Abstract
In Drosophila, aversive associative memory of an odor consists of heterogeneous components with different stabilities. Here we report that Bruchpilot (Brp), a ubiquitous presynaptic active zone protein, is required for olfactory memory. Brp was shown before to facilitate efficient vesicle release, particularly at low stimulation frequencies. Transgenic knockdown in the Kenyon cells of the mushroom body, the second-order olfactory interneurons, revealed that Brp is required for olfactory memory. We further demonstrate that Brp in the Kenyon cells preferentially functions for anesthesia-resistant memory. Another presynaptic protein, Synapsin, was shown previously to be required selectively for the labile anesthesia-sensitive memory, which is less affected in brp knockdown. Thus, consolidated and labile components of aversive olfactory memory can be dissociated by the function of different presynaptic proteins.
Introduction
The fruit fly Drosophila melanogaster can form aversive associative memory of an odor if the odor is paired with electric shock (Quinn et al., 1974; Tully and Quinn, 1985). A single associative training induces memory lasting several hours. It consists of multiple memory components with different stabilities: a stable component can be operationally defined by being resistant to cold anesthesia [i.e., anesthesia-resistant memory (ARM)] (Quinn and Dudai, 1976; Margulies et al., 2005). In contrast to ARM, a labile component is characterized by its susceptibility to cold anesthesia [i.e., anesthesia-sensitive memory (ASM)] (Quinn and Dudai, 1976; Margulies et al., 2005). These memory components are distinct in many physiological characteristics (Tamura et al., 2003; Mery, 2007), such as dependency on reinforcement intensity (Knapek et al., 2010).
To establish odor–shock association, the neuronal signals of these two stimuli must converge in the brain. In Drosophila, this likely happens in the major intrinsic neurons of the mushroom body (i.e., Kenyon cells) (Heisenberg, 2003; Gerber et al., 2004; McGuire et al., 2005). Although both ASM and ARM require the mushroom body, different subsets of Kenyon cells dissociate these components (Isabel et al., 2004; Shuai et al., 2010), implying parallel formation of ASM and ARM with distinct neural circuits. Also at the molecular level, ARM and ASM can be distinguished. For instance, ASM is specifically affected in mutants with the impaired cAMP/protein kinase A (PKA) signaling pathway (e.g., rutabaga, encoding the type I adenylyl cyclase) (Dudai et al., 1988; Folkers et al., 1993; Isabel et al., 2004; Schwaerzel et al., 2007; Shuai et al., 2010). Similarly, Synapsin, a presynaptic vesicle protein essential for the regulation of the reserve pool vesicles, is preferentially required for ASM (Knapek et al., 2010). In contrast, selective modulation of ARM has been shown in the radish mutant (Folkers et al., 1993), augmentation of protein kinase C (PKC) signaling (Drier et al., 2002), or reduced PKA activity in the mushroom body (Horiuchi et al., 2008). The radish gene encodes a functionally unknown protein that has many potential phosphorylation sites for PKA and PKC (Folkers et al., 2006). The involvement of PKA suggests that cAMP/PKA signaling regulates both ASM and ARM (Horiuchi et al., 2008).
To address synaptic determinants of the distinct memory components, we here examine the role of Bruchpilot (Brp). Brp is homologous to the mammalian ELKS/CAST family of active zone proteins, specifically localized to the presynaptic release sites (active zones), in which it is an essential component of the presynaptic dense bodies (T-bars) (Wagh et al., 2006; Fouquet et al., 2009). Downregulation of the Brp protein causes an impairment in the release probability, particularly at low action potential frequencies (Kittel et al., 2006; Wagh et al., 2006; Fouquet et al., 2009). Importantly, vesicle release at neuromuscular synapses under high-frequency stimulation is still possible, yet not complete, after the elimination of Brp (Kittel et al., 2006). Here we examine the role of Brp-dependent neurotransmission in olfactory associative memory by knocking down the brp gene specifically in the Kenyon cells of the mushroom body. We demonstrate that the requirement of Brp is preferential to ARM.
