Abstract
While deep learning methods have demonstrated performance comparable to human readers in tasks such as computer-aided diagnosis, these models are difficult to interpret, do not incorporate prior domain knowledge, and are often considered as a “black-box.” The lack of model interpretability hinders them from being fully understood by end users such as radiologists. In this paper, we present a novel interpretable deep hierarchical semantic convolutional neural network (HSCNN) to predict whether a given pulmonary nodule observed on a computed tomography (CT) scan is malignant. Our network provides two levels of output: 1) low-level semantic features; and 2) a high-level prediction of nodule malignancy. The low-level outputs reflect diagnostic features often reported by radiologists and serve to explain how the model interprets the images in an expert-interpretable manner. The information from these low-level outputs, along with the representations learned by the convolutional layers, are then combined and used to infer the high-level output. This unified architecture is trained by optimizing a global loss function including both low- and high-level tasks, thereby learning all the parameters within a joint framework. Our experimental results using the Lung Image Database Consortium (LIDC) show that the proposed method not only produces interpretable lung cancer predictions but also achieves significantly better results compared to using a 3D CNN alone.
Keywords: Lung nodule classification, lung cancer diagnosis, Computed tomography, deep learning, convolutional neural networks, model interpretability
1. Introduction and Background
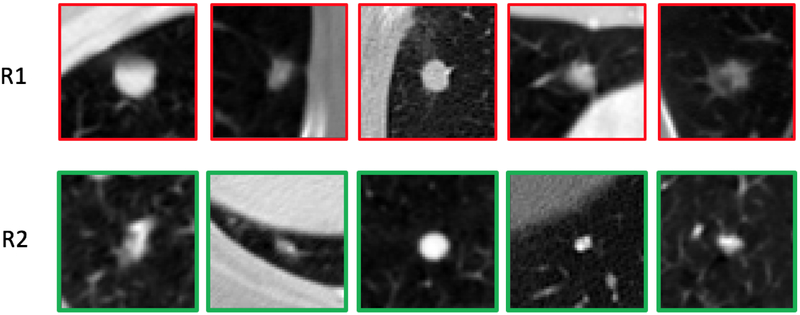
Lung cancer is the leading cause of cancer mortality worldwide (Torre et al., 2016; Shen et al., 2017a). Computed tomography (CT) imaging is increasingly being used to detect and characterize pulmonary nodules with the purpose of diagnosing lung cancer earlier. The National Lung Screening Trial (NLST) (Team et al., 2011) in the United States demonstrated a 20% lung cancer mortality reduction in high-risk subjects who underwent screening using low-dose CT relative to plain chest radiography. Based on the findings of the NLST, the United States Preventative Services Task Force (USPSTF) recommends low-dose CT lung cancer screening for current and former smokers aged 55–80 with a smoking history of at least 30 pack-years, or former smokers having quit within the past 15 years (ten Haaf et al., 2017). However, the potential consequences of implementing lung cancer screening is an increase in false positive screens that result in unnecessary medical, economic, and psychological costs. Indeed, some studies indicate that the false positive rate for low-dose CT is upwards of 20%. Moreover, detection rates vary among less experienced radiologists, particularly in subtle cases, as interpretation heavily relies on past experience (Zhao et al., 2013). Figure 1 illustrates examples of malignant (top row, R1) and benign (bottom row, R2) nodules. The visual appearance of these nodules is highly varied with subtle differences in size, shape, and texture, underscoring the challenge faced by radiologists in differentiating between the two categories.
Figure 1:
Illustrations of malignant and benign nodules: R1 are malignant nodules; R2 are benign nodules.
In response, computer-aided diagnosis (CADx) systems (Armato et al., 2003; Shen et al., 2015a; Duggan et al., 2015; Firmino et al., 2016; Amir & Lehmann, 2016; Huang et al., 2017) have been explored to help distinguish between malignant from benign nodules in small nodules (Huang et al., 2017). While architectures may vary, contemporary lung nodule CADx systems typically consist of modules that perform: 1) image reconstruction and enhancement (image pre-processing); 2) identification and segmentation of nodule candidates (candidate generation); 3) characterization and filtering of nodule candidates (false positive reduction); and 4) classification of each candidate as benign or malignant (diagnosis). For example, Armato et al. (Armato et al., 2003) segmented the lung nodule using multilevel thresholding techniques; extracted morphological and gray-level features; and classified nodules as benign or malignant using linear discriminant analysis. Zinovev et al. (Zinovev et al., 2011) employed both texture and intensity features using belief decision trees and a multi-label approach to perform lung nodule classification. Way et al. (Way et al., 2009) segmented lung nodules using k-means clustering, combined nodule surface features together with texture and morphological features, and used linear discriminant analysis to diagnose malignant lung cancers. However, these approaches achieve variable performance because: 1) nodules are inherently difficult to segment due to the range of nodule morphology and potential overlap with surrounding structures (e.g., chest wall, vessels) (Shen et al., 2015b); and 2) extracted features vary due to differences in segmentation results and acquisition parameters (Shen et al., 2017b; Piedra et al., 2016). Thus, using segmented regions may lead to inaccurate features that are subsequently used as inputs into downstream classifiers (Shen et al., 2017b). Another critical question raised by this type of CADx design is how to define the “optimal” subset of features that can best encode characteristics of the lung nodule (Ciompi et al., 2015). The optimal feature set is dependent on the characteristics of the dataset and methods used to train the model, which lead to models that perform well on their training dataset but not other datasets.
To overcome these issues, deep learning methods (Shen et al., 2015b, 2017b; Ciompi et al., 2015; Kumar et al., 2015; Hua et al., 2015), particularly convolutional neural networks (CNNs), have recently been used for lung nodule classification, with promising results. These deep learning models adaptively learn the optimal representation in a fully data-driven way, taking raw image data as input without relying on a priori nodule segmentation masks or handcrafted features. For instance, Kumar et al. (Kumar et al., 2015) first trained an unsupervised deep autoencoder to extract latent features from 2D CT patches. These extracted deep features were then used together with decision trees to predict lung cancer. Similarly, Hua et al. (Hua et al., 2015) employed supervised techniques with a deep belief network and CNN, outperforming methods that use scale-invariant feature transform (SIFT) features and local binary patterns (LBP) (Farag et al., 2011); and using fractal analysis (Lin et al., 2013). Ciompi et al. (Ciompi et al., 2015) used pre-trained CNN models to classify candidates as peri-fissural nodules (PFNs) or non-PFNs. Deep features were extracted from the pre-trained model for three 2D image patches in axial, coronal, and sagittal views. An ensemble of the deep features and bag-of-frequency features were then used to train supervised binary classifiers for the PFN classification task. Shen et. al. (Shen et al., 2015b) designed a multi-scale CNN using 3D nodule patches at three different resolutions to perform the lung cancer diagnosis task. This work is further extended in (Shen et al., 2017b) by adding a multi-crop pooling strategy to improve model performance. Markedly, these cited works use deep learning as a “black-box” and do not attempt to explain what representations have been learned or why the model generates a given prediction. This low degree of interpretability arguably hinders target end users, such as radiologists, from understanding how the models work and ultimately impedes model adoption for clinical usage. As discussed in (Jorritsma et al., 2015), interpretability is critical in facilitating radiologist-CADx interactions by providing transparent and trustworthy predictions.
