Abstract
G protein-coupled receptors (GPCRs) represent important drug targets, as they regulate pivotal physiological processes and they have proved to be readily druggable. Natural products have been and continue to be amongst the most valuable sources for drug discovery and development. Here, we surveyed small molecules and (poly-)peptides derived from plants, animals, fungi, and bacteria, which modulate GPCR signaling. Among naturally occurring compounds, peptides from plants, cone-snails, snakes, spiders, scorpions, fungi, and bacteria are of particular interest as lead compounds for the development of GPCR ligands, since they cover a chemical space, which differs from that of synthetic small molecules. Peptides, however, face challenges, some of which can be overcome by studying plant-derived compounds. We argue here that the opportunities outweigh the challenges.
G Protein-Coupled Receptors and Natural Products: A Magic Dyad
In the beginning of the 19th century, the German pharmacist Friedrich Sertürner extracted opium (i.e., the dried latex of the poppy Papaver somniferum) and isolated for the first time a pharmacologically active compound from a plant [1]. The most active opium component, which he named morphine, became a precedent for the exploration of natural products for medicinal purposes. Since then, many drugs from plants and other organisms, including fungi, bacteria, and animals, have been discovered. In fact, these compounds were instrumental for the emerging discipline of pharmacology. This can be gauged from the nomenclature of receptors: acetylcholine receptors, for instance, are still classified based on their prototypical agonists, nicotine (from the herbaceous plant Nicotiana tabacum) and muscarine (from the mushroom Amanita muscaria). Because of their very large structural and chemical diversity, natural products still continue to be powerful and invaluable sources of compounds with drug-like properties (see Glossary) [2]. For instance, a recent systematic survey demonstrated that between 1981 and 2014, 34% of the 1562 new drugs approved by the FDA were derived from natural products and their derivatives [3].
Glossary.
Aquaretic effect: increase in urinary volume with no loss of electrolytes.
Cyclic cystine-knot motif: conserved structural motif of cyclotides comprising a head-to-tail cyclic backbone and a cystine-knot in which an embedded ring formed by two disulfide bonds is threaded by a third disulfide bond.
Drug-like properties: specific characteristics of a given molecule such as size, shape, or solubility shared with other molecules, which are considered as precursors of drugs (lead compounds).
EC50/IC50: measure of ligand potency; it defines the ligand concentration that produces 50% of the maximum effect (Emax) or reduces the response/binding by 50%, respectively.
Enkephalin-like peptide: peptide that resembles sequence or structure of enkephalin, a neuropeptide that binds to opioid receptors.
Grafting: insertion of a bioactive peptide epitope into a naturally occurring stable peptide scaffold, thereby generating a more stable peptide while retaining biological activity.
Hypereosinophilia: persistent elevation of peripheral blood eosinophilic leukocytes greater than 1.5 × 109 l–1.
Kd/Ki: measure of ligand affinity; it is the equilibrium dissociation constant that indicates the concentration at which 50% of the receptor binding sites are occupied by the ligand.
Kunitz domain: domain of Kunitztype protease inhibitors consisting of about 60 amino acid residues stabilized by three disulfide bonds.
Ligand bias: ligand-dependent selectivity for activating a certain signaling pathway of a receptor relative to a reference (e.g., the endogenous peptide ligand).
Polycystic kidney disease: genetic disorder associated with occurrence of numerous cysts within the kidneys as well as other organs.
Secondary metabolites: biologically active small chemicals produced by microbes or plants, which are not directly required for normal growth, development, and reproduction. They are often involved in interspecies communication or defense.
Sequence diversity: variety of amino acid sequences within peptides or proteins likely evolved due to natural selection.
Structural plasticity: the ability of biomolecules such as peptides or proteins to tolerate amino acid substitutions, insertions, or deletions within the backbone chain that do not change the overall fold.
Taste 2 GPCRs: referred to as T2Rs or TAS2Rs, belong to a family of ~25 human GPCRs that enable perception of bitter taste, or more generally are activated by ‘bitter’ substances, not only in the tongue.
Uterotonic: agents which induce tone and contractions of the uterus muscle.
Venom: mixture of toxic substances produced by an animal for prey capture and defense.
G protein-coupled receptors (GPCRs), also referred to as seven-transmembrane receptors [4], represent the largest family of membrane proteins: the human genome encodes approximately 800 members [5] (i.e., GPCRs account for about 13% of all membrane proteins). The seven-transmembrane core of these receptors, in combination with variable N-terminal extensions, allows for recognition of chemically diverse ligands ranging from ions, small amines and organic acids (including volatile odorants), nucleosides and nucleotides to lipids, peptides, and large proteins. The extracellular ligand binding pocket represents the input side; the helical bundles of the hydrophobic core allow for translating the conformational change induced by ligand binding to the output side on the intracellular face: this results in both G protein-dependent and G protein-independent cellular signals [4,6,7]. Apart from the orthosteric site, where the cognate agonists are bound, GPCRs also harbor additional binding sites, which support allosteric modulation of their activity [5,7]. Because structures of more than 50 distinct GPCRs have been solved, it is possible to combine computational approaches, combinatorial chemistry, and high-content screening to identify new GPCR ligands [8]. It is thus likely that, in the foreseeable future, GPCRs will remain a prime target of approved and marketed drugs [5].
Considering GPCRs as today’s most exploited drug targets and the utmost importance of natural products for drug discovery, in this review we provide an overview of natural products from plants, animals, fungi, and bacteria that have been discovered as GPCR ligands throughout the past seven decades. For the sake of clarity, we define a natural product as any unmodified compound either: (i) isolated from a plant, an animal, a fungus, or a bacterium; or (ii) identified in any of these organisms via in silico approaches, followed by its chemical synthesis and pharmacological characterization. Further, we present a compilation of nature-derived ligands that are available as approved drugs (past and present) acting on GPCRs. Nature-derived peptides cover a chemical space, which is not readily accessible to synthetic combinatorial chemistry. Hence, we will summarize recent findings on nature-derived peptides as GPCR ligands and point out the opportunities provided by plant-derived cysteine-rich peptides, venom-derived peptides from cone-snails, snakes, spiders, and scorpions, as well as peptides from marine fungi and bacteria, as novel drug leads. Finally, we will discuss challenges and opportunities of nature-derived peptides for GPCR ligand discovery. Overall, this review provides a brief historical overview and summary about the discovery of natural products as GPCR ligands and highlights the emerging potential of nature-derived peptides as a toolbox and treasure-trove of GPCR drug discovery and development.
Diversity of Natural GPCR Ligands
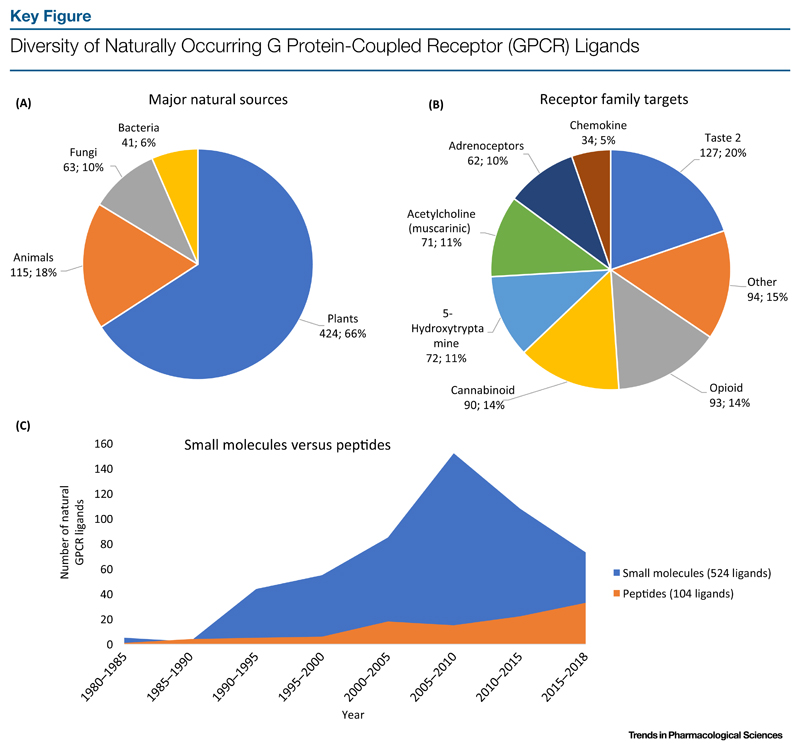
As a starting point for a comprehensive overview of natural products targeting GPCRs, we performed database searches using PubMed and Science Direct to mine the published literature from September 1954 until August 2018. The common Medical Subject Heading terms including natural product, GPCR, plant, bacteria, fungi, and venom in various combinations were used in the database search engines. Our analysis focused on the known GPCRs of all five major families in humans, including class A (rhodopsin), class B (secretin), class C (glutamate), class F (frizzled/taste), and the adhesion receptor family. In total, there are 643 unmodified natural products that act on GPCRs. Figure 1A (Key Figure) illustrates the ratio and number of natural GPCR ligands that have been identified in the four kingdoms of life, namely plants, animals, fungi, and bacteria. Among these, plants represent the most eminent source, accounting for 66% (424) of all nature-derived GPCR ligands (643) (Figure 1A). Furthermore, we determined that secondary metabolites represent the majority of natural GPCR ligands isolated from plants.
Figure 1.
(A) Plants constitute the best source of nature-derived GPCR ligands. (B) Taste 2, aminergic, as well as opioid and cannabinoid GPCRs are leading targets of natural products. (C) Trend in discovery of small molecules (524) versus peptides (104) as nature-derived GPCR ligands from 1980 to 2018. Ligands before 1980 were not included.
Based on published knowledge it is evident that aminergic, opioid, and cannabinoid receptors are primary GPCR targets of natural products (Figure 1B). Additionally, taste 2 GPCRs and its diverse receptors were identified as one of the major families targeted by nature-derived compounds (Figure 1B). However, according to our analysis based on published knowledge, only 119 GPCRs (out of an estimated >800 receptors) have been targeted by natural products. This further highlights that more than 600 GPCRs remain to be discovered as targets of naturally occurring ligands. In this context it is noteworthy to emphasize that small molecules account for the majority of GPCR ligands, which we identified (539; 84%), while 104 ligands (16%) are peptides. Most small molecules were found in plants, whereas animals, fungi, and bacteria represent the most abundant sources of peptide GPCR ligands.