Materials and Methods
Flies and genetic crosses
Stocks were raised at 25°C and 60% humidity with a 12 h light/dark cycle on a standard cornmeal-based food. The knockdown of the Brp protein in the mushroom body was generated using different RNA interference (RNAi) lines for brp under control of upstream activating sequence (UAS) [w−; RNAiB3 (III), w−; RNAiC8 (III), and w−; RNAiB3,C8 (combination of RNAiB3 and RNAiC8) (Wagh et al., 2006)] and GAL4 drivers [w−; OK107 (IV) (Connolly et al., 1996), w−; MB247 (III) (Zars et al., 2000), and w−; c739 (II) (Yang et al., 1995)]. To generate the genetic controls, GAL4 and UAS lines were crossed to w− flies. Two different RNAi constructs (B3 and C8) gave the same phenotype in immunohistochemistry and memory, making an off-target effect unlikely (Echeverri et al., 2006). For the experiment in Figure 3B, flies carrying the GAL4-repressor GAL80 under the control of the MB247 promoter [w−; MB247–GAL80 (II) (Krashes et al., 2007)] and UAS–RNAiB3,C8 were crossed to GAL4 drivers. For brp knockdown in the rutabaga mutant background, females rut2080; UAS–RNAiB3,C8 were crossed to w−; OK107 or w− males. rut2080 is a hypomorphic allele of the rutabaga gene (Pan et al., 2009). To ensure the knockdown, all crosses were set at 25°C and later shifted to 29°C when larvae reached the late third instar. This temperature shift had no significant effect on ARM (data not shown).
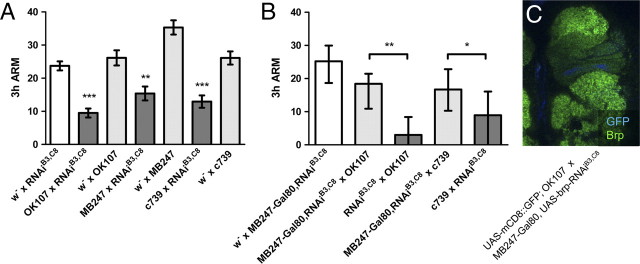
Figure 3.
Bruchpilot function in the mushroom body is necessary for ARM. A, The impairment of ARM by brp–RNAi is consistent with three different drivers for the mushroom body: OK107, MB247, and c739 (n = 18–43). All of these drivers have common expression in the α/β neurons. B, The impaired ARM by brp knockdown is significantly improved with the suppression of transgenic expression in the mushroom body with MB247–GAL80 (n = 14–42). This rescue is not complete, because ARM is significantly lower than the control group without any expression of brp–RNAi (p < 0.05 and p < 0.01 for OK107 and c739, respectively). Because the data are significantly different from the normal distribution, bars and error bars represent median and interquartile range, respectively. C, Expression of Brp in the mushroom body is still reduced with MB247–Gal80 (see also Fig. 1B), while green fluorescent protein (GFP) expression with OK107 is mostly silenced. *p < 0.05; **p < 0.01; ***p < 0.001.
Immunohistochemistry
The brains of female flies were prepared and fixed in 2% paraformaldehyde for 1 h at room temperature. Mouse monoclonal anti-Brp, nc82 (Hofbauer et al., 2009), and Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen) were applied as the primary and secondary antibodies, respectively. Brains for Brp quantification were counterstained with Phalloidin (catalog #R415; Invitrogen). Image stacks of whole-mount brains were taken with a confocal microscope Leica SP1. Projection of the confocal stacks and quantification of the signal intensity were done with NIH ImageJ. Brightness and contrast of images were adjusted with Photoshop (Adobe Systems).
For quantification of the Brp signal, a confocal slice containing the medial lobes of the mushroom body was selected from a stack (see Fig. 1). The average pixel intensity values in the experimental (the γ lobe of the mushroom body) and the adjacent control region [anterior inferior medial protocerebrum (aimpr)] was measured by the “Histogram” function in NIH ImageJ. The signal of the experimental region was normalized by the control region of an identical confocal slice. The typical size of the region of interest was ∼1000–2500 pixels.
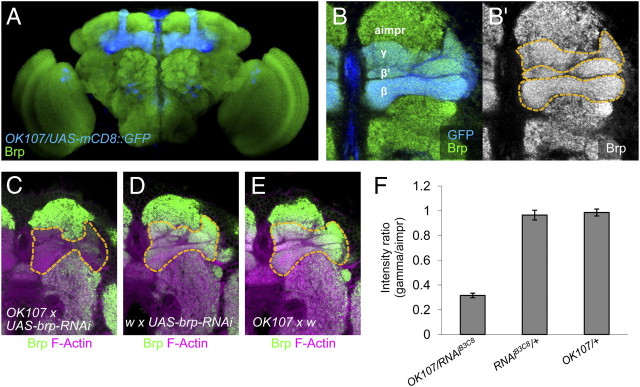
Figure 1.
Verification of the knockdown of Brp in the Kenyon cells of the mushroom body. A, Expression of Brp (green) and OK107 (blue) in the frontal view of the whole-mount fly brain (projection, dorsal up). Brp is ubiquitously expressed in the brain and localized to the neuropile regions. B, A magnified confocal slice in which the different lobes of the mushroom body (γ, β′, and β) and the adjacent neuropile (aimpr) are readily discernible. C–E, Magenta, Filamentous actin (F-actin); green, Brp. C–E, Knockdown by brp–RNAiC8 using OK107 (C) and genetic controls (D, E). Brp and F-actin are visualized in green and magenta, respectively. F, Intensity ratio of Brp signals in the γ neurons of the Kenyon cells and the adjacent control region (aimpr). After the knockdown with brp–RNAiB3C8, the Brp signal in the mushroom body was significantly reduced to ∼30% (p < 0.001, n = 8–17). GFP, Green fluorescent protein.