A number of radiologist-interpreted features derived from CT scans have been considered influential when assessing the malignancy of a lung nodule (Kim et al., 2015; Erasmus et al., 2000). These features are referred to as semantic features in this study. Examples of such semantic features include nodule spiculation, lobulation, consistency (texture), and shape. Although qualitative in nature, studies have shown that these semantic features can be char acterized numerically using low-level image features (Kaya & Can, 2015). Hancock et al. (Hancock & Magnan, 2016) demonstrated that machine learning can achieve high prediction accuracy for lung cancer malignancy using only semantic features as inputs. In addition, semantic features are intuitive to radiologists and are moderately robust against perturbations in image resolution and reconstruction kernel. An opportunity exists to incorporate these semantic features into the design of deep learning models, combining the advantages of both.
In this study, we propose a novel interpretable hierarchical semantic convolutional neural network (HSCNN) to predict whether a nodule is malignant in CT images. The HSCNN takes the raw CT image cubes centered at nodules as input and generates two levels of outputs. The first predictive level provides intermediate outputs in terms of diagnostic semantic features, while the second level represents the final lung nodule malignancy prediction score. Jump connections are employed to feed the information learned from the first level semantic features to the final malignancy prediction. As such, our first level outputs provide explanations about what the HSCNN model has learned from the raw image data and correlates semantic features with the specific malignancy prediction; it also provides additional information to improve the final malignancy prediction task through the jump connections. This entire model is trained by minimizing a global cost function, where both first- and second-level task losses are included.
The contributions of this paper are threefold:
We describe an approach to build a radiologist-interpretable deep convolution neural network. The intermediate outputs from the model give predictions of diagnostic semantic features associated with the final classification, helping to explain the prediction. To the best of our knowledge, this is the first example of a network architecture emphasizing the interpretability of the results.
We provide a hierarchical design that integrates both semantic features and deep features to predict malignancy. Shared convolution modules in the HSCNN are used to learn generalizable features across tasks. The information learned for each specific low-level semantic feature is then fed into the final high-level malignancy prediction task.
We present a new global cost function to train the whole model jointly, taking both first- and second-level outputs into consideration simultaneously. The new objective function concurrently handles data imbalance issues for both tasks.
The remainder of this paper is organized as follows. In Section 2, we describe the dataset used in this study and the proposed HSCNN model. In Section 3, we present results and compare the proposed method with a traditional 3D CNN. In Sections 4 & 5, we discuss the findings and limitations of the work.
2. Materials and Methods
2.1. Lung Image Database Consortium Dataset
The Lung Image Database Consortium image collection (LIDC-IDRI) (Armato et al.,2011) is a publicly available dataset, which we used to train and test our proposed methods. LIDC-IDRI contains both screening and diagnostic CT scans collected from 7 academic centers and 8 medical imaging companies. Inclusion criteria for CT scans were: 1) having a collimation and reconstruction interval no greater than 3 mm; and 2) each scan approximately containing no more than 6 lung nodules with the longest dimension ranging from 3–30 mm, as determined by a cursory review during case selection at the originating institution (Armato et al., 2011). For the whole dataset, the slice thicknesses varied from 0.6 to 5 mm, and the in-plane pixel size varied from 0.461 to 0.977 mm. LIDC-IDRI contains 1,018 cases (representing 1,010 different patients, 8 patients having 2 distinct scans); each case consists of at least one CT scan and associated eXtensible Markup Language (XML) file, containing nodule annotations made by up to four human readers following a two-phase image annotation process. Pixel-level 3D contour segmentations, assessment of nodule likelihood for malignancy, and interpretation of eight nodule characteristics were provided for nodules ≥ 3 mm. We considered the following eight nodule characteristics as semantic features: calcification, subtlety, lobulation, sphericity, internal structure, margin, texture, spiculation, and malignancy. Each feature was rated from 1 to 5 or 6 by each reader. Table 1 lists the description and definitions for each of the labels from (McNitt-Gray et al., 2007).
Table 1:
Detailed nodule characteristics labels in LIDC dataset.
| Semantic Feature | Description | Ratings |
|---|---|---|
| Malignancy | Likelihood of malignancy | 1. Highly unlikely 2. Moderately unlikely 3. Indeterminate 4. Moderately suspicious 5. Highly suspicious |
| Margin | How well defined the margins are | 1. Poorly defined 2. 3. 4. 5. Sharp |
| Sphericity | Three dimensional shape in terms of roundness | 1. Linear 2. 3. Ovoid 4. 5. Round |
| Subtlety | Difficulty of detection relative to surround | 1. Extremely subtle 2. Moderately subtle 3. Fairly subtle 4. Moderately obvious 5. Obvious |
| Spiculation | Degree of exhibition of spicules | 1. Marked 2. 3. 4. 5. None |
| Radiographic solidity (texture) | Internal texture (consistency) of nodule | 1. Non-solid 2. 3. Part Solid 4. 5. Solid |
| Calcification | Presence and pattern of calcification | 1. Popcorn 2. Laminated 3. Solid 4. Non-central 5. Central 6. Absent |
| Internal structure | Expected internal composition of the nodule | 1. Soft tissue 2. Fluid 3. Fat 4. 5. Air |
| Lobulation | The presence and degree of lobulation of the nodule margin | 1. Marked 2. 3. 4. 5. None |
2.2. Our Usage of the LIDC dataset
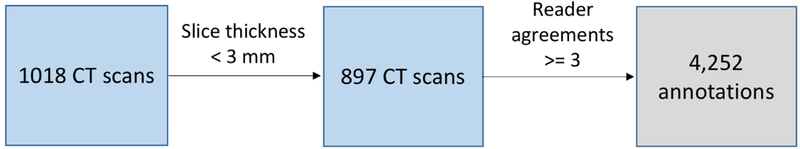
A nodule could be associated with up to 4 annotations, depending on how many of the readers demarcated the nodule.We used a list provided in (A. P. Reeves, A. M. Biancardi, 2011) to determine which annotations referred to the same nodule. Only nodules identified by at least three radiologists were included in this study. CT scans with slice thickness larger than or equal to 3 mm were also excluded. Figure 2 summarizes the inclusion criteria for this study, resulting in the inclusion of 4,252 nodule annotations. Each annotation was considered independently (e.g., an object marked by all four radiologists as a nodule was considered as four independent nodules) to maximize the use of available annotations and to follow the convention used in prior studies (Clark et al., 2013; Hancock & Magnan, 2016; Froz et al., 2017). Uniform labels for each feature were assigned to all annotations that referred to the same nodule. As shown in Table 1, the LIDC annotation process employed one ordinal feature (likelihood of malignancy) and four semantic features (margin, sphericity, nodule subtlety, and texture (consistency)). Scores for these five nodule characteristics were binarized by averaging the scores for each nodule as in (Shen et al., 2015b) and then binarizing each feature: average scores between 1–3 were assigned Label 0 while 4–5 were assigned Label 1. Label 0 typically indicated a benign nodule, poorly defined margin, lesser roundness, poor conspicuity between nodule and surroundings, and a non-solid (ground-glass-like) consistency. Conversely, Label 1 more typically denoted a malignant nodule, sharp margins, higher sphericity, high conspicuity between nodule and surroundings, and solid consistency. Calcification was handled differently: annotations were made using a categorical scale from 1 to 6. Here, nodules with averaged ratings of 6 were labeled as absent of calcification pattern (Label 1); all other ratings represented the presence of calcification (Label 0).
