Abstract
Purpose of review
There are over 100 serotypes of human enteroviruses, which cause a spectrum of illnesses, including meningitis, encephalitis, paralysis, myocarditis and rash. Increasing incidence of hand-foot-and-mouth disease in the Asia-Pacific and recent outbreaks of enterovirus-associated disease, such as severe respiratory illness in the United States in 2014, highlight the threat of these viruses to human health.
Recent findings
We describe recent outbreaks of human enteroviruses and summarise knowledge gaps regarding their burden, spectrum of diseases and epidemiology.
Summary
Reported outbreaks of respiratory, neurological, skin and eye diseases associated with human enteroviruses have increased in frequency and size in recent years. Improved molecular diagnostics and genetic sequence analysis are beginning to reveal the complex dynamics of individual serotypes and genotypes, and their contribution to these outbreaks. However, the biological mechanisms underlying their emergence and transmission dynamics remain elusive. They are likely to involve changes in the virus, such as fitness, antigenicity, virulence or tropism, and in the human population, such as levels of sanitation and of homo- and heterotypic immunity. Improvements in surveillance, serological surveys and detailed genetic and antigenic characterisation of viral populations would help to elucidate these mechanisms. This will be important for the design of outbreak control and vaccine development strategies.
Keywords: Enterovirus, epidemiology, hand-foot-and-mouth disease, outbreak, vaccine
Introduction
Enteroviruses (EVs, Picornaviridae family) are a genus of small, non-enveloped, single-stranded RNA viruses. They have high mutation rates and undergo frequent recombination. The genome region encoding their capsid protein VP1 (which contains the majority of neutralization epitopes) correlates well with serotypes and molecular methods use it for serotyping [1]. Molecular typing has largely replaced neutralization tests and is now considered the gold standard. More than 100 enterovirus serotypes infect humans, and are currently classified into four species, A to D, according to molecular and antigenic properties (Table 1, Figure 1). This large group of viruses includes polioviruses, coxsackieviruses A and B, echoviruses, and various serotypes identified more recently simply called “enterovirus” and sequentially numbered (starting at 68).
Table 1. Human enterovirus serotypes. Updated from[2].
| EV species | Serotypes |
|---|---|
| Species A | Coxsackievirus A 2−8, 10, 12, 14, 16, Enterovirus 71, 76, 89−91, 114, 119−121 |
| Species B | Coxsackievirus A 9, Coxsackievirus B 1−6, Echovirus 1−7, 9, 11−21, 24−27, 29−33, Enterovirus 69, 73−75, 77−88, 93, 97, 98, 100, 101,106, 107 |
| Species C | Poliovirus 1−3, Coxsackievirus A 1, 11, 13, 17, 19−22, 24, Enterovirus 95, 96, 99, 102, 104, 105, 109, 116−118 |
| Species D | Enterovirus 68, 70, 94, 111 |
Figure 1.
Schematic representation of the phylogenetic relationship between representative serotypes of the four species of human enteroviruses based on VP1 sequences reported in GenBank. A multiple sequence alignment was generated using Clustal Omega. A Neighbour-Joining tree was then created from this alignment, and visualised using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Branch lengths are not to scale. Accession numbers were as follows: CV-A6, AF081297; CV-A10, AF081300; CV-A24, AF081311; E6; AF081321; E11, AF081326; E30, AF081340; EV-68, AF081348; EV-71, JN168790; EV-70, D00820 (nucleotides 2425–3339); PV1, V01149 (nucleotides 2480–3385); and CV-A16, U05876 (nucleotides 2446–3336). CV-A: coxsackievirus A; E: echovirus; EV: enterovirus; PV: poliovirus.
Most human enteroviruses enter the body via the alimentary or respiratory tract and spread by both fecal-oral and respiratory routes, although a few exceptions to the usual modes of transmission exist [2]. Rhinoviruses, which are closely related to enteroviruses, have recently been classified within the genus Enterovirus, but they have unique biological and physical properties that cause them to be restricted to the upper respiratory tract [2]. Rhinoviruses are beyond the scope of this review, as are polioviruses, which are the target of a major eradication initiative.
Enterovirus infections can result in a wide range of clinical diseases, from minor febrile illness and rash to severe, sometimes fatal, conditions, including meningitis, encephalitis, paralysis and myocarditis (Table 2). Although most infections are subclinical, sporadic or regular outbreaks of disease are common worldwide, and can lead to significant morbidity and mortality. In particular, non-polio enteroviruses are recognised as an emerging cause of neurological diseases, which more often arise as complications of mild infections. This has become more evident during recent years. For example, hand-foot-and-mouth disease (caused by EV-A71 and CV-A16, among others) has been increasing in incidence in the Asia-Pacific, and despite polio having been eliminated from most countries, the burden of acute flaccid paralysis (AFP) remains high, with many of those cases associated with non-polio enteroviruses [3].
Table 2. Diseases caused by enteroviruses. Based on data from [2]. CV-A: coxsackievirus A; CV-B: coxsackievirus B; EV: enterovirus; E: echovirus; PV: poliovirus.
