosci.org as supplemental material). In both cases, the sequence of stimulus presentation was without effect on test performance in WT (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) as well as in Sap47156 (data not shown).
Statistical analyses.
In a conservative approach, we used nonparametric analyses throughout; comparisons of values against zero, i.e., chance level, were made with one-sample sign tests. All comparisons were significantly different from chance, unless mentioned otherwise. Comparisons between multiple or two genotypes were done with Kruskal–Wallis or Mann–Whitney U tests, respectively. We corrected the level of significance in cases where multiple comparisons were made by dividing the p level of 0.05 by the number of comparisons made (Bonferroni correction) to maintain an experiment-wide judgement error rate of 5%. Shared letters above boxes indicate that groups behave not significantly different, whereas significant differences between groups are indicated by different letters above boxes. Data are displayed as box plots with the middle line indicating the median and box boundaries and whiskers the 25, 75, 10, and 90% quantiles, respectively. Analyses were performed with Statistica 7.0 (StatSoft) on a PC.
Results
Ultrastructural localization of SAP47
After confirming the genomic status of WT and the outcrossed Sap47156 mutant (Fig. 1A,B), we determined the ultrastructural localization of SAP47 at presynaptic terminals of WT larval motor neurons (Fig. 1C). Of 1631 gold particles in sections from 18 different presynaptic terminals, 87.5% were located within 30 nm of a synaptic vesicle (SV). Only 8.5% of the particles did not have a vesicle within that distance. We considered 4% of the gold particles detected outside boutons as unspecific background. Interestingly, SAP47 did not appear to be integral to the synaptic vesicle membrane because SAP47 was found in the soluble fraction of brain homogenate [Arnold et al. (2004), their Fig. 2], and glycerol density gradient centrifugation separated SAP47 from known integral synaptic vesicle membrane proteins [such as CSP: Umbach et al. (1994) and Arnold et al. (2004) (their Fig. 3)].
Figure 3.
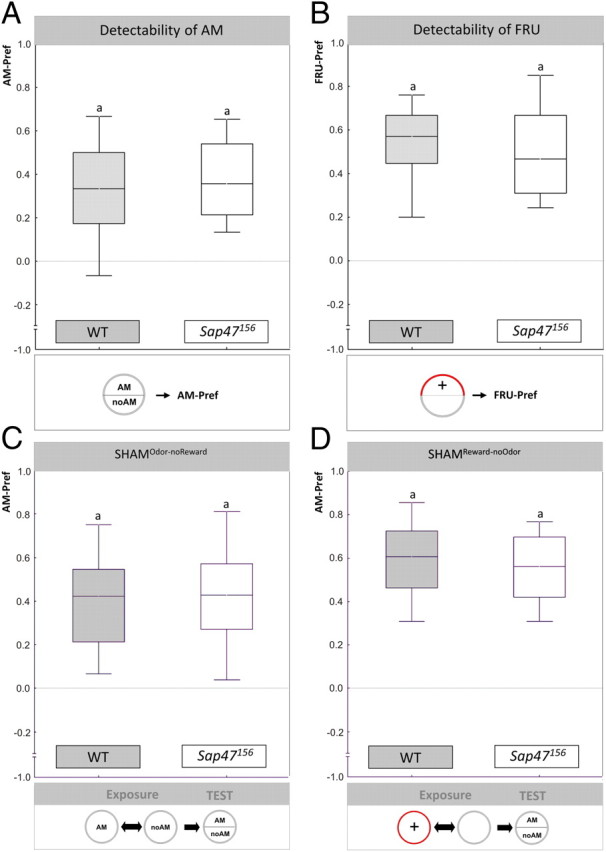
Behavioral controls: Sensory and motor ability are normal in Sap47156. In A and B, the behavior of experimentally naive animals toward the to-be-associated stimuli is compared between WT and Sap47156 mutants; in C and D, behavior toward the odorant is compared after animals had undergone “sham-training.” Insets in the figure illustrate the experimental regimen. A, Behavior toward AM in experimentally naive larvae: WT and Sap47156 show the same level of preference for amyl acetate. N = 42, 44. B, Behavior toward FRU in experimentally naive larvae: WT and Sap47156 show the same level of preference for fructose. N = 53, 53. C, Behavior toward AM after sham training that involves exposure to the odorant, but not to the reward (SHAMOdor-noReward). After such treatment, the Sap47156 mutants and the WT larvae show the same level of AM preference. N = 39, 42. D, Behavior toward AM after sham training that involves exposure to the reward, but not to the odorant (SHAMReward-noOdor). The Sap47156 mutants and the WT larvae show the same level of AM preference also after this kind of treatment. N = 50, 48. Shared lettering above plots signifies p > 0.05 in Mann–Whitney U tests.
Thus, SAP47 is associated with synaptic vesicles, but is not an integral part of the synaptic vesicle protein complement.