It is interesting to note that at least 16 FDA-approved drugs (past and present) that target GPCRs, are natural products (Table 1); this list is largely a compilation to provide an overview. Among those drugs, exendin-4 isolated from the Gila monster (Heloderma suspectum) is an example of a nature-derived peptide drug (BYETTA®). It was introduced in 2005 to treat diabetes mellitus type 2 and acts as an agonist of the glucagon-like peptide-1 receptor [9].
Table 1. Natural Products Acting on GPCRs that are Approved (Past and Present) by the FDAa.
| Drug name | Source | Targets | Medical use | Drug type |
|---|---|---|---|---|
| Atropine | Plant (Atropa belladonna) |
ACM1–ACM5 | Control of heart rate; antidote for organophosphate poisoning; cycloplegia; mydriasis | Small molecule (alkaloid) |
| Caffeine | Plant (Coffea arabica) |
AA1R, AA2AR, AA2BR | Central nervous system stimulant; infant apnea | Small molecule (alkaloid) |
| Cannabidiol | Plant (Cannabis sativa) |
GPR55b | Seizures associated with epilepsy: Lennox-Gastaut and Dravet syndromes | Small molecule (cannabinoid) |
| Codeine | Plant (Papaver somniferum) |
OPRD, OPRK, OPRM | Analgesic | Small molecule (alkaloid) |
| Ergonovine | Fungus (Claviceps purpurea) |
ADRA1A | Antihemorrhagic | Small molecule (peptide-alkaloid) |
| Ergotamine | Fungus (C. purpurea) |
5HT1B, 5HT1D, ADRA1B, ADRA1D, ADRA2A, ADRA2B, ADRA2C | Antimigraine | Small molecule (peptide-alkaloid) |
| Ephedrine | Plant (Ephedra sinica) |
ADRA1A | Hypotension | Small molecule (alkaloid) |
| Exendin-4 | Animal; Gila monster (Heloderma suspectum) |
GLP1R | Diabetes mellitus type 2 | Peptide |
| Hyoscyamine | Plant (Hyoscyamus niger) |
ACM1, ACM2 | Antispasmodic | Small molecule (alkaloid) |
| Morphine | Plant (P. somniferum) |
OPRD, OPRK, OPRM | Analgesic; opioid addiction | Small molecule (alkaloid) |
| Pilocarpine | Plant(Pilocarpus microphyllus) | ACM1–ACM3 | Glaucoma; dry mouth | Small molecule (alkaloid) |
| Pseudoephedrine | Plant (E. sinica) |
ADA1A, ADA2A | Anti-allergic | Small molecule (alkaloid) |
| Scopolamine | Plant (Datura stramonium) |
ACM1 | Motion sickness | Small molecule (alkaloid) |
| Tetrahydro-cannabinol | Plant (C. sativa) |
CNR1, CNR2 | Analgesic: neuropathic pain; restless legs syndrome | Plant extractc |
| Theophylline | Plant (Camellia sinensis) |
AA1R, AA2AR, AA2BR, AA3R | Bronchodilator; anti-asthmatic | Small molecule (alkaloid) |
| Yohimbine | Plant(Pausinystalia yohimbe) | 5HT1A, 5HT1B, 5HT1D, 5HT2B, 5HT5A, 5HT7R, ADRA2A, ADRA2B, ADRA2C | Erectile dysfunction | Small molecule (alkaloid) |
We list natural ligands identified as drugs in the Drugs@FDA databasei as well as GPCRdbii, which are considered to function via GPCRs (data extracted in January 2019). GPCRs are listed using the protein name according to UniProt. For more details on GPCR nomenclature, see the IUPHAR/BPS Guide to Pharmacology.
The exact mechanism of action is unknown. Antiseizure activity of cannabidiol is probably mediated by multiple seven-transmembrane receptors, ion channels, and neurotransmitter transporters, however GPR55 is suggested to play an important role (see [73] for a recent review).
The approved form of this drug is an extract. This has been exemplarily included, however, please note that there may be other pharmacological mixture that exist as approved drugs, which are not listed.
Nature-Derived Peptides as GPCR Ligands
Naturally occurring small molecules have had an important role in the history of GPCR drug discovery. In many instances, this can be traced to their superior drug-like properties, most notably high stability and good oral bioavailability [10]. Besides, small molecules are associated with further advantages, such as low production costs and lipophilicity, a characteristic that confers them the ability to penetrate cells and cross membranes. Small molecules currently dominate the drug market mainly for these reasons [5,11]. However, several drawbacks limit their usefulness in drug development. For instance, limited target specificity and hence increased probability for off-target effects remain a persistent problem [12]. By contrast, over the past few years peptides have gained remarkable interest and significance in drug discovery. They are recognized as reliable alternatives for small molecules, owing to their high selectivity and low toxicity [11]. In addition, peptides may be metabolized and cleared without accumulation in body tissues, thereby minimizing the occurrence of side effects [13]. This explains why there are currently more than 60 peptides approved as drugs, over 150 peptides under clinical investigation for a variety of indications [13], and many more in preclinical development.
It is safe to conclude that naturally occurring small molecules will remain important templates for GPCR drug discovery. However, our analysis demonstrates that the discovery of small molecules reached an apparent peak in 2010 (Figure 1C). By contrast, starting from 1980, the number of identified peptide GPCR ligands has been continuously rising, with a notable boost between 2015 and 2018 as nature-derived peptides were starting to be recognized for their potential in drug development. We predict this trend will continue with the help of emerging peptide-mining and -chemistry technologies and because of the steadily increasing importance of peptides for drug development.
An exhaustive list of 103 GPCR-targeting nature-derived peptides, including their natural sources, targets, mode-of-action, and structural properties is illustrated in Table 2 [one peptide GPCR ligand, exendin-4 has already been approved (Table 1)]. This number would be considerably larger if one were to consider human endogenous or protein-embedded GPCR ligands that become activated by proteases, for instance upon viral infection (Box 1). Representative examples of these nature-derived peptide classes will be discussed in detail in the sections below.
Table 2. Nature-Derived Peptides Acting as GPCR Ligands.
| Peptide name | Source | Targets | Mode-of-action | Peptide structure | Refs |
|---|---|---|---|---|---|
| BE-18257B | Bacteria (Streptomyces misakiensis) |
EDNRA | Competitive antagonist | Cyclic pentapeptide | [74] |
| Duramycin | Bacteria (Streptomyces alboflavus) |
CXCR3 | Antagonist | 19-Amino acid (aa) cyclic thiopeptide | [75] |
| RES-701-1, 701-2, 701-3, and 701-4 | Bacteria (Streptomyces sp.) |
EDNRB | Antagonist | 16-aa cyclic peptides | [76] |
| SP 1, 2, 6-9 | Bacteria (Streptococcus suis, Bacillus cereus, Psychromonas ingrahamii, Shewanella baltica, Desulfotomaculum reducens, and Borrelia burgdorferi) |
FPR1, FPR2 | Agonist | N-formylated signal hexapeptides | [77] |
| Brintonamides C, D, and Ea | Cyanobacteria (Oscillatoria sp.) |
CXCR7, CCR10, OXTR, TACR2, SSTR3 | Agonist, antagonist | Linear hexapeptides | [39] |
| CJ-15,208 | Fungus (Ctenomyces serratus) |
OPRK, OPRM, OPRD | Antagonist | Cyclic tetrapeptide | [78] |
| Cyclosporin-A and –H | Fungus (Tolypocladium inflatum) |
FPR1 | Antagonist | Cyclic undecapeptides | [79,80] |
| Endolide A and Bb | Fungus (Stachylidium sp.) |
V1AR 5HT2B | n.d. | Cyclic N-methylated and 3-(3-furyl)-alanine-containing tetrapeptides | [38] |
| SCH-378161, –217048, –378199, and –378167 | Fungus (taxonomically unidentified) |
NK2R | Antagonist | Cyclic nonadepsipeptides | [81] |
| Rubiscolins 5 and 6 | Plant (Spinacia oleracea) |
OPRD | Agonist | Rubisco-derived linear penta-and hexapeptides | [82] |
| Kalata B7 | Plant (Oldenlandia affinis) |
OXTR, V1AR | Partial agonist | 29-aa cyclic cystine-knot peptide; three disulfide bonds (I–IV, II–V, III–VI) | [17] |
| Caripe 8 | Plant (Carapichea ipecacuanha) |
CRFR1 | Antagonist | 31-aa cyclic cystine-knot peptide; three disulfide bonds (I–IV, II–V, III-VI) | [23] |
| Cyclopsychotride A | Plant (Psychotria longipes) |
NTR1 | Antagonist | 31-aa cyclic cystine-knot peptide; three disulfide bonds (I–IV, II–V, III–VI) | [83] |
| Soymorphins 5, 6, and 7 | Plant (Glycine max) |
OPRM | Agonist | β-conglycinin-derived linear penta-, hexa-, and heptapeptides | [84] |
| Barettin and 8,9-dihydrobarettinc | Sponge (Geodia baretti) |
5HT2A, 5HT2C, 5HT4R | n.d. | Brominated cyclodecapeptides | [85] |
| Polydiscamides B, C, and D | Sponge (Ircinia sp.) |
SNSR | Agonist | 13-aa depsipeptides | [86] |
| ρ-TIAd | Cone-snail (Conus tulipa) |
