Abstract
Background
The major timothy grass pollen allergen Phl p 5 belongs to the most potent allergens involved in hay fever and asthma.
Objective
This study characterized immune-dominant IgE- and T-cell–recognition sites of Phl p 5.
Methods
Seven peptides, P1 to P7 with a length of 31 to 38 amino acids that spanned the Phl p 5 sequence, were synthesized, characterized by circular dichroism spectroscopy, and tested for IgE reactivity, basophil activation, and T-cell reactivity. Carrier-bound peptides were studied for their ability to induce IgG antibodies in rabbits which recognize Phl p 5 or cross-reactive allergens from different grass species. Peptide-specific antibodies were tested for the capability to inhibit IgE reactivity to Phl p 5 and allergen-induced basophil activation of patients with allergy.
Results
The peptides exhibited no secondary structure and showed no IgE reactivity or relevant allergenic activity, indicating that Phl p 5 IgE epitopes are conformational. Except for P3, peptide-specific IgG antibodies blocked IgE binding to Phl p 5 of patients with allergy and cross-reacted with temperate grasses. IgE inhibition experiments and molecular modeling identified several clustered conformational IgE epitopes on the N- as well as C-terminal domain of Phl p 5. P4, which stimulated the strongest T-cell and cytokine responses in patients, was not part of the major IgE-reactive regions.
Conclusion
Our study shows an interesting dissociation of the major IgE- and T-cell–reactive domains in Phl p 5 which provides a basis for the development of novel forms of immunotherapy that selectively target IgE or T-cell responses.
Keywords: Allergy, grass pollen allergen, peptides, epitopes
Approximately 50% of patients with allergy experience grass pollen-induced allergic symptoms that range from rhinoconjunctivitis (ie, hay fever) to severe asthma attacks.1–4 Grass pollen has, therefore, been recognized since 1880 as a major allergen source.5 In addition, the first allergen-specific immunotherapy was performed with grass pollen preparations in 1911.6 Today, allergen-specific immunotherapy is well established as a clinically effective treatment for allergy.7–9 New approaches are based on genetically modified recombinant allergens with reduced allergenic activity,10–13 T-cell epitope-containing peptides for selective targeting of allergen-specific T cells,14 and carrier-bound allergen peptides for selective induction of protective allergen-specific IgG responses.15,16
Group 5 allergens belong to the most frequent and potent grass pollen allergens which, because of their release in the form of submicronic respirable pollen particles, are implicated in asthma attacks.3,17–20 The complete 3-dimensional structure of group 5 allergens has not yet been solved, but the structures of the N- and C-terminal domains of 2 isoforms, Phl p 5a and Phl p 5b, from timothy grass pollen showed that these domains consist each of anti-parallel 4 helix bundles.21,22 Both the N-23 and C-terminal domains of group 5 allergens20,24,25 are described as containing IgE-binding sites that still need to be characterized in detail.
Different earlier T-cell epitope mapping studies for group 5 allergens performed with T-cell clones and lines report highly discrepant results for the location of major T-cell epitopes.26–29 Therefore, little is known about the immune dominance of certain T-cell epitopes.
Here, we used Phl p 5 peptide-specific IgG antibodies for IgE competition experiments as an approach for the mapping of conformational IgE epitopes of group 5 allergens.30,31 For this purpose we synthesized 7 peptides of 31 to 38 amino acids that cover large parts of the Phl p 5 sequence to raise peptide-specific antibodies. With the synthetic peptides the presence of sequential (ie, continuous) IgE epitopes of group 5 allergens was investigated, and peptide-specific antibodies were used for inhibiting the IgE binding to Phl p 5 from a large number of patients with grass pollen allergy and to search for immunedominant conformational/discontinuous IgE epitopes. Furthermore, we studied the ability of the individual peptide-specific IgG antibodies and mixes thereof to inhibit Phl p 5-induced basophil activation to learn about the clonal distribution of patients’ IgE responses. In parallel, we used the synthetic peptides to study lymphocyte proliferation and cytokine responses in peripheral blood mononuclear cells (PBMCs) from patients with grass pollen allergy to identify immunedominant T-cell epitopes.