Sap47156 is a protein-null mutant allele
At neuromuscular junctions of muscle pair 6/7 and in preparations of the whole larval brain viewed under the confocal microscope, the Sap47156 strain did not show any SAP47 immunoreactivity, whereas in WT, presynaptic terminals and the complete neuropil region, respectively, were strongly stained (Fig. 1D,E) (used antibody: nc46). SAP47 was also expressed in the cephalic chemosensory system [dorsal organ (DO), terminal organ (TO), and the Bolwig organ] of WT third-instar larvae (Fig. 1F). Furthermore, we could not detect any SAP47 signal on a Western blot for Sap47156, whereas WT showed the expected (Reichmuth et al., 1995; Funk et al., 2004; Hofbauer et al., 2009) strong band at 47 kDa with both monoclonal antibodies used (Fig. 1G) [nc46 with its epitope FSGLTNQFTS, which was within the Sap47156 deletion; nb200 with its epitope QQAKHF, which is downstream, C terminal of the Sap47156 deletion (Hofbauer et al., 2009)]. One of the heavier bands at ∼62 kDa was seen only with the nc46 antibody; this band, however, is typically weaker and more variable in Western blots, potentially because of temporal and/or local specificity of expression (Funk et al., 2004) (see also Figs. 1G, 6B, 7B).
Figure 6.
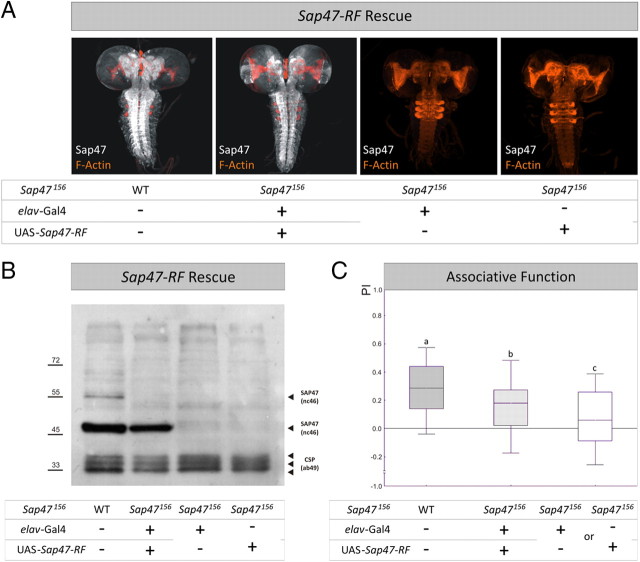
SAP47 rescue with 47 kDa isoform. A, Whole mounts. In contrast to WT and rescue larvae, the elav-Gal4; Sap47156 driver-control and the UAS-Sap47-RF; Sap47156 effector-control strains show no SAP47 expression. Phalloidin is used to visualize F-actin (orange). B, Western blot. In contrast to WT, the elav-Gal4; Sap47156 driver-control and the UAS-Sap47-RF; Sap47156 effector-control strains show no SAP47 expression; the rescue larvae show an obvious SAP47 band of 47 kDa, as is to be expected from the coding region used for the UAS-Sap47-RF transgene; a higher isoform is only detectable in WT. Anti-SAP47: nc46; anti-CSP: ab49 (loading control). C, Expression of the PF isoform of SAP47 partially rescues impairment in associative function. Associative function is reduced to ∼50% of control level (WT) in genetic controls, using elav-Gal4; Sap47156 as driver- and UAS-Sap47-RF; Sap47156 as effector-strain. Both controls perform equally well and are therefore pooled as genetic controls (supplemental Fig. 8B, available at www.jneurosci.org as supplemental material). The experimental group shows higher associative performance compared to genetic controls, but does not reach wild-type level. N = 68, 69, 136. Different lettering above plots signifies p < 0.05/2 in Mann–Whitney U tests. The PREF scores underlying all PI values are documented in supplemental Figure 5 (available at www.jneurosci.org as supplemental material).
Figure 7.
