Abstract
Correlated light and electron microscopy (CLEM) has become a popular technique for combining the protein-specific labeling of fluorescence with electron microscopy, both at room and cryogenic temperatures. Fluorescence applications at cryo-temperature have typically been limited to localization of tagged protein oligomers due to known issues of extended triplet state duration, spectral shifts, and reduced photon capture through cryo-CLEM objectives. Here, we consider fluorophore characteristics and behaviors that could enable more extended applications. We describe how dialkylcarbocanine DiD and its autoquenching by resonant energy transfer can be used to distinguish the fusion state of a lipid bilayer at cryo-temperatures. By adapting an established fusion assay to work under cryo-CLEM conditions, we identified areas of fusion between influenza virus-like particles and fluorescently labeled lipid vesicles on a cryo-EM grid. This result demonstrates that cryo-CLEM can be used to localize functions in addition to tagged proteins, and that fluorescence autoquenching by resonant energy transfer can be incorporated successfully into cryo-CLEM approaches. In the case of membrane fusion applications, this method provides both an orthogonal confirmation of functional state independent of the morphological description from cryo-EM and a way to bridge room-temperature kinetic assays and the cryo-EM images.
Keywords: CLEM, FRET, resonant energy transfer, membrane fusion, cryo-EM
Introduction
Correlated light and electron microscopy (CLEM) provides its users both the specificity of fluorescence microscopy and the resolution of electron microscopy (EM), and has now become an established technique with many variations (Thomas Muller-Reichert & Paul Verkade, 2017). Early applications typically involved room-temperature light microscopy on unfixed samples to observe event progression, quickly followed by resin embedding and evaluation of the sample by EM (Bright, Gratian, & Luzio, 2005). More recently, samples are often chemically fixed or frozen prior to visualization by light microscopy. Room-temperature applications tolerate better fluorescence optics but limit the EM to resin-embedded samples; conversely, cryo-temperature applications allow the full power of cryo-EM but at the expense of the numerical aperture of the fluorescence objective (Kaufmann, Hagen, & Grünewald, 2014). CLEM has been widely applied to study membrane remodeling and virus-host cell interactions (Kopek et al., 2012; Kukulski et al., 2012a; Lebrun et al., 2014; Martinez et al., 2014; Li et al., 2014; Romero-Brey et al., 2015; Bykov et al., 2016). In these cases, the object of interest is too small or rare to be easily found without relying on fluorescence microscopy, while the structure is smaller than the point spread function of the light microscope and must therefore rely on EM for its characterization.
Despite its optical drawbacks, interest and applications for cryo-fluorescence microscopy (cryo-FM) are growing due to its potential to correlate with cryo-EM; the advantages of vitrification as a means of rapid, chemical-free sample fixation; and the possibility of reduced photobleaching at low temperatures (Schwartz et al., 2007; Sartori et al., 2007; Wolff et al., 2016). One area where cryo-fluorescence is still lacking is the expansion to applications that draw on unique dye photophysics such as Förster Resonance Energy Transfer (FRET) and stochastic photobleaching. Such approaches are rarely used at cryo-temperatures due to optical limitations and potential temperature dependence in the behavior of the dyes. This has functionally limited cryo-CLEM to being a method for targeting a fluorophore-tagged molecule for cryo-EM imaging.
During a reaction or process in which no protein is added or lost, localization-based methods will not give a distinguishable signal change in cryo-CLEM. Such processes include lipid membrane events like fusion and fission. In a process like endocytosis individual proteins can be tagged to organize images in time (Kukulski et al., 2012a). In contrast, fluorescence co-localization of two fluorophores in apposing membranes cannot be used to determine whether two membranes in tight apposition are exchanging lipids or not, or exactly when a hemifusion stalk becomes a fusion pore. In such cases a chemical reporter is necessary to define the point at which reactions begin (Blumenthal et al., 2002; Kreye, Malsam, & Söllner, 2008; Floyd et al., 2008).
In this study, we have adapted a standard lipid-mixing assay to cryo-CLEM, employing an application of autoquenching based on resonant energy transfer (RET). By doing so, we apply cryo-CLEM to localize a functional event rather than a protein, drawing upon the expanded fluorophore uses that have been mostly neglected in cryo-CLEM applications. We use the model system of influenza virus fusion, which has been well-studied separately by fluorescence kinetics (Floyd et al., 2008) and cryo-EM and tomography (Gui et al., 2016; Chlanda et al., 2016). The use of the cryo-fluorescence signal to localize a function allows distinction between apposition (lipid unmixed) and hemifusion or fusion (lipid mixed), allowing targeted imaging of fusion events and permitting more accurate assignment of micrographs into their appropriate functional state.
Materials and Methods
Fluorophore Stock Preparation
Octadecyl Rhodamine B Chloride (R18, Thermo Fisher Scientific) was solubilized in 100% ethanol at roughly 10 mg/mL and stored at -80 C in glass vials sealed with Teflon tape for up to 1 month. DiIC18(5) solid (DiD, 1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindodicarbocyanine, 4-Chlorobenzenesulfonate Salt, Thermo Fisher Scientific) was solubilized in 100% ethanol at roughly 5 mM and stored at -20 C in glass vials sealed with Teflon tape. The exact concentration was determined by measuring absorbance in methanol (1 μL stock in 1.5 mL methanol), using the manufacturer’s extinction coefficient under these conditions.
