Summary
The billions of proteins inside a eukaryotic cell are organized among dozens of sub-cellular compartments, within which they are further organized into protein complexes. The maintenance of both levels of organization is crucial for normal cellular function. Newly made proteins that fail to be segregated to the correct compartment or assembled into the appropriate complex are defined as orphans. In this review, we discuss the challenges faced by a cell of minimizing orphaned proteins, the quality control systems that recognize orphans, and the consequences of excess orphans for protein homeostasis and disease.
Introduction
The advent of methods to observe the interior of cells by electron microscopy in the 1950’s revealed far more compartmentalization than was apparent previously using light microscopy with vital dyes and stains (Porter, 1956). This led to the definition of many membrane-bound organelles in the cell, and our first views of many non-membrane bound structures such as the ribosome (Palade, 1955). In parallel with direct visualization, biochemical fractionation was able to isolate these various morphologically identifiable structures (Claude, 1943, 1946). The structures were soon shown to have distinct functions (De Duve, 1965) and eventually associated with specific subsets of cellular proteins that are often part of larger complexes with defined composition and stoichiometry. These studies initiated modern cell biology, a major goal of which is to understand how intracellular organization is generated and maintained to facilitate cellular function.
The most extensive efforts have been aimed at determining how newly made proteins are segregated to the appropriate organelle (Blobel, 1980; Wickner and Schekman, 2005). Although the accuracy of intracellular protein targeting was usually assumed to be high, one could anticipate that it cannot be perfect. As detailed below, it is increasingly clear that the recognition and disposal of mis-targeted copies of a protein is an important facet of achieving effective net segregation. Similarly, the assembly of proteins into complexes is likely to be imperfect (Harper and Bennett, 2016), necessitating the degradation of unassembled components. Thus, the prompt degradation of proteins that fail to be correctly localized or assembled is critical to the maintenance of intracellular organization. We refer to proteins that are terminally separated from their correct location or partners as orphans.
In this review, we discuss our understanding of how cells recognize orphans and selectively route them for degradation. We begin by providing an accounting of the eukaryotic proteome and the extent of its subcellular organization into compartments and complexes. After defining the challenges to achieving a well organized proteome, we consider successively the mechanisms cells use to identify mis-localized and mis-assembled proteins. We end with a consideration of how pathways for the quality control of orphaned proteins impact cellular physiology and disease.
Organization of the eukaryotic proteome
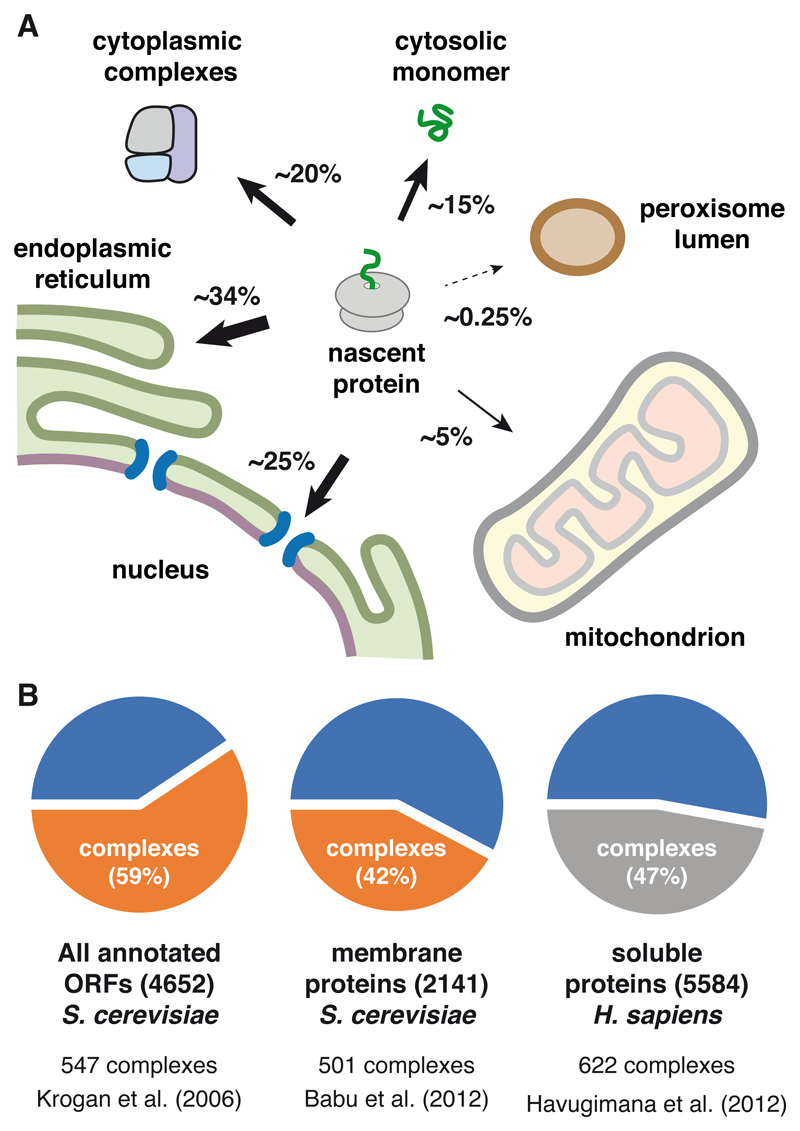
Bioinformatic tools to predict protein location based on known trafficking signals (Emanuelsson et al., 2007), together with increasingly thorough proteome-wide analytic tools (Itzhak et al., 2016; Mulvey et al., 2017), provide estimates of how many proteins are delivered to non-cytosolic destinations. In parallel, systematic analyses of protein complexes (Babu et al., 2012; Gavin et al., 2006; Krogan et al., 2006) and their relative abundances (Kulak et al., 2014) inform on what proportion of nascent proteins need additional assembly. These studies show that the vast majority of newly made proteins are destined for a different location, assembled with other cellular factors, or both (Fig. 1).
Figure 1. Accounting of Protein Localization and Assembly.
(A) The approximate percentages of human genes whose proteins are destined for various intracellular compartments are shown.
(B) Data from three proteomic studies indicating that roughly half of all proteins in yeast and mammals are in protein complexes.
Of the 20,341 reference proteins of the human genome (UniProt Consortium, 2018), ~7000 are targeted to the endoplasmic reticulum for eventual residence in the endomembrane system, nuclear envelope, plasma membrane, outside the cell, and peroxisomal membrane. ~1000 proteins are destined for mitochondria, ~50 for the peroxisomal lumen, and a relative handful for more specialized structures such as lipid droplets. Around ~5000 proteins operate primarily in the nucleus, although the entry and exit of many of these are often dynamic and regulated. Thus, ~65% of genes encode for proteins that must be recognized for selective trafficking to a membrane-enclosed compartment (Fig. 1A).
Analysis of protein interactions by mass spectrometry across the yeast proteome (Gavin et al., 2006; Krogan et al., 2006) indicates that over half of all proteins may be in stable multi-protein complexes (Fig. 1B). While the proportion is somewhat less for membrane proteins (Babu et al., 2012), this may be due to the additional challenge of retaining interactions during detergent solubilization. Although data on the human proteome is less complete, analysis of the relatively abundant cytosolic proteins in HeLa and HEK293 cells indicate that a little less than half of them are in stable complexes (Havugimana et al., 2012). This means that of the ~35% of proteins that do not traffic to another destination (see above), fewer than half are unaccompanied by partners. Thus, only ~15% of genes encode proteins that simply fold and function without first requiring sub-cellular localization and assembly.
When abundance, cell type, and growth rate are taken into account, there are many circumstances where the vast majority of synthesized proteins must be localized or assembled. For example, in highly secretory cell types such as pancreatic exocrine cells or antibody-secreting plasma cells, nearly all ribosomes are synthesizing proteins destined for the ER (Brewer and Hendershot, 2005; Pfeffer et al., 2016). In reticulocytes, almost all protein synthesis is dedicated to alpha and beta globin which assemble into a heterotetramer (Benz and Forget, 1974). Even in a less specialized cell type making a diverse proteome, the cell’s most abundant proteins are localized (e.g., histones to the nucleus) or part of complexes (e.g., tubulin, ribosomes, and proteasomes).
These considerations indicate that even if the rate of failure for localization or assembly is less than 0.1%, the absolute number of polypeptides that must be recognized by quality control is high. As argued below, the actual failure rates are likely to be a few percent, imposing a substantial constitutive burden on quality control pathways. Based on these estimates, we propose that orphans are the major source of substrates for most cellular quality control pathways in normal unstressed cells. Thus, a mechanistic framework for their selective recognition and degradation is of substantial importance to understanding how protein homeostasis is maintained in cells to avoid various disease states.
Challenges to accurate protein localization
The fundamental principle underlying protein segregation to organelles is the recognition of signal sequences within a nascent protein by targeting factors that specify the appropriate destination (Blobel, 1980). With few exceptions, the targeting signal for a particular organelle is not a specific sequence. Instead, it is typically specified by more general properties such as hydrophobicity, length, and charge (von Heijne, 1995). Thus, the set of proteins that are recognized by a targeting factor have signals that can differ substantially in sequence as long as they share the relevant underlying biochemical feature(s).
While molecular recognition of a distinct sequence can be extremely specific, recognition of a diverse set of sequences with loosely shared features will necessarily be limited in specificity. The actual rate of failure has been studied only cursorily, and is best understood for secretory protein segregation to the ER. Very early studies anecdotally noted that the signal sequences of some proteins are more efficient than others in cell-free translocation assays. It was later appreciated that not only do signals differ in their relative efficiencies (Kim et al., 2002; Levine et al., 2005), but also in their requirements for translocation machinery (Fons et al., 2003; Ng et al., 1996; Voigt et al., 1996).
Mammalian cell culture experiments comparing different ER signal sequences showed that even the best signals fail ~5% of the time (Levine et al., 2005; Rane et al., 2004). In mouse brain, it has been estimated that the efficient and well-characterized signal from prolactin fails ~1-2% of the time, while the more average signal from prion protein fails ~5% of the time (Rane et al., 2010). These in vitro, cell culture, and in vivo experiments all indicate that failure rates of protein segregation to the ER are on the order of 1-10%. Although similar measurements of mitochondrial import efficiency remain to be performed, the comparable diversity of signal sequences (von Heijne, 1995) and analogous mechanisms of recognition suggest that failure rates may be comparable.