| Disease | Serotypes commonly implicated |
|---|---|
| CNS infections | |
| Acute flaccid paralysis | PVs, EV-A71 |
| Aseptic meningitis | PVs, CV-Bs, CV-A5, 7, 9, 16, E4, 6, 9, 11, 14, 16, 25, 30, 31, EV-A71 |
| Encephalitis | PVs, E6, 9, 17, 21, CV-A2, 9, CV-B1, EV-A71 |
|
Heart disease (acute and chronic myocarditis, dilated cardiomyopathy) |
CV-Bs |
| Eye infections | |
| Acute haemorrhagic conjunctivitis | CV-A24v, EV-D70 |
| Skin rashes | |
| Hand-foot-and-mouth disease | CV-A 4−6, 9, 10, 16, CV-B2, 5, EV-A71 |
| Herpangina | CV-As, CV-Bs, E6, 9, 11, 16, 17, 22, 25, EV-A71 |
|
Respiratory infections (common cold, rhinitis, pharyngitis, pneumonia, bronchiolitis and others) |
CV-As, CV-Bs, EV-D68, Es |
| Muscle disease | |
| Epidemic pleurodynia | CV-Bs, CV-A4, 6, 9, 10, E1, 6, 9, 16, 19 |
| Acute and chronic inflammatory | CVs (in experimental animals) |
| myopathies | |
| Pancreatitis | CV-Bs |
| Diarrhea | CV-As, Es |
| Undifferentiated febrile illness | PVs, CV-Bs |
| Neonatal infections (including prenatal, natal and postnatal infections) | CV-Bs, Es |
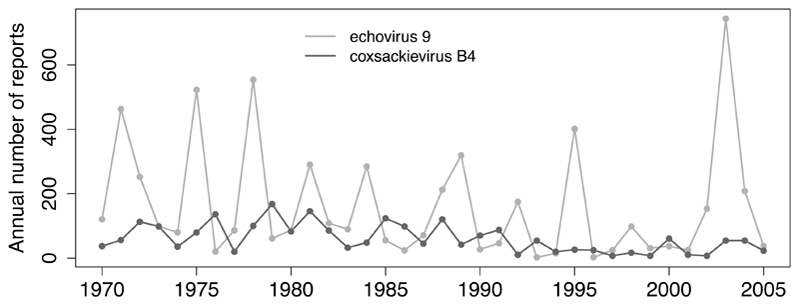
Enteroviruses often show seasonal patterns of incidence in both temperate and tropical climates, although seasonality is typically more marked in the former, with infections more common in summer and early autumn [2]. Passive laboratory surveillance of clinical isolates reveals different patterns of circulation over time and by geographic location for individual serotypes. For example, surveillance in the USA [4] recorded large and periodic outbreaks followed by years of relative quiescence for some serotypes (e.g. echovirus 9 exhibits large outbreaks every 3–5 years), whilst other serotypes have endemic patterns with relatively stable circulation (e.g. coxsackievirus B4), and others irregular patterns (Figure 2).
Figure 2.
Number of annually reported echovirus 9 and coxsackievirus B4 detections in the United States. Based on data from [4].
Enterovirus surveillance relies on passive, voluntary, laboratory-based systems, where participating laboratories report enterovirus detection to a reference centre (e.g. in the USA, the UK, the Netherlands, France [4,5,6,7]). Although enterovirus surveillance may in theory provide information relevant to the detection of outbreaks and changing patterns of infection, the burden of disease and the emergence of new strains, in practice these systems suffer a number of important biases. First, most enterovirus-associated diseases are not notifiable (except AFP), such that only a small proportion of all cases of a clinical illness are tested. Second, methods for virus identification have changed during the last decade (cell culture has been superseded by molecular methods). Third, an increasing number of enterovirus-positive specimens remain untyped because many laboratories do not perform molecular typing and samples may not be submitted to reference centres with this capacity. Fourth, laboratory methods used to test specimens are not standardised (e.g. use of different primers). Finally, in a context of an outbreak, the number of reports is likely to increase due to enhanced surveillance.
Recent outbreaks of disease, such as hand-foot-and-mouth disease (HFMD) in the Asia-Pacific and the outbreak of severe respiratory illnesses (caused by EV-D68) last year in the USA [8,9], have raised basic and applied questions about the mechanisms determining their emergence and transmission dynamics. Here, we review the epidemiology of recent outbreaks, progress towards the development of vaccines to control such outbreaks, and outstanding research questions that are important from a public health perspective.
Outbreak of severe respiratory illness in the USA in 2014
Enterovirus D68 (EV-D68) has biological features similar to rhinoviruses (e.g. acid lability and optimal growth at 33ºC [10]) and has long been exclusively associated with respiratory diseases. First isolated in 1962 from four children with pneumonia and bronchiolitis in California [11], only sporadic cases were reported until the late 2000s. In the USA, only 26 cases were notified between 1970 and 2005 [4].
During the last decade, several clusters of relatively small size (no more than 40 cases) of respiratory illnesses associated with EV-D68 occurred in Asia [12,13,14,15], Europe [12,16,17] and the USA [12,18], mostly affecting young children aged 0–4 years. The clinical respiratory manifestations ranged from mild to severe, sometimes fatal. Most clusters showed an atypical late seasonality compared to other enteroviruses with a peak in autumn, instead of summer [12]. The apparent increase in the number of detections was first thought to be related to improvements in detection techniques, since older tests could not detect EV-D68 or misidentified it as a rhinovirus [12]. However, retrospectively tested samples from the Netherlands and Japan confirmed the increase [13,16], which was also accompanied by an expansion in genetic diversity in the late 2000s [16,19]. Currently, three co-circulating clades of EV-D68 have been described, which probably diversified during the mid to late 1990s [13,16,19]. These clades have recently been shown to have different antigenic properties [20*].
A number of biological mechanisms have been hypothesized for the observed global increase in EV-D68 detections over the last decade. For example, amino acid changes in the VP1 compared to the Fermon reference strain that have resulted in altered antigenic properties may have permitted immune escape [16,20*]. Alternatively, EV-D68 may have become more virulent as a result of nucleotide deletions in the 5’ untranslated region [19].
In 2014, the largest outbreak to date of severe respiratory illness associated with EV-D68 occurred in the USA. The first cases were reported in August 2014 from Missouri and Illinois, and by January 2015 more than 1,150 cases across the country had been confirmed [8,9]. Of note, almost all the cases were children and a considerable proportion had a previous history of asthma or wheezing [8,9].
Coinciding with the 2014 outbreak in the USA, there was an unusual number of reports of children who developed a sudden onset of acute weakness in arms or legs similar to that caused by polioviruses; this is now referred to as acute flaccid myelitis (AFM) [21]. The first cluster was reported in Colorado, started in August 2014 and involved 12 children [22,23]. The total number of AFM reports by mid-April 2015 was 118 from 34 states in the USA and most cases were children <18 years of age [21,24]. Interestingly, during the two years preceding the EV-D68 outbreak, an unusual number (23 in total) of “polio-like” illness were also reported in California [25], among which some had a clinical and radiologic picture similar to the AFM cases described since August 2014 [25]. Most AFM patients reported a respiratory and/or febrile illness preceding the onset of the neurological symptoms [22,24] and some tested positive for EV-D68 in nasopharyngeal samples (5/11 in Colorado [22], 8/41 tested at CDC [24], 2/23 in California [25]). Although the virus has not been detected in the CSF of patients [22,24] (in fact, EV-D68 has only been isolated from CSF in two cases in the literature, once in 2005 [4] and once in 2008 [26]), these observations raised the question of an association between the outbreak of EV-D68 respiratory disease and those cases of neurologic illness. A new phylogenetic study has strengthened that putative association [27*]. It showed that EV-D68 strains from AFM cases belonged to a recent evolutionary cluster, clade B1, which emerged in 2010. The same study identified six single nucleotide polymorphisms (SNPs) specific to clade B1, some of which matched with SNPs in polioviruses and enterovirus D70 (which has previously been linked with AFP) and might have conferred neurovirulence to EV-D68 [27*].