Vesicle Preparation
POPC vesicles (Figures 1-2)
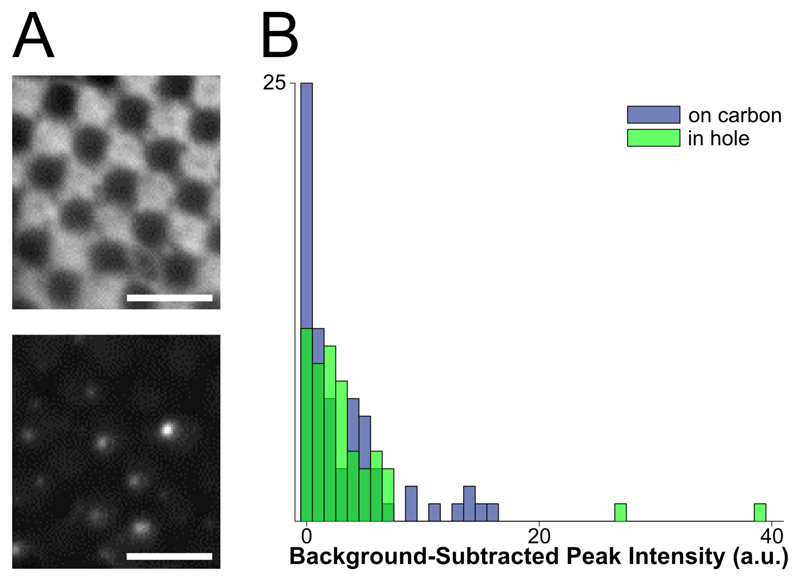
Figure 1. Assessment of fluorophores for labelling vesicles on cryo-EM grids.
A: top: cryo-FM image of POPC vesicles labeled with 4% R18, showing a lawn of fluorescence intensity staining the carbon of the EM grid. Bottom: cryo-FM image of POPC vesicles labeled with 1% DiD, displaying punctate behavior. Scale bars = 5 μm. B: quantification of intensities for clearly separated vesicles labelled with 5% DiD, sorted by location in holes or on carbon.
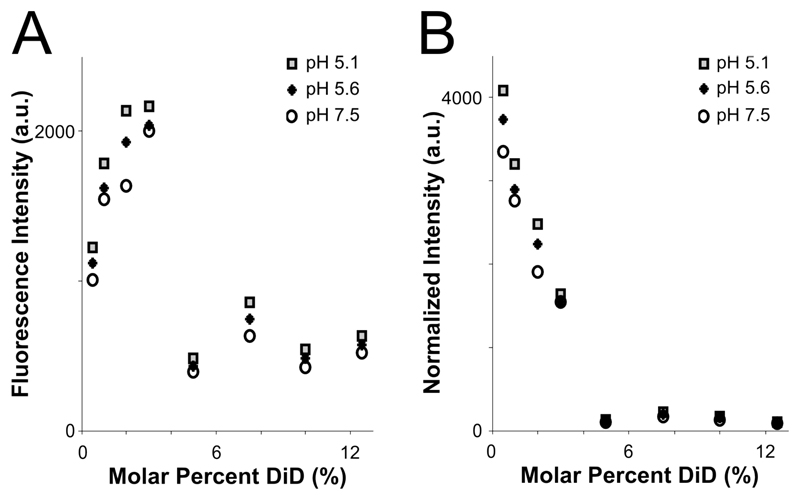
Figure 2. Fluorophore quenching at room temperature.
A: Fluorescence intensity of vesicles with varying lipid concentration, measured in a plate reader. Even as the total fluorophore concentration of the sample increases, autoquenching at 5% DiD results in lower total fluorescence intensity. B: Results from A, normalized to equal fluorophore concentration (varying total lipid concentration). Autoquenching is linear from 0.5-5% DiD, at which point the dye is fully quenched.
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (Avanti Lipids) was dissolved in chloroform at 1 mg/mL concentration, then dried into a thin film in a round-bottom flask by evaporation using nitrogen gas. This thin film rested overnight in a room-temperature dessicator. Following evaporation, the POPC film was incubated in TNE buffer (50 mM Tris-HCL pH 7.4, 100 mM NaCl, 0.1 mM EDTA) for 30 minutes at 4 C, then subjected to 7 cycles of freeze/thaw by alternating baths in an acetone/dry ice mixture and warm water. The resulting lipid mixture was extruded for 21 passes through a membrane with a pore size 50-200 nm depending upon the experiment (Avanti Lipids). Average vesicle size and quality were confirmed by dynamic light scattering (Zetasizer, Malvern Panalytical).
For Figure 1B, a mixture of 90% POPC / 10% POPS was used to decrease the clumping tendency of fully POPC vesicles, allowing better quantification of fluorescence intensity from separated vesicles on the EM grid.
For DiD-labeled vesicles, the desired amount of fluorophore was incorporated into the chloroform/methanol mixture immediately prior to drying. For R18-labeled vesicles, fluorophore was added to a glass autosampler vial with the extruded vesicle mixture and stirred for 30 minutes at room temperature. Following labeling, aggregates and any unincorporated fluorophore were removed by passing vesicles through two consecutive 0.5 mL 7K MWCO Pierce Zeba columns, following manufacturer’s protocols.
Lipid vesicles for hemifusion experiments (Figures 3-5)
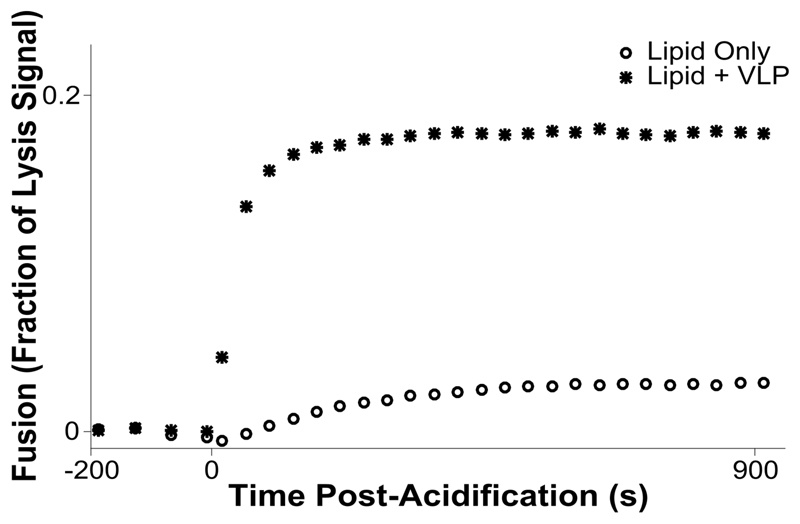
Figure 3. Kinetic assay for hemifusion between influenza VLPs and 5% DiD liposomes.