In addition to intrinsic failure of localization under normal conditions, acute organelle stress has been shown to impair protein import into the ER and mitochondria (Kang et al., 2006; Wright et al., 2001). The step that fails appears to involve the actual translocation reaction across the membrane, albeit by different mechanisms. In the case of ER translocation, it is thought that chaperones in the lumen bind partially translocated polypeptides to prevent their back-sliding (Brodsky et al., 1995; Matlack et al., 1999). Engagement of these chaperones with misfolded proteins during ER stress reduces their availability for driving translocation. This is thought to be a protective mechanism to minimize the load of misfolded proteins in the ER during stress (Kang et al., 2006), but results in an increased number of mis-localized proteins orphaned from the ER.
In the case of mitochondria, import of many proteins relies on the electrochemical gradient across the inner membrane (Wiedemann and Pfanner, 2017). Stress conditions that impair this gradient will result in reduced translocation. At least one protein whose import is impaired initiates a stress response (Nargund et al., 2012), while the general increase in mislocalized proteins seems to initiate a compensatory increase in cytosolic degradation capacity (Wang and Chen, 2015; Wrobel et al., 2015). Other cellular states, such as cell division, may also result in temporary attenuation of mitochondrial protein import (Harbauer et al., 2014; Schmidt et al., 2011; Weidberg and Amon, 2018). Thus, cells active in protein synthesis are constantly challenged with proteins orphaned in the cytosol due to a combination of intrinsic and stress-dependent failures in segregating proteins to the ER and mitochondria. The extent of import failure for proteins that function exclusively in the nucleus (e.g., histones) remains poorly studied, and may further contribute to orphans.
Recognition of mislocalized protein orphans
A number of early observations indicated that mis-localized proteins in the cytosol are degraded. First, rare human mutations that disrupt the function of a signal peptide result in lower overall expression levels (Cassanelli et al., 1998; Karaplis et al., 1995; Seppen et al., 1996). Second, proteasome inhibition led to the appearance of a non-glycosylated form of prion protein (Ma and Lindquist, 2001), which later proved to be generated by stabilization of the non-targeted population (Chakrabarti et al., 2011; Drisaldi et al., 2003; Rane et al., 2004). Third, inhibition of ER translocation by small molecules (Besemer et al., 2005; Garrison et al., 2005) or acute ER stress (Kang et al., 2006) resulted in rapid proteasome-dependent degradation of the non-translocated protein. While these findings showed that mislocalized proteins are degraded in the cytosol, the factors involved were not known.
Various studies had informally noted that cell-free translation of a protein intended for the ER results in the protein’s ubiquitination. This observation was exploited to identify factors needed for recognition and ubiquitination of the mislocalized protein (Hessa et al., 2011). These experiments led to Bag6 (also called Bat3 or Scythe), a large, widely conserved cytosolic protein capable of recognizing mislocalized proteins. A role for Bag6 in quality control was consistent with earlier studies that reported a role in the degradation of newly synthesized polypeptides prematurely released from the ribosome with puromycin (Minami et al., 2010). A ubiquitin-like (UBL) domain in Bag6 associates with RNF126, a ubiquitin ligase that works with the E2 enzyme UbcH5 to poly-ubiquitinate substrates associated with Bag6 (Rodrigo-Brenni et al., 2014). Knockdown of either Bag6 or RNF126 partially impairs degradation of proteins that fail to be successfully imported into the ER.
The key feature of mislocalized proteins that are recognized by Bag6 proved to be precisely the elements that are also recognized by the targeting and insertion machinery: transmembrane domains (TMDs) and signal sequences (Fig. 2). Thus, deleting these hydrophobic domains precludes Bag6 recognition and ubiquitination despite the fact that the resulting protein is still mis-localized and presumably unable to fold (Hessa et al., 2011). It is instead apparently recognized by cytosolic quality control pathways that recognize protein misfolding. Indeed, enforced mislocalization by deletion of a signal sequence in yeast results in recognition by the general chaperone Hsp70 (Park et al., 2007).
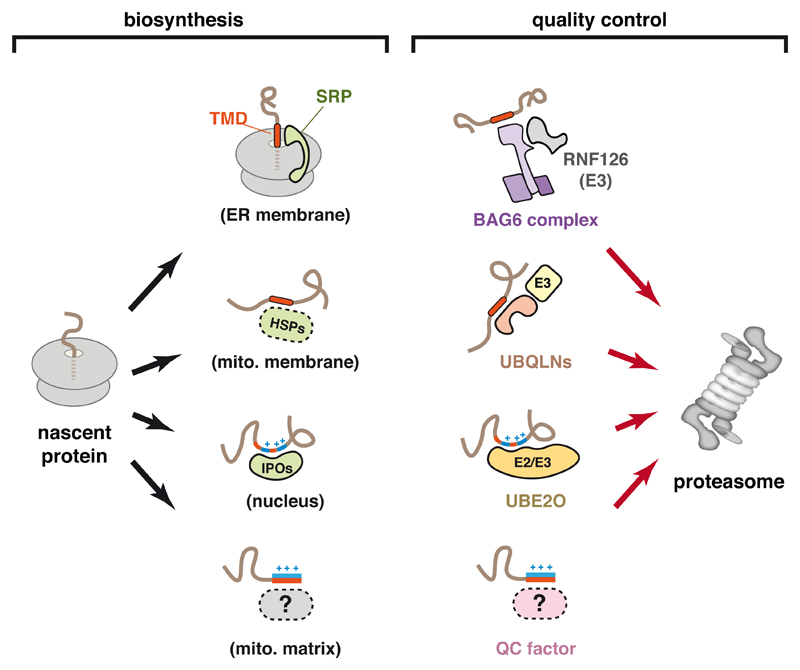
Figure 2. Recognition Factors for Protein Localization and Mislocalization.
Sequence features in a nascent protein can be recognized by various factors that mediate localization to the destinations indicated in parentheses (left side). Not all factors or destinations are shown. Orange indicates hydrophobicity, and blue indicates basic residues. SRP, signal recognition particle; IPO, importin family member; HSP, heat shock protein. The right side shows that the same sequence features used by biosynthesis factors can also be recognized by quality control factors that ultimately lead to degradation at the proteasome.
Analogous studies of a mitochondrial membrane protein showed that prior to its insertion into mitochondria, it associates with members of the Ubiquilin (UBQLN) family (Itakura et al., 2016). Structure-function analysis of UBQLN1 identified a middle (M) domain that specifically associates with the TMDs of membrane proteins in the cytosol. Unlike substrate interaction with Bag6 (Shao et al., 2017), the UBQLN1 association is dynamic (Itakura et al., 2016). However, if the interaction is prolonged as would be the case if import into mitochondria fails, the membrane protein becomes ubiquitinated in a UBQLN-dependent manner. This ubiquitination is mediated by a yet-unidentified ligase that is recruited to the ubiquitin-associating (UBA) domain in UBQLN. Although other members of the UBQLN family (mammals have four) have been less well studied, they have similar M-domains that may also interact with TMDs of mislocalized proteins (Itakura et al., 2016; Suzuki and Kawahara, 2016). It is attractive to speculate that their specificities are somewhat different from each other to collectively cover the wide range of sequences that define TMDs. Indeed, Bag6 appears to prefer more hydrophobic TMDs than UBQLN1 (Itakura et al., 2016). Additional studies are needed to investigate the substrate ranges and molecular basis of specificity of these factors.
Unlike membrane proteins whose exposed TMDs provide a distinctive cue for recognition of mislocalization, the soluble proteins of membrane-bound compartments more closely resemble cytosolic proteins. In the case of proteins destined for the ER, signal sequences are sufficiently similar to TMDs that they seem to be recognized by Bag6 (Hessa et al., 2011). Soluble proteins destined for mitochondria or the nucleus have amphipathic or basic signals, respectively. Whether dedicated factors recognize persistent residence of these signals in the cytosol for degradation remains largely unexplored. One candidate factor for this role is UBE2O, a hybrid E2-E3 enzyme that was recently shown to recognize and ubiquitinate ribosomal proteins in the cytosol (Nguyen et al., 2017; Yanagitani et al., 2017). Under normal circumstances, newly made ribosomal proteins are recognized by dedicated nuclear import factors (Jakel and Görlich, 1998), delivered to the nucleolus, and assembled with rRNA into pre-ribosomal subunits (Peña et al., 2017). It appears that failure of nuclear import leads to recognition by UBE2O, multi-mono-ubiquitination, and proteasomal degradation.
Although the precise sequence features recognized by UBE2O are unclear, it appears to be the juxtaposition of hydrophobic and basic residues (Yanagitani et al., 2017). This is noteworthy because it is similar in some ways to both nuclear import (Jakel and Görlich, 1998) and mitochondrial import signals (von Heijne, 1995), as well as a common feature of nucleic acid-binding proteins (Hentze et al., 2018; Nelson, 1995). Ribosomal protein interactions with UBE2O and nuclear import factors appears to be mutually exclusive (Yanagitani et al., 2017), suggesting that UBE2O may interact with the nuclear import signal. It will therefore be interesting to determine whether this is a general property of UBE2O, and whether other nucleic acid binding proteins, such as histones, subunits of the signal recognition particle (SRP), and mitoribosomal proteins, are recognized by UBE2O when they are orphaned in the cytosol.
Thus, the cytosol appears to be patrolled by a set of factors that can interact with signal sequences and TMDs on the one hand, and ubiquitin ligases on the other (Fig. 2). These factors collectively recognize proteins that expose organelle targeting domains, an indicator that targeting may have failed, and mediate their tagging for degradation. It is noteworthy that the substrate specificity of these factors, while likely to be rather broad, appears to be especially well suited to the same elements recognized by protein targeting factors such as SRP and TRC40 (also called Get3), which mediate membrane protein targeting to the ER (Shao and Hegde, 2011), and Importins 5 and 7, which mediate nuclear import of ribosomal proteins (Jakel and Görlich, 1998). In order to not interfere with targeting, Bag6, UBQLNs, and UBE2O must act after substrates have attempted targeting, the mechanisms of which are discussed later. Even though these factors collectively recognize a broad range of orphans mislocalized to the cytosol, additional quality control machinery that recognize nuclear and mitochondrial targeting signals may remain to be discovered.