Behavioral experiments
All flies were collected and transferred to fresh food vials at least 24 h before the experiments. All behavioral experiments were performed at room temperature (22 ± 1°C) with a relative humidity of 78 ± 2%. Mixed populations of males and females aged for 1–6 d were measured and used to calculate the performance indices (PIs), except for the experiments in Figure 4, where the sexes have been separately counted after the test and only males were used for calculation because of the crossing design.
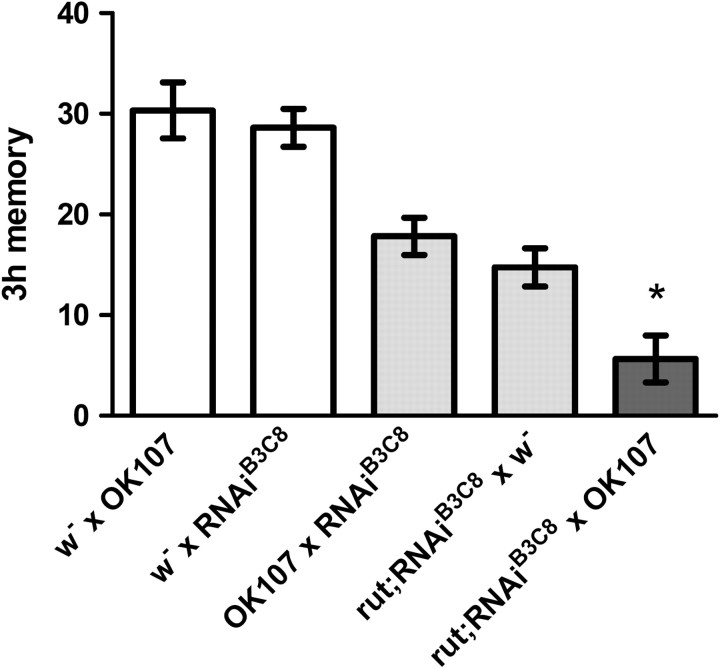
Figure 4.
Additive memory impairment in the rutabaga mutant and brp knockdown. Three hour memory of flies expressing brp–RNAi is significantly lower than the controls both in the wild-type and rut mutant backgrounds (n = 34–57; p < 0.05). Unlike the other groups, memory of brp knockdown in the rut background was not significantly different from zero (p > 0.05, one-sample t test).
Olfactory learning.
A standard differential olfactory conditioning with two odorants was used (Tully and Quinn, 1985; Knapek et al., 2010). In a subsequent test in a T-maze, flies were allowed to choose between the two odors for 120 s. For memory retention, flies were kept in empty vials in darkness between training and test. To correct a learning-independent preference to one of the two odors, two separate groups of flies were trained reciprocally: one group received odor A with shock (A+) and B without (B−), and the other group received odor B with shock (B+) and A without (A−). A learning index was then calculated as the mean preference of these two reciprocally trained groups (Tully and Quinn, 1985). To rule out non-associative effects caused by the reinforcement order, in half of the experiments, the first presented odor was punished (A+, B− and B+, A−) and, in the other half, the second presented odor was punished (A−, B+ and B−, A+).
For all learning experiments (except Fig. 4), 4-methylcyclohexanol (Fluka) diluted 1:80 and 3-octanol (Fluka) diluted 1:100 were presented in odor cups with a diameter of 14 mm. For Figure 4, benzaldehyde (VWR International) diluted 1:1000 in a 16 mm cup was used instead of 4-methylcyclohexanol, because the odor response of experimental groups was significantly impaired with 4-methylcyclohexanol. Paraffin oil (Fluka) was used for dilution. Twelve pulses of electric shock (90 V direct current) were applied with an interpulse interval of 5 s (Tully and Quinn, 1985).
To apply cold anesthesia, vials containing trained flies were put into an ice-floating water bath at given time points after training. After 2 min, the flies were transferred to a vial at room temperature and kept there until the test. They typically recovered from the anesthesia within ∼30 s. To verify the sufficiency of 2 min cold anesthesia, we extended anesthesia to 10 min but did not find additional decrement in short-term memory (STM) (data not shown; procedure as in Fig. 2D).