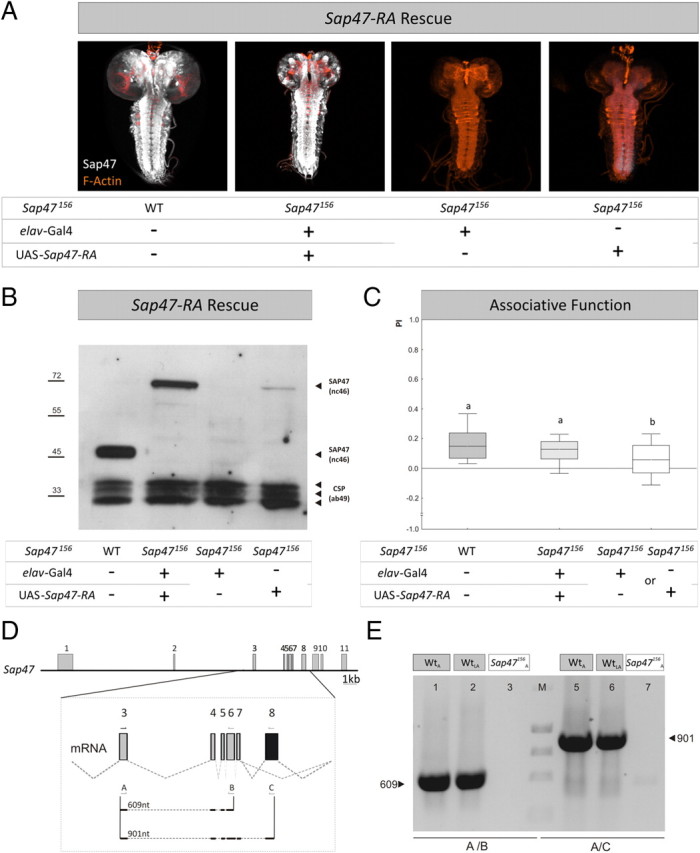
SAP47 full-length rescue. A, Whole mounts. In contrast to WT and rescue larvae, the elav-Gal4; Sap47156 driver-control shows no SAP47 expression, whereas there is a weak leaky expression detectable at the caudal tip of the ventral nerve cord of the UAS-Sap47-RA; Sap47156 effector-control strain. Phalloidin is used to visualize F-actin (orange). B, Western blot. The WT shows the prominent band at 47 kDa, whereas the rescue strain shows the band of the full-length PA isoform of SAP47 of ∼70 kDa, as to be expected from the coding region used for the UAS-Sap47-RA element. No SAP47 signal is detectable in the driver-control, but a weak leaky expression is seen in the UAS-Sap47-RA; Sap47156 effector-control strain, corresponding to the expression seen in the whole mount at the caudal tip of the ventral nerve cord (A). Anti-SAP47: nc46; anti-CSP: ab49 (loading control). C, Expression of the full-length isoform of SAP47 fully rescues associative function. Driver and effector-control are impaired in associative function to ∼50% of control level (WT), using elav-Gal4; Sap47156 as driver- and UAS-Sap47-RA; Sap47156 as effector-control. Both controls perform equally well and are therefore pooled as genetic controls (supplemental Fig. 8C, available at www.jneurosci.org as supplemental material). Larvae expressing the full-length cDNA of SAP47 perform as good as WT. N = 40, 40, 80. Different lettering above plots signifies p < 0.05/2 in Mann–Whitney U tests. The PREF scores underlying all PI values are documented in supplemental Figure 6 (available at www.jneurosci.org as supplemental material). D, Gene structure of SAP47 in WT larvae. Shown is the gene structure of Sap47 with exons and introns in wild type (exons gray, see also Fig. 1A). Eight transcripts are annotated, five longer and 3 shorter transcripts (Flybase: http://flybase.bio.indiana.edu). All of the longer transcripts contain exon 8, whereas this exon is spliced out in all of the short transcripts, indicated by black color in the magnification of the mRNA from exon 3 to exon 8. This situation allows differentiating the long from the short transcripts. The arrows and letters indicate binding sites of primers used for PCR in E and size of the expected PCR products of 609 nt using primer pair A and B and of 901 nt using primer pair A and C. E, PCR. After isolation of total RNA from 100 WT adult flies (WTA), 200 WT third-instar larvae (WTLA), and 100 adult SAP47156 flies (SAP47156A) and producing Sap47 cDNAs, the primer pair A/B generates a 609 nt fragment only in WT. Also using primer pair A/C, a 901 nt fragment is generated in only WT, and notably in both larvae and adults, indicating that long isoforms of SAP47 are expressed. Note that, on the protein level, these longer isoforms are often hard to detect (see Western blot in B).
Thus, also at the larval stage, Sap47156 qualifies as a protein-null mutant (for adult Drosophila: Funk et al., 2004).
Sap47156 larvae are defective in associative function
We next tested whether Sap47156 mutant larvae are impaired in associative function. In an odorant–reward associative learning experiment, we found significant performance indices for both Sap47156 (Fig. 2A) (one-sample sign test: p < 0.05/2; N = 35) and WT (Fig. 2A) (one-sample sign tests: p < 0.05/2; N = 35), arguing that associative faculties in principle are available to both genotypes. However, associative function in Sap47156 was reduced to ∼50% of WT levels (Fig. 2A) (p < 0.05, U = 408.5; sample sizes as above). The same defect was observed for a two-odor differential conditioning paradigm (Fig. 2B; see legend for statistics) as well as for another deletion allele, which is also a protein-null mutant (Fig. 2C; see legend for statistics).