Proteins orphaned to the wrong organelle
The TMDs of membrane proteins targeted to the ER and mitochondria are very similar in their biophysical properties (Guna and Hegde, 2018), consistent with their eventual residence in a lipid bilayer. For many of these proteins, the TMD(s) serve as the main or sole targeting signal. Due to the similarities among these TMDs, membrane proteins can be routed to the wrong organelle. How cells deal with this problem is only partially understood.
The simplest and best-studied case involves tail-anchored (TA) membrane proteins (Hegde and Keenan, 2011). These proteins contain a single TMD close to the C-terminus which serves as the sole targeting signal. TA proteins destined for the mitochondria tend to have slightly less hydrophobic TMDs (Wattenberg et al., 2007) and often have downstream basic residues in the C-terminal tail (Horie et al., 2002). Nevertheless, there is substantial overlap in their properties and a clear risk of mis-targeting.
The best-characterized TA targeting pathway to the ER is known as the TRC (TMD recognition complex) pathway in mammals and the homologous GET (guided entry of TA proteins) pathway in yeast (Hegde and Keenan, 2011). Deletion of GET pathway components in yeast results in mislocalization to the cytosol (often as protein aggregates) and mitochondria (Jonikas et al., 2009; Schuldiner et al., 2008). A membrane-embedded AAA-ATPase Msp1 in the mitochondrial outer membrane is thought to be involved in removing TA proteins mislocalized to mitochondria (Chen et al., 2014; Okreglak and Walter, 2014). A sub-population of Msp1 is also found in the peroxisomal membrane, where it is thought to serve a similar role (Weir et al., 2017). Hence, the levels of mislocalized TA proteins in mitochondria or peroxisomes increases when Msp1 (or its human homolog ATAD1) is deleted. As this stabilization is seen even in wild type cells containing an intact TRC/GET pathway (Chen et al., 2014), mislocalization must be a constitutive problem in cells and tissues, consistent with the challenges of achieving precise targeting specificity.
The mechanistic basis of Msp1 function in clearance of mislocalized protein substrates is not well understood. Purified Msp1 reconstituted into synthetic liposomes was shown to drive ATP-dependent extraction of a co-reconstituted TA protein to the cytosol (Wohlever et al., 2017). The ATPase domain of Msp1, which faces the cytosol, was shown to be a hexamer containing a central pore. Because mutations within the pore impair the TA protein extraction activity, it has been posited that the substrate is pulled through the pore into the cytosol (Wohlever et al., 2017). Such a model would be consistent with the mechanism of action of other AAA protein unfoldases for whom translocation through the central pore is strongly supported (Sauer and Baker, 2011). How the substrate is kept in a soluble form after extraction, how it is ubiquitinated, and how it is delivered to the proteasome all remain unknown. One possibility is that cytosolic factors such as Bag6 or UBQLNs may mediate these downstream steps similarly to how they recognize and degrade cytosolic orphans generated by failed targeting. Consistent with this concept, Bag6 has been shown to maintain the solubility of membrane proteins extracted from the ER until their degradation at the proteasome (Wang et al., 2011).
The basis of substrate selection by Msp1 is also only partially understood. Studies of the peroxisomal TA protein Pex15 have provided initial insights into this key issue (Weir et al., 2017). Pex15 is normally found in complex with Pex3. It was observed that excess Pex15 in peroxisomes is degraded in an Msp1-dependent manner, while Pex15 in complex with Pex3 evades Msp1 (Weir et al., 2017). As discussed in detail later, orphans of multi-protein complexes are often targets for quality control via recognition of the regions that are normally shielded by interaction partner(s). It appears that Msp1 may use such a mechanism to recognize orphaned Pex15, and perhaps other substrates (Fig. 3).
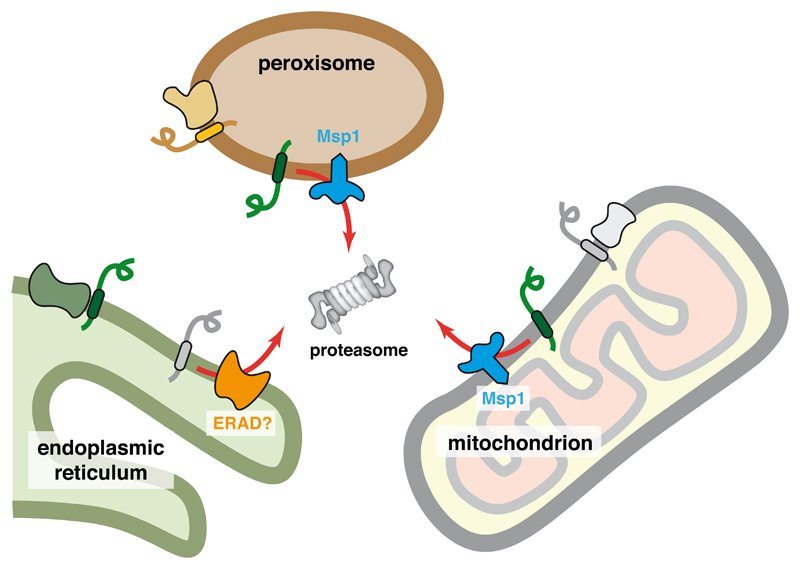
Figure 3. Recognition of Protein Mislocalization at Organelles.
Membrane protein insertion into the wrong organelle can be recognized by its lack of association with an interaction partner. In mitochondria and peroxisomes, the hexameric ATPase Msp1 is involved in extracting such mislocalized proteins to the cytosol for degradation by the proteasome. In the ER, the specific machinery for mislocalized proteins has not been studied, but probably involves known ER-associated degradation (ERAD) pathways. The molecular basis of recognition is not known, but may involve the same features that facilitate assembly into protein complexes, explaining why successful assembly would prevent recognition by quality control factors.
This finding suggests that when Pex15 is mis-targeted to mitochondria, its recognition by Msp1 for degradation is due to the absence of Pex3. Whether all Msp1 substrates are normally part of larger complexes whose absence cues their recognition remains to be determined. Furthermore, it is unclear whether Msp1 directly recognizes substrates or uses adaptors. In this context, it is noteworthy that Msp1 associates with Cis1, a protein that is upregulated by excessive mitochondrial import failure and plays a yet unknown role in clearance of failed import products (Weidberg and Amon, 2018). Whether Cis1 is involved in identifying substrates for delivery to Msp1 remains unclear. Thus, both the specific features of mis-targeted proteins that are recognized and the factors that mediate recognition remain to be elucidated in molecular terms.
The factor(s) that play the analogous role to Msp1 in the ER are unknown. It has recently been shown that mitochondrial TA proteins can be mis-targeted to the ER, particularly under over-expression conditions (Vitali et al., 2018). Furthermore, the ribosome-associated chaperone-like protein NAC appears to antagonize SRP and limit its promiscuous recognition of nascent mitochondrial proteins (Gamerdinger et al., 2015; Wiedmann et al., 1994). Thus, when NAC levels are reduced, mitochondrial proteins can be detected at the ER. Although these examples of ER mis-targeting are seen under perturbed conditions, it is likely that a low level of mis-targeting occurs in normal cells. How these mis-targeted proteins are recognized and degraded is unclear. One possibility is that known ER-associated degradation pathways are involved (Vembar and Brodsky, 2008). This problem warrants investigation. Tools to induce excessive ER mis-targeting should facilitate this line of inquiry.
Challenges to assembly of protein complexes
After segregating proteins to the correct compartment, most proteins must additionally assemble with one or more partners (typically other proteins, but also RNAs). The assembly of multi-protein complexes imposes three major challenges to the cell. First, the components of the complex must be synthesized in the appropriate stoichiometry. Second, the unassembled subunits of a complex must avoid inappropriate interactions until their assembly. Third, partners must find each other within a crowded cell. As with protein localization, limitations to the overall efficiency of achieving these essential steps in complex assembly results in orphans that must be recognized and degraded.
Direct analysis of translation rates by sequencing of ribosome-protected footprints (ribosome profiling) indicate that cells typically express subunits of a multi-protein complex at close to the stoichiometry found in the final complex (Li et al., 2014). A combination of several mechanisms contributes to this concordance. First, the genes for proteins that function together are frequently organized in a single operon in prokaryotes (Ames and Martin, 1964). This allows the transcript levels to necessarily be increased and decreased in unison. Differences in the order of genes in an operon and their respective Shine-Dalgarno sequences can presumably further tune their relative levels of translation (Bonde et al., 2016; Lim et al., 2011).
In eukaryotes, gene order (Dávila López et al., 2010) and the similarities of promoters engaged by the same transcription factors (Lee et al., 2009) can also coordinate transcript levels of protein subunits. Beyond this, further fine tuning of mRNA half-lives and translation rates via untranslated regions and codon optimality probably contribute to expression at the desired stoichiometry. For example, the 5’ end of mRNAs encoding ribosomal proteins and many translation factors contain terminal oligo-pyrimidine (TOP) sequences (Meyuhas and Kahan, 2015). These sequences allow the translation of these mRNAs to be repressed and de-repressed in concert, thereby maintaining their approximately stoichiometry. Nevertheless, these mechanisms are all subject to stochasticity (Munsky et al., 2012), meaning that the inevitable imbalances in synthesis levels generate orphans. It is reasonable to assume that achieving expression stoichiometry to greater than 90% precision is very challenging given the inherent noisiness of gene expression. For complexes with many subunits, such as ribosomes and proteasomes, matching expression levels may be particularly difficult.
The mechanisms by which proteins are assembled into complexes has been studied in a number of contexts including ribosome biogenesis in the nucleolus (Peña et al., 2017), haemoglobin assembly in pre-erythrocytes (Feng et al., 2004; Kihm et al., 2002), and histone assembly (Hammond et al., 2017). Each of these processes is distinctive in their use of specific chaperones and assembly factors. The main underlying principle from these studies is that unassembled subunits of a complex are held temporarily by an assembly factor until it is displaced by the appropriate interacting subunit. In many cases, the precise order of protein complex assembly is important and under selective pressure to be maintained, presumably to minimize mis-assembled products (Marsh et al., 2013). However, the efficiency of these processes is unknown in most cases, especially in vivo, so their relative contributions to the generation of orphans remains to be determined.