ADRA1A; ADRA1B; ADRA1D | Competitive and non-competitive antagonist | C-terminally amidated 19-aa peptide; two disulfide bonds (I-III;II-IV) | [29,30] |
| Contulakin-G | Cone-snail (Conus geographus) |
NTR1 | Agonist | N-terminal pyroglutamate 16-aa O-linked glycopeptide | [87] |
| Conopressin T, S and Ge | Cone-snail (C. tulipa, Conus striatus, and C. geographus) |
V1AR, OXTR, V1BR | Antagonist, partial agonist | C-terminally amidated nonapeptides; one disulfide bond | [88,89] |
| τ-CnVA and LiC32 | Cone-snail (Conus consor and Conuslividus) |
SSR3 | Antagonist | C-terminally amidated 14- and 15-aa peptides; two disulfide bonds (I–III, II–IV) | [90] |
| Conorphin-T | Cone-snail (Conus textile) |
OPRK | Agonist | C-terminally amidated nonapeptide; two disulfide bonds (I–II, III–IV) | [91] |
| BulA | Cone-snail (Conus bullatus) |
LPAR6 | Competitive antagonist | C-terminally amidated 13-aa peptide; two disulfide bonds (I–III,II–IV) | [92] |
| Vc1.1, RgIA, AuIB, Vc1.2, and PeIA | Cone-snail (Conus victoriae, Conus regius, Conus aulicus, and Conus pergrandis) |
GABR1, GABR2 | Allosteric modulator | C-terminally amidated 14- and 16-aa peptides; two disulfide bonds (I–III, II–IV) | [27] |
| Helokinestatin | Gila monster (Heloderma suspectum) and Mexican beaded lizard (Heloderma horridum) |
BKRB2 | Antagonist | Proline-rich decapeptide | [93] |
| Inotocinf | Black garden ant (Lasius nigert) |
OXTR, V1AR, V1BR, and V2R | Agonist,allosteric modulator | C-terminally amidated nonapeptide | [71] |
| Apamin | Bee (Apis mellifera) |
ACM2 | Agonist | 18-aa cyclic peptide; two disulfide bonds (I–III, II–IV) | [94] |
| THR6-BK | Wasp (Polybia occidentalis) |
BKRB2 | Agonist | Linear nonapeptide | [95] |
| NLP-24 | Nematode (Caenorhabditis elegans) |
OPRK, OPRM | Agonist | C-terminally amidated pentapeptide | [96] |
| α-Latrotoxin | Spider (Latrodectus tredecimguttatus) |
ADGRL1 | Agonist | 128-kDa cysteine-rich polypeptide | [35] |
| δ-CNTX-Pn1a | Spider (Phoneutria nigriventer) |
CNR1, OPRM, OPRD | Agonist | 48-aa peptide; five disulfide bonds (I–V, II–III, IV–VI, VII–VIII, IX– | [36] |
| BmK-YA | Scorpion (Buthus martensii) |
OPRM, OPRK, OPRD | Agonist | C-terminally amidated linear octapeptide | [37] |
| TsHpt-I | Scorpion (Titys serrulatus) |
BKRB2 | Agonist | 25-aa linear proline-rich peptide | [97] |
| Bv8 | Frog (Bombina variegata) |
PKR1, PKR2 | Agonist (Bombina variegata) | 77-aa peptide; five disulfide bonds (I–V, II–III, IV–VI, VII–VIII, IX–X) | [89] |
| Kinestatin | Frog (Bombina maxima) |
BKRB2 | Antagonist | C-terminally amidated linear nonapeptide | [98] |
| Deltorphin-1 and –2 and dermorphin | Frog (Phyllomedusa sauvagii) |
OPRM, OPRD | Agonist | C-terminally amidated linear heptapeptides | [99,100] |
| Mambaqauretin-1 | Snake (Dendroaspis angusticeps) |
V2R | Competitive antagonist | 57-aa Kunitz-domain peptide;three disulfide bonds (I–VI, II–IV,III–V) | [34] |
| WTX | Snake (Naja kaouthia) |
ACM1-ACM5 | Allosteric modulator | 66-aa peptide; five disulfide bonds (I–V, II–III, IV–VI, VII–VIII, IX–X) | [101] |
| γ-Bungarotoxin | Snake (Bungarus multicinctus) |
ACM2 | n.d. | 68-aa peptide; five disulfide bonds (I–V, II–III, IV–VI, VII–VIII, IX–X) | [102] |
| ρ-Da1a and ρ-Da1bg | Snake (D. angusticeps) |
ADRA1A, ADRA2A, DRD3 | Non-competitive antagonist | 66-aa peptides; four disulfide bonds (I–III, II–IV, V–VI, VII–VIII) | [103,104] |
| Sarafotoxins m, b and i3 | Snake (Atractaspis irregularis) |
EDNRB | Agonist | 20-aa, 24-aa and 23-aa peptides; two disulfide bonds (I–IV, II–III) | [105] |
| β-Cardiotoxin | Snake (Ophiophagus hannah) |
ADRB1, ADRB2 | Antagonist | 63-aa peptide; four disulfide bonds (I–III, II–IV, V–VI, VII–VIII) | [106] |
| α-Cobratoxin | Snake (Naja naja kaouthia) |
ACM4 | Agonist | 71-aa peptide; five disulfide bonds (I–V, II–III, IV–VI, VII–VIII, IX–X) | [107] |
| MT-MIα | Snake (Micrurus lemniscatus) |
ACM1–ACM5 | Antagonist | Cysteine-rich peptide; 12-aa partial sequence available only | [108] |
| MT1 and MT2h | Snake (D. angusticeps) |
ACM1, ACM4, ADRA2B | Agonist, antagonist, allosteric modulator | 65 and 66-aa peptides, fourdisulfide bonds (I–III, II–IV, V–VI,VII–VIII) | [103,109] |
| MT3 (m-4 toxin) and MT6i | Snake (D. angusticeps) |
ACM1, ACM4, ADRA1A, ADRA2A, ADRA2C | Antagonist, non-competitive antagonist | 65-aa peptides, four disulfide bonds (I–III, II–IV, V–VI, VII–VIII) | [89,103] |
| MT4 and MT5j | Snake (D. angusticeps) |
ACM1, ACM2, ACM4, ADRB2 | Antagonist | 65 and 66-aa peptides, four disulfide bonds (I–III, II–IV, V–VI,VII–VIII) | [89,110] |
| MT7 (m-1 toxin) | Snake (D. angusticeps) |
ACM1 | Negative allosteric modulator | 65-aa peptide; four disulfide bonds (I–III, II–IV, V–VI, VII–VIII) | [103] |
| MT-α and MT-βk | Snake (Dendroaspis polylepis) |
ACM1, ACM2, ACM3, ACM4, ACM5, ADRA2B | Antagonist | 65 and 66-aa peptides, four disulfide bonds (I–III, II–IV, V–VI,VII–VIII) | [110,111] |
| Bj-PRO-7a | Snake (Bothrops jararaca) |
ACM1 | Agonist | Proline-rich linear heptapeptide | [112] |
| BM14 | Snake (B. multicinctus) |
ACM2 | n.d. | 82-aa peptide; five disulfide bonds (I–V, II–III, IV–VI, VII–VIII, IX–X) | [113] |
| Sarafotoxins a, c, and S6c | Snake (Atractaspis engaddensis) |
EDNRA, EDNRB | Agonist | 21-aa peptides; two disulfide bonds (I–IV, II–III) | [114,115] |
| Bibrotoxin | Snake (Atractaspis bibronii) |
EDNRB | Agonist | 21-aa peptide; two disulfide bonds (I–IV, II–III) | [116] |
| MIT1 | Snake (D. polylepis) |
PKR1, PKR2 | Agonist | 81-aa peptide; five disulfide bonds (I–V, II–III, IV–VI, VII–VIII, IX–X) | [89] |
| Crotalphine | Snake (Crotalus durissus terrificus) |
OPRK | Agonist | 14-aa peptide; one disulfide bond | [117] |
| MTLP-1 | Snake (N. kaouthia) |
ACM1–ACM5 | n.d. | 65-aa peptide; four disulfide bonds (I–III, II–IV, V–VI, VII–VIII) | [118] |
| Pep 1-8 | Bovine | T2R4 | Antagonist | Bovine protein-derived peptides | [119] |
| β-Lactotensin | Bovine | NTR2 | Agonist | Bovine protein-derived peptide | [120] |
GPCRs are listed using the protein name according to UniProt. For more details please refer to the IUPHAR/BPS Guide to Pharmacology. n.d. refers to not determined (ligand’s mode-of-action).
Brintoamides C, D, and E are agonists of CXCR7. Brintonamide C is further an antagonist of SSTR3 and TACR2. Brintonamide D antagonizes CCR10, OXTR, SSTR3, and TACR2. Brintonamide E acts as an antagonist of CCR10, OXTR, and TACR2.
Endolide A targets V1AR, while endolide B is selective for 5HT2BR.
Barettin binds to 5HT2A, 5HT2C, and 5HT4R, whereas 8,9-dihydrobarettin is selective for 5HT2C.
ρ-TIA noncompetitively antagonizes ADRA1B, while it antagonizes ADRA1A and ADRA1D in a competitive manner.
Conopressin S was shown to bind to V1AR, V1BR, and OXTR, while conopressin G was demonstrated to bind to OXTR. Their mechanism of action is not determined.
Inotocin shows the ability to bind to human OXTR, V1AR, and V1BR but it acts as a full agonist on human V1BR, an allosteric modulator on human V2R, and an antagonist on human V1AR.
ρ-Da1a was reported to be a noncompetitive antagonist on ADRA1A whereas ρ-Da1b functions as an antagonist on ADRA2A and DRD3 in a noncompetitive manner.
The mode-of-action of MT1 and MT2 is controversial. Several studies reported agonistic, antagonistic, competitive, or allosteric properties of these polypeptides. MT2 selectively binds to ACM1. Their mechanisms of action are summarized in [121].
Venom-derived MT3 shows affinity towards ACM4, ADRA1A, ADRA2A, and ADRA2C. It was reported as a highly potent and selective antagonist of ACM4. Synthetic form of MT3 was demonstrated to be a competitive antagonist of ACM4 as well as highly potent on α-adrenoceptors, in particular ADRA1A, ADRA2A, and ADRA2C. Herein, a noncompetitive antagonism was suggested. MT6 is proposed as an isotoxin of MT3 and its primary sequences have not been determined. It shows selectivity towards ACM1, but its mode-of-action is unknown.
MT4 was shown to be an antagonist of ACM1, ACM2, and ADRAB2. In contrast, MT5 binds to ACM1 and ACM4 but its mechanism of action is not known.
Venom-derived MT-α binds to muscarinic receptors; synthetic MT-α does not have muscarinic activity but rather antagonistic properties on ADRA2B. MT-β shows binding affinity towards ACM1, ACM3, and ACM4 but its detailed mode-of-action is unknown.
Box 1. Human Protein-Embedded Peptide GPCR Ligands Activated by Proteases.