Figure 2:
Lung nodule inclusion criteria.
The feature “internal structure” was overwhelmingly annotated as soft tissue, thus provided little discriminative information (Hancock & Magnan, 2016) and was excluded from our analysis. Moreover, the Cancer Imaging Archive (TCIA) reported that an indeterminate subset of cases in the dataset were inconsistently annotated with respect to spiculation and lobulation (The Cancer Imaging Archive, 2017). As such, we did not consider these two features in our model. Finally, it should be noted that biopsy-confirmed diagnoses of the nodules are not known. For the purposes of this work, the likelihood of malignancy served as the proxy for truth. Table 2 summarizes the generation of the binary labels from LIDC rating scales as described above. Table 3 lists the data counts for each label of the nodule characteristics.
Table 2:
Summary of generating binary labels from LIDC rating scales for nodule characteristics.
| Nodule characteristics | Label 0 | Label 1 |
|---|---|---|
| Malignancy | Scale 1 – 3 Benign |
Scale 4 – 5 Malignant |
| Sphericity | Scale 1 – 3 Lesser roundness |
Scale 4 – 5 High degree of roundness |
| Margin | Scale 1 – 3 Poorly defined margin |
Scale 4 – 5 Sharp margin |
| Subtlety | Scale 1 – 3 Poor contrast between nodule and surroundings |
Scale 4 – 5 High contrast between nodule and surroundings |
| Texture | Scale 1 – 3 Non-solid internal density |
Scale 4 – 5 Solid internal density |
| Calcification | Scale 1 – 5 Present of calcification |
Scale 6 Absent of calcification |
Table 3:
Label counts for nodule characteristics.
| Nodule characteristics | Label 0 (#) | Label 1 (#) | Total (#) |
|---|---|---|---|
| Malignancy | 3212 | 1040 | 4252 |
| Sphericity | 2304 | 1948 | 4252 |
| Margin | 1640 | 2612 | 4252 |
| Subtlety | 1570 | 2682 | 4252 |
| Texture | 518 | 3734 | 4252 |
| Calcification | 496 | 3756 | 4252 |
2.3. Data Preprocessing
The LIDC dataset contains a heterogeneous set of scans obtained using various acquisition and reconstruction parameters. To normalize pixel values, all CT scans were first transformed to Hounsfield (HU) scales using the information in the DICOM (Digital Imaging and Communication in Medicine) series header and converted to a range of (0, 1) from (−1000, 500 HU). A 3D patch sized 40 × 40 × 40 mm were extracted for each candidate. Each patch was centered around the candidate. 40 mm was chosen so that all candidates would be fully contained in the patch as the largest nodules in our subset were 30 mm in diameter. We then rescaled each patch to a fixed size of pixels in all three dimensions, resulting in isotropic cubes for all cases. During preprocessing, we retained the original relative nodule size information within each patch with the belief that nodule size is informative in subsequent prediction tasks.
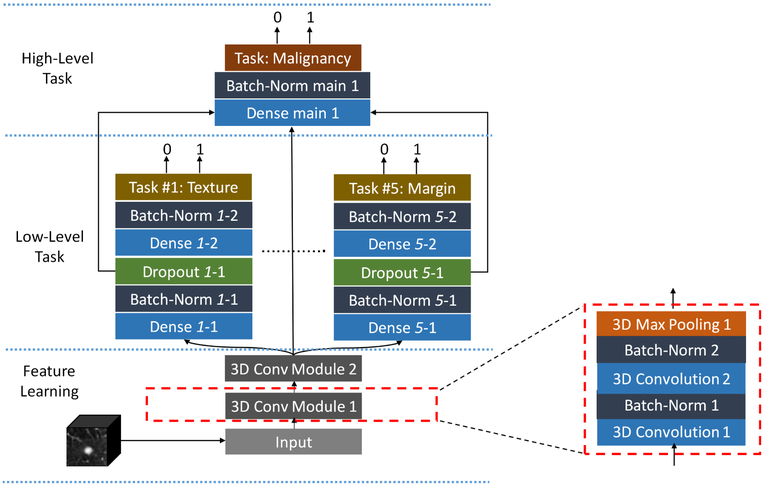
2.4. Hierarchical Semantic Convolutional Neural Network
The proposed HSCNN utilizes a 3D patch capturing the lung nodule as input and outputs two levels of predictions, as shown in Figure 3. This architecture comprises three parts: 1) a feature learning module; 2) a low-level task module; and 3) a high-level task module. The feature learning module adaptively learns the image features that are generalizable across different tasks. The low-level task predicts five semantic diagnostic features: margin, texture, sphericity, subtlety, and calcification. The high-level task incorporates information from both the generalizable image features and the low-level tasks to produce an overall prediction of lung nodule malignancy.
Figure 3:
Model architecture of the hierarchical semantic convolutional neural network.
The feature learning module (Figure 3, feature learning) consists of two convolution module blocks where each block shares the same structure and contains two stacked 3D convolution layers followed by batch normalization and one 3D average pooling layer. Each convolution layer has a kernel size of 3×3×3. These layers perform the convolution operation on input feature maps along all three dimensions of the input cube to produce an output feature map defined by:
| (1) |
where fj and fi are the jth output feature map and ith input feature map, respectively. And cj is the jth convolution kernel and * represents the 3D convolution operation between the convolution kernel and input feature map. bj is the jth bias corresponding to the jth convolution kernel. After convolution, batch normalization is applied to all output feature maps to accelerate the training process and reduce the internal covariate shift by normalizing the feature maps (Ioffe & Szegedy, 2015). Rectified linear units (ReLUs) (Krizhevsky et al., 2012) are used as the nonlinear activation functions to take the output from batch normalization. 16 feature maps are used for both convolution layers in the first convolution module, and 32 feature maps are adopted for both convolution layers in the second convolution module. A 3D max pooling layer is used in the end for each convolution module block to progressively reduce the spatial size of the feature maps to reduce the number of parameters and control for overfitting. This layer is defined as:
| (2) |
where x (the row index), y (the column index), and z (the depth index) start from zero. Here, s is the stride size (downscale factor) and d is the size of the max pooling window. We employ a pooling window size of d = (2, 2, 2) and stride size of s = (2, 2, 2). This design downsamples the input feature maps by a factor of 2 across all three cube dimensions. This pooling layer has no learnable parameters.
After the last convolutional module, output features are fed simultaneously into the lowand high-level task modules. The low-level task module (Figure 3, low-level task) consists of five branches, each with the same architecture, representing a distinct semantic feature (i.e., texture, margin, sphericity, subtlety, or calcification). A fully-connected layer (densely-connected) is the major basic building block for each of these branches. One fully-connected layer connects each input unit to each output unit, designed to capture correlations from all input feature units to the output. Batch normalization and dropout techniques are both used to control model overfitting. The dropout method randomly removes connections between input and output units during network training to prevent units from co-adapting too much (Srivastava et al., 2014). Two fully-connected layers are employed before the final binary prediction with 256 neurons and 64 neurons for the first and second layer, respectively.