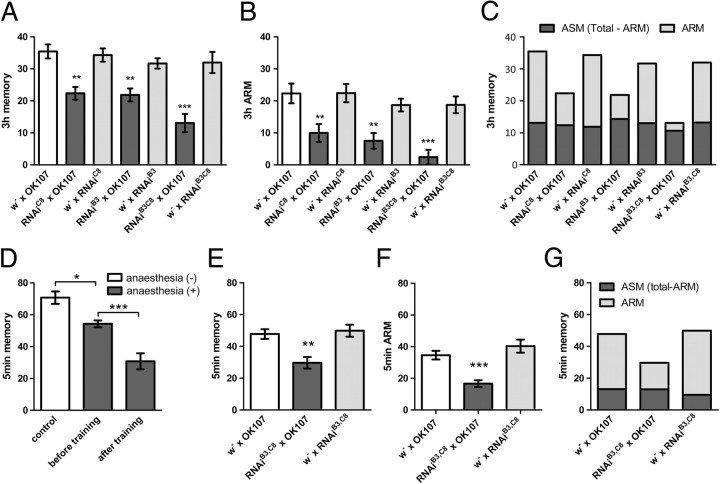
Figure 2.
Bruchpilot is selectively required for ARM. A, brp knockdown in the mushroom body with three different UAS–brp–RNAi lines driven by OK107 leads to a significantly impaired aversive olfactory memory tested at 3 h after training (n = 15–24). B, ARM of the same genotypes. The impairment is similar to that in total 3 h memory that is the sum of ARM and ASM (A) (n = 17–29). C, The score of ASM (calculated by subtracting ARM from total 3 h memory) is similar in all groups (dark gray), whereas ARM is reduced in the experimental groups (light gray). This indicates that ARM is selectively impaired by brp knockdown. Same data as in A and B are represented. D, Five minute memory of wild-type Canton-S flies consists of ASM and ARM (n = 8). Cold anesthesia was applied either before or directly after training. E, brp knockdown with OK107 significantly impairs aversive olfactory STM tested at 5 min after training (n = 13). Cold anesthesia was applied before training (see also D). F, Short-term ARM tested 5 min after training is similarly impaired as the total STM shown in E (n = 13–14). Cold anesthesia was applied directly after training (see also D). G, The score of short-term ASM (calculated by subtracting short-term ARM from the total 5 min memory) is similar in all groups (dark gray), whereas ARM (light gray) is reduced in the experimental group. This indicates that ARM is preferentially impaired by brp knockdown. Same data as in E and F are represented. *p < 0.05; **p < 0.01; ***p < 0.001; see also Materials and Methods.
Avoidance tests.
Flies were tested for responsiveness to electric shock and the odors in the same T-maze assay as used for learning experiments. For electric shock responsiveness, flies were given 1 min to choose between an electrified (12 pulses of the indicated voltage) and a non-electrified arm of the T-maze. From each experiment, the number of flies choosing the electrified tube (N+) and the non-electrified one (N−) were counted, and a PI (avoidance) was calculated as the difference of N− and N+ divided by the total number of flies. A positive value indicates avoidance from the shock, and PI of zero indicates no response.
To test the responsiveness to olfactory cues, flies were given 2 min to choose between two arms of the T-maze: one scented with the respective odor used for conditioning and the other one unscented. A PI was calculated in the same way as shock avoidance.
Statistics
The significance level in each experiment was set to 5%. Data were always first tested for normal distribution and homogeneity of variance with Shapiro–Wilk test, followed by Bonferroni's correction and Bartlett's test, respectively. If none of these assumptions were violated, parametric comparisons (one-way ANOVA, followed by Bonferroni's-corrected post hoc tests) were applied. If data were not normally distributed or had significantly different variances, nonparametric statistical tests were performed; medians were compared with Kruskal–Wallis test, followed by the post hoc pairwise test (Dunn's test). All statistical calculation was done using the software Prism (GraphPad Software). Bars and error bars represent means and SEM, respectively, in all figures if not stated otherwise. *p < 0.05; **p < 0.01; ***p < 0.001. Throughout the report, p values refer to the more conservative result of the two respective statistical comparisons (e.g., OK107/brp–RNAi vs brp–RNAi/+ and OK107/+).
Results
Bruchpilot is preferentially required for anesthesia-resistant memory
Brp is a ubiquitously expressed active zone protein of Drosophila synapses (Fig. 1A). Because a null mutant for brp is not viable to adulthood (Kittel et al., 2006), we suppressed Brp expression in a targeted group of cells with a transgenic RNA interference approach (Wagh et al., 2006). For this, we used the OK107 GAL4 driver for targeting brp RNAi to the majority of mushroom body Kenyon cells (Fig. 1A) (Connolly et al., 1996; Aso et al., 2009), because the output synapses of these neurons are thought to undergo associative plasticity (Heisenberg, 2003; Gerber et al., 2004). We verified the knockdown in Kenyon cells by quantifying Brp protein levels with immunohistochemistry. The Brp signal in the mushroom body of knockdown flies was significantly reduced to ∼30% of genetic controls (Fig. 1F) (p < 0.001, n = 8–17). The knockdown by OK107 did not cause a conspicuous morphological change of the mushroom body. Residual Brp in the lobes was expected given that many presynaptic terminals of extrinsic neurons of the mushroom body should not be affected by OK107 (Johard et al., 2008; Tanaka et al., 2008). Therefore, reduction by 70% is likely a conservative estimation of the downregulation of Brp in Kenyon cells. Different RNAi constructs that target distinct regions of the brp mRNA (brp–RNAiC8 and brp–RNAiB3) caused similar silencing effects (data not shown) (Wagh et al., 2006). Thus, Brp can be locally downregulated in the presynaptic terminals of the Kenyon cells.