Vesicles and VLPs are incubated in a 37 C plate reader, and fusion is stimulated by acidification to pH 5.1 at time = 0 s. The acidification results in hemifusion between VLPs and liposomes, dequenching the dye as it is distributed into a larger lipid pool.
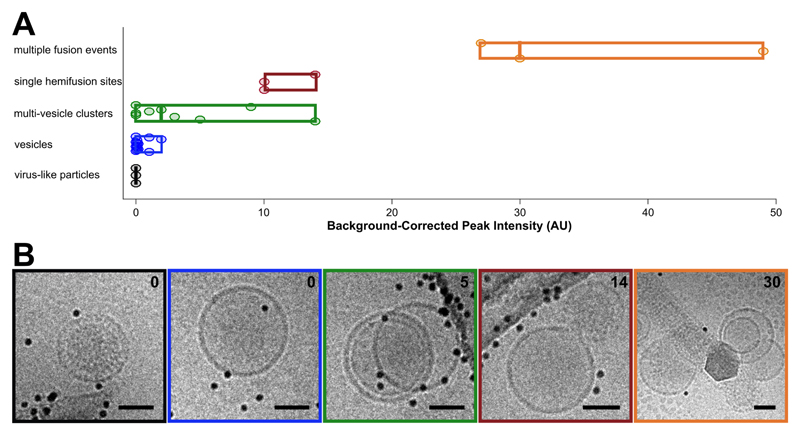
Figure 5. Categorization of events by cryo-CLEM.
A: Fluorescence intensities for vesicles and virus-like particles. Events are classified visually. Circular points are single events, boxes display ranges per category, and mid-lines represent the median value for that category. B: Examples of events for each of the classes in A. Box colors correspond to category colors in A, and baseline-subtracted peak intensities are reported in the upper right hand corner of each image. Scale bars = 50 nm.
The lipid composition for these vesicles was adapted from (Chlanda et al., 2016). A mixture of 47% POPC / 13% POPE / 35% cholesterol / 5% total ganglioside (Avanti Lipids) was prepared in a mixture of roughly 1 mg/mL in 2 parts chloroform / 1 part methanol; 5% DiD was then added to this mixture prior to drying into a thin film overnight as above. Lipid vesicles were prepared by extrusion as above, using a buffer of 10 mM HEPES / 50 mM sodium citrate / 150 mM NaCl at pH 7.5 (Gui et al., 2016) and a 100 nm pore size. Vesicle preparations were filtered by spin column as above. Vesicle concentration was estimated by DiD absorbance at 645 nm, assuming the 5% molar composition.
Vesicle Size Assessment
We prepared a comparison of vesicle sizes by DLS on a Zetasizer (Malvern Panalytical). Briefly, 100 μL of an unlabeled vesicle preparation (nominally 1 mg/mL) were measured for 3 sets of 13 reads. By this method, a 100 nm membrane produced vesicles with a mean diameter of 163.4 nm, with polydispersity of 0.144 (roughly 62 nm standard deviation).
We then imaged this vesicle preparation on a cryo-EM grid. A mixture of 1.5 μL vesicle preparation and 1.5 μL 10 nm gold fiducial marker was pipetted onto a C-Flat 2/2-300C EM grid, blotted in a Vitrobot (Mark 2, offset -4, 3 s blot time, >85% humidity, 15 C), and plunged into liquid ethane. The resulting grid was imaged on a FEI Tecnai T12 microscope at 1.25 nm pixel size (9800x microscope magnification setting), and vesicle sizes were measured using Amira software. We found that the vesicles on the grid had a mean diameter of 88 ± 36 nm.
We believe that this marked difference in vesicle sizes is due to systematic biases in both instruments. Briefly, DLS can over-estimate a vesicle diameter based on charge and hydration shells, and any transient vesicle-vesicle association might also result in larger vesicle sizes without severely increased polydispersity. On the other hand, we expect that large lipid vesicles are selected against during cryo-EM imaging, possibly due to increased contact with the blotting paper, greater exposure to the air-water interface, or preferential localization to areas of thick ice.
Buffer Titration for pH Control
We chose a HEPES-citrate dual-buffer system (10 mM HEPES / 50 mM sodium citrate / 150 mM NaCl) to control pH over a greater range (Gui et al., 2016). pH is adjusted by addition of acidic buffer to the neutral buffer containing the sample, thus avoiding osmotic stress. A titration curve was measured using a pH electrode, and was found to be linear between pH 4.6 – 6.1. For these experiments, buffers at pH 7.5 and pH 3 were used. The linear regression variables are an intercept of pH 6.4 (due to non-linearity outside the 10-50% range) and a slope of -0.036 pH units per percent pH 3 buffer.
Virus-Like Particle (VLP) Preparation
VLPs were prepared by transfection of influenza A proteins into HEK cells by adapting a previously described protocol (Chlanda et al., 2015). PCAGGS plasmids for influenza A hemagglutinin (A/Hong Kong/1/68, HA), neuraminidase (A/Singapore/1/57, NA), and matrix proteins 1 (A/Hong Kong/1/68, M1) and 2 (A/Hong Kong/1/68, M2) were transfected into 1*106 HEK 293T cells in serum-free DMEM, with 2.2 μg HA, 2.2 μg NA, 4.4 μg M1, and 1.1 μg M2 DNA per plate and using 30 μL FuGene according to manufacturer’s instructions (Promega Corporation). Following transfection, a final concentration of 100 mU/μL exogenous NA (New England BioLabs, P0720S) was added to the plates to facilitate VLP release.