To find out whether reduced associative ability is secondary to sensory or motor impairments, we tested whether Sap47156 mutants and WT differ in their behavior toward the to-be-associated stimuli. We did not find any difference between the genotypes in the behavior of experimentally naive larvae to amyl acetate (Fig. 3A) (p > 0.05; U = 827.0; N = 42, 44). Within each genotype, we found approach toward the odorant (Fig. 3A) (one-sample sign tests: p < 0.05/2 in both cases; sample sizes as above). This argues that animals from both genotypes are able to detect amyl acetate, are attracted by it, and do not differ in those kinds of motor ability that are necessary to behaviorally express their attraction toward this odorant. With respect to the reinforcer, a trend toward lower response levels to the fructose reward in the Sap47156 mutants remained far from being statistically significant, although sample size and scatter seems permissive to pick up such an effect (Fig. 3B) (p = 0.21; U = 1207.5; N = 53, 53) (for two additional datasets confirming this lack of effect, see supplemental Fig. 7A,B, available at www.jneurosci.org as supplemental material). Specifically, animals from both genotypes showed a clear preference for fructose (Fig. 3B) (one-sample sign tests: p < 0.05/2 in both cases; sample sizes as above).
These behavioral control procedures are state of the art in the neurogenetics of Drosophila learning. We have, however, recently argued that such testing of behavior in experimentally naive animals is not sufficient to conclude that an apparent associative defect is indeed due to reduced associative faculties (Michels et al., 2005; Knapek et al., 2010). That is, any associative training procedure obviously requires exposure to the to-be-associated stimuli, i.e., to both the odorant and the reward. Odor exposure is often found to reduce odor preferences in larval Drosophila (Boyle and Cobb, 2005) [see discussion in Colomb et al. (2007) and Gerber and Stocker (2007)]. If in the Sap47156 mutants such a decrease in preference would be particularly strong, this could feign an “associative” defect. Following Michels et al. (2005), we therefore ran a “sham-training” control, which involved exactly the same treatment as during a learning experiment, except that the reward was omitted (SHAMOdor-noReward). After such treatment, we found that both genotypes behaved toward the odorant in the same way (Fig. 3C) (p > 0.05, U = 743.0, N = 39, 42), in that both genotypes were attracted to the odorant (Fig. 3C) (one-sample sign tests: p < 0.05/2 in both cases; sample sizes as above). Regarding reward exposure, we in turn run a second kind of “sham training” (Michels et al., 2005), which again mimicked the learning protocol, except that this time the odorants were omitted (SHAMReward-noOdor). Also after this kind of treatment, behavior toward the odorant did not differ between genotypes (Fig. 3D) (p > 0.05; U = 1066.5, N = 50, 48), in that both genotypes were attracted by the odorant (Fig. 3D) (one-sample sign tests: p < 0.05/2 in both cases; sample sizes as above).
Thus, Sap47156 mutants have defects in associative function that do not seem to be due to any task-relevant sensory or motor impairment, or to an altered sensitivity to any nonassociative effect of odor exposure, reward exposure, satiety, or handling.
Synaptic transmission is intact but short-term plasticity is distorted in Sap47156 mutant larvae
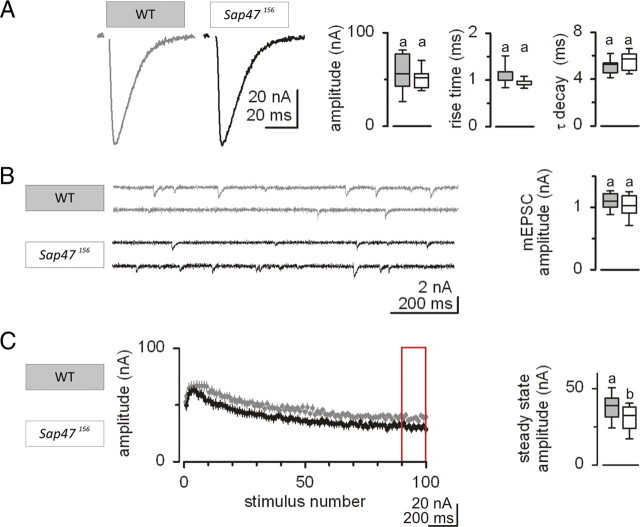
Consistent with normal locomotion, basal synaptic transmission at the neuromuscular junction was not altered in Sap47156. That is, in voltage-clamp recordings, both evoked and spontaneous miniature EPSCs (eEPSCs and mEPSCs, respectively) were unaltered in Sap47156 mutants compared to WT (Fig. 4A,B). Furthermore, evoked and spontaneous synaptic potentials were unaltered in current-clamp recordings, too (supplemental Fig. 9, available at www.jneurosci.org as supplemental material). However, genotypes differed in short-term plasticity during high-frequency stimulation (100 pulses at 60 Hz): Sap47156 mutants showed stronger depression of synaptic transmission during sustained stimulation as quantified by the steady-state amplitude, which was significantly lower in Sap47156 mutants than in WT (Fig. 4C) (p < 0.05, U = 64; N = 15, 15).