The high-level task module (Figure 3, high-level task) predicts whether the nodule is malignant. This module combines the output features from the feature learning module and each of the low-level task branches as its input. As shown in Figure 3, the output feature maps from the last convolution module are used, along with the output from the last second fully-connected layer of each subtask branch. This design makes the final prediction utilize the basic features learned from the shared convolution modules and forces the convolution blocks to extract representations that are generalizable across all tasks. It also makes use of the information learned from each related semantic subtask to ultimately infer nodule malignancy. The last fully-connected layer in each subtask branch is trained to extract representations more specific to the corresponding subtask compared to the second to last fully-connected layer. Thus, the second to last layer of the subtask branch is chosen to provide less specific but salient information for the final malignancy prediction task. The concatenated features are inputted into a fully-connected layer with 256 neurons, followed by a batch normalization operation before the final malignancy prediction.
To jointly optimize the HSCNN during network training, a global loss function is proposed to maximize the probability of predicting the correct label for each task by:
| (3) |
where N is the total number of training samples and i indicates the ith training sample. j is the jth subtask and j ∈ [1, 5]. λj is the weighting hyperparameter for the jth subtask. Lj,I represents the loss for sample i and task j. LM,i is the loss for the malignancy prediction task for the ith sample. Each loss component is defined as weighted cross entropy loss by:
| (4) |
where yi is true label for the ith sample (xi, yi). Here, yi equals 0 or 1. fyi,j is the prediction score of the true class yi for task j and fyn,j represents a prediction score for class yn. We use ωyi,j to represent the weight of class yi for task j. The use of !yi,j is important because the labels are imbalanced in all the tasks and ωyi,j is helpful in reducing the training bias introduced by such data imbalance. Specifically, ωyi,j weights each class loss proportional to the reciprocal of the class counts in the training data. For instance, ωyi=0,j = Nyi=1,j/(Nyi=0,j + Nyi=1,j) and ωyi=1,j = Nyi=0,j/(Nyi=0,j + Nyi=1,j). Nyi=1,j represents the total count of samples in the training data for task j, where the true class label equals 1. The global loss function is minimized during the training process by iteratively computing the gradient of Lglobal over the learnable parameters and updates the parameters through back-propagation. During training, model learnable parameters are initialized using the Xavier algorithm (Glorot & Bengio, 2010) and are updated using the Adam stochastic optimization algorithom (Kingma & Ba, 2014).
2.5. Training
We performed model training, validation, and testing using 897 LIDC cases, selected as described in Section 2.2. A 4-fold cross validation study design was employed to obtain the final assessment of the model performance. Within each fold, we split these cases into four subsets, where each subset had a similar number of nodules. 2 subsets are used for training, 1 subset for validation, and 1 subset for holdout testing. The validation set is used to tune the hyperparameters and test set is employed as external holdout to report the final model performance. Each subset is used as the test set once during the cross validation. This design ensures that the test set is independent of model training and parameter optimization and should reflect the true model performance without information leakage. We note that earlier studies in (Shen et al., 2015b, 2017b; Kumar et al., 2015; Hua et al., 2015) only use training and validation splits during the cross validation process, without consideration for holdout test sets. Such designs arguably have information leakage and may overestimate model performance.
To better control for model overfitting, 3D data augmentation was applied during the training process. Data augmentation artificially inflates the dataset by using label-preserving transforms to generate more data examples and is considered as a model regularization scheme (Krizhevsky et al., 2012). One or more random operations were applied on each training dataset to generate artificial samples. The spatial affine operations used in this study included translating the position of the nodule within 4 mm or flipping the 3D nodule cube along one of the three axes. The translation limit was set to 4 mm to ensure that the boundaries of the largest nodules were captured properly in the 3D cube (40 × 40 × 40 mm).
3. Experimental Results
This section first describes how we trained the models. We compare our model to a traditional 3D CNN model and other state-of-art methods. We also evaluate the accuracy of semantic feature predictions, providing illustrations of correct and incorrect predictions.
3.1. Model Training
Models were trained for 300 epochs during each fold of cross-validation. After 100 epochs of training, the model loss on the validation set became stable. The best model for each fold was chosen to be the one that achieved the lowest malignancy prediction loss on the validation dataset. Only the independent test dataset was used to calculate end model performance. An online augmentation scheme was employed during model training: during each training epoch, additional artificially created training samples were generated by randomly picking one or multiple augmentation operations, as described in Section 2.5. The same augmentation process was also applied to the validation dataset. To capture a majority of nodule morphology while reducing the input data dimensions, the input nodule cube size was set to be 52 × 52 × 52 voxels. The learning rate was set to be 0.001. The convolution kernel size, number of feature maps, pooling window size, downscale factor, and number of neurons for each fully-connected layer were reported in Section 2.4. The choices of these parameters have been commonly used, as shown in (Krizhevsky et al., 2012; Simonyan & Zisserman, 2014). The hyperparameters presented in Equation 3 were chosen by using a randomized coarse-to-fine grid search with the validation dataset in the first 20 epochs of each fold (Bergstra & Bengio, 2012).
The proposed HSCNN model was implemented in Python 2.7 with TensorFlow (Abadi et al., 2016) and the Keras toolkit (Chollet et al., 2015). All experiments were performed on a server with 6-core Intel Xeon E5–2630 processor, 32GB memory, and one NVIDIA TITAN Xp GPU (12GB on-board memory). The training of one HSCNN model took about 5 hours for 300 epochs.
3.2. Malignancy Prediction Results
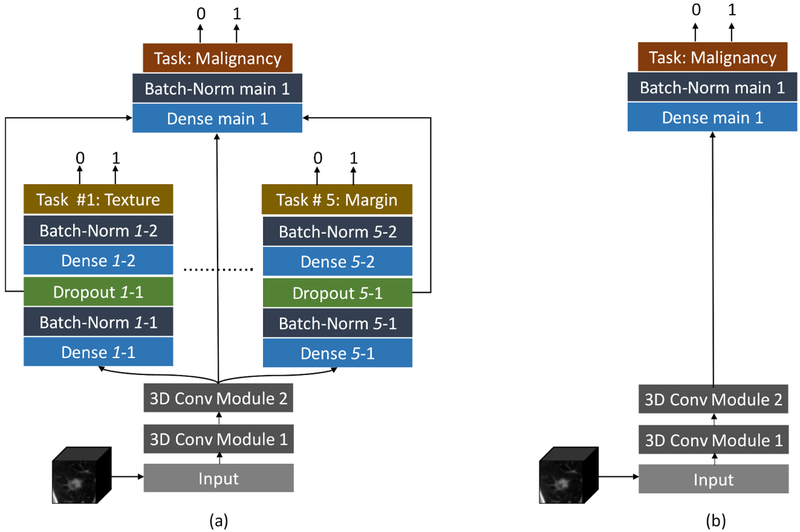
To evaluate and compare the HSCNN performance on lung nodule malignancy prediction, a 3D convolutional neural network (3D CNN) was implemented as a baseline model, shown in Figure 4b. This 3D CNN uses the same feature learning and high-level task modules as the HSCNN but do not include the low-level subtask module. The baseline model was trained and evaluated using the same 4-fold cross validation process and with the same data splitting for each fold (using the same randomization seed).