After 48 hours, the VLPs were treated with trypsin to cleave the HA subunits (Chlanda et al., 2016). Inhibited trypsin and cellular debris were removed by centrifugation at 900 xG for 10 minutes; VLPs were then harvested by centrifugation over a 32.5% sucrose cushion (SW40i rotor, 30k RPM, 90 minutes). VLP pellets were overlaid with citrate-HEPES buffer (above), allowed to rest overnight, and then resuspended gently with wide-mouthed pipet tips. VLP preparations were cleaned of excess sucrose and soluble proteins by a 4-hour dialysis against citrate-HEPES buffer using a 300 MWCO membrane, and gently concentrated in Microcon 100 MWCO concentrators (EMD Millipore) using a rotor speed of no more than 900 xG. VLPs prepared in this manner retained their mixed morphology (spheres and long filaments).
VLP concentrations were approximated by monitoring accessible surface protein concentration using a Bio-Rad Protein Assay according to manufacturer’s instructions.
Room-Temperature Fluorophore Controls
To assess the quenching behavior of DiD, labeled 1 mg/mL POPC vesicle preparations with varying molar percentages of DiD: 0.5, 1, 2, 3, 5, 7.5, 10, and 12.5 mol%, and used buffers at pH from 5.1-7.5 (the pH range relevant for endosomal escape). Vesicle preparations were mixed 1:1 with citrate-HEPES buffer and measured in a Synergy HT plate reader to monitor total brightness relative to fluorophore content (BioTek, monochromator excitation 644 nm, emission 670 nm, background correction 500 nm).
Fusion Assay
All VLP preparations were assayed for fusion activity on a BioTek Synergy H1 plate reader. In all fusion experiments, fluorophore was incorporated into the liposomes due to the highly polymorphic nature of VLPs from the filamentous virus strain used here, but the protocol could be adapted to label homogeneous influenza particles. 80 μL fusion mix was prepared using 50 μM target membrane lipid, pH 7.5 HEPES-citrate buffer, and a volume of VLPs corresponding to 0.014 mg accessible protein. The mix was prepared in the wells of a 96-well plate, which was then loaded immediately into the plate reader. After a five-minute incubation at 37 C, the mix was acidified to pH 5.1 by injection of 45 μL of pH 3 HEPES-citrate buffer. The plate was shaken for 15 s to mix, followed by reads every 30 s for 20 minutes (620/40 excitation, 680/30 emission, gain 35). Finally, the plate was removed and 0.5% Triton X-100 was added to the wells to lyse the vesicles and VLPs. Assays were normalized for total DiD concentration by subtracting the pre-acidification intensity and dividing by the intensity following lysis with 0.5% Triton X-100, which corresponds to complete dequenching.
Cryo-EM/cryo-CLEM Grid Preparation
For the quenched lipid-only grid (Figure 1B), fluorescent signals were of roughly even intensity across the grid, making fiducial-free alignments difficult as vesicles appeared identical to each other by both EM and FM. We therefore used fluorescent fiducial markers to assist with high-precision correlation (Kukulski et al., 2012b; Schorb et al., 2017). Briefly, a purchased stock solution of 50 nm Tetraspec beads was sonicated for 5 minutes in a waterbath sonicator, and Protochip C-Flat grids (2/2, 300 mesh) were glow-discharged for 30 s at ~30 mA. The tetraspec solution was diluted 1:50 in PBS, and 10 μL drops were incubated on the grids for 15 minutes. Grids were back-side blotted, rinsed with two washes of deionized water, and allowed to dry prior to use.
All cryo-EM/cryo-CLEM grids were frozen using the protocol from the Vesicle Size Assessment section above.
For hemifusion grids, vesicles and VLPs were prepared as above, mixed, and incubated on ice at relative concentrations of 0.16 mg/mL accessible viral protein and 0.12 mM target membrane lipid for roughly 30 minutes. This provided ample time for VLPs and vesicles to adhere. The tubes were then brought to room temperature and acidified to pH 5.1 for 60 s, followed by addition of 10 nm Protein A-coated gold fiducial markers (Cell Microscopy core, University Medical Center Utrecht). The sample was then pipetted onto grids and frozen as above. The total time between acidification and freezing was roughly 2.5 minutes, in keeping with the kinetics of the hemifusion plateau at room temperature.
Cryo-Fluorescence Microscopy and Intensity Analysis
Cryo-fluorescence microscopy was performed on Leica DM 1200 or DM6 FS cryo-CLEM microscopes running LAS X software and an OrcaFlash 4.0 V2 SCMOS camera (Hamamatsu Photonics). Excitation intensities were adjusted as needed to avoid devitrification from extended high-intensity illumination; exposure times were chosen to optimize dynamic range without saturating the camera. Typically, white transmitted light exposures were 80-100 ms, green channel imaging (480/40, 505, 527/30 nm excitation, dichroic, emission filters, Tetraspecs) used 30% intensity with aperture 4 and exposure time of 0.8-1 s, red channel imaging (560/40, 585, 630/76 nm excitation, dichroic, emission filters, R18) used 17% intensity for 25 ms, and far-red channel imaging (620/60, 660, 700/75 nm excitation, dichroic, emission filters, DiD) used 17% intensity with aperture 4 and exposure times of 14 ms (hemifusion, Figure 4) or 150 ms (quenched vesicles, Figure 1B). Grids were focus-mapped using built-in software functions, and imaged in Z-stacks of 10-12 slices and ~1 μm step sizes.