The CXC-motif-chemokine receptor 4 (CXCR4), originally discovered in 1996 as a co-receptor required for the entry of the human immunodeficiency virus type 1 (HIV-1) [59], and its endogenous chemokine CXCL12 are involved in the regulation of cellular migration and homing processes, which underlie organogenesis, hematopoiesis, and immune responses [60,61]. Aberrant activation of CXCR4 is observed in cancer, autoimmune diseases, and atherosclerosis [60–62]. Accordingly, several synthetic CXCR4 ligands have been developed to block CXCR4 [63]. Plerixafor/AMD3100 is an example of a clinically approved CXCR4 antagonist [64]. Münch et al. recently discovered a novel endogenous CXCR4 ligand by screening a human hemofiltrate-derived peptide library [65,66]. This strategy led to the identification of EPI-X4, a 16-amino acid peptide fragment that arises from an albumin protein precursor that is cleaved by proteases such as cathepsin D and E under acidic conditions, for instance upon viral infection. Functional and binding studies demonstrated that EPI-X4 acts as an antagonist on CXCR4 by competing with CXCL12. Accordingly, EPI-X4 inhibits Ca2+-mobilization and receptor internalization. Furthermore, this endogenous antagonist inhibits migration and invasion of cancer cells along a CXCL12 gradient. This observation suggests that EPI-X4 has anti-invasive and antimetastatic properties [66]. Structural studies reveal that EPI-X4 binds to the second extracellular loop of the receptor, thereby presumably impeding envelope protein glycoprotein 120 (gp120) attachment and HIV-1 entry [66,67]. In addition, EPI-X4 was shown to induce mobilization of stem cells and suppress infiltration of immune cells into the lung in a mouse model of acute allergic airway hypereosinophilia [66]. Interestingly, EPI-X4 was detected in the urine of patients suffering from inflammatory kidney diseases. Thus, EPI-X4 may represent a biomarker for chronic kidney diseases and related disorders [66,67]. These studies describe a novel strategy to identify endogenous and ‘natural’ GPCR ligands, namely by exploring peptide libraries derived from human body fluids.
Plant-Derived Cyclotides as Starting Points for GPCR Drug Discovery
Cyclotides are disulfide-rich plant peptides characterized by a head-to-tail cyclized backbone and six conserved cysteine residues, which form three knotted disulfide bonds. This unique topology, referred to as cyclic cystine-knot motif, provides them with a tightly packed 3D fold resulting in exceptional stability against thermal, chemical, and enzymatic degradation [14,15]. Hitherto, cyclotides have been identified in several plant families, including violet (Violaceae), coffee (Rubiaceae), cucurbit (Cucurbitaceae), pea (Fabaceae), potato (Solanaceae), and grass (Poaceae) [16]. Cyclotides exhibit manifold bioactivities, such as uterotonic [17] and immunomodulatory properties [14]. Their endogenous function appears to be as plant defense molecules: they modulate insect GPCRs [18] and exhibit antiherbivore effects towards plant pests [19]. Intriguingly, a single species can express over 150 distinct cyclotides and the number of cyclotides to be discovered in plants has been predicted to exceed 150 000 [16]. They comprise one of the most abundant classes of ribosomally synthesized peptides in plants [20] and display substantial structural plasticity and sequence diversity around the conserved cystine-knot motif [16,21]. Owing to their outstanding stability, cyclotides constitute interesting starting points for peptide-based GPCR drug discovery.
In 1994, preliminary experiments with cyclopsychotride A isolated from a tropical Psychotria species demonstrated the ability of cyclotides to interfere with neurotensin receptor binding [83]. The field of cyclotide GPCR ligand discovery received a breakthrough by Koehbach et al., who provided an evidence-based explanation for the use of cyclotide plants as traditional uterotonic medicine [17]. A bioactivity-guided fractionation approach was used to analyze an herbal extract from the African medicinal plant Oldenlandia affinis. This led to the identification of peptide-enriched fractions, which stimulated contractions of human uterine smooth muscle cells. Peptidomics analysis of these fractions allowed the isolation of kalata B7 (kB7) as active compound. This cyclic peptide was shown to bind to human oxytocin (OXT) and arginine-vasopressin (AVP) V1A receptor [17] with an affinity in the low μM range. Additional experiments confirmed that kB7 acts as a partial agonist of the OXT receptor (OXTR) (EC50 = 12 μM) and V1AR (EC50 = 4 μM). To gain more insights into receptor–ligand interaction, the structure of kB7 was determined by nuclear magnetic resonance spectroscopy. This uncovered high similarity between loop 3 of kB7 and human OXT, explaining the properties of kB7 to act as a GPCR agonist. Since cyclotides are larger and bulkier than the nonapeptides OXT and AVP, four OXT-like peptides were designed by using the structure of loop 3 of kB7 as a template. This approach yielded the nonapeptide [G5,T7,S9]-OXT with improved affinity (Ki = 218 nM) and increased potency as a full agonist with an EC50 of 145 nM [17]. Intriguingly, the plant-inspired ligand had improved receptor subtype selectivity for the human OXTR over its three AVP receptor counterparts (V1AR, V1BR, and V2R). The development of selective ligands is of relevance in this area: lack of receptor selectivity limits their use as therapeutic drugs or chemical probes [22]. This study provided a proof-of-concept that plant-derived cyclotides can be exploited as templates for peptide-based GPCR ligand design [17]. Knowing that cyclotides are capable of modulating GPCRs of class A family, in a more recent study Fahradpour and colleagues explored modulatory properties of cyclotides, isolated from an ipecac root extract (Carapichea ipecacuanha), on corticotropin releasing factor type I receptor (CRFR1), a prototypical class B GPCR [23]. Herein, they provided an ipecac root extract that antagonized CRFR1 signaling (IC50 = 2.0 μg ml–1), which was subjected to bioactivity-guided fractionation to isolate cyclotides responsible for the observed CRFR1 antagonism. Further pharmacological analysis of cyclotide-enriched fractions resulted in isolation and sequencing of seven cyclotides, referred to as caripe peptides, of which caripe 8 had the most pronounced antagonistic effect [23]. This study reported for the first time the ability of cyclotides to modulate class B GPCR signaling and highlighted potential of ipecac root-derived cyclotides as useful tools and templates to design and develop antagonists that target the CRFR1 [23].
GPCR Peptide Ligands Derived from Cone-Snails and Snakes
Many animals produce venoms that are unique sources of naturally occurring peptides that have evolved to cover a large repertoire of pharmacological properties [24]. Cone-snails produce a strikingly diverse collection of peptides, which are referred to as conopeptides or conotoxins [25]. Due to their small size, stability, and amenability for synthesis, these (often) disulfide-rich peptides constitute valuable drug leads [26]. The majority of conotoxins target ion channels; only a minor portion (i.e., 14 venom peptides) act on GPCRs [27,28]. For instance, the conopeptide ρ-TIA was identified as an allosteric modulator acting on the α1B adrenoceptor (ADRA1B) [29]. Pharmacological analysis of this peptide isolated from the crude venom of fish-hunting Conus tulipa revealed a unique mechanism of action; ρ-TIA noncompetitively antagonized ADRA1B (IC50 = 2 nM) [29]. In a follow-up study, Chen et al. further explored the pharmacological profile of ρ-TIA in radioligand binding assays and observed a competitive antagonism at the α1A- (ADRA1A; IC50 = 18 nM) and α1D-(ADRA1D; IC50 = 25 nM) adrenoceptors, suggesting that ρ-TIA might be exploited as a template for rational design of highly selective adrenoceptor ligands [30]. Other examples of conotoxin-derived GPCR ligands have been reported in recent years. These include conorphin-T, a κ-opioid receptor (OPRK) ligand (Ki = 80.4 nM; EC50 = 9.8 μM), conopressin-T, a ligand with nanomolar affinity for human OXTR (Ki = 100 nM), and V1AR (Ki = 319 nM) and contulakin-G, a neurotensin receptor 1 (NTR1) agonist (EC50 = 960 nM) (summarized in [27]). Intriguingly, conotoxins further target gamma aminobutyric acid (GABA) B receptors, an attractive therapeutic target for pain management, although the exact mechanism remains unclear. Initial studies revealed that conotoxins, in particular Vc1.1 and RgIA, inhibit N-type calcium channels Cav2.2 and Cav2.3 via a voltage-independent mechanism probably mediated by Gαi/o subunit of GABAB receptors and c-Src tyrosine kinase activity [27]. By contrast, subsequent studies reported that Vc1.1 and RgIA do not compete with orthosteric ligands such as baclofen or GABA, suggesting an allosteric interaction to a currently unknown binding site [27]. Hence, further studies are required to elucidate a precise mode-of-action of GABAB receptor-provoked Cav inhibition mediated by conotoxins.
Snakes also produce potent peptides that act as natural GPCR ligands. In our analysis we identified published information of about 30 snake peptides that target GPCRs. For example, mamba snake venom contains toxins that can modulate GPCRs by distinct modes-of-action: MT7 peptide is a negative allosteric modulator of the M1 muscarinic receptor [31]; ρ-Da1a and ρ-Da1b antagonize α1A- and α2A-adrenoceptors, respectively [32,33]. Recently, a peptide of 57-amino acids, which targets a GPCR, was identified in the venom of a green mamba [34]: this peptide, termed mambaquaretin-1, exhibited nanomolar affinity for the V2R (Ki = 2.8 nM) but was inactive on nine cardiac ion channels and 155 additional GPCRs. mambaquaretin-1 antagonized V2R-dependent cAMP production (Ki = 12 nM), β-arrestin-1 mobilization (Ki = 110 nM), and mitogen-activated-protein kinase phosphorylation (Ki = 210 nM) in a competitive manner. Interestingly, mambaquaretin-1 belongs to the family of proteins comprising a Kunitz domain; it exerts its inhibitory action on the V2R via its first loop (in the same manner that aprotinin inhibits trypsin). Injection of mambaquaretin-1 in rodents resulted in an aquaretic effect (i.e., enhanced urine outflow and decreased urine osmolality). Moreover, in a juvenile model of polycystic kidney disease, mambaquaretin-1 inhibited progression of cysts. This highlights its potential usefulness for the treatment of polycystic kidney disease [34].