Figure 4:
Framework comparison between proposed HSCNN and baseline 3D CNN. (a) The proposed HSCNN architecture; (b) a baseline 3D CNN architecture. The baseline model has the same structure as the HSCNN but without the low-level semantic task component.
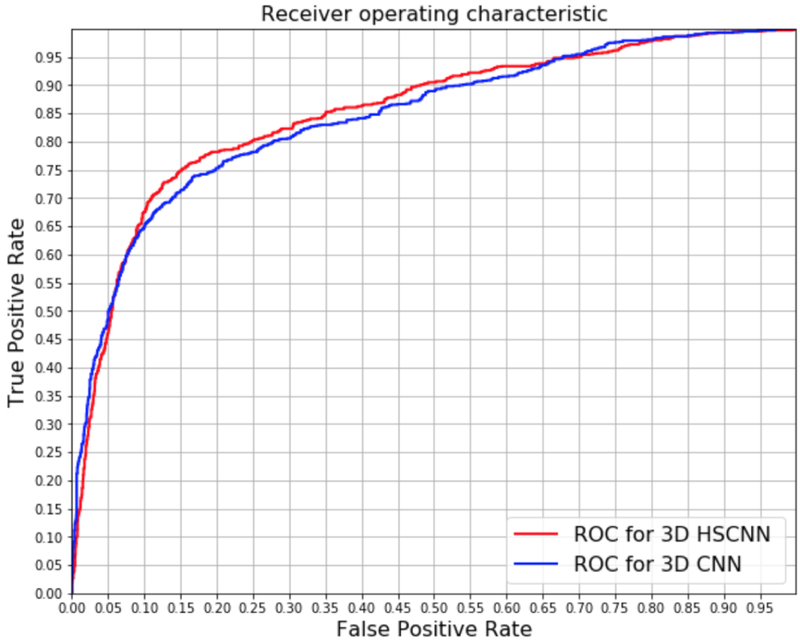
Figure 5 shows the receiver operating characteristic (ROC) curve plots comparing HSCNN versus 3D CNN performance. These plots represent the intuitive trade-off between sensitivity and specificity. By visual inspection of the ROC curves, HSCNN performs better than the traditional 3D CNN model. The area under the ROC curve (AUC) quantitatively compares the overall performance of a classification model and is frequently used as a metric to access performance in nodule classification (Shen et al., 2017b; Ciompi et al., 2015; Hancock & Magnan, 2016; Clark et al., 2013; Froz et al., 2017). Table 5 summarizes the mean AUC score, accuracy, sensitivity, and specificity for both models. The HSCNN model achieved a mean AUC 0.856, mean accuracy 0.842, mean sensitivity 0.705 and mean specificity 0.889; while the 3D CNN model achieved a mean AUC 0.847, mean accuracy 0.834, mean sensitivity 0.668 and mean specificity 0.889. Both ROC plots and metric assessments show that the proposed HSCNN achieved better performance for malignancy prediction compared with the conventional 3D CNN approach.
Figure 5:
Receiver operating characteristic curve comparison: HSCNN versus 3D CNN. The AUC of 3D HSCNN is significantly higher than 3D CNN according to a paired T-test as shown in Table 5.
Table 5:
Paired T-Test summarizes for AUC scores between HSCNN and 3D CNN model on test set of each fold. CI represents for confidence interval.
| Test Fold | HSCNN AUC |
3D CNN AUC |
AUC Difference (HSCNN - 3D CNN) |
Paired T-Test |
|---|---|---|---|---|
| Fold 1 | 0.878 | 0.869 | 0.009 | P-value=0.005, Mean_difference=0.009, CI = [0.0051, 0.0129] |
| Fold 2 | 0.813 | 0.807 | 0.006 | |
| Fold 3 | 0.874 | 0.862 | 0.012 | |
| Fold 4 | 0.860 | 0.851 | 0.009 |
To assess the statistical significance of model performance improvements, we conducted a paired sample t-test to evaluate the mean differences in AUC scores between the HSCNN and 3D CNN model. Group 1 consists of the AUC score of the HSCNN model for each holdout test fold during the cross validation. Group 2 consists of the corresponding AUC score for the 3D CNN for the same fold. The null hypothesis is that the mean difference of AUC score between these two models equals 0. Table 5 summarizes the AUC scores for these groups and results of a paired t-test. The test obtained a p-value of 0.005 and confidence interval of [0.0051, 0.0129], thus rejecting the null hypothesis and indicating that the HSCNN model achieved a statistically significantly better AUC relative to the 3D CNN. The mean improvement of the AUC score was 0.009. This finding demonstrates that adding a low-level task component on an existing CNN structure may improve the prediction of malignancy in a lung nodule.
We also compared our results with other deep learning models for lung nodule malignancy prediction that utilized the LIDC dataset reported in literature to date in Table 6. Kumar et al. (Kumar et al., 2015) developed a deep autoencoder-based model with 4,323 nodules of the LIDC dataset, achieving model accuracy of 0.7501. Hua et al. (Hua et al., 2015) presented a CNN model and deep belief network (DBN) model. Both models were trained and validated using 2,545 lung nodule samples from LIDC. The CNN model had specificity of 0.787 and sensitivity 0.737; and the DBN model obtained specificity of 0.822 and sensitivity 0.734. In (Shen et al., 2015b), Shen et al. used a model based on multi-scale 3D CNN. Developed with 1,375 LIDC nodule samples, the average accuracy is reported above 0.84 with different configurations. In (Shen et al., 2017b), Shen et al. extended this multi-scale model using a multi-crop approach and achieved accuracy of 0.839, 0.8636, and 0.8714 with 340, 1030 and 1375 nodules of LIDC, respectively. All of these previously reported methods were evaluated with only training and validation data splits without an independent holdout test dataset as discussed in Section 2.5. Generally, our model achieved better or similar performances compared with these reported methods. However, direct comparison of these models is difficult given that each model was trained and tested on different subsets of the LIDC dataset.
Table 6:
Comparison with other current deep learning models.
| Method | Nodules | Hold-out Test | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|---|---|
| Deep Auto-encoder (Kumar et al., 2015) |
4,323 | No | - | - | 0.7501 | - |
| CNN (Hua et al., 2015) |
2,545 | No | 0.737 | 0.787 | - | - |
| DBN (Hua et al., 2015) |
2,545 | No | 0.734 | 0.822 | - | - |
| Multi-scale CNN (Shen et al., 2015b) |
1,375 | No | - | - | 0.84 | - |
| Multi-crop CNN (Shen et al., 2017b) |
1,375 | No | - | - | 0.8714 | - |
| Proposed | 4,252 | Yes | 0.705 | 0.889 | 0.842 | 0.856 |
3.3. Semantic Feature Prediction Results and Model Interpretability
Table 7 presents the classification performance for each of the low-level tasks (i.e., semantic features). We achieved mean accuracy of 0.908, 0.725, 0.719, 0.834 and 0.552; mean AUC score of 0.930, 0.776, 0.803, 0.850 and 0.568; mean sensitivity of 0.930, 0.758, 0.673, 0.855 and 0.552; and mean specificity of 0.763, 0.632, 0.796, 0.636 and 0.554 for calcification, margin, subtlety, texture, and sphericity, respectively. These results suggest that the HSCNN model is able to learn feature representations that are predictive of semantic features while simultaneously achieving high performance in predicting nodule malignancy.