Figure 4.
Electrophysiology. A, Evoked EPSCs. Representative traces of EPSCs evoked by 0.1 Hz nerve stimulation. The peak amplitude, 10–90% rise time, and decay time constant of EPSCs were not significantly altered in Sap47156 mutants (white, N = 8) compared to WT (gray, N = 9). B, Spontaneous EPSCs. Sample traces of spontaneous mEPSCs. The amplitude of miniature EPSCs was not significantly different in Sap47156 mutants and WT. C, Short-term depression quantified by steady-state amplitude. To the left, EPSC amplitudes (mean and SEM as error bars) evoked by 60 Hz stimulations (100 pulses) are displayed; the Sap47156 mutants showed stronger depression (white, N = 15) than WT (gray, N = 15). This is quantified for the red boxed area, showing that the steady-state amplitude was significantly reduced in Sap47156 mutants compared to WT. Shared lettering above plots signifies p > 0.05, different lettering p < 0.05 in Mann–Whitney U tests.
Thus, we conclude that in Sap47156 mutant larvae basic synaptic transmission is intact, but short-term plasticity is distorted.
RNAi-mediated knockdown of SAP47 phenocopies the defect in associative function
To independently test for the requirement of SAP47 for associative function, we knocked down SAP47 by RNA interference (Smith et al., 2000; Kalidas and Smith, 2002). We used a UAS-RNAi-SAP47 fly strain as the effector strain and elav-Gal4 as the driver strain. As these transgenic Drosophila strains are in the white mutant background (w1118; this is necessary to monitor for a possible loss of the transgenic constructs), we first tested for SAP47 expression in WT and w1118 on a Western blot as well as for associative function in these two strains. In both respects, the w1118 mutation was without phenotype (Fig. 5A,A′): SAP47 levels appeared normal (Fig. 5A) and both genotypes showed associative performance indices (Fig. 5A′) (one-sample sign tests: p < 0.05/2; N = 16, 16) at equal levels (Fig. 5A′) (p = 0.66, U = 109; sample sizes as above) (see also Yarali et al., 2009). Therefore, data obtained with transgenic strains in our paradigm can be interpreted without reference to white function.
After confirming the effectivity of the RNAi-mediated knockdown of SAP47 on a Western blot (Fig. 5B), we thus could move on to test for the effect of this knockdown on associative function. Given that the transgenic driver and effector control strains showed equal levels of SAP47 as well as of associative function (supplemental Fig. 8A, available at www.jneurosci.org as supplemental material) (p = 0.79; U = 153; N = 19, 17), behavioral data were pooled for subsequent analyses. Compared to WT, performance indices in the knockdown group were reduced by ∼50% (Fig. 5B′) (p < 0.05/2, U = 199; N = 32, 19), as was the case when compared to controls (Fig. 5B′) (p < 0.05/2, U = 186; N = 19, 36; the Kruskal–Wallis ANOVA across all three groups yielded: p < 0.05; H = 8.58; df = 2; sample sizes as above).
We therefore conclude that an RNAi-mediated reduction of SAP47 causes an impairment in associative function similar to the one seen in the Sap47156 null mutant.
The associative defect in the Sap47156 mutant is rescued by transgenic SAP47 expression
For a rescue of the defect in associative function of the Sap47156 mutant, we used the driver strain elav-Gal4; Sap47156 crossed to UAS-Sap47-RF; Sap47156 as effector strain to transgenically restore expression of the 47 kDa PF isoform of the protein broadly throughout the larval brain (Fig. 6A,B). Because no significant difference in associative function was found between the driver and effector control (supplemental Fig. 8B, available at www.jneurosci.org as supplemental material) (p = 0.59; U = 2188; N = 67, 69), these were pooled for subsequent analysis. It turned out that rescue larvae were significantly better in associative function than the control larvae in the Sap47156 mutant background (Fig. 6C) (p < 0.05/2, U = 3668; N = 69, 136), but rescue larvae did not quite reach WT levels in associative performance indices (Fig. 6C) (p < 0.05/2; U = 1655; N = 68, 69) (the Kruskal–Wallis test across all three genotypes yielded the following: p < 0.05; H = 32.49; df = 2; sample sizes as above). This suggests that transgenic expression of the 47 kDa PF isoform of SAP47 partially rescues the Sap47156 mutant learning defect.