GPCR Peptide Ligands from Arachnids: Scorpions and Spiders
Venoms from spiders and scorpions are best known for their action on ion channels; however, they also contain peptides, which act on GPCRs. For instance, α-latrotoxin isolated from the black widow spider of the genus Latrodectus is a large polypeptide toxin (128 kDa) that binds to the latrophilin 1 receptor (ADGRL1), a member of the class B family of GPCRs [35], with high affinity (Kd = 0.54 nM). Additionally, δ-CNTX-Pn1a, a peptide from Phoneutria nigriventer spider venom, induced antinociception in in vivo pain models by activating cannabinoid 1 (CNR1) as well as the μ-opioid receptor (OPRM) and the OPRK [36]. Further, BmK-YA, identified as an enkephalin-like peptide by screening venom extracts of Asian scorpion Buthus martensii, activates mammalian opioid receptors with 6.8- and 12-fold increased selectivity for δ-opioid receptor (OPRD) over OPRM and OPRK, respectively. BmK-YA is a full agonist of OPRD with an EC50 of 2.5 μM [37].
GPCR Peptide Ligands Derived from Marine Fungi and Bacteria
Marine-derived fungi have further proven to be a rich source of biologically active peptides that might be exploited for development of novel GPCR-based drug leads. Recently, in the study of Almeida et al., the marine-derived fungus Stachylidium sp. was isolated from the sponge Callyspongia flammea, and the culture on a biomalt medium supplemented with sea salt enabled the isolation of endolide A and B [38]. These unusual cyclic tetrapeptides containing an N-methylation and a very rare 3-(3-furyl)-alanine moiety were studied in radioligand binding assays. Here, endolide A was demonstrated to bind to the V1AR (Ki = 7.04 μM), whereas endolide B showed an affinity to the serotonin 5HT2B receptor (Ki = 0.77 μM). Intriguingly, endolide B is selective for the 5HT2B receptor, exhibiting no affinity towards ten other serotonin receptor subtypes [38].
Additionally, marine cyanobacteria have received increasing attention in recent years as another rich source of bioactive peptides with diverse activities. Al-Awadhi et al. isolated five novel linear hexapeptides, termed brintonamides A–E, from a marine cyanobacterial sample [39]. Following chemical synthesis and their structural determination highlighting major differences in the N terminus, they were screened in a panel of 241 GPCR targets to uncover their cellular activities. Brintonamides A and B were inactive at all tested GPCRs, highlighting the importance of the hydroxy group at the N terminus for activity. By contrast, brintonamide C, associated with an N,N-Me2-Phe residue at the N terminus, activated C-X-C chemokine receptor type 7 (CXCR7) (EC50 = 10.5 μM), while it antagonized somatostatin receptor 3 (SSTR3) and tachykinin receptor 2 (TACR2) with similar IC50 values, 6.1 nM and 5.5 nM, respectively. The cis and trans isomers, brintonamides D and E, containing a cinnamic acid at the N terminus showed moderate agonistic/antagonistic activities. The trans isomer brintonamide D was active at CXCR7 (EC50 = 4 μM), OXTR (IC50 = 6.8 μM), SSTR3 (IC50 = 3.1 μM), TACR2 (IC50 = 1.8 μM), and C–C chemokine receptor type 10 (CCR10), at which it exhibited the highest potency with respect to the other brintonamides (IC50 = 440 nM) [39]. Compared with brintonamide D, the cis isomer brintonamide E was inactive at SSTR3, while it was similarly potent at four other GPCRs, indicating that the trans configuration of brintonamides is important for maintaining activity against SSR3. Given the role of CCR10 in cancer progression and metastasis, this study further revealed that the most potent CCR10 antagonist, brintonamide D, is capable of inhibiting proliferation and migration of breast cancer cells in a CCR10-dependent manner [39]. Taken together, these examples demonstrate that peptides isolated from venoms of cone-snails, snakes, spiders, and scorpions, as well as peptides derived from marine fungi and bacteria, constitute important and rich natural sources for the discovery of novel GPCR ligands. Their remarkable structural and functional diversity make them valuable templates for the development of novel GPCR ligands, with potential drug lead-like properties.
Concluding Remarks and Future Perspectives
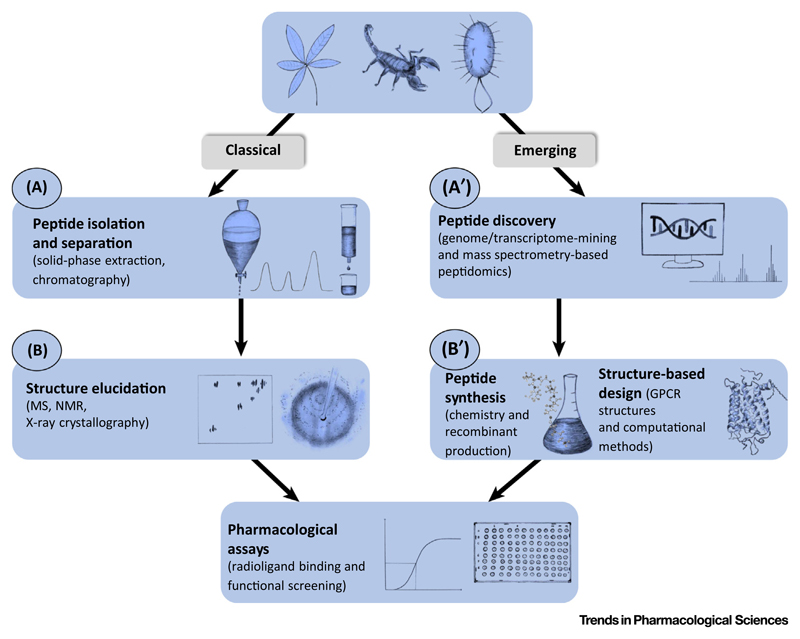
The classical workflow for nature-derived drug discovery starts with sample extraction and isolation. Samples from various organisms are crude extracts that contain the peptide(s) of interest, usually in a low concentration, in a complex mixture with other biological products [16]. The peptide GPCR ligands are extracted by extensive fractionation and purification, for instance using solid-phase extraction or chromatography (Figure 2A). The isolated peptides are then subjected to structure elucidation methods such as mass spectrometry, nuclear magnetic resonance spectroscopy, or X-ray crystallography (Figure 2B). Finally, the purified peptides are screened in pharmacological ligand binding and functional assays using cells or tissues expressing (endogenous or heterologous) GPCRs [40] (Figure 2). This enables analysis of affinity and functional GPCR responses, such as second messenger production or arrestin recruitment.
Figure 2.
Classical and Emerging Approaches of Peptide Drug Discovery to Identify Novel G Protein- Coupled Receptor (GPCR) Ligands. Classical approaches used for the isolation and characterization of nature-derived peptides from various sources include three major steps: (A) isolation and purification methods (e.g., solid-phase extraction and chromatography), and (B) structure elucidation by mass spectrometry (MS), nuclear magnetic resonance (NMR) spectroscopy, and X-ray crystallography. Emerging discovery strategies of peptides will focus on (A′) genome- and transcriptome-mining, and mass spectrometry-based peptidomics, (B′) structure-based design utilizing GPCR structures and computational methods, as well as (B′) improved peptide synthesis using chemistry methods and recombinant production. Both the classical and emerging approaches will be followed by pharmacological assays by ligand binding and functional screening (high-throughput if applicable).
There are multiple challenges and obstacles associated with the classical workflow that must be overcome to develop naturally occurring peptides into clinically relevant drugs. For instance, extensive separation techniques and structural elucidation methods are time-consuming and labor-intensive. In addition, the cost of peptide production is high when compared with small molecules. Currently, the production of peptides relies on solid-phase peptide synthesis, which is limited by the peptide length, and it cannot achieve production scales typical of organic synthesis. In addition, oral bioavailability of peptides is, in general poor and they do not readily cross membranes, which limits their biodistribution. Due to a lack of stability, peptides are in many instances rapidly degraded in biological fluids. These drawbacks and limitations must be overcome to make a convincing case for their suitability in drug development [41].
Rapid advances in the field of peptide-based drug discovery and development have progressed to manage some of these challenges. For instance, the time-consuming and laborintensive isolation procedures can be accelerated by relying on the rapidly growing number of publicly available genome- and transcriptome data (e.g., 1KPiii and 1KITEiv) (Figure 2A′). In fact, in silico mining is a powerful tool in identifying novel and potentially bioactive peptides [42]. Limitations associated with in silico mining, such as inaccurate prediction of open-reading frames or determination of post-translational modifications, can be improved by using it in combination with mass spectrometry-based peptidomics [16,43,44]. Further, we believe that advances in GPCR structural biology and computational methods (Box 2) will greatly improve discovery and design of nature-derived peptide ligands (Figure 2B′). Progress in peptide synthesis (i.e., improved automated workflows and native chemical ligation strategies) now enables production of longer peptides of high quality [45,46]. In addition, nature-derived peptides can be produced by recombinant expression in plants or microbes (Figure 2B′) [15,47,48].
Box 2. GPCR Structures and Computational Methods to Discover and Characterize Nature-Derived Peptide Ligands.
Advances in GPCR structural biology and computational methods have provided new momentum for the field of GPCR ligand discovery [5,68]. High-resolution GPCR structures determined by X-ray crystallography or nuclear magnetic resonance spectroscopy increased our understanding of GPCR structure and function [5,68]. Recently, cryo-electron microscopy was used to elucidate the active-state structure of the human glucagon-like peptide 1 receptor (GLP-1R) in complex with exendin-P5 (ExP5) and a Gs heterotrimer [69]. ExP5, an analogue of exendin-4 (see Table 1 in main text), is a potent G protein-biased, selective agonist of GLP-1R [70]. Major differences between the endogenous ligand glucagon-like peptide 1 (GLP-1) and ExP5-bound GLP-1R complexes were discovered in transmembrane helix 1, extracellular portions of helices 6 and 7, and extracellular loop 3, suggesting that these structural features are important for ligand bias [69]. Additionally, ExP5-mediated GLP-1R activation not only induces conformational changes of Gαs, but also increases the G protein activation rate via distinct flexibility of helix 5 and intracellular loop 3 [69]. These observations provide valuable insights into ligand bias that may be exploited for design of novel peptide-based therapeutics that target GLP-1R [69]. The availability of GPCR structures may further be used as a template for homology modelling and peptide–receptor interaction studies. For instance, Di Giglio et al. leveraged this approach to elucidate the pharmacology of inotocin, an ant-derived neuropeptide [71]. Structural models of several OXT- and AVP-type receptors were utilized to identify conserved sequence positions responsible for peptide binding, selectivity, and function [71]. Considering the conserved residues of the peptide-binding cavity it was possible to explain the binding and functional properties of inotocin, and a synthetic D-arginine analog, which might be useful for design of selective agonists and antagonists [71]. In addition, emerging computational methods facilitate rational design of peptides by using nature-derived peptides as valuable starting points. Recently, Bhardwaj et al. reported accurate de novo design of conformationally restricted peptides [72]. They designed 18–47 residue constrained peptides as either: (i) genetically encodable disulfide-rich peptides, (ii) synthetic disulfide crosslinked peptides with non-canonical sequences, or (iii) heterochiral cyclic peptides associated with non-canonical sequences. Each of these peptide categories demonstrated stability against thermal and chemical denaturation and all structures that were experimentally determined compared well with the computational designed models [72]. This approach has great potential for design and development of novel peptide-based GPCR drugs.