Recent proteomic pulse-chase experiments indicate a substantial constitutive burden of unassembled orphans in cultured cells (McShane et al., 2016). In this study, kinetic analysis of protein degradation found ~10% of proteins are degraded non-exponentially, with a proportion (ranging from 10-70%) being degraded immediately after synthesis. Approximately 70% of these non-exponentially degraded proteins are subunits of known multi-protein complexes, suggesting that the rapidly degraded populations are those polypeptides that failed assembly. As this analysis identified only the most abundant proteins, it appears that cells must constantly identify and degrade unassembled orphans.
Recognition of unassembled cytosolic protein orphans
It has been apparent for many decades that unassembled orphans of multi-protein complexes are selectively degraded. The earliest appreciation of this phenomenon comes from the study of haemoglobin, a hetero-tetramer normally formed of two α and two β subunits (Perutz et al., 1960). Mutations in haemoglobin that markedly reduce its unusually long half life were correlated with an increased tendency of tetramer dissociation into subunits and decreased binding to heme (Jacob et al., 1968; Rieder, 1974). Many years later, studies of the multi-subunit T-cell receptor (TCR) showed that the alpha subunit is rapidly degraded when expressed without its interaction partners (Lippincott-Schwartz et al., 1988). Conversely, it is well appreciated that knockdown or knockout of one subunit of a multi-protein complex often destabilizes the other subunits. Thus, in numerous contexts and cellular compartments, cells have mechanisms to selectively identify orphaned subunits and target them for degradation.
Two general mechanisms can explain how orphans are recognized. First, some proteins may not be able to achieve a stable folded state in the absence of its interaction partner. This instability would result in its unfolding and recognition by general quality control pathways that monitor protein folding. Second, a subunit interface that is normally shielded in the intact complex is recognized by a factor which ultimately leads to degradation. Factors for both mechanisms have been identified in different biological contexts.
The most challenging multi-subunit complex to assemble in cells is probably the ribosome. Its biogenesis from 80 proteins and pre-rRNA involves dozens of dedicated assembly factors, numerous processing reactions of both proteins and rRNA, a variety of post-translational modifications, and multiple transport steps across the nuclear envelope (Peña et al., 2017). Individual ribosomal proteins have long been known to be refractory to over-expression, suggesting efficient mechanism(s) for degrading excess copies (Abovich et al., 1985; Warner et al., 1985). Despite the fact that ribosomes have a very long half-life, ribosomal proteins are a major set of proteins that accumulate when proteasome activity is diminished (Mayor et al., 2005, 2007). This suggests that ribosomal proteins may be a major source of orphans in actively growing cells.
As discussed above, a ribosomal protein orphaned at the step of import into the nucleus is recognized in the cytosol by UBE2O (Yanagitani et al., 2017). After nuclear import, failure at the step of assembly with rRNA is recognized by a different mechanism. In yeast, the large nuclear-localized E3 ligase Tom1 was shown to interact with unassembled ribosomal proteins and mediate their ubiquitination (Sung et al., 2016). The sites of ubiquitination suggest that Tom1 probably interacts with basic regions that would ordinarily interact with rRNA. These regions are inaccessible in intact ribosomes, which are not recognized by Tom1. HUWE1 is the closest homolog of Tom1 in mammals. Its knockdown partially increases the levels of over-expressed uL24/RPL26, but the basis for this effect was not explored in sufficient depth to conclude a direct effect (Sung et al., 2016).
In separate studies however, it was observed that HUWE1 is required for efficient degradation of farnesyltransferase alpha (FTNA) and UBL4A when their respective binding partners are absent (Xu et al., 2016). Although interaction studies were not performed, HUWE1 presumably recognizes the exposed interfaces of these and other proteins to mediate their ubiquitination. Proteomic analysis identified 72 candidate HUWE1 substrates (although curiously, not ribosomal proteins), almost all of which are known subunits of multi-protein complexes. Most of these candidate substrates operate in the nucleus, yet HUWE1 appears to function in the cytosol. Thus, HUWE1 may identify orphans that fail localization, assembly, or both. The molecular features shared by HUWE1 clients and the basis of their recognition remain to be addressed.
In addition to its role in recognizing mislocalized ribosomal proteins discussed above, UBE2O can also recognized folded but unassembled proteins. This conclusion comes from the observation that in vitro and in vivo, UBE2O is needed for efficient ubiquitination and degradation of unassembled α-globin (Nguyen et al., 2017; Yanagitani et al., 2017). The interface of α-globin that interacts with β-globin is usually shielded by α -haemoglobin stabilizing protein (AHSP) until assembly with β -globin (Feng et al., 2004, 2005) (Fig. 4A). Mutations in α-globin that impair interaction with AHSP (and β -globin) are more effectively recognized and ubiquitinated by UBE2O (Yanagitani et al., 2017). This indirectly suggests that UBE2O recognizes the same region as AHSP. This interface is basic and hydrophobic (Fig. 4B), which is similar to other established UBE2O targets such as unassembled ribosomal proteins (Nguyen et al., 2017; Yanagitani et al., 2017). Thus, orphan α -globin appears to be identified for degradation by its exposed assembly interface.
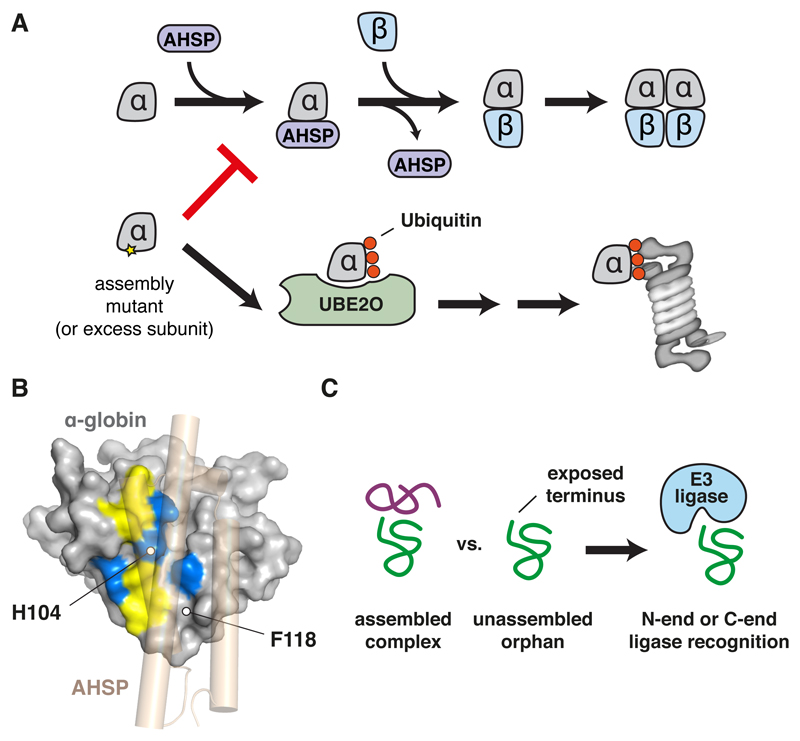
Figure 4. Assembly Interfaces Serve as Degradation Signals.
(A) Schematic of hemoglobin assembly in which the α subunit is temporarily shielded by α-hemoglobin-stabilizing protein (AHSP) until it assembles with the β subunit. Free α subunit, generated due to excess or a mutation that prevents assembly, is instead recognized by UBE2O for ubiquitination and targeting to the proteasome for degradation.
(B) The assembly interface of α-globin showing hydrophobic (yellow) and basic (blue) surfaces. AHSP is shown in transparent tan to illustrate how this interface is shielded. The positions of two assembly mutants that cause human anemia are indicated. UBE2O is known to bind composite basic/hydrophobic peptides, suggesting that the interface is recognized via these features when assembly fails.
(C) An exposed N or C terminus with a destabilizing residue can be shielded in a correctly assembled complex (left) but exposed as an unassembled orphan. N-end and C-end E3 ubiquitin ligases that recognize such residues would then selectively target the orphan for degradation.
Notably, UBE2O is highly up-regulated during erythrocyte development concordantly with increased globin production (Wefes et al., 1995). Whether this up-regulation is a programmed event in preparation for the inevitable increase in substrate load, or a response to increased orphans remains to be determined. In support of the latter possibility, acute induction of protein misfolding has been shown to cause UBE2O upregulation in cultured cells (Miyazaki et al., 2015). Structure-function analysis of UBE2O indicates that it has two major substrate interaction domains with different binding properties (Nguyen et al., 2017; Yanagitani et al., 2017). Identifying the client ranges of these domains and the structural basis of their interactions remains an important goal.
The general concept of orphan recognition via exposure of a normally buried interface has been exploited by other ubiquitination factors. The N-end rule pathways of substrate ubiquitination recognize certain residues at an exposed N-terminus and selectively target these proteins for degradation (Tasaki et al., 2012). One mechanism involves recognition of N-terminal acetylation by the ubiquitin ligase CNOT4 (Shemorry et al., 2013). Several multi-protein complexes appear to contain N-acetylated subunits whose N-terminus is shielded by other subunits in the intact complex. Failure to assemble into a complex (or complex disassembly) exposes the N-terminal degron, resulting in ubiquitination by an N-end rule ubiquitin ligase (Fig. 4C). Examples include the Hcn1 subunit of the APC/C complex and the Cog1 subunit of the yeast COG complex.
It is possible that other N-end rule ligases exploit a similar mechanism to recognize different unassembled proteins. For example, the N-end rule ubiquitin ligase Ubr1 was shown to be needed for efficient clearance of unassembled Fas2, a subunit of the fatty acid synthase complex (Scazzari et al., 2015). Whether Ubr1 uses its N-end rule recognition domain or uses an adaptor such as Hsp70 to identify unassembled Fas2 is unclear at present. Hsp70 is needed for Ubr1-dependent Fas2 ubiquitination, supporting the latter hypothesis. Presumably, an interface of Fas2 shielded by its interaction partner Fas1 is exploited for its recognition by chaperones, which over time recruits a ubiquitin ligase to initiate degradation. The recent discovery of a “C-end rule” (Koren et al., 2018; Lin et al., 2018) raises the possibility that exposure of a normally-shielded C-terminus might also be exploited by cells to recognize orphans (Fig. 4C). This idea remains to be investigated.