Table 7:
Classification performance for semantic feature predictions.
| Semantic Features |
Accuracy (SD) | AUC (SD) | Specificity (SD) | Sensitivity (SD) |
|---|---|---|---|---|
| Calcification | 0.908 (0.050) | 0.930 (0.034) | 0.763 (0.092) | 0.930 (0.067) |
| Margin | 0.725 (0.049) | 0.776 (0.033) | 0.632 (0.109) | 0.758 (0.091) |
| Subtlety | 0.719 (0.019) | 0.803 (0.015) | 0.796 (0.045) | 0.673 (0.044) |
| Texture | 0.834 (0.086) | 0.850 (0.042) | 0.636 (0.199) | 0.855 (0.108) |
| Sphericity | 0.552 (0.027) | 0.568 (0.015) | 0.554 (0.076) | 0.552 (0.095) |
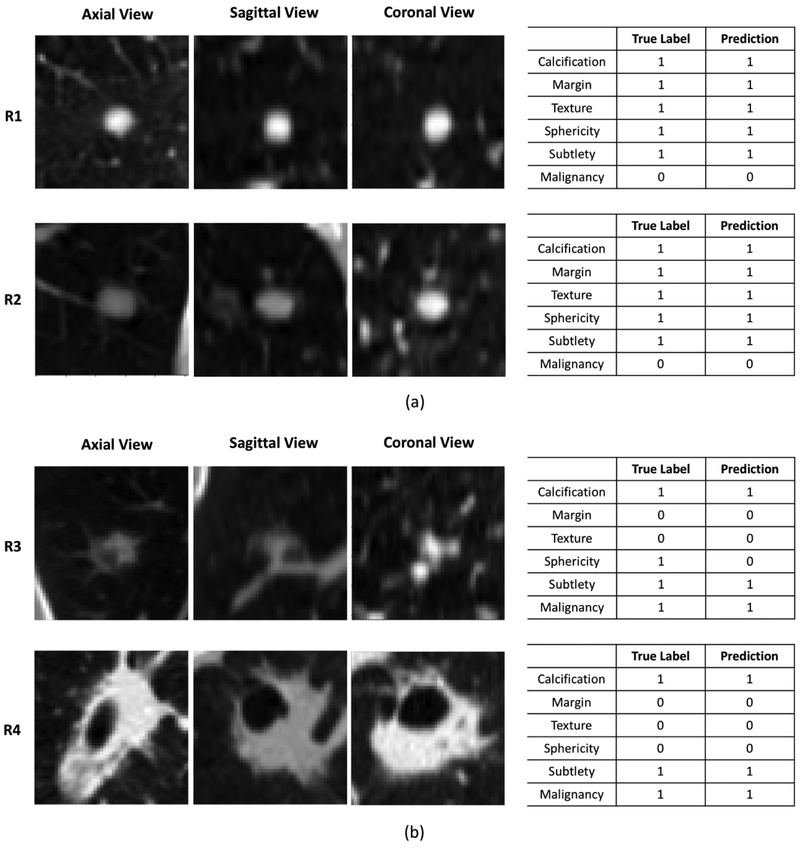
Figure 6 demonstrates the interpretability of the HSCNN model by visualizing the central slices of the 3D nodule patches in axial, coronal, and sagittal projections while presenting the predicted interpretable semantic labels along with the malignancy classification results. Figure 6a–R1 shows that the HSCNN model classifies the lung nodule as benign (the true label is also benign). This decision correlated to predictions of this nodule as having no calcification, sharp margins, roundness, obvious contrast between nodule and surroundings, and solid consistency. The predictions of these five semantic characteristics are the same as the true label and corresponds to our knowledge about benign lung nodules. Compared to a 3D CNN malignancy prediction model, the HSCNN provides more insight for interpreting its predictions. Similarly, in Figure 6b–R3, the proposed model predicts the lung nodule as malignant (true label is also malignant). Different from the benign case, the HSCNN model predicts this nodule having poorly defined margins, ground glass consistency, and non-round shape. This partly explains why the HSCNN makes a malignancy classification with such nodule characteristics corresponding to our expert knowledge about typical malignant nodules. We note that the sphericity predictions made by the model are different from the true label. This result is explained by the fact that while the nodule has a more regular round shape in axial view, the shape is actually more elongated in the two other projections, as shown in Figure 6b–R3.
Figure 6:
Illustrating the HSCNN model interpretability: lung nodule central slices, interpretable semantic feature prediction and malignancy prediction. R1, R2, R3 and R4 are four different nodules. (a) Central slices of axial, coronal and sagittal view of two benign nodule samples; true and predicted labels for interpretable semantic features and malignancy. (b) Central slices of axial, coronal and sagittal view of two malignant nodule samples; true and predicted labels for interpretable semantic features and malignancy.
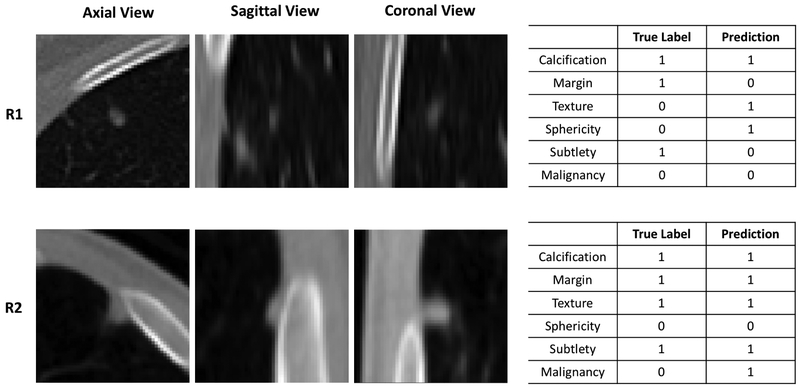
Figure 7 shows two representative cases where the HSCNN fails to predict either one or more semantic features or cancer malignancy. Figure 7–R1 shows that the HSCNN model classifies the lung nodule correctly as benign but incorrectly for four semantic features of this nodule (margin, texture, sphericity and subtlety). In Figure 7–R2, the HSCNN model incorrectly classifies the lung nodule as malignant (the true label is benign). However, all semantic features of this nodule are predicted correctly. These two cases present the situation where the correctness are inconsistent between the malignancy and semantic predictions. Section 4 provides further discussion about how the HSCNN model can be augmented with more semantic features.
Figure 7:
Example cases where the HSCNN model incorrectly predicts semantic features or cancer malignancy. R1 and R2 are two different nodules. R1: This case has four incorrect semantic feature predictions yet a correct malignancy prediction. R2: This case has all correct semantic predictions yet an incorrect malignancy prediction.