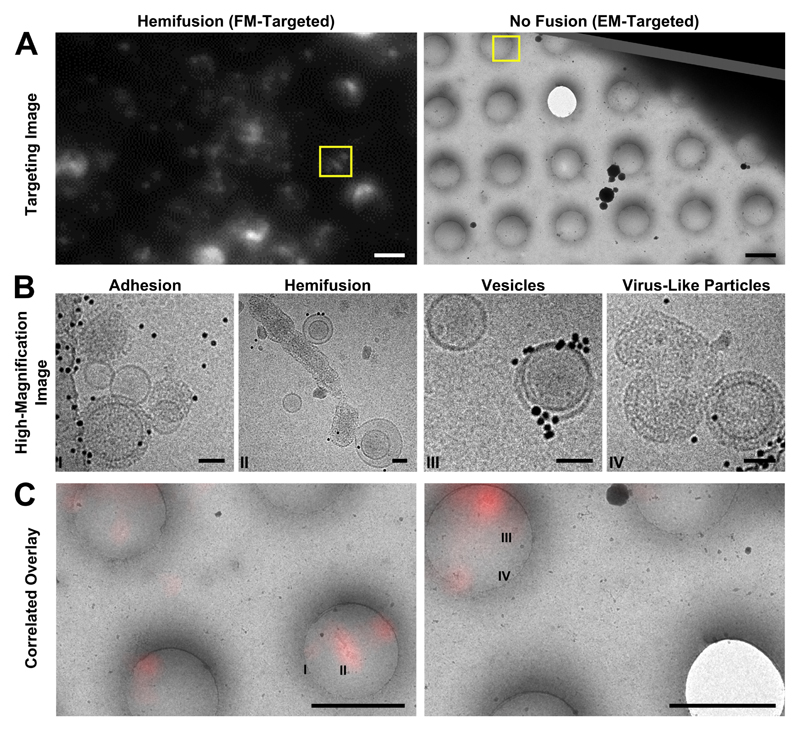
Figure 4. Cryo-CLEM images of different types of on-grid events and their corresponding fluorescence signals.
A: Targeting images are used to define points of interest (yellow boxes). Left: fluorescence-based targeting of potential hemifusion sites. Right: EM-based targeting of VLPs and vesicles irrespective of fluorescence. Scale bars = 2 μm. B: High-magnification micrographs are taken within the targeted areas. Scale bars = 50 nm. C: During post-processing, fluorescence and EM maps are correlated with greater precision, and fluorescence intensities are quantified for all imaging locations. Roman numerals correspond to the high-magnification images in B. Scale bars = 2 μm.
Images of grid squares of interest were built using FIJI’s built-in Max Intensity projection algorithm (Schindelin et al., 2012; Schneider, Rasband, & Eliceiri, 2012). Full-grid maps for correlating on-the-fly were generated with the Fiji Stack Focuser plug-in.
Spot intensities were quantified in FIJI using the built-in 3D Surface Plot feature. Briefly, the maximum peak intensity was recorded for each location, searching within the area of accuracy predicted by the correlation software and informed by visible chromatic offsets in the cryo-CLEM. The local background was determined by the plateau nearest the peak in cases where multiple peaks were present; otherwise the local background was determined by averaging along the edge of the visualization box (extended to reach a plateau when necessary), using the same surface as the point of interest. Points of interest that were near to contamination (ice) or areas of bright fluorescence without lipid/protein density (from fluorescent crystalline ice or suspected reflections) were avoided. Reported peak intensities are of the background-subtracted maximum of the peak center.
Cryo-Electron Microscopy and Correlation with Cryo-Fluorescence Microscopy
Cryo-EM grids were imaged on an FEI Tecnai T12 microscope (hemifusion) or FEI Tecnai F20 microscope (quenched DiD vesicles) running SerialEM. Grids were mapped in full at 150X nominal magnification, which was correlated with the white transmitted light cryo-FM map using SerialEM map registration functions (Schorb et al., 2017).
From this point, workflows followed different paths depending upon the goal of the experiment. For quenched lipid grids where each vesicle’s intensity needed to be quantified, it was necessary to image at a magnification where each individual vesicle would be visible irrespective of fluorescence. Therefore, grid squares of interest were imaged by a 5x5 montage at 6000X nominal magnification on the F20 microscope, and all alignment with cryo-fluorescence images was performed during post-processing using Matlab cryo-CLEM correlation scripts (Kukulski et al., 2012b).
For hemifusion grids, we sought to both prepare maps for later correlation and to image bright and dim spots at higher magnification. Grid squares of interest were mapped at 1200X nominal magnification and aligned on-the-fly at the T12 microscope using SerialEM registration points based on visible grid square features and defects. This rapid, fiducial-free approach provided an estimated 0.2-0.3 μm precision for the correlation, which was sufficient for the hemifusion sample. (While higher magnification and alignment with fiducials would provide more precise correlation, this is time-consuming and was found to be unnecessary for this sample.) The fluorescence map location was used to direct the stage to the expected position, at which point a 5000X nominal magnification image was used to center the object closest to the expected position. The image was then collected with 30,000X nominal magnification (0.37 nm pixel size, ~10 e/A2, -5 μm defocus). Matlab cryo-CLEM scripts were again used in post-processing to correlate all remaining points and confirm that the higher-magnification images truly corresponded to the fluorescence they were targeting. Final correlation accuracy varied by square and ranged from roughly 60-200 nm.
Results
Fluorophore selection criteria and controls
Assays that monitor membrane fusion are usually based on tracking lipid mixing, content release, or both. In lipid mixing assays, a fluorophore is typically embedded in one membrane; upon fusion, the fluorophore is diluted in the expanded membrane and gives a fluorescence signal, typically dequenching. Quenching can occur by multiple mechanisms, including collision (dynamic quenching), formation of a dark complex (static quenching), or resonant energy transfer (RET), and can be a product of contact with the fluorophore itself (autoquenching) or with other small molecules (Joseph Lakowicz, 2006). While room-temperature, solution-based applications can draw from a wide variety of molecules and mechanisms, cryo-FM places specific demands on fluorophores and their photophysical behavior.
There are many considerations for choosing a fluorophore for cryo-CLEM applications. At cryo-temperatures (-180 to -135 C), macromolecular diffusion will not occur. The cryo-EM grid has an unprotected surface that can nonspecifically bind or activate fluorophores, increasing background signal. Cryo-temperatures have strong effects on fluorophores: excitation and emission spectra can shift or change in width (Moerner & Orrit, 1999; Creemers et al., 2000); fluorophores can spend large portions of time trapped in the triplet state (Zondervan et al., 2003; Kaufmann et al., 2014); and some photo-switchable fluorophores fail to work as expected or experience decreased quantum yields (Faro et al., 2010; Chang et al., 2014). Finally, due to the optical and experimental limitations of cryo-FM, the labeled sample is visualized with a low-numerical aperture objective and a low enough illumination intensity to avoid devitrification.