Recognition of unassembled protein orphans at the ER
Each of the examples of unassembled protein degradation discussed thus far occur in the cytosol. Analogous processes operate in the ER membrane and lumen, and probably also different mitochondrial compartments. In the ER, the most extensively studied multi-protein assemblies have been the immunoglobulins and T-cell receptor (TCR). TCRα and CD3δ, two subunits of the TCR, have been widely used as models for ERAD (Bonifacino and Lippincott-Schwartz, 1991; Bonifacino et al., 1989; Huppa and Ploegh, 1997; Lippincott-Schwartz et al., 1988). Both subunits are single-spanning transmembrane proteins that are recognized for degradation via their TMDs (Bonifacino et al., 1991; Cosson et al., 1991; Manolios et al., 1990). The prevailing model is that the orphaned TMDs of TCRα and CD3δ are recognized by the membrane-embedded regions of a ER-resident ubiquitin ligase complex centered around Hrd1 (Kikkert et al., 2004). Although the structural basis of their recognition remains unknown, it depends critically on charged residues within their TMDs that are shielded by other subunits in the assembled TCR. A similar mode of recognition by Hrd1 was also shown for other integral membrane substrates such as Hmg2 (Sato et al., 2009).
Based on these examples, one might speculate that orphans of multi-protein complexes assembled via their TMDs are recognized by the exposure of polar residues within the lipid bilayer. This would provide a relatively universal cue for unassembled proteins without relying on sequence-specific recognition. In this light, it is intriguing that the recently determined cryo-EM structure of Hrd1 shows a hydrophilic pocket that might serve as the site of recognition of such orphaned TMDs (Schoebel et al., 2017). Even though the functional role of this putative binding region of Hrd1 remains to be investigated experimentally, the model is consistent with previous observations that mutations within this hydrophilic region of Hrd1 affect recognition of some integral membrane ERAD substrates (Sato et al., 2009).
Although recognition of orphaned TMDs within the membrane has long been presumed, experimental evidence for this idea is generally lacking. An alternative idea is that orphaned TMDs, due to their exposed polar or charged residues, are unstable in the lipid bilayer and translocate into the ER lumen (Feige and Hendershot, 2013). This results in their recognition by lumenal chaperones, particularly BiP, which are proposed to deliver them for degradation. Successful assembly would prevent this translocation, providing an explanation for why orphans are selectively degraded. Although both models ultimately involve Hrd1 (or another ubiquitin ligase complex) mediating retrotranslocation into the cytosol for degradation, one posits recognition in the ER lumen while the other involves recognition in the membrane.
Although the two views have yet to be reconciled, one attraction of the lumenal recognition model is its similarity to how misassembled lumenal protein complexes might be monitored. One example of protein complex assembly in the ER lumen is immunoglobulins typically built from two heavy and two light chains (Huber et al., 1976). After translocation into the ER lumen, the first heavy chain immunoglobulin domain (CH1) binds to the chaperone BiP in an unfolded and reduced state (Bole et al., 1986; Haas and Wabl, 1983; Hendershot et al., 1987; Vanhove et al., 2001). CH1 can only fold upon interaction with light chain (Feige et al., 2009). Hence, in the absence of light chains, heavy chains are retained in the ER and are eventually dislocated to the cytosol to be degraded by the proteasome (Mancini et al., 2000). Even though the assembly process varies between different immunoglobulin isotypes (Baumal et al., 1971), BiP-mediated retention of heavy chains in the ER appears to be universal (Hendershot et al., 1987). Prolonged association with BiP, whether of orphan heavy chains or TCR subunits, may result in delivery to ERAD machinery via adaptors such as specific J-proteins (Shen and Hendershot, 2005).
Distinguishing intermediates from orphans
Proteins should be recognized as orphans to be degraded only after reasonable attempt(s) at localization and assembly have failed. Thus, the biosynthesis machinery should have higher priority for access to nascent chains than the quality control machinery, but this priority should be time-limited. The mechanistic basis of how priority and timing are determined is important to understand because this critical decision balances promiscuous degradation of normal biosynthetic intermediates versus excessive persistence of potentially toxic and aggregation-prone orphans.
One mechanism by which priority can be conferred is by spatial segregation of the biosynthetic and quality control factors. The best illustration of this principle is SRP interaction with the ribosome. The majority of eukaryotic proteins destined for the ER are recognized co-translationally by SRP (Chartron et al., 2016; Costa et al., 2018; Keenan et al., 2001). This interaction is facilitated by the ability of SRP to interact with the ribosome such that the signal sequence binding domain is positioned precisely at the ribosomal exit tunnel (Halic et al., 2004; Voorhees and Hegde, 2015). Not only is SRP thought to sample ribosomes for the presence of an emerging signal, but it may be stabilized there while the signal is still inside the ribosomal tunnel (Berndt et al., 2009; Voorhees and Hegde, 2015).
In this manner, nascent secretory and membrane proteins would not have an opportunity to engage quality control factors such as Bag6 or UBQLNs unless SRP recognition has failed. Thus, despite its ~5-10 fold lower abundance (Kulak et al., 2014), SRP nevertheless has priority due to its precise localization at the ribosome. Once a protein has been released from the ribosome, SRP’s relatively low abundance and poor capacity to bind proteins in solution precludes its interference with other processes.
At a later step in biogenesis, secretory and membrane proteins are translocated into the ER through the ribosome-associated Sec61 translocation channel (Rapoport et al., 2017). Any proteins that selectively associate, even dynamically, with Sec61 will get priority for interaction with the nascent chain over those that do not. Indeed, processing enzymes such as oligosaccharyl transferase and signal peptidase enjoy such priority (Braunger et al., 2018), as might the chaperone BiP [via its recruitment by the translocon component Sec63 (Brodsky et al., 1995)] and Calnexin (Lakkaraju et al., 2012). In this manner, biosynthetic factors could be given priority before degradation factors such as Hrd1.
A second mechanism of conferring priority is to use a combination of abundance and fast binding. This appears to be how newly made tail-anchored proteins are prioritized for ER targeting ahead of proteasomal degradation. Recent studies show that among the three TMD binding factors involved in TA protein targeting, SGTA is the fastest and first interactor (Shao et al., 2017). Its competitive advantage may be further increased by its interaction with the Bag6 complex or Hsp70, both of which can associate with the ribosome. TA proteins can transfer from SGTA to TRC40 (Mock et al., 2015; Shao et al., 2017)by a mechanism that does not require release of TA protein into the bulk cytosol (Shao et al., 2017). Loading on TRC40 is a commitment to targeting because TA protein dissociation is very slow relative to the rate of targeting. TA proteins have a limited time to complete these events as dictated by their off-rate from SGTA. Dissociation from SGTA permits an opportunity to be captured by Bag6, which recruits an E3 ligase for substrate ubiquitination (Rodrigo-Brenni et al., 2014). Very slow dissociation from Bag6 means TA binding to this factor is effectively a commitment for degradation. Thus, triage by multiple factors of seemingly identical specificity can be accomplished simply by their differential on- and off-rates combined with committed downstream events such as membrane insertion or ubiquitination (Shao et al., 2017).
The final mechanism is based on an intrinsically slow reaction followed by a specific commitment step. This is best exemplified by glycoprotein quality control in which a specific irreversible trimming event of an N-linked glycan generates a product that is recognized by a lectin coupled to quality control pathways (Caramelo and Parodi, 2015; Tannous et al., 2015). The trimming event is carried out by intrinsically slow mannosidases, effectively placing a time limit on protein maturation attempts. As predicted by this model, over-expression of the mannosidase accelerates the rate of degradation (Hosokawa et al., 2001).
An analogous mechanism of a slow enzymatic reaction linked to a commitment step seems to be used for some orphans in the cytosol. Membrane proteins bound to UBQLNs are not initially committed for degradation (Itakura et al., 2016). Instead, their dynamic release and re-binding provides opportunities at successful insertion. However, a relatively slow ubiquitination step seems to be the key commitment step. When ubiquitin is added to the substrate, a UBA domain in UBQLNs binds to ubiquitin, preventing substrate release and thereby ending insertion attempts (Itakura et al., 2016). Presumably, the time allowed for insertion is dependent on the affinity of the ligase for UBQLNs, its abundance, and speed of ubiquitination. Identifying the ligase should allow these aspects of the model to be tested.
Can failures be detected co-translationally?
The systems for detection and degradation of orphans discussed so far all operate post-translationally. This is consistent with the idea that biosynthesis must necessarily have an opportunity to succeed before a polypeptide is deemed an orphan. However, many biosynthetic reactions are initiated or occur co-translationally: recognition by SRP, the initial stages of polypeptide folding, various modifications, and even multi-subunit assembly. It is therefore possible that cells have evolved mechanisms to detect orphans co-translationally via failure of a decisive co-translational biosynthetic step. Early detection is advantageous because minimizing the time an aberrant polypeptide resides in a cell reduces the risk of inappropriate interactions, aggregation, and other adverse consequences. Although co-translational orphan detection has not been established unambiguously, two examples are noteworthy.
The first example concerns an autoregulatory mechanism to control β-tubulin expression. As noted already, α- and β -tubulin form a constitutive complex (Feit et al., 1971). In the presence of excess unpolymerized tubulin subunits, synthesis of both α-tubulin and β - tubulin polypeptides is reduced (Cleveland et al., 1981). One regulatory mechanism involves selective degradation of β-tubulin mRNA (Pittenger and Cleveland, 1985). This degradation is strictly dependent on translation, and more specifically, on the first four amino acids in the nascent polypeptide (Bachurski et al., 1994; Gay et al., 1989). Remarkably, a monoclonal antibody that binds these four residues abolished translation-dependent mRNA degradation (Theodorakis and Cleveland, 1992). These findings led to the idea that co-translational association of nascent β-tubulin with some yet-unidentified factor (which is not α-tubulin) is needed to escape mRNA degradation.
In the intervening time since these findings, various pathways of mRNA decay have been characterized, including ones dependent on ribosome stalling (Shoemaker and Green, 2012). Thus, one attractive model is that a limiting assembly factor binds nascent β-tubulin, the absence of which leads to translation arrest and mRNA degradation. Assuming that this factor is displaced upon assembly with α-tubulin (or tubulin incorporation into microtubules), β-tubulin orphans would sequester the assembly factor and lead to mRNA decay. In this way, the production of orphan β-tubulin would be tightly restricted via a co-translational detection mechanism.