The problem with peptide stability can be tackled in different ways: peptide cyclization [15], introduction of unnatural amino acids, or N-methylation may reduce enzymatic degradation and potentially increase oral bioavailability. In addition, linear peptides can be stabilized by molecular grafting using cyclic disulfide-rich peptides as scaffolds [15,49]. This approach has been successfully applied for design and development of peptide-based GPCR ligands [50–53]. These grafted peptides have been mainly generated by solid-phase peptide synthesis, however synthesis of such constrained peptides with three disulfide bonds is challenging: folding problems may occur leading to non-native connectivity of disulfide bonds [15]. Several additional strategies have been introduced to enhance membrane permeability of peptides, including peptide delivery to the brain: they rely on nature-inspired cell-penetrating and shuttle peptides, which can readily cross biological barriers [54,55]. Finally, conjugation of peptides with small molecules or antibodies allows for the development of compounds with improved pharmacological properties, including efficacy, safety, and tolerability [56]. Peptide–drug conjugates are an important class of oncologic imaging probes and therapeutics; for instance, chlorotoxin, a scorpion-derived peptide conjugated to a fluorescent dye is an excellent example of how nature-derived peptides might serve to target cancer [57].
GPCRs continue to represent a class of privileged drug targets, as evident from 475 FDA-approved drugs targeting 108 unique GPCRs [5]. Natural products from plants, animals, fungi, and bacteria have historically played an important role in drug discovery [3,58]. In the recent past, many natural products have been appreciated as a largely untapped source of GPCR ligands. An analysis of the published literature exemplified that there are now approximately 600 nature-derived GPCR ligands, most of which are small molecules isolated from plants. There are at least 16 FDA-approved drugs (15 small molecules and 1 peptide) derived from natural products that target various GPCRs (Table 1). While small molecules continue to play an important role in GPCR drug discovery, nature-derivedpeptides(Table 2) aregaining momentum asimportant compound class for GPCR ligand discovery since they cover a distinct chemical space. Challenges of nature-derived peptide drug development can be overcome by the vast array of new technologies. Hence, we anticipate that nature-derived peptides will provide new opportunities for GPCR drug discovery and development (see Outstanding Questions).
Outstanding Questions.
Can we develop more accurate genome-mining tools/software to predict coding sequences of nature-derived peptides?
Is homology-based modeling an alternative to labor intensive methods to define the native 3D structure of nature-derived peptides?
Can we improve synthesis and folding yields (chemistry and recombinant) to lower production costs (scale-up) in comparison with organic synthesis of small molecules?
Will it be possible to systematically increase the oral bioavailability of peptides without negatively affecting efficacy or tolerability of the peptide drugs?
Can we rely on nature-derived peptide scaffolds to improve stability, oral bio-availability, and biodistribution of active peptide epitopes, including delivery of peptide drugs into the brain?
Can we further utilize nature-derived peptides as (bivalent) ligands and chemical probes to study biased signaling and receptor oligomerization of GPCRs?
Are nature-derived peptides suitable ligands for GPCR structural biology to provide novel insight into GPCR signaling?
Will it be possible to optimize existing computational methods to advance the field of structure-based peptide design (e.g., docking)?
Could nature-derived peptides be used as a template for de novo design of GPCR peptide drug leads?
Highlights.
Natural products have been and continue to be an important source of GPCRs ligands.
Over 600 natural GPCR ligands have been isolated from plants, animals, fungi, and bacteria; they predominantly target aminergic, opioid, cannabinoid, and taste 2 receptors.
At least 16 FDA-approved drugs targeting GPCRs are natural products, mainly small molecules from plants.
Nature-derived peptides isolated from bacteria, fungi, plants, and venomous animals, such as cone-snails, snakes, spiders, and scorpions are an emerging compound class for GPCR ligand discovery. They represent valuable starting points for GPCR drug development.
New technologies in peptide discovery and peptide chemistry allow for reliable identification of numerous nature-derived peptides and their synthesis to advance pharmacological screening, lead discovery and optimization, and eventually clinical applications.
Acknowledgments
The authors would like to thank Bernhard Retzl, Fabiola Emser, and Martin Olalde Quintanar for help with database searches and drawings. Research in the laboratory of C.W.G is funded by the Austrian Science Fund (FWF) through project I3243.
Footnotes
Disclaimer Statement
None to be declared.
References
- 1.Brownstein MJ. A brief history of opiates, opioid peptides, and opioid receptors. Proc Natl Acad Sci U S A. 1993;90:5391–5393. doi: 10.1073/pnas.90.12.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues T, et al. Counting on natural products for drug design. Nat Chem. 2016;8:531–541. doi: 10.1038/nchem.2479. [DOI] [PubMed] [Google Scholar]
- 3.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 4.Pierce KL, et al. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 5.Hauser AS, et al. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruber CW, et al. Ligand-based peptide design and combinatorial peptide libraries to target G protein-coupled receptors. Curr Pharm Des. 2010;16:3071–3088. doi: 10.2174/138161210793292474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wacker D, et al. How ligands illuminate GPCR molecular pharmacology. Cell. 2017;170:414–427. doi: 10.1016/j.cell.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumari P, et al. Emerging approaches to GPCR ligand screening for drug discovery. Trends Mol Med. 2015;21:687–701. doi: 10.1016/j.molmed.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Andersen A, et al. Glucagon-like peptide 1 in health and disease. Nat Rev Endocrinol. 2018;14:390–403. doi: 10.1038/s41574-018-0016-2. [DOI] [PubMed] [Google Scholar]
- 10.Di L. Strategic approaches to optimizing peptide ADME properties. AAPS J. 2015;17:134–143. doi: 10.1208/s12248-014-9687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craik DJ, et al. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 12.Ho TT, et al. The polypharmacology of natural products. Future Med Chem. 2018;10:1361–1368. doi: 10.4155/fmc-2017-0294. [DOI] [PubMed] [Google Scholar]
- 13.Lau JL, Dunn MK. Therapeutic peptides: historical perspectives, current development trends, and future directions. Bioorg Med Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 14.Thell K, et al. Oral activity of a nature-derived cyclic peptide for the treatment of multiple sclerosis. Proc Natl Acad Sci U S A. 2016;113:3960–3965. doi: 10.1073/pnas.1519960113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang CK, Craik DJ. Designing macrocyclic disulfide-rich peptides for biotechnological applications. Nat Chem Biol. 2018;14:417–427. doi: 10.1038/s41589-018-0039-y. [DOI] [PubMed] [Google Scholar]
- 16.Hellinger R, et al. Peptidomics of circular cysteine-rich plant peptides: analysis of the diversity of cyclotides from Viola tricolor by transcriptome and proteome mining. J Proteome Res. 2015;14:4851–4862. doi: 10.1021/acs.jproteome.5b00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koehbach J, et al. Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design. Proc Natl Acad Sci U S A. 2013;110:21183–21188. doi: 10.1073/pnas.1311183110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keov P, et al. Discovery of peptide probes to modulate oxytocin-type receptors of insects. Sci Rep. 2018;8 doi: 10.1038/s41598-018-28380-3. 10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weidmann J, Craik DJ. Discovery, structure, function, and applications of cyclotides: circular proteins from plants. J Exp Bot. 2016;67:4801–4812. doi: 10.1093/jxb/erw210. [DOI] [PubMed] [Google Scholar]
- 20.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craik DJ, et al. Ribosomally-synthesised cyclic peptides from plants as drug leads and pharmaceutical scaffolds. Bioorg Med Chem. 2018;26:2727–2737. doi: 10.1016/j.bmc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Muttenthaler M, et al. Subtle modifications to oxytocin produce ligands that retain potency and improved selectivity across species. Sci Signal. 2017;10 doi: 10.1126/scisignal.aan3398. eaan3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fahradpour M, et al. Cyclotides isolated from an ipecac root extract antagonize the corticotropin releasing factor type 1 receptor. Front Pharmacol. 2017;8:616. doi: 10.3389/fphar.2017.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prashanth JR, et al. Pharmacological screening technologies for venom peptide discovery. Neuropharmacology. 2017;127:4–19. doi: 10.1016/j.neuropharm.2017.03.038. [DOI] [PubMed] [Google Scholar]
- 25.Gao B, et al. Cone snails: a big store of conotoxins for novel drug discovery. Toxins. 2017;9 doi: 10.3390/toxins9120397. E397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mir R, et al. Conotoxins: structure, therapeutic potential and pharmacological applications. Curr Pharm Des. 2016;22:582–589. doi: 10.2174/1381612822666151124234715. [DOI] [PubMed] [Google Scholar]
- 27.Daniel JT, Clark RJ. G-protein coupled receptors targeted by analgesic venom peptides. Toxins. 2017;9 doi: 10.3390/toxins9110372. E372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadeghi M, et al. Analgesic conopeptides targeting G protein-coupled receptors reduce excitability of sensory neurons. Neuropharmacology. 2017;127:116–123. doi: 10.1016/j.neuropharm.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 29.Sharpe IA, et al. Two new classes of conopeptides inhibit the alpha1 adrenoceptor and noradrenaline transporter. Nat Neurosci. 2001;4:902–907. doi: 10.1038/nn0901-902. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, et al. Subtype-selective noncompetitive or competitive inhibition of human alpha1-adrenergic receptors by rho-TIA. J Biol Chem. 2004;279:35326–3533. doi: 10.1074/jbc.M403703200. [DOI] [PubMed] [Google Scholar]