Given these constraints, the fluorophores with the best potential for the study of membrane fusion by cryo-CLEM would autoquench, and could be embedded in either target or fusogen-containing membrane. The signal must be bright, and the change upon lipid mixing should be strong enough to be visible without extensive processing (a near-binary dark-bright transition). Based on these criteria, we selected two dyes for testing: octadecyl rhodamine B (R18) and the dialkylcarbocanine DiD. Fluorophores were loaded into a 1 mg/mL preparation of extruded POPC vesicles at concentrations below those required for full quenching, plunge-frozen onto EM grids, and imaged in a Leica cryoCLEM system. R18 was tested in 100 nm POPC vesicles at a 4% concentration. In these grids, the R18 appears to have partitioned onto the carbon support layer of the EM grid and entered a high-fluorescence state at that location, leading to an unsuitably high background (Figure 1A, top). In contrast to the R18, cryo-FM images of 200 nm POPC vesicles containing 1% DiD show fluorescent punctae scattered over the carbon and holes (Figure 1A, bottom).
After confirming that DiD labeling produced punctate behavior in cryo-FM, we sought to determine the intensity change that could be expected from DiD dequenching upon lipid mixing. Room-temperature fusion assays are often baseline-corrected and normalized, allowing the use of fluorophores with modest intensity changes; however, such processing is error-prone in cryo-FM due to intensity variability by ice thickness, stray reflections, and a lack of negative and positive control areas within the imaging square for establishing baselines. For cryo-CLEM, therefore, both a dark base state and a large intensity change upon fusion are required.
To assess the quenching behavior of DiD, we prepared POPC vesicles labeled with varying molar percentages of DiD, and measured fluorescence emission under steady excitation in a room-temperature plate reader (see Methods). The total fluorescence of the vesicle samples rose from 0.5% to 3% DiD, then dropped to a lower intensity plateau for concentrations 5% -12.5% with little fluorescence gain from fluorophore added past 5% (Figure 2A). Importantly, the total fluorescence in the cuvette was lower for the higher-DiD concentrations, where the sample is quenched, than for the dequenched samples, even when the total amount of fluorophore was an order of magnitude larger. We conclude that if the molar percentage of DiD is above roughly 5%, then variation in total fluorophore amount due to heterogeneity in vesicle or virion sizes will be small in comparison with the large intensity shifts that would occur if the dye dequenches due to dilution upon fusion with a target membrane (Figure 2). However, intensities in post-fusion states could vary more widely based on the initial fluorophore amount.
When measurements were normalized according to total fluorophore concentration (as determined by absorbance at 645 nm), a linear decrease in fluorophore brightness from 0.5% to 5% was visible, with a dark plateau at concentrations 5% and above, and a total intensity difference of roughly 10-fold between 5% and 0.5% DiD (Figure 2B). The effect of pH was minimal compared to that of concentration (Figure 2). In the context of a fusion sample, fusion between a 5% DiD vesicle and a virion of similar size, both smaller than the point-spread function of the cryo-FM, would result in a roughly 10-fold increase in fluorescence at that location (Figure 2B). Other size relationships would be dependent upon both the dilution factor of the DiD upon fusion according to target surface area, and distribution of fluorescence intensity over the imaging area of the fused membranes.
We next assessed the consistency of small, quenched DiD vesicles that would be appropriate for fusion assays. A 1 mg/mL POPC/POPS suspension was labeled with 5% DiD, extruded through 50 nm pores, and plunge-frozen onto grids containing 50 nm tetraspec beads. Grids were imaged in cryo-FM as above. Grid squares of interest were then imaged on an FEI Tecnai F20 electron microscope, correlated with the cryo-FM images (Kukulski et al., 2012b), and fluorescence pixel intensities were quantified for each lipid vesicle within the squares of interest (Figure 1B).
These cryo-CLEM results confirmed that the fluorescence intensity of the quenched DiD liposomes is consistently low, and is not affected by adhesion to the carbon grid surface (Figure 1B). No intensity corrections were made for vesicle size variation or multiple lamellae, neither of which can be assessed prior to using the cryo-FM image for targeting EM imaging. In this context, the narrow intensity distribution in Figure 1B shows that variation in the total amount of fluorophore present in a spot does not result in strong signal heterogeneity in the quenched state, consistent with full quenching occurring at the same fluorophore concentration as at room-temperature (Figure 2).
Localizing membrane fusion and hemifusion events in cryo-CLEM
We elected to use influenza virus-like particles (VLPs) to induce fusion; the influenza fusion mechanism is well-studied and VLP samples are well-suited for the concentration and thickness constraints of EM. VLPs were prepared by transfecting HEK 293T cells with plasmids containing the influenza A proteins hemagglutinin, M1, M2, and neuraminidase as in (Chlanda et al., 2015). VLPs were harvested by centrifugation through a sucrose cushion, after which their mixed morphology (filaments and spheres) was maintained, and assessed for fusogenicity (Figure 3). Target vesicles were prepared by extruding a 1 mg/mL suspension of 44.7% POPC, 12.3% POPE, 33.2% cholesterol, 4.8% total ganglioside, and 5% DiD in HEPES-citrate buffer through 100 nm pores (membrane composition adapted from (Chlanda et al., 2016)).
We prepared EM grids to assess fusion localization by cryo-CLEM. VLPs and quenched-DiD vesicles were incubated on ice at relative concentrations of 0.16 mg/mL accessible viral protein and 0.12 mM target membrane lipid to allow VLP-vesicle adhesion. The tube contents were then acidified at pH 5.1 for 60 s at room temperature, followed by addition of 10 nm gold fiducial markers and plunge-freezing in liquid ethane. The total time between acidification and freezing was roughly 2.5 minutes, at which time the lipid mixing reaches a plateau at room temperature (Figure 3).