To see whether rescue expression of the full-length cDNA of SAP47 would yield a full rescue of associative function, we crossed elav-Gal4; Sap47156 as driver strain to the UAS-Sap47-RA; Sap47156 effector strain. Expression of the full-length PA isoform of SAP was restored throughout the larval brain (Fig. 7A,B). Again, no significant difference in associative function was found between driver and effector control (supplemental Fig. 8C, available at www.jneurosci.org as supplemental material) (p = 0.27; U = 686; N = 40, 40), so these were pooled for subsequent analysis. Larvae expressing SAP47-PA performed significantly better than control larvae in the Sap47156 mutant background (Fig. 7C) (p < 0.05/2, U = 1155; N = 40, 80). Indeed, these rescue larvae reached WT levels of associative function (Fig. 7C) (p = 0.18; U = 661; N = 40, 40) (the Kruskal–Wallis test across all three genotypes yielded: p < 0.05; H = 16.5; df = 2; sample sizes as above). Please note a tendency for overall low associative performance indices in this experiment; this is within the normal range of variation of behavioral experiments and underlines the necessity to train and test all genotypes to be compared statistically in parallel, as was done throughout this study. We further note a weak leaky expression in the effector control detectable on the Western blot (Fig. 7B). This expression is at the caudal tip of the ventral nerve cord (Fig. 7A, rightmost panel), a region not previously implicated in learning and memory, and indeed is inconsequential for associative function (see rightmost plot in Fig. 7C, and the trend for lower associative performance indices in the effector than in the driver control: supplemental Fig. 8C, available at www.jneurosci.org as supplemental material). In any event, given that the full-length PA isoform cannot be detected in Western blots of WT larvae, potentially because of local restriction of expression, we wondered whether longer isoforms of SAP47 are expressed in WT larvae at all. This is indeed the case, as suggested by PCR (Fig. 7D,E).
We therefore conclude that SAP47 functions in associative behavioral plasticity.
Discussion
We report that the SAP47 protein is widely expressed in the neuropil regions of the larval brain (Fig. 1E). The protein is associated with synaptic vesicles (Fig. 1C), but is not an integral part of the synaptic vesicle membrane [Mastrogiacomo et al., 1994; Umbach et al., 1994; Arnold et al., 2004 (their Fig. 3)]. The Sap47156 mutation leads to a total absence of the SAP47 protein in larvae (Fig. 1D,E,G) (for adult Drosophila: Funk et al., 2004). Lack of SAP47 entails a ∼50% reduction in odorant–reward associative ability as compared to WT (Fig. 2A); both strains had been adjusted for genetic background by extensive outcrossing. Importantly, this associative defect appears to be specific on the behavioral level because sensory and motor capacity regarding the to-be-associated stimuli is intact, be it in terms of naive responsiveness or in terms of the ability to behave toward the learned odorant at the moment of test (Fig. 3).
To independently verify this impairment, we tested Sap47156 larvae also in a two-odor version of the conditioning paradigm (AM vs OCT) and found a similar reduction in associative function (Fig. 2B). Also, another deletion allele (Sap47201) shows the same phenotype in associative function as Sap47156 (Fig. 2C). When SAP47 was transgenically knocked down by RNAi (Fig. 5B,B′), we found an ∼50% reduction of associative function, similar to that caused by the Sap47156 or the Sap47201 deletion (Fig. 2). From these results, and considering the outcrossing regimen for the Sap47156 mutant, it seems reasonable to attribute the learning defect upon deletions in the Sap47 gene to a lack of the SAP47 protein, rather than to spurious differences in genetic background (see also below).
We note that the associative defect in all cases (in Sap47156, Sap47201, and RNAi knockdown larvae, as well as in the genetic controls in both rescue experiments) is partial, arguing that there are SAP47-independent mechanisms to support associative function in our paradigm. Alternatively, there could be hitherto unknown Sap47-like genes in the fly genome; however, we do not see any SAP47 protein in the mutants, with either antibody used (Figs. 1D,E,G, 6A,B) (SAP47 expression in the effector control of Fig. 7, A and B, is due to weak and mnemonically inconsequential leaky expression from the used effector construct), and the Drosophila genome does not contain any obvious SAP47 sequence homolog. In this regard, SAP47 is similar to Synapsin, the lack of which also entails a reduction of associative performance indices in our paradigm by ∼50% (Michels et al., 2005). Both proteins are also similar regarding their association with synaptic vesicles without being an integral part of the vesicular protein complement (Hilfiker et al., 1999; Arnold et al., 2004). Whether and how these proteins interact and whether their learning phenotypes are additive remains to be investigated.
Maybe most importantly, transgenic expression of the full-length SAP47 protein from the elav-Gal4 driver fully rescues associative function in the Sap47156 mutant (Fig. 7), providing compelling evidence for a function of SAP47 in behavioral associative plasticity.
With respect to the physiological mechanism of SAP47 function, we analyzed basic transmission as well as short-term plasticity at the larval neuromuscular junction, the only cellular site in Drosophila that at present is routinely amenable to such analyses. The pronounced synaptic depression during sustained bursts of neuronal activity is consistent with a hypothesis that SAP47 contributes to the recruitment of vesicles to the release site (Hallermann et al., 2010). Although the plasticity processes that underlie odorant-taste learning and memory likely happen within the central brain (Gerber and Stocker, 2007), previous extrapolations between behavioral and synaptic plasticity at the neuromuscular synapses have been surprisingly successful [e.g., regarding the cAMP-PKA cascade (Kidokoro et al., 2004; Ueda and Wu, 2009)]. Thus, the kind of distortion of short-term plasticity observed here may well be the cause for impaired associative function on the behavioral level (Abbott and Regehr, 2004; Rothman et al., 2009).