Large-scale epidemics of acute haemorrhagic conjunctivitis
Two enteroviruses are the main etiological agents of explosive and large-scale outbreaks of acute haemorrhagic conjunctivitis (AHC): enterovirus D70 (EV-D70) and an antigenic variant of coxsackievirus A24 (CV-A24v). Both strains spread mainly via direct or indirect contact with eye secretions and both have been found in stool, suggesting that the fecal-oral route is also a mode of transmission [28,29]. A tropism of CV-A24v for the airways has also been suggested, which makes respiratory transmission also likely [30]. Although most cases resolve without sequelae after a 5–7 day course, microbial superinfection can occur and neurological complications such as “polio-like” paralysis have been reported during AHC outbreaks of EV-D70 [29,31]; however, the virus was not isolated from CSF [29,31] and cases were reported from settings where polio was still endemic, leaving the causal link uncertain.
The first documented outbreak of AHC due to EV-D70 occurred in Ghana in 1969 [32,33], and during 1970 and 1971 the virus spread globally, producing the first AHC pandemic and affecting millions of people [33,34]. Since the mid-1990s, no more outbreaks due to EV-D70 have been documented [35,36,37].
CV-A24v was first isolated in 1970 in Singapore [38] and remained restricted to South-East Asia and India until 1985. Since then, it has been responsible for outbreaks worldwide (e.g. Cuba 2008–2009 [28]; China 2010 [39]; India 2010 [40]; Japan 2011 [41]; Thailand 2014 [42]), including two pandemics: the first started in 1985 [43,44] and the second in 2002 [45]. Phylogenetic analyses of CV-A24v allowed the identification of four genotypes (GI to GIV): GI and GII included isolates from the 1970s, GIII from the late 1980s and early 1990s (i.e. including the first pandemic), and GIV from the second pandemic and others more recently identified [28,39,40,42,46,47]. This suggests a turnover of CV-A24v genotypes: new genetic variants rapidly spread globally and then fade out to be replaced by new variants.
With some exceptions, outbreaks of AHC seem to principally affect tropical and subtropical regions during the hot, rainy seasons and some countries have experienced several outbreaks during the last decades (e.g. Singapore, Thailand, Brazil, China, India). A recent study from Thailand suggests that CV-A24v has been continuously circulating at least since 2002, producing large and periodic epidemics every 2–3 years interspersed with minor outbreaks and sporadic cases [42]. Older studies showed that infected individuals develop neutralizing antibodies that last only for a few years [48,49], which could explain the patterns observed in Thailand, where extensive outbreaks may occur when population immunity falls below a certain threshold.
The size of AHC outbreaks and their rapid spread impose a significant burden on primary healthcare systems and normal activities of affected countries may be partly paralyzed (e.g. schools, companies, sports) [50]. Because no treatment (either vaccines or antiviral drugs) is currently available, control measures are reduced to limiting the spread of the virus (e.g. encouraging hand-washing and increased hygiene, and alerts via radio and media channels).
Hand-foot-and-mouth disease in the Asia-Pacific region
Hand-foot-and-mouth disease (HFMD) is a common, typically mild and self-limiting illness that mainly affects young children under 5 years of age (clinical manifestations include fever, a rash with blisters on hands, feet and buttocks, and vesicles in the mouth). The most common etiological agents are enterovirus A71 (EV-A71) and coxsackievirus A16 (CV-A16). Approximately 30% of hospitalised HFMD patients with EV-A71 infection develop more severe disease including neurological symptoms (e.g. aseptic meningitis, encephalitis, acute flaccid paralysis) and cardiopulmonary complications [51,52], which in some cases can be fatal. These severe outcomes are very unusual with CV-A16 [51]. EV-A71 is currently considered the most neurotropic enterovirus beyond polioviruses and there is a considerable literature on it (for reviews see [53,54,55]).
Only small outbreaks and sporadic cases of EV-A71 were reported throughout the world between the early 1970s and the late 1990s, with the exception of two large outbreaks, one in Bulgaria in 1975 [56] and one in Hungary in 1978 [57], both associated with a high mortality. During the last 10–20 years, major outbreaks of HFMD have occurred in the Asia-Pacific, becoming an important public health problem. This has led to increasing attention from health authorities, declaration of HFMD as a notifiable disease and the introduction of case-based surveillance systems [58]. Unprecedented outbreaks occurred in 1997 in Sarawak, Malaysia (at least 34 deaths [59,60]) and in 1998 in Taiwan (total number of cases estimated at around 1.5 million, >120,000 cases reported, among which >400 had severe diseases and 78 died [60,61]). Large-scale outbreaks have subsequently been reported in Malaysia, Taiwan, Singapore, China, Hong Kong, Japan, Republic of Korea, Vietnam and Cambodia. In China, from 2008 to 2014, >10 million cases of HFMD and >3,000 deaths were reported [62], with 80% of severe cases and 93% of fatal cases due to EV-A71 [63**].
Although outbreaks tend to occur in warmer months, significant geographical variability in the epidemic patterns of HFMD has been observed. A study based on surveillance data from China showed that the periodicity of the outbreaks and the timing of the peak varied with latitude [63**]: northern provinces had one peak in June, (as observed in Japan [64]), whereas southern provinces had two peaks, around May and October (as observed in the South of Vietnam [65]). Other countries, however, such as Hong Kong and Singapore, have observed changes over time in the periodicity of the peaks [66,67,68,69]. Although seasonal variation has been suggested to be associated with climatic factors [63**], other factors, like changes in the circulating and predominant serotypes (some of which could circulate or peak earlier than others) could also be involved (e.g. in Singapore, changes in the dominant serotype coincided with changes in the annual number of peaks [66]).
Temporal changes in the circulating and predominant serotypes have been observed in many places. For example, EV-A71 seems to have a periodicity of 3–4 years in Japan [64], and 3 years in Malaysia [70], whereas in China it circulated widely every year from 2008 to 2014 [62,63**]. Coxsackievirus A6 (CV-A6) has become a predominant serotype in many places during the last 2 to 5 years, often replacing EV-A71 and CV-A16 as the main cause of HFMD: e.g. Japan [64], Singapore [67,68], and different regions in China [71,72,73,74,75,76]. In a few places, coxsackievirus A10 (CV-A10) has also been recently reported as a predominant serotype [77,78]. Some of the CV-A6 and CV-A10 strains currently circulating in the Asia-Pacific are likely to have originated in Europe, where they caused outbreaks of atypical HFMD (e.g. associated with nail shedding [79,80]).