4. Discussion
We present a HSCNN model that incorporates domain knowledge into the model architecture design, predicting semantic nodule characteristics along with the primary task of nodule malignancy diagnosis. Five semantic features were considered: calcification, margin, subtlety, texture, and sphericity. Our results in Section 3.3 suggest that the HSCNN model is capable of providing accurate predictions of semantic descriptors while simultaneously classifying nodule malignancy. The semantic labels are useful in interpreting the model’s predictions. Moreover, Section 3.2 shows that our HSCNN architecture improves model performance over a 3D CNN architecture.
There are some limitations to this study. Our semantic labels did not include those of known higher association with malignancy, such as nodule size, margin spiculation, lobulation, and anatomic location, which have previously been reported as informative (McWilliams et al., 2013; Swensen et al., 1997). In the case of lobulation and spiculation, known labeling errors in the LIDC dataset made them unsuitable for our use. Additionally, semantic labels are subject to moderate inter-reader variability; performance might be enhanced by limiting semantic labels to those on which there is high reader agreement. Third, the malignancy labels provided in the LIDC dataset do not reflect pathological diagnosis but rather, suspicion levels of the interpreting radiologists. Finally, the original semantic features have scales of 5 or 6; binarizing the labels may lose some of the semantic information. Changing the threshold for binary classification would also affect results. Our rationale for binary labels in this case was to overcome data sparsity, where the number of cases labeled for certain scales might be very small compared with the other scales (e.g., only 11 cases are labeled as linear for sphericity out of total 4252 cases). Moreover, analysis shows that inter-reader agreement is much lower for 5 or 6 scales compared with the proposed binary labels. Thus, binary labeling helps to reduce labeling noise caused by inter-reader variability. These limitations may be circumvented by training on large datasets that have been systematically annotated using a shared lexicon that includes discriminating features.
Several improvements can be investigated as part of future work. First, further optimization of the network architecture to achieve higher prediction performance can be performed. For instance, densely connected designs (Huang et al., 2016) and residual designs (He et al., 2016) could be used to potentially improve model performance. Given limitations in computational power, not all designs were optimally searched; we will investigate these as part of future work. Second, as our HSCNN model facilitates interpretation of the utility of each semantic feature in predicting malignancy, the model can be fine-tuned by domain experts by weighting more discriminating features in difficult cases. Third, exploration of more granular or continuous labels for semantic features could be performed. Information of each semantic label’s distributions could be incorporated into the model’s design to boost performance. Fourth, our HSCNN architecture could be easily extended to incorporate additional semantic features. However, too many low-level subtasks (e.g., more than 20) would make model convergence more difficult. Thus, improving the model scalability should be studied in future works. Not all combinations of semantic labels may co-occur. Therefore, this observation could be employed to improve the model design. Finally, the inputs of current models were the 3D cubes centered at each nodule with all background pixel intensities. Background objects such as the lung walls of the juxtapleural nodules might prevent the model from learning useful information for the classification task. A possible future work is to explore feeding the deep learning model with nodular versus perinodular regions as two distinct separated inputs for each input data.
5. Conclusion
In this paper, we have developed a novel radiologist-interpretable HSCNN model for predicting lung cancer in CT-detected indeterminate nodules. This model is able to simultaneously predict nodule malignancy while classifying five nodule semantic characteristics, including calcification, margin, subtlety, texture, and sphericity of nodules. These diagnostic semantic features predictions are intermediate outputs associated with the final malignancy prediction, and are useful to explain the diagnosis prediction. Information from each low-level semantic feature prediction is incorporated into the malignancy prediction task by employing jump connections. This framework is able to enforce the shared basic convolution modules in the HSCNN to learn features that are generalizable across tasks. This unified model is trained by minimizing a joint global loss function, where the losses of both malignancy and semantic feature prediction tasks are incorporated. Extensive experiments and statistical tests show that the proposed HSCNN model is able to significantly improve the classification performance for nodule malignancy prediction and the semantic characteristics predictions have improved the model interpretability. This trained model could also serve as a lung nodule semantic feature generator.
Table 4:
Results comparison: HSCNN versus 3D CNN.
| Model | AUC (SD) | Accuracy (SD) | Sensitivity (SD) | Specificity (SD) |
|---|---|---|---|---|
| 3D CNN | 0.847 (0.024) | 0.834 (0.022) | 0.668 (0.040) | 0.889 (0.022) |
| HSCNN | 0.856 (0.026) | 0.842 (0.025) | 0.705 (0.045) | 0.889 (0.022) |
Highlights.
A novel interpretable hierarchical deep learning model for lung cancer diagnosis
Network design incorporates semantic features that are intuitive to radiologists
A single joint network predicts interpretable features and malignancy
Architecture maintains prediction accuracy while improving model interpretability
Acknowledgement
The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study. Research reported in this publication was partly supported by the National Cancer Institute of the National Institutes of Health under award number R01 CA210360, the Center for Domain-Specific Computing (CDSC) funded by the NSF Expedition in Computing Award CCF-0926127, and the National Science Foundation under Grant No. 1722516. Computing resources were provided by the NIH Data Commons Pilot and a donation of a TITAN Xp graphics card by the NVIDIA Corporation. The content is solely the responsibility of the authors and does not necessarily represent the official views of sponsor agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
None Declared.
Contributor Information
Shiwen Shen, Email: shiwenshen@engineering.ucla.edu.
Simon X Han, Email: simon.x.han@ucla.edu.
Denise R Aberle, Email: daberle@mednet.ucla.edu.
Alex A Bui, Email: buia@mii.ucla.edu.
William Hsu, Email: whsu@mednet.ucla.edu.