We next addressed the function of Brp in the mushroom body for olfactory aversive memory. The knockdown in the Kenyon cells using OK107 significantly impaired memory measured at 3 h after training (p < 0.01) (Fig. 2A). Three hour memory consists of both labile and consolidated memory (i.e., ASM and ARM, respectively) (Quinn and Dudai, 1976; Margulies et al., 2005). To measure ARM, we disrupted ASM by applying cold anesthesia and tested flies with brp knockdown in the mushroom body. In fact, ARM was significantly impaired (Fig. 2B) (p < 0.01). The extent of the ARM impairment was similar to that of the total 3 h memory (Fig. 2C). Thus, ARM but not ASM seems to be affected at 3 h after training by brp knockdown. The brp–RNAiC8 and brp–RNAiB3 as well as the double insertion showed the same phenotype in ARM (Fig. 2), making it difficult to explain the observed memory phenotype by an off-target effect (Echeverri et al., 2006). Moreover, brp knockdown did not lead to a significant reduction of avoidance of electric shock or odors compared with the corresponding controls (Table 1).
Table 1.
| Shock avoidance [median (25/75 percentile)] | 3-Octanol avoidance [mean (SEM)] | 4-Methylcyclohexanol avoidance [mean (SEM)] | |
|---|---|---|---|
| w− × RNAiB3C8 | 66.14 (58.75/80.62) | 36.80 (3.75) | 56.62 (3.44) |
| OK107 × RNAiB3C8 | 64.68 (56.22/70.41) | 24.36 (5.70) | 31.27 (5.86) |
| w− × OK107 | 70.03 (67.98/77.29) | 32.46 (6.60) | 25.31 (5.06) |
| MB247 × RNAiB3C8 | 69.70 (62.50/73.24) | 49.15 (8.00) | 58.96 (5.11) |
| w− × MB247 | 66.35 (61.79/71.96) | 33.73 (5.45) | 40.27 (6.04) |
| c739 × RNAiB3C8 | 61.90 (48.15/62.96) | 32.33 (5.79) | 41.05 (5.78) |
| w− × c739 | 62.07 (53.78/65.99) | 29.11 (6.05) | 35.46 (6.78) |
None of the experimental groups are significantly different from their two respective controls, except for the comparison between OK107 × RNAiB3C8 and w− × RNAiB3C8 in the 4-methycyclohexanol avoidance (p < 0.05). n = 7–10.
Cold anesthesia directly after associative training has been shown to disrupt all memory when tested hours later (Quinn and Dudai, 1976; Margulies et al., 2005). Therefore, ARM is built gradually after training, and STM consists mainly of ASM (Quinn and Dudai, 1976; Margulies et al., 2005). However, we found that application of the same cold anesthesia directly after training does not erase all memory if it is tested for STM (Fig. 2D). Interestingly, this anesthesia-resistant STM is significantly decreased during the knockdown of Brp in the mushroom body (Fig. 2E,F) (p < 0.01). The knockdown impaired ARM and total 5 min memory to a similar extent (Fig. 2G).
Altogether, these results suggest a preferential requirement of Brp in the mushroom body for ARM, without impairing sensitivity to the tested odors and shock.
Bruchpilot is required in the mushroom body
Because most GAL4 driver lines have additional expression outside of the investigated structures (Aso et al., 2009), we tried to confirm the specificity of the RNAi effect in the mushroom bodies with two different strategies: (1) brp knockdown induced by different GAL4 drivers with overlapping expression in the Kenyon cells, and (2) blocking of GAL4 transactivation in the mushroom body using MB247–GAL80.
In parallel to the knockdown with OK107, we examined two other mushroom body drivers: MB247 with strong expression in the α/β and γ neurons (Zars et al., 2000) and c739 with strong expression in the α/β neurons (Yang et al., 1995). These three drivers do not apparently have common GAL4 expression outside the mushroom body (Aso et al., 2009). With all three drivers, brp knockdown caused a significant defect in ARM (Fig. 3A) (p < 0.001, p < 0.01, and p < 0.001 for OK107, MB247, and c739, respectively). Given common expression in the α/β neurons in the three drivers, the impairment of ARM is consistent with the requirement of the α/β neuron output for ARM (Isabel et al., 2004).