Concerns that large-scale outbreaks of HFMD may occur in India have arisen, mainly because of the close proximity to China and other conditions that could favour the rapid spread of the virus, including high population density and poor hygiene and sanitation [81]. Nevertheless, only small-scale outbreaks have occurred to date. Why the disease seems to be mainly restricted to the Asia-Pacific remains unknown.
HFMD vaccine development
Owing to the growing threat of HFMD in countries in the Asia-Pacific and the lack of effective antiviral therapies, efforts to develop an EV-A71 vaccine have recently been accelerated. The successful use of inactivated and live-attenuated poliovirus vaccines as part of the Global Polio Eradication Initiative emphasizes the potential to achieve safe and durable protective immunity against enterovirus infection. A variety of EV-A71 vaccine candidates have been evaluated in preclinical studies, including inactivated [82], live-attenuated [83], virus-like particle [84], subunit [85], and DNA vaccines [85]. Among these, formalin-inactivated whole-virus vaccines are the most advanced, and five candidates developed in mainland China, Taiwan and Singapore have entered clinical evaluation [86]. Recently, phase III randomized, double-blind, placebo-controlled trials were conducted in China for three of these vaccines [87,88,89]. Each trial involved the administration of two doses of alum-adjuvant inactivated EV-A71 vaccine or placebo at an interval of 28 days to more than 10,000 children aged 6–35 months [88,89] or 6–71 months [87]. Surveillance for EV-A71-associated disease (including HFMD, CNS disease and other EV-A71-associated sequelae) was then performed for up to 1 year. Each vaccine displayed an acceptable safety profile, and protective efficacy against EV-A71-associated HFMD ranged between 90.0% and 97.4%, raising the possibility that an EV-A71 vaccine will soon be licensed in China.
Despite this recent progress, several hurdles to the introduction of a vaccine against HFMD remain. Co-administration studies are required to test for potential interactions with other vaccines included in the Expanded Program on Immunization, while further studies are merited to determine whether the vaccines are efficacious in children <6 months of age. The cross-protective efficacy of the EV-A71 vaccines is also uncertain. To date, the vaccines that have entered clinical evaluation have been based on locally dominant EV-A71 genotypes (C4 in China, B4 in Taiwan and B2 in Singapore). Although antibodies induced by a given EV-A71 strain exhibit cross-neutralizing activity against other genotypes [90], this may not extend to all other EV-A71 strains [91]. Moreover, vaccinating against EV-A71 tackles just one cause of HFMD. The EV-A71 vaccines showed no efficacy against HFMD caused by CV-A16 or other serotypes during the recent phase III trials [87,88]. In the longer term, the development of multivalent vaccines targeting EV-A71, CV-A16 and other enteroviruses associated with HFMD may be desirable. Preclinical studies of multivalent HFMD vaccines have commenced [92*].
Conclusion
Focusing on a few examples of disease outbreaks, we have illustrated various clinical syndromes caused by non-polio enteroviruses, the complexity of their circulation dynamics and their capacity to spread rapidly.
The mechanisms of emergence of outbreaks are not known and our understanding is limited by relatively weak surveillance systems, typically based on passive reporting by public health laboratory. A combination of virus-specific, population-level and other external factors is likely to be involved. The evolutionary dynamics of enteroviruses confer them with the capacity to experience rapid phenotypic changes, including transmissibility, antigenicity, virulence and tropism (i.e. spectrum of disease). At the population level, the number of non-immune individuals needs to be above a certain threshold for an outbreak to be triggered, and depends on the number of births and the serological status of the population. The immune status of the population at a given time is related, among others, to the previous exposure of that population to the same or a related virus, the cross-reactivity between different genogroups of a given serotype (and between different serotypes), and the decay of neutralizing antibodies over time. Finally, climatic and environmental variables seem to correlate with the peaks of incidence of disease (e.g. HFMD), which suggests that they might affect the survival of the virus outside the host or the efficiency of transmission, which might differ among serotypes depending on their transmission routes.
Through genome sequencing, molecular epidemiology is helping to track the temporal and spatial circulation of different strains and determine their origin and geographical range. Large-scale genetic analyses have revealed the evolutionary and epidemiological dynamics of some serotypes. Distinct phylogenetic patterns have been described for different serotypes or even for different clades within a serotype (e.g. coxsackievirus B5 [93]). In some cases, ladder-like trees similar to influenza A have been obtained, consistent with continuous selection for new antigenic variants (e.g. echovirus 9 [94]). In other cases, co-circulation of several lineages has been observed, suggesting different selective forces or greater geographic isolation (e.g. echovirus 6 [95]). It has also been reported that peaks in incidence of some serotypes have been preceded by the emergence of recombinant groups (e.g. echovirus 9 [94]).
Unfortunately, molecular analyses alone do not allow the biological mechanisms determining the patterns of circulation of individual serotypes to be elucidated, and in particular, the role of immunity remains poorly understood. However, molecular epidemiological studies combined with investigations of antigenic changes and serological surveys have the potential to provide novel insights towards better understanding the occurrence of outbreaks. This would also allow elucidation of the mechanisms acting across genogroups or serotypes, which could explain the periodic changes in predominant variants. For instance, one study with isolates of EV-A71 from Taiwan showed using phylogenetic analyses that every large outbreak between 1998 and 2008 was associated with a genotypic change, alternating between B and C (B1, C2, B4, C4, B5), and then using antigenic cartography showed that the predominant strain in every major outbreak was antigenically distinct from the predominant strain in the previous outbreak [96].
Understanding the contribution of strain- or serotype-specific and heterotypic population immunity to the patterns of circulation of non-polio enteroviruses may help explain and predict outbreaks of disease. Ultimately this may lead to improved vaccination strategies, including the selection of appropriate vaccine strains.
Key points.
-
-
Non-polio enteroviruses are an emerging cause of neurological diseases, predominantly arising as complications of mild, self-limiting infections.
-
-
The full spectrum of illnesses caused by individual serotypes remains unclear; e.g. EV-D68 has long been associated with respiratory diseases but is now suspected to also be responsible for acute flaccid myelitis.