Methods
Immunization of rabbits with KLH-coupled peptides, reactivity of rabbit antipeptide antibodies with Phl p 5, and natural allergens from different grass species
New Zealand white rabbits were immunized with each of the keyholelimpet hemocyanin (KLH)-conjugated peptides (200 μg/injection) or with complete rPhl p 5 with the use of Freunds complete and incomplete adjuvans (Charles River, Kisslegg, Germany). IgG reactivity was tested by direct ELISA.32
Inhibition of allergic patient’s IgE binding to Phl p 5 by peptide-specific IgG
ELISA inhibition experiments were done as described, using 1 μg/mL rPhl p 5 for coating, a 1:250 dilution of each of the rabbit antisera, and 1:10 diluted sera from patients with grass pollen allergy. Detection of bound IgE antibodies and calculation of inhibition of IgE binding was done as described previously.32
Allergen-induced upregulation of CD203c and CD63 expression on patient’s basophils and inhibition of allergen-induced CD203c upregulation by allergen-specific antibodies
Heparinized blood samples (100 μL) from patients with grass pollen allergy were incubated with serial dilutions of rPhl p 5 (10−4 to 10 μg/mL), an equimolar mix of Phl p 5-derived peptides, an equimolar mix of KLH-conjugated Phl p 5 peptides, a monoclonal anti-IgE antibody (Immunotech, Marseille, France; 1 μg/mL), or buffer alone (PBS) for 15 minutes at 37°C.
In CD203c inhibition experiments serial dilutions of rPhl p 5 or an unrelated allergen, that is, birch pollen allergen, Bet v 1 (negative control) (10−1 to 10−4 μg/mL), were preincubated with a 1:100 diluted mix of rabbit antipeptide antisera or a mix of the corresponding preimmune sera before incubation with the blood samples.30
Measurement of CD203c upregulation was done as described.33
Lymphocyte proliferation assays, CFSE staining, and multiple cytokine measurements
PBMCs were isolated from patients with grass pollen allergy and were cultured as described previously.34 Cells were stimulated with different concentrations of each of the synthetic peptides, mixes of the 7 peptides (each peptide 1.25, 0.6, 0.3, and 0.15 μg per well), and equimolar concentrations of rPhl p 5 for comparison. Endotoxin concentrations were determined in each of the tested preparations with the use of the Limulus Amebocyte Lysate QCL-1000 kit (BioWhittaker, Walkersville, Md). The endotoxin concentrations of the peptides and of Phl p 5 (measured at a concentration of 10 μg/mL) were below 1 EU/mL (data not shown). For control purposes 4 U of IL-2 per well (Boehringer, Mannheim, Germany) or medium alone were tested. Cultures were done in triplicates, and results are shown as mean cpm values ± SD.34
For carboxyfluorescein diacetate succinimidyl ester (CFSE) staining experiments, cells were labeled with CFSE (Invitrogen, Oslo, Norway) by incubating 5 × 106 cells (triplicates) in a solution of 2.5 μmol/L CFSE in PBS for 10 minutes at 37°C, washed, and then cultured with rPhl p 5 (5 μg/well), the individual peptides or a peptide mix (0.6 μg/well per peptide), medium alone (negative control), or Dynabeads that contained anti-CD3 and anti-CD28 (positive control) (3 μL/well; Invitrogen) for 7 days at 37°C in Ultra Culture serum-free medium (Lonza, Verviers, Belgium). Then cells were centrifuged and resuspended in 50 μL of blocking buffer (PBS, 0.01% wt/vol NaN3, 1% wt/vol BSA, 10% mouse serum) for 20 minutes on ice, centrifuged, and finally stained with phycoerythrin/cyanine 7-labeled mouse monoclonal IgG1 anti-human CD3, an IgG1 isotype control, 7-amino-actinomycin D (7-AAD), phycoerythrin-labeled mouse monoclonal IgG2 anti-human CD4, and the corresponding isotype control (Biolegend, San Diego, Calif) diluted in fluorescence-activated cell sorting buffer (PBS, 0.01% wt/vol NaN3, 1% wt/vol BSA) in dark on ice for 20 minutes and analyzed on a Cytomics FC 500 (Beckman Coulter, Fullerton, Calif). Cells were gated according to forward and sideward scatter, dead cells were excluded by 7-AAD staining, and gating was on CD4 cells. After subtraction of results for medium, mean percentages ± SDs of CD4 cells that proliferate in response to each of the agonists are shown.