The second example concerns the consequences of failed co-translational recognition by SRP. It has been observed that knockdown of SRP or mutating a signal peptide such that it cannot be recognized by SRP causes a reduction in the corresponding mRNA and protein (Karamyshev et al., 2014). This effect was dependent on Argonaute2, which was also observed to interact with the SRP-deficient nascent chain at the ribosome (Karamyshev et al., 2014). These findings suggest a model in which co-translational failure of a hydrophobic sequence to be recognized by SRP triggers Argonaute2-dependent mRNA decay.
These two examples illustrate a potentially important principle (Fig. 5). If an interaction critical for biogenesis occurs co-translationally, its failure would necessarily result in an orphan. There may be mechanisms to detect such failures co-translationally, which would provide the cellular quality control systems access to both the nascent polypeptide and associated mRNA. Although still poorly studied, there is evidence that many protein complexes may initiate assembly during translation (Duncan and Mata, 2011; Williams and Dichtl, 2018). If such co-translational interactions were coupled to translation elongation, the mRNA decay pathways downstream of ribosome stalling could be exploited to fine-tune the balanced expression of subunits and minimize the generation of orphans. Such an early-detection system may represent one of the physiologic roles of ribosome-associated quality control pathways.
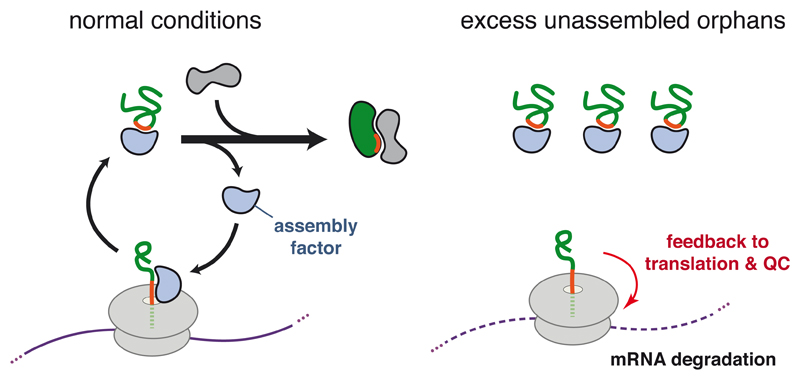
Figure 5. Hypothetical Model for Co-translational Monitoring of Protein Assembly.
A nascent polypeptide emerging from the ribosome is recognized by an assembly factor, which is recycled when assembly occurs correctly. When assembly fails, the assembly factor is titrated by excess unassembled orphans (right side). The unavailability of this nascent chain binding protein at the ribosome is proposed to trigger ribosome-associated quality control pathway via effects on translation (red arrow), perhaps via factors (not shown) that bind to the sequence motif normally recognized by the assembly factor.
Orphan-related pathologies
As with other types of quality control such as protein misfolding or processing, excessive production of orphans can be dominantly detrimental to cellular homeostasis. This is observed in the disease phenotypes of various inherited mutations. Rare mutations have been described in the signal sequences of a wide range of proteins (Cassanelli et al., 1998; Karaplis et al., 1995; Seppen et al., 1996). In most cases, the mutation disrupts the hydrophobic core of the signal to reduce or eliminate its targeting function. This would lead to an increase of orphans in the cytosol of cells expressing the mutant protein. Accordingly, the phenotypes are typically dominant and tissue-specific to the most highly expressing cell type. This suggests that the increase in orphans, not simply a loss-of-function, is the basis of cellular damage. In mouse studies of the non-essential prion protein (PrP), introduction of a version containing a signal sequence that is only ~50% efficient led to a dominant neurodegenerative phenotype (Rane et al., 2008). Thus, over long times in a complex organism, even a modest increase in the load of mis-localized orphans in the cytosol can be detrimental.
Perhaps the most common set of diseases involving orphans are the Thalassemias, a group of hereditary blood diseases characterized by anemia, hemolysis, and various downstream consequences (Olivieri, 1999; Piel and Weatherall, 2014). These diseases result when one or more of the alleles encoding α- or β -globin is deficient, resulting in imbalanced synthesis of haemoglobin subunits. Mutations in the α-β interface that impair assembly of haemoglobin also cause rare variants of anemia related to the Thalassemias (Clarke and Higgins, 2000; Kohne, 2011). While a reduced level of mature haemoglobin is certainly a major contributor to the pathogenesis of these diseases, substantial evidence indicates that the unpaired globin proteins are dominantly toxic in many ways. For example, unpaired α- or β-globin has been suggested to generate increased reactive oxygen species, form intracellular aggregates, or interfere with other cellular processes such as assembly of a membrane cytoskeleton (Weatherall JD, 2001). As noted above, unassembled α-globin is selectively recognized, ubiquitinated, and degraded by UBE2O. However, this does not seem to be the only degradation pathway, as some α-globin ubiquitination was observed even in UBE2O knockout cells (Nguyen et al., 2017). Furthermore, the pathway of unpaired β-globin degradation remains unknown. These may prove to be important facets of the pathophysiology of the Thalassemias.
Given the detrimental consequences of orphans, it is noteworthy that aneuploidy is a prominent feature in large proportion of cancers (Rajagopalan and Lengauer, 2004). As the genes for different subunits of multi-protein complexes are often on different chromosomes, aneuploidy would necessarily unbalance expression. Experiments in yeast have shown that duplication of a single chromosome in an otherwise haploid cell is detrimental, while an additional chromosome in diploid cell is less so (Dephoure et al., 2014; Dodgson et al., 2016; Oromendia et al., 2012). This suggests that proteome imbalance may underlie the fitness cost, an idea consistent with the observed protein homeostasis defects seen in aneuploid yeast. Experiments in human cells induced to be aneuploid support the findings in yeast (Ben-David et al., 2014; Stingele et al., 2012; Williams et al., 2008). Yet, aneuploid cancer cells paradoxically grow unrestrained. This might imply that they manage to grow robustly despite aneuploidy, not because of it. Consistent with this idea, many cancer cells have higher levels of the major cytosolic chaperone Hsp70 (Murphy, 2013), elevated proteasome activity (Chen and Madura, 2005) and increased autophagy (Singh et al., 2018). It is also noteworthy that UBE2O is amplified in many cancers, while reduction of UBE2O in different mouse cancer models provides a benefit by attenuating tumor growth (Liang et al., 2017; Vila et al., 2017). Thus, understanding the mechanistic basis of orphan recognition and degradation may provide new therapeutic opportunities in a number of common diseases ranging from Thalassemias to cancers.
Acknowledgements
We thank Hegde lab members for useful discussions. Work in the authors’ lab is supported by the UK Medical Research Council (MC_UP_A022_1007 to RSH).
References
- Abovich N, Gritz L, Tung L, Rosbash M. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3429–3435. doi: 10.1128/mcb.5.12.3429. TL - 5. VN-re. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN, Martin RG. Biochemical Aspects of Genetics: The Operon. Annu Rev Biochem. 1964;33:235–258. doi: 10.1146/annurev.bi.33.070164.001315. [DOI] [PubMed] [Google Scholar]
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BDM, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- Bachurski CJ, Theodorakis NG, Coulson RM, Cleveland DW. An amino-terminal tetrapeptide specifies cotranslational degradation of beta-tubulin but not alpha-tubulin mRNAs. Mol Cell Biol. 1994;14:4076–4086. doi: 10.1128/mcb.14.6.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumal R, Potter M, Scharff MD. Synthesis, assembly, and secretion of gamma globulin by mouse myeloma cells. 3. Assembly of the three subclasses of IgG. J Exp Med. 1971;134:1316–1334. doi: 10.1084/jem.134.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Arad G, Weissbein U, Mandefro B, Maimon A, Golan-Lev T, Narwani K, Clark AT, Andrews PW, Benvenisty N, et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat Commun. 2014;5 doi: 10.1038/ncomms5825. 4825. [DOI] [PubMed] [Google Scholar]
- Benz EJ, Forget BG. The biosynthesis of hemoglobin. Semin Hematol. 1974;11:463–523. [PubMed] [Google Scholar]
- Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci U S A. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Harant H, Wang S, Oberhauser B, Marquardt K, Foster CA, Schreiner EP, de Vries JE, Dascher-Nadel C, Lindley IJD. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436:290–293. doi: 10.1038/nature03670. [DOI] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci U S A. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986;102:1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde MT, Pedersen M, Klausen MS, Jensen SI, Wulff T, Harrison S, Nielsen AT, Herrgård MJ, Sommer MOA. Predictable tuning of protein expression in bacteria. Nat Methods. 2016;13:233–236. doi: 10.1038/nmeth.3727. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Lippincott-Schwartz J. Degradation of proteins within the endoplasmic reticulum. Curr Opin Cell Biol. 1991;3:592–600. doi: 10.1016/0955-0674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Suzuki CK, Lippincott-Schwartz J, Weissman AM, Klausner RD. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J Cell Biol. 1989;109:73–83. doi: 10.1083/jcb.109.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Shah N, Klausner RD. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunger K, Pfeffer S, Shrimal S, Gilmore R, Berninghausen O, Mandon EC, Becker T, Förster F, Beckmann R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science. 2018;360:215–219. doi: 10.1126/science.aar7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JW, Hendershot LM. Building an antibody factory: a job for the unfolded protein response. Nat Immunol. 2005;6:23–29. doi: 10.1038/ni1149. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo JJ, Parodi AJ. A sweet code for glycoprotein folding. FEBS Lett. 2015;589:3379–3387. doi: 10.1016/j.febslet.2015.07.021. [DOI] [PubMed] [Google Scholar]
- Cassanelli S, Bertolini S, Rolleri M, De Stefano F, Casarino L, Elicio N, Naselli A, Calandra S. A“de novo” point mutation of the low-density lipoprotein receptor gene in an Italian subject with primary hypercholesterolemia. Clin Genet. 1998;53:391–395. doi: 10.1111/j.1399-0004.1998.tb02752.x. [DOI] [PubMed] [Google Scholar]
- Chakrabarti O, Rane NS, Hegde RS. Cytosolic aggregates perturb the degradation of nontranslocated secretory and membrane proteins. Mol Biol Cell. 2011;22:1625–1637. doi: 10.1091/mbc.E10-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Hunt KCL, Frydman J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016;536:224–228. doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K. Increased Proteasome Activity, Ubiquitin-Conjugating Enzymes, and eEF1A Translation Factor Detected in Breast Cancer Tissue. Cancer Res. 2005;65:5599–5606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- Chen Y-C, Umanah GKE, Dephoure N, Andrabi SA, Gygi SP, Dawson TM, Dawson VL, Rutter J. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 2014;33:1548–1564. doi: 10.15252/embj.201487943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke GM, Higgins TN. Laboratory Investigation of Hemoglobinopathies and Thalassemias: Review and Update. Clin Chem. 2000;46 [PubMed] [Google Scholar]