-
-
Some enteroviruses spread worldwide very quickly, as illustrated by the pandemics of acute haemorrhagic conjunctivitis caused by CV-A24v, with a rapid turnover of genetic variants that emerge, spread globally and then vanish being replaced by new variants.
-
-
Patterns of circulation of individual serotypes, including changes in the predominant ones, are complex and how serotype-specific and heterotypic immunity affects those is not well understood; e.g. why have EV-A71 and CV-A16 been replaced by CV-A6 as the main causative agent of HFMD in several countries in recent years?
-
-
Three inactivated candidate vaccines against EV-A71 have completed phase III trials in China and could be licensed soon.
Acknowledgements
We would like to thank M. Steven Oberste, Mark A. Pallansch and Miren Iturriza-Gomara for helpful discussions on the epidemiology of enteroviruses.
Financial support and sponsorship. MPS is supported by a Sir Henry Wellcome Fellowship funded by the Wellcome Trust [106073/Z/14/Z]. EPKP is funded by the UK Medical Research Council (MRC). NCG is funded by the Gates Foundation, MRC and World Health Organisation.
Footnotes
Conflicts of interest. None.
References
* of special interest
** of outstanding interest
- 1.Oberste MS, Maher K, Kilpatrick DR, et al. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pallansch MA, Oberste MS, Whitton JL. Enteroviruses: Polioviruses, Coxsackieviruses, Echoviruses, and Newer Enteroviruses. In: Knipe DM, Howley P, editors. Fields Virology 2. 6th ed. Lippincott Williams & Wilkins; 2013. [Google Scholar]
- 3.Rao CD, Yergolkar P, Shankarappa KS. Antigenic diversity of enteroviruses associated with nonpolio acute flaccid paralysis, India, 2007-2009. Emerg Infect Dis. 2012;18:1833–1840. doi: 10.3201/eid1811.111457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khetsuriani N, Lamonte-Fowlkes A, Oberste S, et al. Enterovirus surveillance--United States, 1970-2005. MMWR Surveill Summ. 2006;55:1–20. [PubMed] [Google Scholar]
- 5.Antona D, Leveque N, Chomel JJ, et al. Surveillance of enteroviruses in France, 2000-2004. Eur J Clin Microbiol Infect Dis. 2007;26:403–412. doi: 10.1007/s10096-007-0306-4. [DOI] [PubMed] [Google Scholar]
- 6.Kadambari S, Bukasa A, Okike IO, et al. Enterovirus infections in England and Wales, 2000-2011: the impact of increased molecular diagnostics. Clin Microbiol Infect. 2014;20:1289–1296. doi: 10.1111/1469-0691.12753. [DOI] [PubMed] [Google Scholar]
- 7.van der Sanden SM, Koopmans MP, van der Avoort HG. Detection of human enteroviruses and parechoviruses as part of the national enterovirus surveillance in the Netherlands, 1996-2011. Eur J Clin Microbiol Infect Dis. 2013;32:1525–1531. doi: 10.1007/s10096-013-1906-9. [DOI] [PubMed] [Google Scholar]
- 8.CDC. Enterovirus D68. [31 May 2015];2015 Available from: http://www.cdc.gov/non-polio-enterovirus/about/ev-d68.html.
- 9.Midgley CM, Jackson MA, Selvarangan R, et al. Severe respiratory illness associated with enterovirus D68 - Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep. 2014;63:798–799. [PMC free article] [PubMed] [Google Scholar]
- 10.Oberste MS, Maher K, Schnurr D, et al. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol. 2004;85:2577–2584. doi: 10.1099/vir.0.79925-0. [DOI] [PubMed] [Google Scholar]
- 11.Schieble JH, Fox VL, Lennette EH. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol. 1967;85:297–310. doi: 10.1093/oxfordjournals.aje.a120693. [DOI] [PubMed] [Google Scholar]
- 12.CDC. Clusters of acute respiratory illness associated with human enterovirus 68--Asia, Europe, and United States, 2008-2010. MMWR Morb Mortal Wkly Rep. 2011;60:1301–1304. [PubMed] [Google Scholar]
- 13.Ikeda T, Mizuta K, Abiko C, et al. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol Immunol. 2012;56:139–143. doi: 10.1111/j.1348-0421.2012.00411.x. [DOI] [PubMed] [Google Scholar]
- 14.Imamura T, Fuji N, Suzuki A, et al. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis. 2011;17:1430–1435. doi: 10.3201/eid1708.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaida A, Kubo H, Sekiguchi J, et al. Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan. Emerg Infect Dis. 2011;17:1494–1497. doi: 10.3201/eid1708.110028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meijer A, van der Sanden S, Snijders BE, et al. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology. 2012;423:49–57. doi: 10.1016/j.virol.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Piralla A, Baldanti F, Gerna G. Phylogenetic patterns of human respiratory picornavirus species, including the newly identified group C rhinoviruses, during a 1-year surveillance of a hospitalized patient population in Italy. J Clin Microbiol. 2011;49:373–376. doi: 10.1128/JCM.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokarz R, Kapoor V, Wu W, et al. Longitudinal molecular microbial analysis of influenza-like illness in New York City, May 2009 through May 2010. Virol J. 2011;8:288. doi: 10.1186/1743-422X-8-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tokarz R, Firth C, Madhi SA, et al. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura T, Okamoto M, Nakakita S, et al. Antigenic and receptor binding properties of enterovirus 68. J Virol. 2014;88:2374–2384. doi: 10.1128/JVI.03070-13. [*This article provided experimental evidence that antigenic changes may explain the increase in the number of detections of EV-D68 in the late 2000s] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CDC. Summary of Findings: Investigation of Acute Flaccid Myelitis in U.S. Children, 2014-15. [31 May 2014];2014 Available from: http://www.cdc.gov/ncird/investigation/viral/2014-15/investigation.html.