To summarize, this study is the first to identify a behavioral and synaptic function of the phylogenetically conserved SAP47 protein: our results indicate that SAP47 is required for normal short-term synaptic plasticity at the neuromuscular junction as well as for normal levels of associative behavioral plasticity. Given that molecular determinants of behavioral and synaptic plasticity in invertebrates have repeatedly turned out to be shared with mammals (Pittenger and Kandel, 2003; Davis, 2005; Keene and Waddell, 2007), this may be an inspiring finding.
Footnotes
This work was supported by grants from the Deutsche Forschungsgemeinschaft [Collaborative Research Centre 554 Arthropod Behavior (A 10 and A 2 to B.G. and E.B., respectively), Collaborative Research Centre TR 58 Fear, Anxiety, Anxiety Disorders (A6 to B.G.), Collaborative Research Centre 581 Molecular Models for Diseases of the Nervous system (B 21 to E.B.), and International Research Training Group 1156 Synaptic and Behavioral Plasticity (to B.G, E.B., and B.M.)]; T.S. was supported by grants of the Excellence Initiative of the German Federal and State Governments to the Graduate School of Life Sciences, University of Würzburg. B.G. is a Heisenberg Fellow of the Deutsche Forschungsgemeinschaft. We are grateful to Scott Waddell, Bert Klagges, and our colleagues from the Würzburg and Leipzig laboratories for comments on the manuscript, and to Katharina Gerber, Katja Tschirner, and Anne Haberberger for help with the experiments. We thank N. Gendre for providing Figure 1F.
References
- Abbott LF, Regehr WG. Synaptic computation. Nature. 2004;431:796–803. doi: 10.1038/nature03010. [DOI] [PubMed] [Google Scholar]
- Arnold C, Reisch N, Leibold C, Becker S, Prüfert K, Sautter K, Palm D, Jatzke S, Buchner S, Buchner E. Structure-function analysis of the cysteine string protein in Drosophila: cysteine string, linker, and C-terminus. J Exp Biol. 2004;207:1323–1334. doi: 10.1242/jeb.00898. [DOI] [PubMed] [Google Scholar]
- Boyle J, Cobb M. Olfactory coding in Drosophila larvae investigated by cross-adaptation. J Exp Biol. 2005;208:3483–3491. doi: 10.1242/jeb.01810. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Colomb J, Grillenzoni N, Stocker RF, Ramaekers A. Complex behavioral changes associated to odour exposure in Drosophila larvae. Anim Behav. 2007;73:587–594. [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M. Expression of Drosophila mushroom body mutations in alternative genetic backgrounds: a case study of the mushroom body miniature gene (mbm) Proc Natl Acad Sci U S A. 1996;93:9875–9880. doi: 10.1073/pnas.93.18.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diegelmann S, Zars M, Zars T. Genetic dissociation of acquisition and memory strength in the heat-box spatial learning paradigm in Drosophila. Learn Mem. 2006;13:72–83. doi: 10.1101/lm.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Huber S, Buchner E, Bork P. BSD: a novel domain in transcription factors and synapse-associated proteins. Trends Biochem Sci. 2002;27:168–170. doi: 10.1016/s0968-0004(01)02042-4. [DOI] [PubMed] [Google Scholar]
- Funk N, Becker S, Huber S, Brunner M, Buchner E. Targeted mutagenesis of the sap47 gene of Drosophila: flies lacking the synapse associated protein of 47 kDa are viable and fertile. BMC Neurosci. 2004;5:16. doi: 10.1186/1471-2202-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B, Stocker RF. The Drosophila larva as a model for studying chemosensation and chemosensory learning: a review. Chem Senses. 2007;32:65–89. doi: 10.1093/chemse/bjl030. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR. Type I repressors of P element mobility. Genetics. 1993;135:81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallermann S, Heckmann M, Kittel RJ. Mechanisms of short-term plasticity at neuromuscular active zones of Drosophila. HFSP J. 2010;4:72–84. doi: 10.2976/1.3338710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel T, Michels B, Neuser K, Schipanski A, Kaun K, Sokolowski MB, Marohn F, Michel R, Heisenberg M, Gerber B. The carrot, not the stick: appetitive rather than aversive gustatory stimuli support associative olfactory learning in individually assayed Drosophila larvae. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2005;191:265–279. doi: 10.1007/s00359-004-0574-8. [DOI] [PubMed] [Google Scholar]