The EM grids were then subjected to cryo-FM, following the same workflow as with the control DiD vesicle sample above (Figure 1B). Cryo-FM imaging was performed using a ten-fold lower exposure time appropriate for the high-intensity, dequenched punctae; we therefore expected a very low signal from the quenched vesicles. Brightfield and fluorescence maps of the EM grid were correlated with EM micrographs applying established SerialEM-based correlation protocols during data collection on an FEI Tecnai T12 microscope (Mastronarde, 2005; Schorb et al., 2017).
Selected grid squares were imaged under low-dose, low-magnification conditions at which VLPs and vesicles could be identified (3500-6000X nominal magnification), allowing later evaluation of the full grid square by correlating the lower-resolution cryo-FM and cryo-EM images in post-processing (Kukulski et al., 2012b). Because we expected lipid mixing during fusion to dequench the DiD and yield bright cryo-FM signals, we targeted bright- and medium-intensity punctae for higher-magnification EM imaging using a SerialEM-based correlation protocol at the cryo-EM (Figure 4A-B) (Schorb et al., 2017). This combination of approaches allows targeted high-resolution acquisitions at spots of interest as well as lower-resolution evaluation of both targeted and untargeted areas during post-processing (Figure 4A-C).
After examining the overlay of fluorescence signal with the electron micrographs, we observed that the brightest spots on the grid corresponded to aggregates or clusters of VLPs and vesicles with multiple fusion sites (Figure 5A). The intermediate intensity spots typically contained putative fusion events, as well as the most intense quartile of vesicle clusters (Figure 5). Importantly, none of the intermediate or bright fluorescence events correlated to unfused VLPs or solitary vesicles, indicating that the DiD remains quenched in the unfused vesicles and that there is no transfer of dye outside of active membrane fusion (Figure 4B-C, Figure 5). Unfused VLPs and solitary vesicles gave very low fluorescent signals (Figure 5A). We note that it may not be appropriate to interpret the relative numbers of fused and unfused samples from an on-grid experiment, since fused and unfused samples could be differently affected by blotting conditions; however, relative numbers could be compared between parallel experiments. Considering all trends together, if all bright and intermediate fluorescence events were targeted, all hemifusion and fusion sites should be contained in the EM dataset, along with a small population of cluster events that can be screened following acquisition.
Discussion and Future Prospects
In this study, we demonstrated that the established DiD-based fusion assay (Rust et al., 2004; Gui et al., 2016) can be incorporated into a cryo-CLEM workflow to specifically locate, target and image membrane fusion sites. By using the same fluorescence-based lipid mixing indicator as room-temperature kinetic assays (Figure 3), this method also provides a way to link the EM-based ultrastructural imaging with orthogonal kinetic and functional information. Furthermore, our use of DiD, which employs an RET-based autoquenching mechanism (Benchimol et al., 2013), confirms that RET-based fluorescence applications can function at cryo-temperatures under favorable conditions. Of these applications, autoquenching is particularly well-suited for cryo-CLEM because the use of a single fluorophore decreases the likelihood of spectral shifts strongly affecting the overlap integral for RET. RET applications with many dye molecules present, such as membrane fusion, are also optimal: the number of fluorophores is likely sufficient to allow the random orientation of the fluorophore dipoles to approximate the tumbling seen at room temperature, and fluorescent emission will be strong despite cryo-temperature limitations such as extended triplet state occupation, gentle illumination intensity or low numerical aperture.
A lipid mixing indicator has many applications. One natural extension of this method will be to load DiD into viruses to locate rare fusion events within eukaryotic endosomes by cryo-CLEM. DiD is known to incorporate readily into influenza virus membranes and experience concentration-dependent autoquenching at room temperature, and fusion inside room-temperature eukaryotic cells has already been tracked using this method (Lakadamyali et al., 2003; Rust et al., 2004; Gui et al., 2016). In this case, detection of lipid mixing would offer a clear advantage over traditional colocalization approaches. If a virus is contained within an endosome, the virus and endosome would give a positive colocalization signal independent of fusion state, while lipid mixing would be specific to hemifusion and fusion.
For the study of membrane fusion in vitro, our cryo-CLEM method also provides an orthogonal signal for lipid mixing, which cannot always be visually distinguished from membrane apposition in cryo-EM images. Furthermore, the use of fluorescence dequenching to study influenza fusion allows correlation between an image and previous kinetics studies based on similar fluorescence signals (Floyd et al., 2008), ensuring that the definition of fusion state is consistent between the kinetic and structural methods.
In conclusion, we have developed a method for localizing a specific function on a cryo-EM grid, using an application of RET in cryo-CLEM. This development allows localization of lipid mixing rather than a protein of interest in cryo-FM, which can be used either to target this site for cryo-EM imaging, or to provide an orthogonal evaluation of the fusion state.
Acknowledgements
This research was technically supported by the Electron Microscopy Core Facility and the Protein Expression and Purification Core Facility at the European Molecular Biology Laboratory (EMBL, Heidelberg), and the EM facility at the MRC Laboratory of Molecular Biology. We thank M. Schorb, W. Hagen, W. Kukulski, E. Lemke, G. Paci, O. Avinoam, and Y. Bykov for helpful discussions and technical assistance with the Leica cryo-CLEM system, and M. Clarke for assistance with VLP preparations. This project was funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC-2014-CoG 648432 – MEMBRANEFUSION) to JAGB. JAGB is an inventor on filed patents that have been licensed by Leica Microsystems and commercialized in the Leica cryo-CLEM system used in this study.
Footnotes
Author Contributions
L.A.M. performed the research and data analysis. L.A.M. and J.A.G.B. designed research, interpreted results and wrote the manuscript.