- 22.Messacar K, Schreiner TL, Maloney JA, et al. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet. 2015;385:1662–1671. doi: 10.1016/S0140-6736(14)62457-0. [DOI] [PubMed] [Google Scholar]
- 23.Pastula DM, Aliabadi N, Haynes AK, et al. Acute neurologic illness of unknown etiology in children - Colorado, August-September 2014. MMWR Morb Mortal Wkly Rep. 2014;63:901–902. [PMC free article] [PubMed] [Google Scholar]
- 24.CDC. Notes from the field: acute flaccid myelitis among persons aged </=21 years - United States, August 1-November 13, 2014. MMWR Morb Mortal Wkly Rep. 2015;63:1243–1244. [PMC free article] [PubMed] [Google Scholar]
- 25.Ayscue P, Van Haren K, Sheriff H, et al. Acute flaccid paralysis with anterior myelitis - California, June 2012-June 2014. MMWR Morb Mortal Wkly Rep. 2014;63:903–906. [PMC free article] [PubMed] [Google Scholar]
- 26.Kreuter JD, Barnes A, McCarthy JE, et al. A fatal central nervous system enterovirus 68 infection. Arch Pathol Lab Med. 2011;135:793–796. doi: 10.5858/2010-0174-CR.1. [DOI] [PubMed] [Google Scholar]
- 27.Greninger AL, Naccache SN, Messacar K, et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012-14): a retrospective cohort study. Lancet Infect Dis. 2015;15:671–682. doi: 10.1016/S1473-3099(15)70093-9. [*The results of this article strengthened the causal link between EV-D68 and AFM. They showed that EV-D68 sequences isolated from AFM cases clustered in a new clade that emerged in 2010, and identified a few SNPs specific to that clade that were also present in other neurotropic enteroviruses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonseca MC, Sarmiento L, Resik S, et al. Isolation of Coxsackievirus A24 variant from patients with hemorrhagic conjunctivitis in Cuba, 2008-2009. J Clin Virol. 2012;53:77–81. doi: 10.1016/j.jcv.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Kono R, Miyamura K, Tajiri E, et al. Virological and serological studies of neurological complications of acute hemorrhagic conjunctivitis in Thailand. J Infect Dis. 1977;135:706–713. doi: 10.1093/infdis/135.5.706. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson EC, Jamshidi F, Johansson SM, et al. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J Virol. 2008;82:3061–3068. doi: 10.1128/JVI.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wadia NH, Wadia PN, Katrak SM, et al. A study of the neurological disorder associated with acute haemorrhagic conjunctivitis due to enterovirus 70. J Neurol Neurosurg Psychiatry. 1983;46:599–610. doi: 10.1136/jnnp.46.7.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee S, Quarcoopome CO, Apenteng A. Unusual type of epidemic conjunctivitis in Ghana. Br J Ophthalmol. 1970;54:628–630. doi: 10.1136/bjo.54.9.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirkovic RR, Kono R, Yin-Murphy M, et al. Enterovirus type 70: the etiologic agent of pandemic acute haemorrhagic conjunctivitis. Bull World Health Organ. 1973;49:341–346. [PMC free article] [PubMed] [Google Scholar]
- 34.Kono R. Apollo 11 disease or acute hemorrhagic conjunctivitis: a pandemic of a new enterovirus infection of the eyes. Am J Epidemiol. 1975;101:383–390. doi: 10.1093/oxfordjournals.aje.a112106. [DOI] [PubMed] [Google Scholar]
- 35.Maitreyi RS, Dar L, Muthukumar A, et al. Acute hemorrhagic conjunctivitis due to enterovirus 70 in India. Emerg Infect Dis. 1999;5:267–269. doi: 10.3201/eid0502.990212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shulman LM, Manor Y, Azar R, et al. Identification of a new strain of fastidious enterovirus 70 as the causative agent of an outbreak of hemorrhagic conjunctivitis. J Clin Microbiol. 1997;35:2145–2149. doi: 10.1128/jcm.35.8.2145-2149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchio E, Yamazaki K, Ishikawa H, et al. An epidemic of acute haemorrhagic conjunctivitis caused by enterovirus 70 in Okinawa, Japan, in 1994. Graefes Arch Clin Exp Ophthalmol. 1999;237:568–572. doi: 10.1007/s004170050280. [DOI] [PubMed] [Google Scholar]
- 38.Mirkovic RR, Schmidt NJ, Yin-Murphy M, et al. Enterovirus etiology of the 1970 Singapore epidemic of acute conjunctivitis. Intervirology. 1974;4:119–127. doi: 10.1159/000149850. [DOI] [PubMed] [Google Scholar]
- 39.Wu B, Qi X, Xu K, et al. Genetic characteristics of the coxsackievirus A24 variant causing outbreaks of acute hemorrhagic conjunctivitis in Jiangsu, China, 2010. PLoS One. 2014;9:e86883. doi: 10.1371/journal.pone.0086883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukla D, Kumar A, Srivastava S, et al. Molecular identification and phylogenetic study of coxsackievirus A24 variant isolated from an outbreak of acute hemorrhagic conjunctivitis in India in 2010. Arch Virol. 2013;158:679–684. doi: 10.1007/s00705-012-1520-7. [DOI] [PubMed] [Google Scholar]
- 41.Nidaira M, Kuba Y, Saitoh M, et al. Molecular evolution of VP3, VP1, 3C(pro) and 3D(pol) coding regions in coxsackievirus group A type 24 variant isolates from acute hemorrhagic conjunctivitis in 2011 in Okinawa, Japan. Microbiol Immunol. 2014;58:227–238. doi: 10.1111/1348-0421.12141. [DOI] [PubMed] [Google Scholar]
- 42.Chansaenroj J, Vongpunsawad S, Puenpa J, et al. Epidemic outbreak of acute haemorrhagic conjunctivitis caused by coxsackievirus A24 in Thailand, 2014. Epidemiol Infect. 2015:1–7. doi: 10.1017/S0950268815000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin KH, Wang HL, Sheu MM, et al. Molecular epidemiology of a variant of coxsackievirus A24 in Taiwan: two epidemics caused by phylogenetically distinct viruses from 1985 to 1989. J Clin Microbiol. 1993;31:1160–1166. doi: 10.1128/jcm.31.5.1160-1166.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamura K, Yamashita K, Takeda N, et al. The first epidemic of acute hemorrhagic conjunctivitis due to a coxsackievirus A24 variant in Okinawa, Japan, in 1985-1986. Jpn J Med Sci Biol. 1988;41:159–174. doi: 10.7883/yoken1952.41.159. [DOI] [PubMed] [Google Scholar]