References
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, …, & Kudlur M (2016). Tensorflow: A system for large-scale machine learning. In OSDI (pp. 265–283). volume 16. [Google Scholar]
- Amir GJ, & Lehmann HP (2016). After detection:: The improved accuracy of lung cancer assessment using radiologic computer-aided diagnosis. Academic radiology, 23, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armato SG, Altman MB, Wilkie J, Sone S, Li F, & Roy AS (2003). Automated lung nodule classification following automated nodule detection on ct: A serial approach. Medical Physics, 30, 1188–1197. [DOI] [PubMed] [Google Scholar]
- Armato SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, …, & Kazerooni E (2011). The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Medical physics, 38, 915–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstra J, & Bengio Y (2012). Random search for hyper-parameter optimization. Journal of Machine Learning Research, 13, 281–305. [Google Scholar]
- Chollet F et al. (2015). Keras. [Google Scholar]
- Ciompi F, de Hoop B, van Riel SJ, Chung K, Scholten ET, Oudkerk M, de Jong PA, Prokop M, & van Ginneken B (2015). Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2d views and a convolutional neural network out-of-the-box. Medical image analysis, 26, 195–202. [DOI] [PubMed] [Google Scholar]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, …, & Tarbox L (2013). The cancer imaging archive (tcia): maintaining and operating a public information repository. Journal of digital imaging, 26, 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan N, Bae E, Shen S, Hsu W, Bui A, Jones E, Glavin M, & Vese L (2015). A technique for lung nodule candidate detection in ct using global minimization methods In International Workshop on Energy Minimization Methods in Computer Vision and Pattern Recognition (pp. 478–491). Springer. [Google Scholar]
- Erasmus JJ, Connolly JE, McAdams HP, & Roggli VL (2000). Solitary pulmonary nodules: Part i. morphologic evaluation for differentiation of benign and malignant lesions. Radiographics, 20, 43–58. [DOI] [PubMed] [Google Scholar]
- Farag A, Ali A, Graham J, Farag A, Elshazly S, & Falk R (2011). Evaluation of geometric feature descriptors for detection and classification of lung nodules in low dose ct scans of the chest In Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on (pp. 169–172). IEEE. [Google Scholar]
- Firmino M, Angelo G, Morais H, Dantas MR, & Valentim R (2016). Computer-aided detection (cade) and diagnosis (cadx) system for lung cancer with likelihood of malignancy. Biomedical engineering online, 15, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froz BR, de Carvalho Filho AO, Silva AC, de Paiva AC, Nunes RA, & Gattass M (2017). Lung nodule classification using artificial crawlers, directional texture and support vector machine. Expert Systems with Applications, 69, 176–188. [Google Scholar]
- Glorot X, & Bengio Y (2010). Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the thirteenth international conference on artificial intelligence and statistics (pp. 249–256). [Google Scholar]
- ten Haaf K, Jeon J, Tammemägi MC, Han SS, Kong CY, Plevritis SK, Feuer EJ, de Koning HJ, Steyerberg EW, & Meza R (2017). Risk prediction models for selection of lung cancer screening candidates: A retrospective validation study. PLoS medicine, 14, e1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock MC, & Magnan JF (2016). Lung nodule malignancy classification using only radiologist-quantified image features as inputs to statistical learning algorithms: probing the lung image database consortium dataset with two statistical learning methods. Journal of Medical Imaging, 3, 044504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, & Sun J (2016). Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 770–778). [Google Scholar]
- Hua K-L, Hsu C-H, Hidayati SC, Cheng W-H, & Chen Y-J (2015). Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets and therapy, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu Z, Weinberger KQ, & van der Maaten L (2016). Densely connected convolutional networks. arXiv preprint arXiv:1608.06993, . [Google Scholar]
- Huang P, Park S, Yan R, Lee J, Chu LC, Lin CT, Hussien A, …, & Hales R (2017). Added value of computer-aided ct image features for early lung cancer diagnosis with small pulmonary nodules: A matched case-control study. Radiology, 286, 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioffe S, & Szegedy C (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167, . [Google Scholar]
- Jorritsma W, Cnossen F, & van Ooijen P (2015). Improving the radiologist–cad interaction: designing for appropriate trust. Clinical radiology, 70, 115–122. [DOI] [PubMed] [Google Scholar]
- Kaya A, & Can AB (2015). A weighted rule based method for predicting malignancy of pulmonary nodules by nodule characteristics. Journal of biomedical informatics, 56, 69–79. [DOI] [PubMed] [Google Scholar]
- Kim H, Park CM, Goo JM, Wildberger JE, & Kauczor H-U (2015). Quantitative computed tomography imaging biomarkers in the diagnosis and management of lung cancer. Investigative radiology, 50, 571–583. [DOI] [PubMed] [Google Scholar]
- Kingma DP, & Ba J (2014). Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, . [Google Scholar]
- Krizhevsky A, Sutskever I, & Hinton GE (2012). Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems (pp. 1097–1105). [Google Scholar]
- Kumar D, Wong A, & Clausi DA (2015). Lung nodule classification using deep features in ct images In Computer and Robot Vision (CRV), 2015 12th Conference on (pp. 133–138). IEEE. [Google Scholar]
- Lin P-L, Huang P-W, Lee C-H, & Wu M-T (2013). Automatic classification for solitary pulmonary nodule in ct image by fractal analysis based on fractional brownian motion model. Pattern Recognition, 46, 3279–3287. [Google Scholar]
- McNitt-Gray MF, Armato SG, Meyer CR, Reeves AP, McLennan G, Pais RC, …, & Laderach G (2007). The lung image database consortium (lidc) data collection process for nodule detection and annotation. Academic radiology, 14, 1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, …, & Atkar-Khattra S (2013). Probability of cancer in pulmonary nodules detected on first screening ct. New England Journal of Medicine, 369, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra EAR, Taira RK, El-Saden S, Ellingson BM, Bui AA, & Hsu W (2016). Assessing variability in brain tumor segmentation to improve volumetric accuracy and characterization of change In Biomedical and Health Informatics (BHI), 2016 IEEEEMBS International Conference on (pp. 380–383). IEEE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Bui AA, Cong J, & Hsu W (2015a). An automated lung segmentation approach using bidirectional chain codes to improve nodule detection accuracy. Computers in biology and medicine, 57, 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Han SX, Petousis P, Weiss RE, Meng F, Bui AA, & Hsu W (2017a). A bayesian model for estimating multi-state disease progression. Computers in biology and medicine, 81, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Zhou M, Yang F, Yang C, & Tian J (2015b). Multi-scale convolutional neural networks for lung nodule classification In International Conference on Information Processing in Medical Imaging (pp. 588–599). Springer. [DOI] [PubMed] [Google Scholar]
- Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y, & Tian J (2017b). Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognition, 61, 663–673. [Google Scholar]
- Simonyan K, & Zisserman A (2014). Very deep convolutional networks for large-scale image recognition. CoRR, abs/1409.1556. [Google Scholar]
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I, & Salakhutdinov R (2014). Dropout: A simple way to prevent neural networks from overfitting. The Journal of Machine Learning Research, 15, 1929–1958. [Google Scholar]
- Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, & Edell ES (1997). The probability of malignancy in solitary pulmonary nodules: application to small radiologically indeterminate nodules. Archives of internal medicine, 157, 849–855. [PubMed] [Google Scholar]
- Team NLSTR et al. (2011). Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med, 2011, 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AP, Biancardi AM (2011). The lung image database consortium (lidc) nodule size report. http://www.via.cornell.edu/lidc/. Accessed 2018-06-01.
- The Cancer Imaging Archive (2017). Lung image database consortium -reader annotation and markup - annotation and markup issues/comments. https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI. Accessed 2018-06-01.
- Torre LA, Siegel RL, & Jemal A (2016). Lung cancer statistics In Lung Cancer andPersonalized Medicine (pp. 1–19). Springer. [DOI] [PubMed] [Google Scholar]
- Way TW, Sahiner B, Chan H-P, Hadjiiski L, Cascade PN, Chughtai A, Bogot N, & Kazerooni E (2009). Computer-aided diagnosis of pulmonary nodules on ct scans: Improvement of classification performance with nodule surface features. Medical physics, 36, 3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tan Y, Bell DJ, Marley SE, Guo P, Mann H, …, & Ghiorghiu DC (2013). Exploring intra-and inter-reader variability in uni-dimensional, bi-dimensional, and volumetric measurements of solid tumors on ct scans reconstructed at different slice intervals. European journal of radiology, 82, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovev D, Feigenbaum J, Furst J, & Raicu D (2011). Probabilistic lung nodule classification with belief decision trees In Engineering in medicine and biology society, EMBC, 2011 annual international conference of the IEEE (pp. 4493–4498). IEEE. [DOI] [PubMed] [Google Scholar]