Moreover, addition of MB247–GAL80 significantly improved ARM of the knockdown with OK107 or c739 (Fig. 3B) (p < 0.01 and p < 0.05 for OK107 and c739, respectively), confirming the effect in the mushroom body. ARM in the presence of MB247–GAL80 was significantly lower than the control groups without RNAi (p < 0.05 and p < 0.01 for OK107 and c739, respectively, n = 14–42), suggesting that the effect of MB247–GAL80 may be incomplete. Consistently, we detected a reduction of the Brp signal in the knockdown with OK107 despite MB247–GAL80 (Fig. 3C). Collectively, these two different experiments suggest that ARM requires a Brp-dependent mechanism at the mushroom body synapses, possibly in the α/β neurons.
Additive memory impairment after combining Bruchpilot knockdown with rutabaga mutation
Previous results indicated that another presynaptic protein, Synapsin, is preferentially required for ASM and that its memory phenotype is included in that of the rutabaga mutant, whose ARM is not significantly impaired (Isabel et al., 2004; Knapek et al., 2010). Because Brp is in turn required for ARM, we addressed whether the brp knockdown in the mushroom body would be additive to memory impairment of a rutabaga mutant (Fig. 4). The memory defect of the combination was significantly larger than the impairment of either rutabaga or brp–RNAi alone (p < 0.05) (Fig. 4). Unlike the controls, these flies showed no detectable memory at 3 h (p > 0.05) (Fig. 4). The avoidance of shock and odors (benzaldehyde and 3-octanol) was not significantly impaired in rutabaga; brp–RNAi flies (Table 2). Altogether, these results are in line with the hypothesis that ASM and ARM depend on genetically dissociable molecular components of synaptic vesicle release.
Table 2.
Avoidance of electric shock and odors for the flies used for Figure 4
| Shock avoidance [mean (SEM)] | 3-Octanol avoidance [mean (SEM)] | Benzaldehyde avoidance [mean (SEM)] | |
|---|---|---|---|
| OK107 × RNAiB3C8 | 41.45 (6.94) | 11.51 (4.84) | 26.55 (6.99) |
| rut; RNAiB3C8 × w− | 63.52 (4.14) | −2.88 (6.83) | 28.21 (4.55) |
| rut; RNAiB3C8 × OK107 | 46.55 (6.88) | 13.51 (4.80) | 15.02 (4.33) |
No statistically significant difference among the corresponding groups. n = 12.
Discussion
Bruchpilot, a novel synaptic molecule for ARM
In Drosophila, middle-term olfactory memory after a single training cycle comprises functionally dissociable forms of memory: the labile ASM and the stable ARM (Quinn and Dudai, 1976). In contrast to ASM, the molecular basis of ARM formation is poorly understood. Only a few molecules have been shown to be important for ARM so far (Drier et al., 2002; Folkers et al., 2006; Horiuchi et al., 2008). We demonstrated that Brp in the Kenyon cells of the mushroom body is preferentially required for ARM (Figs. 2, 3). Although there is no apparent developmental defect in the downregulation of Brp (Fig. 1) (Wagh et al., 2006), this study does not specify the requirement for ARM in the adult.
The Brp protein is specifically localized to the active zone at the presynaptic terminals, in which it forms electron dense projections (Kittel et al., 2006; Wagh et al., 2006; Fouquet et al., 2009). Interestingly, the Radish protein that is also required for ARM is highly enriched in the lobes of the mushroom body (Folkers et al., 2006). Thus, Brp and Radish might interact at the active zones to regulate neurotransmission underlying ARM.
Several memory mutants have been shown to have a selective phenotype in ASM. Consistent with the parallel memory formation of ASM and ARM, the brp knockdown and a rutabaga mutation caused an additive memory deficit (Fig. 4). Interestingly, Synapsin is required for ASM preferentially, and the null mutation caused no augmentation of the memory phenotype in the rutabaga single mutant (Knapek et al., 2010). Thus, the complementary forms of memory might recruit differential signaling mechanisms that rely on distinct presynaptic machineries.
In a current model of memory dynamics, ARM gradually develops after training, whereas ASM occupies the largest part of early memory and decays more quickly (Quinn and Dudai, 1976). Although Radish and Brp are selectively required for ARM measured at 3 h after training, flies lacking either of the proteins are impaired also in immediate memory (Fig. 2E) (Folkers et al., 1993; Dubnau and Tully, 1998). By applying cold anesthesia for STM, we found that STM does contain a significant ARM component (Fig. 2D). The consistent requirement of Brp for short-term and 3 h ARM may contribute to a synaptic mechanism of memory that is stable against amnesic treatment.