- 45.Oh MD, Park S, Choi Y, et al. Acute hemorrhagic conjunctivitis caused by coxsackievirus A24 variant, South Korea, 2002. Emerg Infect Dis. 2003;9:1010–1012. doi: 10.3201/eid0908.030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayoub EA, Shafik CF, Gaynor AM, et al. A molecular investigative approach to an outbreak of acute hemorrhagic conjunctivitis in Egypt, October 2010. Virol J. 2013;10:96. doi: 10.1186/1743-422X-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavares FN, Campos Rde M, Burlandy FM, et al. Molecular characterization and phylogenetic study of coxsackievirus A24v causing outbreaks of acute hemorrhagic conjunctivitis (AHC) in Brazil. PLoS One. 2011;6:e23206. doi: 10.1371/journal.pone.0023206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aoki K, Sawada H. Long-term observation of neutralization antibody after enterovirus 70 infection. Jpn J Ophthalmol. 1992;36:465–468. [PubMed] [Google Scholar]
- 49.Goh KT, Ooi PL, Miyamura K, et al. Acute haemorrhagic conjunctivitis: seroepidemiology of coxsackievirus A24 variant and enterovirus 70 in Singapore. J Med Virol. 1990;31:245–247. doi: 10.1002/jmv.1890310313. [DOI] [PubMed] [Google Scholar]
- 50.Finger C. Brazil faces worst outbreak of conjunctivitis in 20 years. Lancet. 2003;361:1714. doi: 10.1016/S0140-6736(03)13391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang LY, Lin TY, Huang YC, et al. Comparison of enterovirus 71 and coxsackie-virus A16 clinical illnesses during the Taiwan enterovirus epidemic, 1998. Pediatr Infect Dis J. 1999;18:1092–1096. doi: 10.1097/00006454-199912000-00013. [DOI] [PubMed] [Google Scholar]
- 52.Ooi MH, Wong SC, Podin Y, et al. Human enterovirus 71 disease in Sarawak, Malaysia: a prospective clinical, virological, and molecular epidemiological study. Clin Infect Dis. 2007;44:646–656. doi: 10.1086/511073. [DOI] [PubMed] [Google Scholar]
- 53.Ooi MH, Wong SC, Lewthwaite P, et al. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 54.Solomon T, Lewthwaite P, Perera D, et al. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 55.Yamayoshi S, Fujii K, Koike S. Receptors for enterovirus 71. Emerg Microbes Infect. 2014;3:e53. doi: 10.1038/emi.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shindarov LM, Chumakov MP, Voroshilova MK, et al. Epidemiological, clinical, and pathomorphological characteristics of epidemic poliomyelitis-like disease caused by enterovirus 71. J Hyg Epidemiol Microbiol Immunol. 1979;23:284–295. [PubMed] [Google Scholar]
- 57.Nagy G, Takatsy S, Kukan E, et al. Virological diagnosis of enterovirus type 71 infections: experiences gained during an epidemic of acute CNS diseases in Hungary in 1978. Arch Virol. 1982;71:217–227. doi: 10.1007/BF01314873. [DOI] [PubMed] [Google Scholar]
- 58.WHO. A Guide to Clinical Management and Public Health Response for Hand, Foot and Mouth Disease (HFMD) 2011. [Google Scholar]
- 59.Cardosa MJ, Krishnan S, Tio PH, et al. Isolation of subgenus B adenovirus during a fatal outbreak of enterovirus 71-associated hand, foot, and mouth disease in Sibu, Sarawak. Lancet. 1999;354:987–991. doi: 10.1016/S0140-6736(98)11032-2. [DOI] [PubMed] [Google Scholar]
- 60.CDC. Deaths Among Children During an Outbreak of Hand, Foot, and Mouth Disease -- Taiwan, Republic of China, April-July 1998. MMWR Morb Mortal Wkly Rep. 1998;47:629–632. [PubMed] [Google Scholar]
- 61.Ho M, Chen ER, Hsu KH, et al. An epidemic of enterovirus 71 infection in Taiwan. N Engl J Med. 1999;341:929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- 62.Liu SL, Pan H, Liu P, et al. Comparative epidemiology and virology of fatal and nonfatal cases of hand, foot and mouth disease in mainland China from 2008 to 2014. Reviews in Medical Virology. 2015;25:115–128. doi: 10.1002/rmv.1827. [DOI] [PubMed] [Google Scholar]
- 63.Xing W, Liao Q, Viboud C, et al. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis. 2014;14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [** This article demonstrated that environmental factors influence the incidence of enterovirus infections. They showed that the seasonality of HFMD varies with latitude and found associations between incidence and precipitation, sunshine, temperature and air pressure.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.NIID. Hand, foot and mouth disease in Japan, 2002-2011. IASR. 2012;33:55–56. [Google Scholar]
- 65.Tu PV, Thao NT, Perera D, et al. Epidemiologic and virologic investigation of hand, foot, and mouth disease, southern Vietnam, 2005. Emerg Infect Dis. 2007;13:1733–1741. doi: 10.3201/eid1311.070632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ang LW, Tay J, Phoon MC, et al. Seroepidemiology of Coxsackievirus A6, Coxsackievirus A16, and Enterovirus 71 Infections among Children and Adolescents in Singapore, 2008-2010. PLoS One. 2015;10:e0127999. doi: 10.1371/journal.pone.0127999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singapore Ministry of Health. Communicable Disease Surveillance Annual Report. 2013 [Google Scholar]
- 68.Singapore Ministry of Health. Communicable Disease Surveillance Annual Report. 2014 [Google Scholar]
- 69.Ma E, Chan KC, Cheng P, et al. The enterovirus 71 epidemic in 2008--public health implications for Hong Kong. Int J Infect Dis. 2010;14:e775–780. doi: 10.1016/j.ijid.2010.02.2265. [DOI] [PubMed] [Google Scholar]
- 70.Podin Y, Gias EL, Ong F, et al. Sentinel surveillance for human enterovirus 71 in Sarawak, Malaysia: lessons from the first 7 years. BMC Public Health. 2006;6:180. doi: 10.1186/1471-2458-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di B, Zhang Y, Xie H, et al. Circulation of Coxsackievirus A6 in hand-foot-mouth disease in Guangzhou, 2010-2012. Virol J. 2014;11:157. doi: 10.1186/1743-422X-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He YQ, Chen L, Xu WB, et al. Emergence, circulation, and spatiotemporal phylogenetic analysis of coxsackievirus A6- and coxsackievirus A10-associated hand, foot, and mouth disease infections from 2008 to 2012 in Shenzhen, China. J Clin Microbiol. 2013;51:3560–3566. doi: 10.1128/JCM.01231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hongyan G, Chengjie M, Qiaozhi Y, et al. Hand, foot and mouth disease caused by coxsackievirus A6, Beijing, 2013. Pediatr Infect Dis J. 2014;33:1302–1303. doi: 10.1097/INF.0000000000000467. [DOI] [PubMed] [Google Scholar]