- Hilfiker S, Pieribone VA, Czernik AJ, Kao H-T, Augustine GJ, Greengard P. Synapsins as regulators of neurotransmitter release. Philos Trans R Soc Lond B Biol Sci. 1999;354:269–279. doi: 10.1098/rstb.1999.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer A, Ebel T, Waltenspiel B, Oswald P, Chen YC, Halder P, Biskup S, Lewandrowski U, Winkler C, Sickmann A, Buchner S, Buchner E. The Wuerzburg hybridoma library against Drosophila brain. J Neurogenet. 2009;23:78–91. doi: 10.1080/01677060802471627. [DOI] [PubMed] [Google Scholar]
- Kalidas S, Smith DP. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–184. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Meth. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Kidokoro Y, Kuromi H, Delgado R, Maureira C, Oliva C, Labarca P. Synaptic vesicle pools and plasticity of synaptic transmission at the Drosophila synapse. Brain Res Brain Res Rev. 2004;47:18–32. doi: 10.1016/j.brainresrev.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Knapek S, Gerber B, Tanimoto H. Synapsin is selectively required for anesthesia-sensitive memory. Learn Mem. 2010;17:76–79. doi: 10.1101/lm.1661810. [DOI] [PubMed] [Google Scholar]
- Lechner HA, Byrne JH. New perspectives on classical conditioning: a synthesis of Hebbian and non-Hebbian mechanisms. Neuron. 1998;20:355–358. doi: 10.1016/s0896-6273(00)80977-0. [DOI] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Mastrogiacomo A, Parsons SM, Zampighi GA, Jenden DJ, Umbach JA, Gundersen CB. Cysteine string proteins: a potential link between synaptic vesicles and presynaptic Ca2+ channels. Science. 1994;263:981–982. doi: 10.1126/science.7906056. [DOI] [PubMed] [Google Scholar]
- Michels B, Diegelmann S, Tanimoto H, Schwenkert I, Buchner E, Gerber B. A role of synapsin for associative learning: the Drosophila larva as a study case. Learn Mem. 2005;12:224–231. doi: 10.1101/lm.92805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuser K, Husse J, Stock P, Gerber B. Appetitive olfactory learning in Drosophila larvae: effects of repetition, reward strength, age, gender, assay type and memory span. Anim Behav. 2005;69:891–898. [Google Scholar]
- Pittenger C, Kandel ER. In search of general mechanisms for long-lasting plasticity: Aplysia and the hippocampus. Philos Trans R Soc Lond B Biol Sci. 2003;358:757–763. doi: 10.1098/rstb.2002.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmuth C, Becker S, Benz M, Debel K, Reisch D, Heimbeck G, Hofbauer A, Klagges B, Pflugfelder GO, Buchner E. The sap47 gene of Drosophila melanogaster codes for a novel conserved neuronal protein associated with synaptic terminals. Brain Res Mol Brain Res. 1995;32:45–54. doi: 10.1016/0169-328x(95)00058-z. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Rothman JS, Cathala L, Steuber V, Silver RA. Synaptic depression enables neuronal gain control. Nature. 2009;457:1015–1018. doi: 10.1038/nature07604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumweber T, Husse J, Gerber B. Innate attractiveness and associative learnability of odors can be dissociated in larval Drosophila. Chem Senses. 2011 doi: 10.1093/chemse/bjq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcho M, Pauls D, Han KA, Stocker RF, Thum AS. The role of dopamine in Drosophila larval classical olfactory conditioning. PLoS One. 2009;4:e5897. doi: 10.1371/journal.pone.0005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang M-B, Stoutjesdijk PA, Green AG, Waterhouse PM. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- Stewart BA, Schuster CM, Goodman CS, Atwood HL. Homeostasis of synaptic transmission in Drosophila with genetically altered nerve terminal morphology. J Neurosci. 1996;16:3877–3886. doi: 10.1523/JNEUROSCI.16-12-03877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Wu CF. Role of rut adenylyl cyclase in the ensemble regulation of presynaptic terminal excitability: reduced synaptic strength and precision in a Drosophila memory mutant. J Neurogenet. 2009;23:185–199. doi: 10.1080/01677060802471726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JA, Zinsmaier KE, Eberle KK, Buchner E, Benzer S, Gundersen CB. Presynaptic dysfunction in Drosophila csp mutants. Neuron. 1994;13:899–907. doi: 10.1016/0896-6273(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Wegener S. Learning and the presynapse in Drosophila melanogaster. Investigating protein-protein interactions and larval learning in fruitflies. 2008 MSc thesis, Universität Würzburg. [Google Scholar]
- Yarali A, Krischke M, Michels B, Saumweber T, Mueller MJ, Gerber B. Genetic distortion of the balance between punishment and relief learning in Drosophila. J Neurogenet. 2009;23:235–247. doi: 10.1080/01677060802441372. [DOI] [PubMed] [Google Scholar]