- 31.Olianas MC, et al. Action of the muscarinic toxin MT7 on agonist-bound muscarinic M1 receptors. Eur J Pharmacol. 2004;487:65–72. doi: 10.1016/j.ejphar.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 32.Palea S, et al. Effects of rho-Da1a a peptidic alpha(1) (A)-adrenoceptor antagonist in human isolated prostatic adenoma and anaesthetized rats. Br J Pharmacol. 2013;168:618–631. doi: 10.1111/j.1476-5381.2012.02231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouget C, et al. Identification of a novel snake peptide toxin displaying high affinity and antagonist behaviour for the alpha2-adrenoceptors. Br J Pharmacol. 2010;161:1361–1374. doi: 10.1111/j.1476-5381.2010.00966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciolek J, et al. Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease. Proc Natl Acad Sci U S A. 2017;114:7154–7159. doi: 10.1073/pnas.1620454114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davletov BA, et al. Isolation and biochemical characterization of a Ca2+-independent alpha-latrotoxin-binding protein. J Biol Chem. 1996;271:23239–23245. doi: 10.1074/jbc.271.38.23239. [DOI] [PubMed] [Google Scholar]
- 36.Emerich BL, et al. Delta-ctenitoxin-Pn1a, a peptide from Phoneutria nigrivente spider venom, shows antinociceptive effect involving opioid and cannabinoid systems, in rats. Toxins. 2016;8:106. doi: 10.3390/toxins8040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. BmK-YA, an enkephalin-like peptide in scorpion venom. PLoS One. 2012;7:e40417. doi: 10.1371/journal.pone.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almeida C, et al. Endolides A and B vasopressin and serotonin-receptor interacting N-methylated peptides from the sponge-derived fungus Stachylidium sp. Org Lett. 2016;18:528–531. doi: 10.1021/acs.orglett.5b03553. [DOI] [PubMed] [Google Scholar]
- 39.Al-Awadhi FH, et al. Discovery, synthesis, pharmacological profiling, and biological characterization of brintonamides A-E, novel dual protease and GPCR modulators from a marine cyanobacterium. J Med Chem. 2018;61:6364–6378. doi: 10.1021/acs.jmedchem.8b00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundstrom K. New winds in GPCR-based drug discovery. Future Med Chem. 2016;8:605–608. doi: 10.4155/fmc-2016-0008. [DOI] [PubMed] [Google Scholar]
- 41.Henninot A, et al. The current state of peptide drug discovery: back to the future? J Med Chem. 2018;61:1382–1414. doi: 10.1021/acs.jmedchem.7b00318. [DOI] [PubMed] [Google Scholar]
- 42.Dang T, Sussmuth RD. Bioactive peptide natural products as lead structures for medicinal use. Acc Chem Res. 2017;50:1566–1576. doi: 10.1021/acs.accounts.7b00159. [DOI] [PubMed] [Google Scholar]
- 43.Skinnider MA, et al. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc Natl Acad Sci U S A. 2016;113:E6343–E6351. doi: 10.1073/pnas.1609014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prashanth JR, et al. Cone snail venomics: from novel biology to novel therapeutics. Future Med Chem. 2014;6:1659–1675. doi: 10.4155/fmc.14.99. [DOI] [PubMed] [Google Scholar]
- 45.Mijalis AJ, et al. A fully automated flow-based approach for accelerated peptide synthesis. Nat Chem Biol. 2017;13:464–466. doi: 10.1038/nchembio.2318. [DOI] [PubMed] [Google Scholar]
- 46.Mende F, et al. Automated Fmoc-based solid-phase synthesis of peptide thioesters with self-purification effect and application in the construction of immobilized SH3 domains. J Am Chem Soc. 2010;132:11110–11118. doi: 10.1021/ja101732a. [DOI] [PubMed] [Google Scholar]
- 47.Gould A, Camarero JA. Cyclotides: overview and biotechnological applications. Chembiochem. 2017;18:1350–1363. doi: 10.1002/cbic.201700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klint JK, et al. Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of E coli. PLoS One. 2013;8:e63865. doi: 10.1371/journal.pone.0063865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang CK, et al. Molecular grafting onto a stable framework yields novel cyclic peptides for the treatment of multiple sclerosis. ACS Chem Biol. 2014;9:156–163. doi: 10.1021/cb400548s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aboye TL, et al. Design of a novel cyclotide-based CXCR4 antagonist with anti-human immunodeficiency virus (HIV)-1 activity. J Med Chem. 2012;55:10729–10734. doi: 10.1021/jm301468k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aboye T, et al. Design of a MCoTI-based cyclotide with angiotensin (1-7)-like activity. Molecules. 2016;21:152. doi: 10.3390/molecules21020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eliasen R, et al. Design, synthesis, structural and functional characterization of novel melanocortin agonists based on the cyclotide kalata B1. J Biol Chem. 2012;287:40493–40501. doi: 10.1074/jbc.M112.395442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong CT, et al. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew Chem Int Ed Engl. 2012;51:5620–5624. doi: 10.1002/anie.201200984. [DOI] [PubMed] [Google Scholar]
- 54.Guidotti G, et al. Cell-penetrating peptides: from basic research to clinics. Trends Pharmacol Sci. 2017;38:406–424. doi: 10.1016/j.tips.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Oller-Salvia B, et al. Blood-brain barrier shuttle peptides: an emerging paradigm for brain delivery. Chem Soc Rev. 2016;45:4690–4707. doi: 10.1039/c6cs00076b. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, et al. Peptide-drug conjugates as effective prodrug strategies for targeted delivery. Adv Drug Deliv Rev. 2017;110–111:112–126. doi: 10.1016/j.addr.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang RR, et al. Beyond the margins: real-time detection of cancer using targeted fluorophores. Nat Rev Clin Oncol. 2017;14:347–364. doi: 10.1038/nrclinonc.2016.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harvey AL, et al. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 59.Feng Y, et al. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 60.Anders HJ, et al. Pathomechanisms: homeostatic chemokines in health, tissue regeneration, and progressive diseases. Trends Mol Med. 2014;20:154–165. doi: 10.1016/j.molmed.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 61.Janssens R, et al. The unique structural and functional features of CXCL12. Cell Mol Immunol. 2018;15:299–311. doi: 10.1038/cmi.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo F, et al. CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 2016;35:816–826. doi: 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- 63.Choi WT, et al. Targeting chemokine receptor CXCR4 for treatment of HIV-1 infection, tumor progression, and metastasis. Curr Top Med Chem. 2014;14:1574–1589. doi: 10.2174/1568026614666140827143541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Larochelle A, et al. AMD3100 mobilizes hematopoietic stem cells with long-term repopulating capacity in nonhuman primates. Blood. 2006;107:3772–3778. doi: 10.1182/blood-2005-09-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Münch J, et al. Discovery of modulators of HIV-1 infection from the human peptidome. Nat Rev Microbiol. 2014;12:715–722. doi: 10.1038/nrmicro3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zirafi O, et al. Discovery and characterization of an endogenous CXCR4 antagonist. Cell Rep. 2015;11:737–747. doi: 10.1016/j.celrep.2015.03.061. [DOI] [PubMed] [Google Scholar]
- 67.Zirafi O, et al. Proteolytic processing of human serum albumin generates EPI-X4, an endogenous antagonist of CXCR4. J Leukoc Biol. 2016;99:863–868. doi: 10.1189/jlb.2MR1115-521RR. [DOI] [PubMed] [Google Scholar]
- 68.Shimada I, et al. GPCR drug discovery: integrating solution NMR data with crystal and cryo-EM structures. Nat Rev Drug Discov. 2019;18:59–82. doi: 10.1038/nrd.2018.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang YL, et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature. 2018;555:121–125. doi: 10.1038/nature25773. [DOI] [PubMed] [Google Scholar]
- 70.Zhang H, et al. Autocrine selection of a GLP-1R G-protein biased agonist with potent antidiabetic effects. Nat Commun. 2015;6 doi: 10.1038/ncomms9918. 8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Giglio MG, et al. Development of a human vasopressin V1a-receptor antagonist from an evolutionary-related insect neuropeptide. Sci Rep. 2017;7 doi: 10.1038/srep41002. 41002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhardwaj G, et al. Accurate de novo design of hyperstable constrained peptides. Nature. 2016;538:329–335. doi: 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Marzo V. New approaches and challenges to targeting the endocannabinoid system. Nat Rev Drug Discov. 2018;17:623–639. doi: 10.1038/nrd.2018.115. [DOI] [PubMed] [Google Scholar]
- 74.Ihara M, et al. An endothelin receptor (ETA) antagonist isolated from Streptomyces misakiensis. Biochem Biophys Res Commun. 1991;178:132–137. doi: 10.1016/0006-291x(91)91789-f. [DOI] [PubMed] [Google Scholar]
- 75.Ondeykal JG, et al. Discovery of structurally diverse natural product antagonists of chemokine receptor CXCR3. Mol Divers. 2005;9:123–129. doi: 10.1007/s11030-005-1296-8. [DOI] [PubMed] [Google Scholar]
- 76.Ogawa T, et al. RES-701-2, -3 and -4, novel and selective endothelin type B receptor antagonists produced by Streptomyces sp I. Taxonomy of producing strains, fermentation, isolation, and biochemical properties. J Antibiot. 1995;48:1213–1220. doi: 10.7164/antibiotics.48.1213. [DOI] [PubMed] [Google Scholar]
- 77.Bufe B, et al. Recognition of bacterial signal peptides by mammalian formyl peptide receptors: a new mechanism for sensing pathogens. J Biol Chem. 2015;290:7369–7387. doi: 10.1074/jbc.M114.626747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saito T, et al. CJ-15,208, a novel kappa opioid receptor antagonist from a fungus, Ctenomyces serratus ATCC15502. J Antibiot. 2002;55:847–854. doi: 10.7164/antibiotics.55.847. [DOI] [PubMed] [Google Scholar]