- Claude A. THE CONSTITUTION OF PROTOPLASM. Science (80-. ) 1943;97:451–456. doi: 10.1126/science.97.2525.451. [DOI] [PubMed] [Google Scholar]
- Claude A. Fractionation of mammalian liver cells by differential centrifugation : II. Experimental procedures and results. J Exp Med. 1946;84:61–89. [PubMed] [Google Scholar]
- Cleveland DW, Lopata MA, Sherline P, Kirschner MW. Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell. 1981;25:537–546. doi: 10.1016/0092-8674(81)90072-6. [DOI] [PubMed] [Google Scholar]
- Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- Costa EA, Subramanian K, Nunnari J, Weissman JS. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science (80-. ) 2018;359:689–692. doi: 10.1126/science.aar3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila López M, Martíne Guerra JJ, Samuelsson T. Analysis of gene order conservation in eukaryotes identifies transcriptionally and functionally linked genes. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson SE, Kim S, Costanzo M, Baryshnikova A, Morse DL, Kaiser CA, Boone C, Amon A. Chromosome-specific and global effects of aneuploidy in Saccharomyces cerevisiae. Genetics. 2016;202:1395–1409. doi: 10.1534/genetics.115.185660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisaldi B, Stewart RS, Adles C, Stewart LR, Quaglio E, Biasini E, Fioriti L, Chiesa R, Harris DA. Mutant PrP Is Delayed in Its Exit from the Endoplasmic Reticulum, but Neither Wild-type nor Mutant PrP Undergoes Retrotranslocation Prior to Proteasomal Degradation. J Biol Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- Duncan CDS, Mata J. Widespread Cotranslational Formation of Protein Complexes. PLoS Genet. 2011;7:e1002398. doi: 10.1371/journal.pgen.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C. The separation and characterization of subcellular particles. Harvey Lect. 1965;59:49–87. [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM. Quality Control of Integral Membrane Proteins by Assembly-Dependent Membrane Integration. Mol Cell. 2013;51:297–309. doi: 10.1016/j.molcel.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feit H, Slusarek L, Shelanski ML. Heterogeneity of tubulin subunits. Proc Natl Acad Sci U S A. 1971;68:2028–2031. doi: 10.1073/pnas.68.9.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Gell DA, Zhou S, Gu L, Kong Y, Li J, Hu M, Yan N, Lee C, Rich AM, et al. Molecular Mechanism of AHSP-Mediated Stabilization of α-Hemoglobin. Cell. 2004;119:629–640. doi: 10.1016/j.cell.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Feng L, Zhou S, Gu L, Gell DA, Mackay JP, Weiss MJ, Gow AJ, Shi Y. Structure of oxidized α-haemoglobin bound to AHSP reveals a protective mechanism for haem. Nature. 2005;435:697–701. doi: 10.1038/nature03609. [DOI] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hanebuth MA, Frickey T, Deuerling E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science (80-. ) 2015;348:201–207. doi: 10.1126/science.aaa5335. [DOI] [PubMed] [Google Scholar]
- Garrison JL, Kunkel EJ, Hegde RS, Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- Gavin A-C, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gay Da, Sisodia SS, Cleveland DW. Autoregulatory control of beta-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci U S A. 1989;86:5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guna A, Hegde RS. Transmembrane Domain Recognition during Membrane Protein Biogenesis and Quality Control. Curr Biol. 2018;28:R498–R511. doi: 10.1016/j.cub.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Hammond CM, Strømme CB, Huang H, Patel DJ, Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18:141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer AB, Opali ska M, Gerbeth C, Herman JS, Rao S, Schonfisch B, Guiard B, Schmidt O, Pfanner N, Meisinger C. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science (80-. ) 2014;346:1109–1113. doi: 10.1126/science.1261253. [DOI] [PubMed] [Google Scholar]
- Harper JW, Bennett EJ. Proteome complexity and the forces that drive proteome imbalance. Nature. 2016;537:328–338. doi: 10.1038/nature19947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12:787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Protein sorting signals: simple peptides with complex functions. EXS. 1995;73:67–76. doi: 10.1007/978-3-0348-9061-8_4. [DOI] [PubMed] [Google Scholar]
- Hendershot L, Bole D, Köhler G, Kearney JF. Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J Cell Biol. 1987;104:761–767. doi: 10.1083/jcb.104.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie C, Suzuki H, Sakaguchi M, Mihara K. Characterization of Signal That Directs C-Tail–anchored Proteins to Mammalian Mitochondrial Outer Membrane. Mol Biol Cell. 2002;13:1615–1625. doi: 10.1091/mbc.01-12-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. A novel ER -mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Deisenhofer J, Colman PM, Matsushima M, Palm W. Crystallographic structure studies of an IgG molecule and an Fc fragment. Nature. 1976;264:415–420. doi: 10.1038/264415a0. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak DN, Tyanova S, Cox J, Borner GH. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5 doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob HS, Brain MC, Dacie JV, Carrell RW, Lehmann H. Abnormal haem binding and globin SH group blockade in unstable haemoglobins. Nature. 1968;218:1214–1217. doi: 10.1038/2181214a0. [DOI] [PubMed] [Google Scholar]
- Jakel S, Görlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S-W, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamyshev AL, Patrick AE, Karamysheva ZN, Griesemer DS, Hudson H, Tjon-Kon-Sang S, Nilsson I, Otto H, Liu Q, Rospert S, et al. Inefficient SRP interaction with a nascent chain triggers a MRNA quality control pathway. Cell. 2014;156:146–157. doi: 10.1016/j.cell.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaplis AC, Lim SK, Baba H, Arnold A, Kronenberg HM. Inefficient membrane targeting, translocation, and proteolytic processing by signal peptidase of a mutant preproparathyroid hormone protein. J Biol Chem. 1995;270:1629–1635. doi: 10.1074/jbc.270.4.1629. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The Signal Recognition Particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Kihm AJ, Kong Y, Hong W, Russell JE, Rouda S, Adachi K, Simon MC, Blobel GA, Weiss MJ. An abundant erythroid protein that stabilizes free alpha-haemoglobin. Nature. 2002;417:758–763. doi: 10.1038/nature00803. [DOI] [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 Is an E3 Ubiquitin Ligase Involved in Degradation of Proteins from the Endoplasmic Reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Mitra D, Salerno JR, Hegde RS. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Kohne E. Hemoglobinopathies: clinical manifestations, diagnosis, and treatment. Dtsch Arztebl Int. 2011;108:532–540. doi: 10.3238/arztebl.2011.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren I, Timms RT, Kula T, Xu Q, Li MZ, Elledge SJ. The Eukaryotic Proteome Is Shaped by E3 Ubiquitin Ligases Targeting C-Terminal Degrons. Cell. 2018 doi: 10.1016/j.cell.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Lakkaraju AK, Abrami L, Lemmin T, Blaskovic S, Kunz B, Kihara A, Dal Peraro M, van der Goot FG. Palmitoylated calnexin is a key component of the ribosome-translocon complex. EMBO J. 2012;31:1823–1835. doi: 10.1038/emboj.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Zemojtel T, Shakhnovich E. Systems-Level Evidence of Transcriptional Co-Regulation of Yeast Protein Complexes. J Comput Biol. 2009;16:331–339. doi: 10.1089/cmb.2008.17TT. [DOI] [PubMed] [Google Scholar]
- Levine CG, Mitra D, Sharma A, Smith CL, Hegde RS. The Efficiency of Protein Compartmentalization into the Secretory Pathway. Mol Biol Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, Weissman JS. Quantifying Absolute Protein Synthesis Rates Reveals Principles Underlying Allocation of Cellular Resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Volk AG, Haug JS, Marshall SA, Woodfin AR, Bartom ET, Gilmore JM, Florens L, Washburn MP, Sullivan KD, et al. Therapeutic Targeting of MLL Degradation Pathways in MLL-Rearranged Leukemia. Cell. 2017;168:59–72.e13. doi: 10.1016/j.cell.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HN, Lee Y, Hussein R. Fundamental relationship between operon organization and gene expression. Proc Natl Acad Sci U S A. 2011;108:10626–10631. doi: 10.1073/pnas.1105692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-C, Yeh C-W, Chen Y-F, Lee T-T, Hsieh P-Y, Rusnac DV, Lin S-Y, Elledge SJ, Zheng N, Yen H-CS. C-Terminal End-Directed Protein Elimination by CRL2 Ubiquitin Ligases. Mol Cell. 2018;70:602–613.e3. doi: 10.1016/j.molcel.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Bonifacino JS, Yuan LC, Klausner RD. Degradation from the endoplasmic reticulum: Disposing of newly synthesized proteins. Cell. 1988;54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- Ma J, Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc Natl Acad Sci. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini R, Fagioli C, Fra aM, Maggioni C, Sitia R. Degradation of unassembled soluble Ig subunits by cytosolic proteasomes: evidence that retrotranslocation and degradation are coupled events. FASEB J. 2000;14:769–778. doi: 10.1096/fasebj.14.5.769. [DOI] [PubMed] [Google Scholar]
- Manolios N, Bonifacino JS, Klausner RD. Transmembrane helical interactions and the assembly of the T cell receptor complex. Science. 1990;249:274–277. doi: 10.1126/science.2142801. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Hernández H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell. 2013;153:461–470. doi: 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of Polyubiquitin Conjugates Reveals That the Rpn10 Substrate Receptor Contributes to the Turnover of Multiple Proteasome Targets. Mol Cell Proteomics. 2005;4:741–751. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative Profiling of Ubiquitylated Proteins Reveals Proteasome Substrates and the Substrate Repertoire Influenced by the Rpn10 Receptor Pathway. Mol Cell Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- McShane E, Sin C, Zauber H, Wells JN, Donnelly N, Wang X, Hou J, Chen W, Storchova Z, Marsh JA, et al. Kinetic Analysis of Protein Stability Reveals Age-Dependent Degradation. Cell. 2016;167:803–815.e21. doi: 10.1016/j.cell.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta - Gene Regul Mech. 2015;1849:801–811. doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Minami R, Hayakawa A, Kagawa H, Yanagi Y, Yokosawa H, Kawahara H. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol. 2010;190:637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y, Chen L, Chu BW, Swigut T, Wandless TJ. Distinct transcriptional responses elicited by unfolded nuclear or cytoplasmic protein in mammalian cells. Elife. 2015;4 doi: 10.7554/eLife.07687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock J-Y, Chartron JW, Zaslaver M, Xu Y, Ye Y, Clemons WM. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proc Natl Acad Sci U S A. 2015;112:106–111. doi: 10.1073/pnas.1402745112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey CM, Breckels LM, Geladaki A, Britovšek NK, Nightingale DJH, Christoforou A, Elzek M, Deery MJ, Gatto L, Lilley KS. Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat Protoc. 2017;12:1110–1135. doi: 10.1038/nprot.2017.026. [DOI] [PubMed] [Google Scholar]