Dissociation of ARM and ASM
If ARM and ASM were formed at the same synapses of Kenyon cells, how could the two synaptic proteins Brp and Synapsin dissociate these different forms of memory? Notably, Brp and Synapsin are meant to be required for distinct components of action potential-evoked vesicle release. In vertebrates, Synapsin has been shown to be particularly important for recruitment of synaptic vesicles from reserve pools at high stimulation frequencies (Pieribone et al., 1995; Rosahl et al., 1995; Gitler et al., 2004; Sun et al., 2006). Consistently, synapsin mutants show normal quantal content (number of synaptic vesicles released per action potential) at moderate action potential frequencies (Pieribone et al., 1995; Rosahl et al., 1995; Gitler et al., 2004; Sun et al., 2006). Similarly, Drosophila Synapsin maintains the reserve pool of vesicles and mediates mobilization of the reserve pool during intense stimulation (Akbergenova and Bykhovskaia, 2007). The brp null mutant in contrast shows decreased quantal contents particularly in response to the first arrival of an action potential (Kittel et al., 2006; Wagh et al., 2006). Vesicle release after subsequently following high-frequency spikes however is less affected, suggesting the importance in vesicle release at low-frequency stimulation (Kittel et al., 2006). The two different modes of neurotransmission (e.g., different release probabilities during high- vs low-frequency stimulation) could therefore differentiate ASM and ARM, even if the traces of these different forms of memory resided in the same synapses of the Kenyon cells.
Alternatively, the memory traces of ASM and ARM could be spatially separated within the same neurons, i.e., localized at different synapse populations. In the lobes, Kenyon cell axons have multiple compartments that are intersected by transverse extrinsic neurons (Tanaka et al., 2008; Aso et al., 2010). We found that brp knockdown in the α/β neurons affected ARM (Fig. 3A). This is consistent with a previous report, in which inhibition of the output of the α/β neurons impaired ARM (Isabel et al., 2004). Interestingly, inhibition of a specific type of dopaminergic neurons that synapse onto another restricted compartment of the β lobe selectively affected ASM (Aso et al., 2010). Thus, associative plasticity underlying ASM and ARM could be formed by stimulating different synapses of the same neurons. This hypothesis may be tested in future by the identification of extrinsic neurons that are specifically required for ARM and corresponding functional imaging of memory traces (Yu et al., 2005; Mao and Davis, 2009; Tomchik and Davis, 2009; Gervasi et al., 2010).
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft [Emmy-Noether Program (H.T.), I-GRK 1156 (S.K.), and Sonderforschungsbereich 554 and Exc 257 (S.S.)] and the Max Planck Gesellschaft. We thank A. B. Friedrich, A. Gruschka, and B. Mühlbauer for technical assistance. We are also grateful to E. Buchner, M. Heisenberg, and A. Yarali for discussion and critical reading of this manuscript.
References
- Akbergenova Y, Bykhovskaia M. Synapsin maintains the reserve vesicle pool and spatial segregation of the recycling pool in Drosophila presynaptic boutons. Brain Res. 2007;1178:52–64. doi: 10.1016/j.brainres.2007.08.042. [DOI] [PubMed] [Google Scholar]
- Aso Y, Grübel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H. The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet. 2009;23:156–172. doi: 10.1080/01677060802471718. [DOI] [PubMed] [Google Scholar]
- Aso Y, Siwanowicz I, Bräcker L, Ito K, Kitamoto T, Tanimoto H. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20:1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Drier EA, Tello MK, Cowan M, Wu P, Blace N, Sacktor TC, Yin JC. Memory enhancement and formation by atypical PKM activity in Drosophila melanogaster. Nat Neurosci. 2002;5:316–324. doi: 10.1038/nn820. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Tully T. Gene discovery in Drosophila: new insights for learning and memory. Annu Rev Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- Dudai Y, Corfas G, Hazvi S. What is the possible contribution of Ca2+-stimulated adenylate cyclase to acquisition, consolidation and retention of an associative olfactory memory in Drosophila. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1988;162:101–109. doi: 10.1007/BF01342707. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prévôt B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Folkers E, Drain P, Quinn WG. Radish, a Drosophila mutant deficient in consolidated memory. Proc Natl Acad Sci U S A. 1993;90:8123–8127. doi: 10.1073/pnas.90.17.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkers E, Waddell S, Quinn WG. The Drosophila radish gene encodes a protein required for anesthesia-resistant memory. Proc Natl Acad Sci U S A. 2006;103:17496–17500. doi: 10.1073/pnas.0608377103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, Sigrist SJ. Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol. 2009;186:129–145. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B, Tanimoto H, Heisenberg M. An engram found? Evaluating the evidence from fruit flies. Curr Opin Neurobiol. 2004;14:737–744. doi: 10.1016/j.conb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Gervasi N, Tchénio P, Preat T. PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron. 2010;65:516–529. doi: 10.1016/j.neuron.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Gitler D, Takagishi Y, Feng J, Ren Y, Rodriguiz RM, Wetsel WC, Greengard P, Augustine GJ. Different presynaptic roles of synapsins at excitatory and inhibitory synapses. J Neurosci. 2004;24:11368–11380. doi: 10.1523/JNEUROSCI.3795-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Hofbauer A, Ebel T, Waltenspiel B, Oswald P, Chen YC, Halder P, Biskup S, Lewandrowski U, Winkler C, Sickmann A, Buchner S, Buchner E. The Wuerzburg hybridoma library against Drosophila brain. J Neurogenet. 2009;23:78–91. doi: 10.1080/01677060802471627. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Yamazaki D, Naganos S, Aigaki T, Saitoe M. Protein kinase A inhibits a consolidated form of memory in Drosophila. Proc Natl Acad Sci U S A. 2008;105:20976–20981. doi: 10.1073/pnas.0810119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel G, Pascual A, Preat T. Exclusive consolidated memory phases in Drosophila. Science. 2004;304:1024–1027. doi: 10.1126/science.1094932. [DOI] [PubMed] [Google Scholar]
- Johard HA, Enell LE, Gustafsson E, Trifilieff P, Veenstra JA, Nässel DR. Intrinsic neurons of Drosophila mushroom bodies express short neuropeptide F: relations to extrinsic neurons expressing different neurotransmitters. J Comp Neurol. 2008;507:1479–1496. doi: 10.1002/cne.21636. [DOI] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- Knapek S, Gerber B, Tanimoto H. Synapsin is selectively required for anesthesia-sensitive memory. Learn Mem. 2010;17:76–79. doi: 10.1101/lm.1661810. [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr Biol. 2005;15:R700–R713. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Deshazer M, Davis RL. Thirty years of olfactory learning and memory research in Drosophila melanogaster. Prog Neurobiol. 2005;76:328–347. doi: 10.1016/j.pneurobio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Mery F. Aging and its differential effects on consolidated memory forms in Drosophila. Exp Gerontol. 2007;42:99–101. doi: 10.1016/j.exger.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Pan Y, Zhou Y, Guo C, Gong H, Gong Z, Liu L. Differential roles of the fan-shaped body and the ellipsoid body in Drosophila visual pattern memory. Learn Mem. 2009;16:289–295. doi: 10.1101/lm.1331809. [DOI] [PubMed] [Google Scholar]
- Pieribone VA, Shupliakov O, Brodin L, Hilfiker-Rothenfluh S, Czernik AJ, Greengard P. Distinct pools of synaptic vesicles in neurotransmitter release. Nature. 1995;375:493–497. doi: 10.1038/375493a0. [DOI] [PubMed] [Google Scholar]
- Quinn WG, Dudai Y. Memory phases in Drosophila. Nature. 1976;262:576–577. doi: 10.1038/262576a0. [DOI] [PubMed] [Google Scholar]
- Quinn WG, Harris WA, Benzer S. Conditioned behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1974;71:708–712. doi: 10.1073/pnas.71.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosahl TW, Spillane D, Missler M, Herz J, Selig DK, Wolff JR, Hammer RE, Malenka RC, Südhof TC. Essential functions of synapsins I and II in synaptic vesicle regulation. Nature. 1995;375:488–493. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Jaeckel A, Mueller U. Signaling at A-kinase anchoring proteins organizes anesthesia-sensitive memory in Drosophila. J Neurosci. 2007;27:1229–1233. doi: 10.1523/JNEUROSCI.4622-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai Y, Lu B, Hu Y, Wang L, Sun K, Zhong Y. Forgetting is regulated through Rac activity in Drosophila. Cell. 2010;140:579–589. doi: 10.1016/j.cell.2009.12.044. [DOI] [PubMed] [Google Scholar]
- Sun J, Bronk P, Liu X, Han W, Südhof TC. Synapsins regulate use-dependent synaptic plasticity in the calyx of Held by a Ca2+/calmodulin-dependent pathway. Proc Natl Acad Sci U S A. 2006;103:2880–2885. doi: 10.1073/pnas.0511300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Chiang AS, Ito N, Liu HP, Horiuchi J, Tully T, Saitoe M. Aging specifically impairs amnesiac-dependent memory in Drosophila. Neuron. 2003;40:1003–1011. doi: 10.1016/s0896-6273(03)00732-3. [DOI] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- Tomchik SM, Davis RL. Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron. 2009;64:510–521. doi: 10.1016/j.neuron.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- Yu D, Keene AC, Srivatsan A, Waddell S, Davis RL. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123:945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]