References
- Muller-Reichert Thomas, Verkade Paul., editors. Correlative Light and Electron Microscopy III. 2017. [Google Scholar]
- Benchimol MJ, Hsu MJ, Schutt CE, Hall DJ, Mattrey RF, Esener SC. Phospholipid/Carbocyanine Dye-Shelled Microbubbles as Ultrasound-Modulated Fluorescent Contrast Agents. Soft Matter. 2013;9:2384–2388. doi: 10.1039/C2SM26900G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R, Gallo SA, Viard M, Raviv Y, Puri A. Fluorescent lipid probes in the study of viral membrane fusion. Chemistry and Physics of Lipids. 2002;116:39–55. doi: 10.1016/s0009-3084(02)00019-1. [DOI] [PubMed] [Google Scholar]
- Bright NA, Gratian MJ, Luzio JP. Endocytic Delivery to Lysosomes Mediated by Concurrent Fusion and Kissing Events in Living Cells. Current Biology. 2005;15:360–365. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Bykov YS, Cortese M, Briggs JAG, Bartenschlager R. Correlative light and electron microscopy methods for the study of virus–cell interactions. FEBS Letters. 2016;590:1877–1895. doi: 10.1002/1873-3468.12153. [DOI] [PubMed] [Google Scholar]
- Chang Y-W, Chen S, Tocheva EI, Treuner-Lange A, Löbach S, Søgaard-Andersen L, Jensen GJ. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nature Methods. 2014;11:737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlanda P, Mekhedov E, Waters H, Schwartz CL, Fischer ER, Ryham RJ, Cohen FS, Blank PS, Zimmerberg J. The hemifusion structure induced by Influenza virus haemagglutinin is determined by physical properties of the target membranes. Nature microbiology. 2016;1 doi: 10.1038/nmicrobiol.2016.50. 16050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlanda P, Schraidt O, Kummer S, Riches J, Oberwinkler H, Prinz S, Kräusslich H-G, Briggs JAG. Structural Analysis of the Roles of Influenza A Virus Membrane-Associated Proteins in Assembly and Morphology. Journal of Virology. 2015;89:8957–8966. doi: 10.1128/JVI.00592-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers TMH, Lock AJ, Subramaniam V, Jovin TM, Völker S. Photophysics and optical switching in green fluorescent protein mutants. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2974–2978. doi: 10.1073/pnas.050365997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro AR, Adam V, Carpentier P, Darnault C, Bourgeois D, de Rosny E. Low-temperature switching by photoinduced protonation in photochromic fluorescent proteins. Photochemical & Photobiological Sciences. 2010;9:254–262. doi: 10.1039/b9pp00121b. [DOI] [PubMed] [Google Scholar]
- Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proceedings of the National Academy of Sciences. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Ebner JL, Mileant A, Williams JA, Lee KK. Visualization and Sequencing of Membrane Remodeling Leading to Influenza Virus Fusion. Journal of Virology. 2016;90:6948–6962. doi: 10.1128/JVI.00240-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz Joseph. Principles of Fluorescence SpectroscopyIII. Springer; 2006. [Google Scholar]
- Kaufmann R, Hagen C, Grünewald K. Fluorescence cryo-microscopy: current challenges and prospects. Current Opinion in Chemical Biology. 2014;20:86–91. doi: 10.1016/j.cbpa.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6136–6141. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye S, Malsam J, Söllner TH. In vitro assays to measure SNARE-mediated vesicle fusion. Methods in Molecular Biology (Clifton, N.J.) 2008;440:37–50. doi: 10.1007/978-1-59745-178-9_3. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Kaksonen M, Briggs JAG. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012a;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JAG. Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods in Cell Biology. 2012b;111:235–257. doi: 10.1016/B978-0-12-416026-2.00013-3. [DOI] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun M, Thelen N, Thiry M, Riva L, Ote I, Condé C, Vandevenne P, Di Valentin E, Bontems S, Sadzot-Delvaux C. Varicella-zoster virus induces the formation of dynamic nuclear capsid aggregates. Virology. 2014;454–455:311–327. doi: 10.1016/j.virol.2014.02.023. [DOI] [PubMed] [Google Scholar]
- Li F, Sewald X, Jin J, Sherer NM, Mothes W. Murine Leukemia Virus Gag Localizes to the Uropod of Migrating Primary Lymphocytes. Journal of Virology. 2014;88:10541–10555. doi: 10.1128/JVI.01104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MG, Snapp E-L, Perumal GS, Macaluso FP, Kielian M. Imaging the alphavirus exit pathway. Journal of Virology. 2014;88:6922–6933. doi: 10.1128/JVI.00592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Moerner WE, Orrit M. Illuminating single molecules in condensed matter. Science (New York, N.Y.) 1999;283:1670–1676. doi: 10.1126/science.283.5408.1670. [DOI] [PubMed] [Google Scholar]
- Romero-Brey I, Berger C, Kallis S, Kolovou A, Paul D, Lohmann V, Bartenschlager R. NS5A Domain 1 and Polyprotein Cleavage Kinetics Are Critical for Induction of Double-Membrane Vesicles Associated with Hepatitis C Virus Replication. mBio. 2015;6:e00759. doi: 10.1128/mBio.00759-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nature Structural & Molecular Biology. 2004;11:567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. Journal of Structural Biology. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorb M, Gaechter L, Avinoam O, Sieckmann F, Clarke M, Bebeacua C, Bykov YS, Sonnen AF-P, Lihl R, Briggs JAG. New hardware and workflows for semi-automated correlative cryo-fluorescence and cryo-electron microscopy/tomography. Journal of Structural Biology. 2017;197:83–93. doi: 10.1016/j.jsb.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. Journal of Microscopy. 2007;227:98–109. doi: 10.1111/j.1365-2818.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- Wolff G, Hagen C, Grünewald K, Kaufmann R. Towards correlative super-resolution fluorescence and electron cryo-microscopy. Biology of the Cell. 2016;108:245–258. doi: 10.1111/boc.201600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondervan R, Kulzer F, Orlinskii SB, Orrit M. Photoblinking of Rhodamine 6G in Poly(vinyl alcohol): Radical Dark State Formed through the Triplet. The Journal of Physical Chemistry A. 2003;107:6770–6776. [Google Scholar]