- Munsky B, Neuert G, Van Oudenaarden A. Using gene expression noise to understand gene regulation. Science (80-. ) 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ME. The HSP70 family and cancer. Carcinogenesis. 2013;34:1181–1188. doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial Import Efficiency of ATFS-1 Regulates Mitochondrial UPR Activation. Science (80-. ) 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson HC. Structure and function of DNA-binding proteins. Curr Opin Genet Dev. 1995;5:180–189. doi: 10.1016/0959-437x(95)80006-9. [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Prado MA, Schmidt PJ, Sendamarai AK, Wilson-Grady JT, Min M, Campagna DR, Tian G, Shi Y, Dederer V, et al. UBE2O remodels the proteome during terminal erythroid differentiation. Science (80-. ) 2017;357:eaan0218. doi: 10.1126/science.aan0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Walter P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci U S A. 2014;111:8019–8024. doi: 10.1073/pnas.1405755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri NF. The β-Thalassemias. N Engl J Med. 1999;341:99–109. doi: 10.1056/NEJM199907083410207. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE. A small particulate component of the cytoplasm. J Biophys Biochem Cytol. 1955;1:59–68. doi: 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-H, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C, Hurt E, Panse VG. Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol. 2017;24:689–699. doi: 10.1038/nsmb.3454. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North ACT. Structure of Hæmoglobin: A three-dimensional fourier synthesis at 5.5-. resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Zimmermann R, Förster F. Organization of the native ribosome-translocon complex at the mammalian endoplasmic reticulum membrane. Biochim Biophys Acta. 2016;1860:2122–2129. doi: 10.1016/j.bbagen.2016.06.024. [DOI] [PubMed] [Google Scholar]
- Piel FB, Weatherall DJ. The α-Thalassemias. N Engl J Med. 2014;371:1908–1916. doi: 10.1056/NEJMra1404415. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Cleveland DW. Retention of autoregulatory control of tubulin synthesis in cytoplasts: demonstration of a cytoplasmic mechanism that regulates the level of tubulin expression. J Cell Biol. 1985;101:1941–1952. doi: 10.1083/jcb.101.5.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter KR. The submicroscopic morphology of protoplasm. Harvey Lect. 1956;51:175–228. [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Rane NS, Yonkovich JL, Hegde RS. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane NS, Kang S-W, Chakrabarti O, Feigenbaum L, Hegde RS. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev Cell. 2008;15:359–370. doi: 10.1016/j.devcel.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane NS, Chakrabarti O, Feigenbaum L, Hegde RS. Signal sequence insufficiency contributes to neurodegeneration caused by transmembrane prion protein. J Cell Biol. 2010;188:515–526. doi: 10.1083/jcb.200911115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Li L, Park E. Structural and Mechanistic Insights into Protein Translocation. Annu Rev Cell Dev Biol. 2017;33:369–390. doi: 10.1146/annurev-cellbio-100616-060439. [DOI] [PubMed] [Google Scholar]
- Rieder RF. Human hemoglobin stability and instability: molecular mechanisms and some clinical correlations. Semin Hematol. 1974;11:423–440. [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Gutierrez E, Hegde RS. Cytosolic Quality Control of Mislocalized Proteins Requires RNF126 Recruitment to Bag6. Mol Cell. 2014;55:227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded Membrane Proteins Are Specifically Recognized by the Transmembrane Domain of the Hrd1p Ubiquitin Ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ Proteases: ATP-Fueled Machines of Protein Destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Scazzari M, Amm I, Wolf DH. Quality control of a cytoplasmic protein complex: Chaperone motors and the ubiquitin-proteasome system govern the fate of orphan fatty acid synthase subunit Fas2 of yeast. J Biol Chem. 2015;290:4677–4687. doi: 10.1074/jbc.M114.596064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, et al. Regulation of Mitochondrial Protein Import by Cytosolic Kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, Baker D, Chambers MG, Su H, Li D, et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature. 2017;548:352–355. doi: 10.1038/nature23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppen J, Steenken E, Lindhout D, Bosma PJ, Elferink RP. A mutation which disrupts the hydrophobic core of the signal peptide of bilirubin UDP-glucuronosyltransferase, an endoplasmic reticulum membrane protein, causes Crigler-Najjar type II. FEBS Lett. 1996;390:294–298. doi: 10.1016/0014-5793(96)00677-1. [DOI] [PubMed] [Google Scholar]
- Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Rodrigo-Brenni MC, Kivlen MH, Hegde RS. Mechanistic basis for a molecular triage reaction. Science. 2017;355:298–302. doi: 10.1126/science.aah6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemorry A, Hwang CS, Varshavsky A. Control of Protein Quality and Stoichiometries by N-Terminal Acetylation and the N-End Rule Pathway. Mol Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SS, Vats S, Chia AY-Q, Tan TZ, Deng S, Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G, et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M-K, Porras-Yakushi TR, Reitsma JM, Huber FM, Sweredoski MJ, Hoelz A, Hess S, Deshaies RJ. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife. 2016;5 doi: 10.7554/eLife.19105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Kawahara H. UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep. 2016;17:842–857. doi: 10.15252/embr.201541402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous A, Pisoni GB, Hebert DN, Molinari M. N-linked sugar-regulated protein folding and quality control in the ER. Semin Cell Dev Biol. 2015;41:79–89. doi: 10.1016/j.semcdb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Sriram SM, Park KS, Kwon YT. The N-End Rule Pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorakis NG, Cleveland DW. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol. 1992;12:791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699–2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove M, Usherwood YK, Hendershot LM. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity. 2001;15:105–114. doi: 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila IK, Yao Y, Kim G, Xia W, Kim H, Kim S-J, Park M-K, Hwang JP, González-Billalabeitia E, Hung M-C, et al. A UBE2O-AMPKα2 Axis that Promotes Tumor Initiation and Progression Offers Opportunities for Therapy. Cancer Cell. 2017;31:208–224. doi: 10.1016/j.ccell.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali DG, Sinzel M, Bulthuis EP, Kolb A, Zabel S, Mehlhorn DG, Figueiredo Costa B, Farkas Á, Clancy A, Schuldiner M, et al. The GET pathway can increase the risk of mitochondrial outer membrane proteins to be mistargeted to the ER. J Cell Sci. 2018;131:jcs211110. doi: 10.1242/jcs.211110. [DOI] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Hegde RS. Structures of the scanning and engaged states of the mammalian srp-ribosome complex. Elife. 2015;4:1–21. doi: 10.7554/eLife.07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Mitra G, Schwindinger WF, Studeny M, Fried HM. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattenberg BW, Clark D, Brock S. An Artificial Mitochondrial Tail Signal/Anchor Sequence Confirms a Requirement for Moderate Hydrophobicity for Targeting. Biosci Rep. 2007;27:385–401. doi: 10.1007/s10540-007-9061-0. [DOI] [PubMed] [Google Scholar]
- Weatherall JD, C J. The thalassaemia syndromes. Blackwell Publ. Ltd; 2001. pp. 132–174. [Google Scholar]
- Wefes I, Mastrandrea LD, Haldeman M, Koury ST, Tamburlin J, Pickart CM, Finley D. Induction of ubiquitin-conjugating enzymes during terminal erythroid differentiation. Proc Natl Acad Sci U S A. 1995;92:4982–4986. doi: 10.1073/pnas.92.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Amon A. MitoCPR—A surveillance pathway that protects mitochondria in response to protein import stress. Science (80-. ) 2018;360 doi: 10.1126/science.aan4146. eaan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir NR, Kamber RA, Martenson JS, Denic V. The AAA protein Msp1 mediates clearance of excess tail-anchored proteins from the peroxisomal membrane. Elife. 2017;6 doi: 10.7554/eLife.28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Protein Translocation Across Biological Membranes. Science (80-. ) 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis Ta, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- Williams NK, Dichtl B. Co-translational control of protein complex formation: a fundamental pathway of cellular organization? Biochem Soc Trans. 2018 doi: 10.1042/BST20170451. BST20170451. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy Affects Proliferation and Spontaneous Immortalization in Mammalian Cells. Science (80-. ) 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlever ML, Mateja A, McGilvray PT, Day KJ, Keenan RJ. Msp1 Is a Membrane Protein Dislocase for Tail-Anchored Proteins. Mol Cell. 2017;67:194–202.e6. doi: 10.1016/j.molcel.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G, Terada K, Yano M, Sergeev I, Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res. 2001;263:107–117. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- Xu Y, Anderson DE, Ye Y. The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation. Cell Discov. 2016;2 doi: 10.1038/celldisc.2016.40. 16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Juszkiewicz S, Hegde RS. UBE2O is a quality control factor for orphans of multiprotein complexes. Science. 2017;357:472–475. doi: 10.1126/science.aan0178. [DOI] [PMC free article] [PubMed] [Google Scholar]