- 74.Huang Y, Zhou Y, Lu H, et al. Characterization of severe hand, foot, and mouth disease in Shenzhen, China, 2009-2013. J Med Virol. 2015 doi: 10.1002/jmv.24200. [DOI] [PubMed] [Google Scholar]
- 75.Tan X, Li L, Zhang B, et al. Molecular epidemiology of coxsackievirus A6 associated with outbreaks of hand, foot, and mouth disease in Tianjin, China, in 2013. Arch Virol. 2015;160:1097–1104. doi: 10.1007/s00705-015-2340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng H, Lu J, Zheng H, et al. The Epidemiological Study of Coxsackievirus A6 revealing Hand, Foot and Mouth Disease Epidemic patterns in Guangdong, China. Sci Rep. 2015;5:10550. doi: 10.1038/srep10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Q, Ding J, Cao J, et al. Epidemiological and etiological characteristics of hand, foot, and mouth disease in Wuhan, China from 2012 to 2013: outbreaks of coxsackieviruses A10. J Med Virol. 2015;87:954–960. doi: 10.1002/jmv.24151. [DOI] [PubMed] [Google Scholar]
- 78.Zhang C, Zhu R, Yang Y, et al. Phylogenetic analysis of the major causative agents of hand, foot and mouth disease in Suzhou city, Jiangsu province, China, in 2012-2013. Emerg Microbes Infect. 2015;4:e12. doi: 10.1038/emi.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bracho MA, Gonzalez-Candelas F, Valero A, et al. Enterovirus co-infections and onychomadesis after hand, foot, and mouth disease, Spain, 2008. Emerg Infect Dis. 2011;17:2223–2231. doi: 10.3201/eid1712.110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osterback R, Vuorinen T, Linna M, et al. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis. 2009;15:1485–1488. doi: 10.3201/eid1509.090438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sarma N. Hand, foot, and mouth disease: current scenario and Indian perspective. Indian J Dermatol Venereol Leprol. 2013;79:165–175. doi: 10.4103/0378-6323.107631. [DOI] [PubMed] [Google Scholar]
- 82.Ong KC, Devi S, Cardosa MJ, et al. Formaldehyde-inactivated whole-virus vaccine protects a murine model of enterovirus 71 encephalomyelitis against disease. J Virol. 2010;84:661–665. doi: 10.1128/JVI.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arita M, Nagata N, Iwata N, et al. An attenuated strain of enterovirus 71 belonging to genotype a showed a broad spectrum of antigenicity with attenuated neurovirulence in cynomolgus monkeys. J Virol. 2007;81:9386–9395. doi: 10.1128/JVI.02856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li HY, Han JF, Qin CF, et al. Virus-like particles for enterovirus 71 produced from Saccharomyces cerevisiae potently elicits protective immune responses in mice. Vaccine. 2013;31:3281–3287. doi: 10.1016/j.vaccine.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 85.Wu CN, Lin YC, Fann C, et al. Protection against lethal enterovirus 71 infection in newborn mice by passive immunization with subunit VP1 vaccines and inactivated virus. Vaccine. 2001;20:895–904. doi: 10.1016/s0264-410x(01)00385-1. [DOI] [PubMed] [Google Scholar]
- 86.Li JX, Mao QY, Liang ZL, et al. Development of enterovirus 71 vaccines: from the lab bench to Phase III clinical trials. Expert Rev Vaccines. 2014;13:609–618. doi: 10.1586/14760584.2014.897617. [DOI] [PubMed] [Google Scholar]
- 87.Li R, Liu L, Mo Z, et al. An inactivated enterovirus 71 vaccine in healthy children. N Engl J Med. 2014;370:829–837. doi: 10.1056/NEJMoa1303224. [DOI] [PubMed] [Google Scholar]
- 88.Zhu F, Xu W, Xia J, et al. Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China. N Engl J Med. 2014;370:818–828. doi: 10.1056/NEJMoa1304923. [DOI] [PubMed] [Google Scholar]
- 89.Zhu FC, Meng FY, Li JX, et al. Efficacy, safety, and immunology of an inactivated alum-adjuvant enterovirus 71 vaccine in children in China: a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2013;381:2024–2032. doi: 10.1016/S0140-6736(13)61049-1. [DOI] [PubMed] [Google Scholar]
- 90.Mao Q, Cheng T, Zhu F, et al. The cross-neutralizing activity of enterovirus 71 subgenotype C4 vaccines in healthy chinese infants and children. PLoS One. 2013;8:e79599. doi: 10.1371/journal.pone.0079599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chou AH, Liu CC, Chang JY, et al. Formalin-inactivated EV71 vaccine candidate induced cross-neutralizing antibody against subgenotypes B1, B4, B5 and C4A in adult volunteers. PLoS One. 2013;8:e79783. doi: 10.1371/journal.pone.0079783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cai Y, Ku Z, Liu Q, et al. A combination vaccine comprising of inactivated enterovirus 71 and coxsackievirus A16 elicits balanced protective immunity against both viruses. Vaccine. 2014;32:2406–2412. doi: 10.1016/j.vaccine.2014.03.012. [* This study provides the foundations for the development of a bivalent vaccine to control HFMD. The inactivated bivalent (EV-A71 and CV-A16) vaccine tested in mice elicited levels of neutralizing antibodies similar to those induced by the monovalent formulations and the antibody titres were sustained at high levels.] [DOI] [PubMed] [Google Scholar]
- 93.Henquell C, Mirand A, Richter J, et al. Phylogenetic patterns of human coxsackievirus B5 arise from population dynamics between two genogroups and reveal evolutionary factors of molecular adaptation and transmission. J Virol. 2013;87:12249–12259. doi: 10.1128/JVI.02075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McWilliam Leitch EC, Cabrerizo M, Cardosa J, et al. Evolutionary dynamics and temporal/geographical correlates of recombination in the human enterovirus echovirus types 9, 11, and 30. J Virol. 2010;84:9292–9300. doi: 10.1128/JVI.00783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cabrerizo M, Trallero G, Simmonds P. Recombination and evolutionary dynamics of human echovirus 6. J Med Virol. 2014;86:857–864. doi: 10.1002/jmv.23741. [DOI] [PubMed] [Google Scholar]
- 96.Huang SW, Hsu YW, Smith DJ, et al. Reemergence of enterovirus 71 in 2008 in Taiwan: dynamics of genetic and antigenic evolution from 1998 to 2008. J Clin Microbiol. 2009;47:3653–3662. doi: 10.1128/JCM.00630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]