- 79.Yan P, et al. The immunosuppressant cyclosporin A antagonizes human formyl peptide receptor through inhibition of cognate ligand binding. J Immunol. 2006;177:7050–7058. doi: 10.4049/jimmunol.177.10.7050. [DOI] [PubMed] [Google Scholar]
- 80.Wenzel-Seifert K, Seifert R. Cyclosporin H is a potent and selective formyl peptide receptor antagonist Comparison with N-t-butoxycarbonyl-L-phenylalanyl-L-leucyl-L-phenylalanyl-L-leucyl-L-phenylalanine and cyclosporins A, B, C, D, and E. J Immunol. 1993;150:4591–4599. [PubMed] [Google Scholar]
- 81.Hedge VR, et al. A family of depsi-peptide fungal metabolites, as selective and competitive human tachykinin receptor (NK2) antagonists: fermentation, isolation, physicochemical properties, and biological activity. J Antibiot. 2001;54:125–135. doi: 10.7164/antibiotics.54.125. [DOI] [PubMed] [Google Scholar]
- 82.Yang S, et al. Rubiscolin, a delta selective opioid peptide derived from plant Rubisco. FEBS Lett. 2001;509:213–217. doi: 10.1016/s0014-5793(01)03042-3. [DOI] [PubMed] [Google Scholar]
- 83.Witherup KM, et al. Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J Nat Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 84.Ohinata K, et al. Soymorphins, novel mu opioid peptides derived from soy beta-conglycinin beta-subunit, have anxiolytic activities. Biosci Biotechnol Biochem. 2007;71:2618–2621. doi: 10.1271/bbb.70516. [DOI] [PubMed] [Google Scholar]
- 85.Hedner E, et al. Brominated cyclodipeptides from the marine sponge Geodia barretti as selective 5 HT ligands. J Nat Prod. 2006;69:1421–1424. doi: 10.1021/np0601760. [DOI] [PubMed] [Google Scholar]
- 86.Feng Y, et al. Polydiscamides B-D from a marine sponge Ircinia sp: as potent human sensory neuron-specific G protein coupled receptor agonists. J Nat Prod. 2008;71:8–11. doi: 10.1021/np070094r. [DOI] [PubMed] [Google Scholar]
- 87.Craig AG, et al. Contulakin-G, an O-glycosylated invertebrate neurotensin. J Biol Chem. 1999;274:13752–13759. doi: 10.1074/jbc.274.20.13752. [DOI] [PubMed] [Google Scholar]
- 88.Dutertre S, et al. Conopressin-T from Conus tulipa reveals an antagonist switch in vasopressin-like peptides. J Biol Chem. 2008;283:7100–7108. doi: 10.1074/jbc.M706477200. [DOI] [PubMed] [Google Scholar]
- 89.Nareoja K, Nasman J. Selective targeting of G-protein-coupled receptor subtypes with venom peptides. Acta Physiol. 2012;204:186–201. doi: 10.1111/j.1748-1716.2011.02305.x. [DOI] [PubMed] [Google Scholar]
- 90.Petrel C, et al. Identification, structural and pharmacological characterization of tau-CnVA, a conopeptide that selectively interacts with somatostatin sst3 receptor. Biochem Pharmacol. 2013;85:1663–1671. doi: 10.1016/j.bcp.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 91.Brust A, et al. Conopeptide-derived kappa-opioid agonists (conorphins): potent, selective, and metabolic stable dynorphin A mimetics with antinociceptive properties. J Med Chem. 2016;59:2381–2395. doi: 10.1021/acs.jmedchem.5b00911. [DOI] [PubMed] [Google Scholar]
- 92.Younis S, Rashid S. Alpha conotoxin-BuIA globular isomer is a competitive antagonist for oleoyl-L-alpha-lysophos-phatidic acid binding to LPAR6; a molecular dynamics study. PLoS One. 2017;12:e0189154. doi: 10.1371/journal.pone.0189154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwok HF, et al. Helokinestatin: a new bradykinin B2 receptor antagonist decapeptide from lizard venom. Peptides. 2008;29:65–72. doi: 10.1016/j.peptides.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 94.de Matos Silva LF, et al. Apamin reduces neuromuscular transmission by activating inhibitory muscarinic M(2) receptors on motor nerve terminals. Eur J Pharmacol. 2010;626:239–243. doi: 10.1016/j.ejphar.2009.09.064. [DOI] [PubMed] [Google Scholar]
- 95.Mortari MR, et al. Inhibition of acute nociceptive responses in rats after i.c.v. injection of Thr6-bradykinin, isolated from the venom of the social wasp, Polybia occidentalis. Br J Pharmacol. 2007;151:860–869. doi: 10.1038/sj.bjp.0707275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheong MC, et al. An opioid-like system regulating feeding behavior in C elegans. Elife. 2015;4:06683. doi: 10.7554/eLife.06683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Verano-Braga T, et al. Structure-function studies of Tityus serrulatus hypotensin-I (TsHpt-I): a new agonist of B(2) kinin receptor. Toxicon. 2010;56:1162–1171. doi: 10.1016/j.toxicon.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 98.Chen T, et al. Kinestatin: a novel bradykinin B2 receptor antagonist peptide from the skin secretion of the Chinese toad, Bombina maxima. Regul Pept. 2003;116:147–154. doi: 10.1016/j.regpep.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 99.Erspamer V, et al. Deltorphins: a family of naturally occurring peptides with high affinity and selectivity for delta opioid binding sites. Proc Natl Acad Sci USA. 1989;86:5188–5192. doi: 10.1073/pnas.86.13.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rossi AC, et al. Opioid receptor binding profile of selected dermorphin-like peptides. Peptides. 1986;7:755–759. doi: 10.1016/0196-9781(86)90091-4. [DOI] [PubMed] [Google Scholar]
- 101.Mordvintsev DY, et al. Weak toxin WTX from Naja kaouthia cobra venom interacts with both nicotinic and muscarinic acetylcholine receptors. FEBS J. 2009;276:5065–5075. doi: 10.1111/j.1742-4658.2009.07203.x. [DOI] [PubMed] [Google Scholar]
- 102.Shiu JH, et al. Solution structure of gamma-bungarotoxin: the functional significance of amino acid residues flanking the RGD motif in integrin binding. Proteins. 2004;57:839–849. doi: 10.1002/prot.20269. [DOI] [PubMed] [Google Scholar]
- 103.Blanchet G, et al. Polypharmacology profiles and phylogenetic analysis of three-finger toxins from mamba venom: case of aminergic toxins. Biochimie. 2014;103:109–117. doi: 10.1016/j.biochi.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 104.Quinton L, et al. Isolation and pharmacological characterization of AdTx1, a natural peptide displaying specific insurmountable antagonism of the alpha1A-adrenoceptor. Br J Pharmacol. 2010;159:316–325. doi: 10.1111/j.1476-5381.2009.00532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mourier G, et al. Pharmacological and structural characterization of long-sarafotoxins, a new family of endothelin-like peptides: Role of the C-terminus extension. Biochimie. 2012;94:461–470. doi: 10.1016/j.biochi.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 106.Rajagopalan N, et al. Beta-cardiotoxin: a new threefinger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity. FASEB J. 2007;21:3685–3695. doi: 10.1096/fj.07-8658com. [DOI] [PubMed] [Google Scholar]
- 107.Zhang L, et al. Alpha-cobratoxin inhibits T-type calcium currents through muscarinic M4 receptor and Gomicron-protein betagamma subunits-dependent protein kinase A pathway in dorsal root ganglion neurons. Neuropharmacology. 2012;62:1062–1072. doi: 10.1016/j.neuropharm.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 108.da Silva DC, et al. Characterization of a new muscarinic toxin from the venom of the Brazilian coral snake Micrurus lemniscatus in rat hippocampus. Life Sci. 2011;89:931–938. doi: 10.1016/j.lfs.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 109.Servent D, et al. Muscarinic toxins. Toxicon. 2011;58:455–463. doi: 10.1016/j.toxicon.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 110.Nareoja K, et al. Adrenoceptor activity of muscarinic toxins identified from mamba venoms. Br J Pharmacol. 2011;164:538–550. doi: 10.1111/j.1476-5381.2011.01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jolkkonen M, et al. Muscarinic toxins from the black mamba Dendroaspis polylepis. Eur J Biochem. 1995;234:579–585. doi: 10.1111/j.1432-1033.1995.579_b.x. [DOI] [PubMed] [Google Scholar]
- 112.Negraes PD, et al. The snake venom peptide Bj-PRO-7a is a M1 muscarinic acetylcholine receptor agonist. Cytometry A. 2011;79:77–83. doi: 10.1002/cyto.a.20963. [DOI] [PubMed] [Google Scholar]
- 113.Chung C, et al. Muscarinic toxin-like proteins from Taiwan banded krait (Bungarus multicinctus) venom: purification, characterization and gene organization. Biol Chem. 2002;383:1397–1406. doi: 10.1515/BC.2002.158. [DOI] [PubMed] [Google Scholar]
- 114.Ducancel F. The sarafotoxins. Toxicon. 2002;40:1541–1545. doi: 10.1016/s0041-0101(02)00159-9. [DOI] [PubMed] [Google Scholar]
- 115.Smith TP, et al. Evidence for the endothelin system as an emerging therapeutic target for the treatment of chronic pain. J Pain Res. 2014;7:531–545. doi: 10.2147/JPR.S65923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Becker A, et al. Bibrotoxin, a novel member of the endothelin/sarafotoxin peptide family, from the venom of the burrowing asp Atractaspis bibroni. FEBS Lett. 1993;315:100–103. doi: 10.1016/0014-5793(93)81142-m. [DOI] [PubMed] [Google Scholar]
- 117.Konno K, et al. Crotalphine, a novel potent analgesic peptide from the venom of the South American rattlesnake Crotalus durissus terrificus. Peptides. 2008;29:1293–1304. doi: 10.1016/j.peptides.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 118.Kukhtina VV, et al. Muscarinic toxin-like proteins from cobra venom. Eur J Biochem. 2000;267:6784–6789. doi: 10.1046/j.1432-1033.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 119.Zhang C, et al. Beef protein-derived peptides as bitter taste receptor T2R4 blockers. J Agric Food Chem. 2018;66:4902–4912. doi: 10.1021/acs.jafc.8b00830. [DOI] [PubMed] [Google Scholar]
- 120.Hou IC, et al. Beta-lactotensin derived from bovine beta-lactoglobulin exhibits anxiolytic-like activity as an agonist for neurotensin NTS(2) receptor via activation of dopamine D(1) receptor in mice. J Neurochem. 2011;119:785–790. doi: 10.1111/j.1471-4159.2011.07472.x. [DOI] [PubMed] [Google Scholar]
- 121.Servent D, Fruchart-Gaillard C. Muscarinic toxins: tools for the study of the pharmacological and functional properties of muscarinic receptors. J Neurochem. 2009;109:1193–1202. doi: 10.1111/j.1471-4159.2009.06092.x. [DOI] [PubMed] [Google Scholar]