Levels of IL-2, IL-4, IL-5, IL-6, IL-10, IL-12, IL-13, IFN-γ, TNF-α, and GM-CSF were measured in supernatant fluids from PBMC cultures which were identically prepared as for the proliferation experiments with the use of the xMAP Luminex fluorescent bead-based technology (Luminex Corp, Austin, Tex) and a human MultiAnalyte Profiling Kit A and B (R&D Systems, Abingdon, United Kingdom). Fluorescent signals were read on a Luminex 100 system (Luminex Corp).
Statistical evaluation
Unpaired Kruskal-Wallis tests were used to assess statistically significant differences for T-cell proliferation and cytokines. A P value ≤ .05 was considered statistically significant.
Results
Characterization of synthetic peptides derived from solvent accessible areas of Phl p 5a
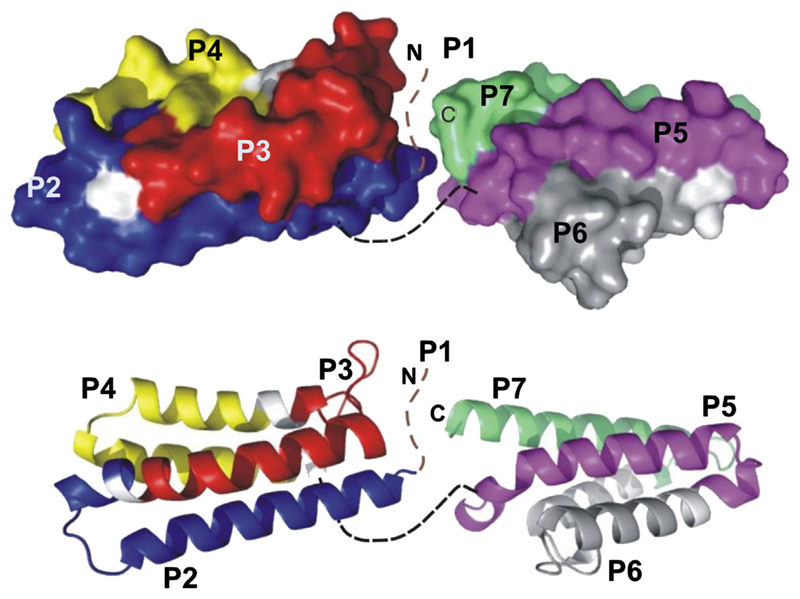
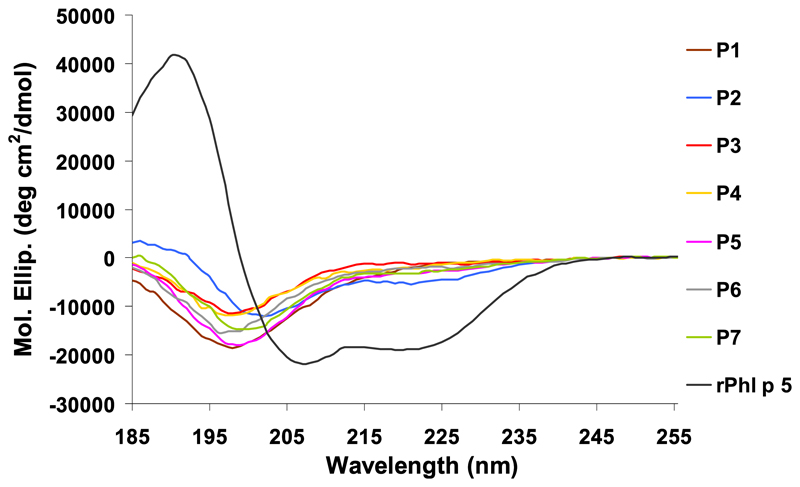
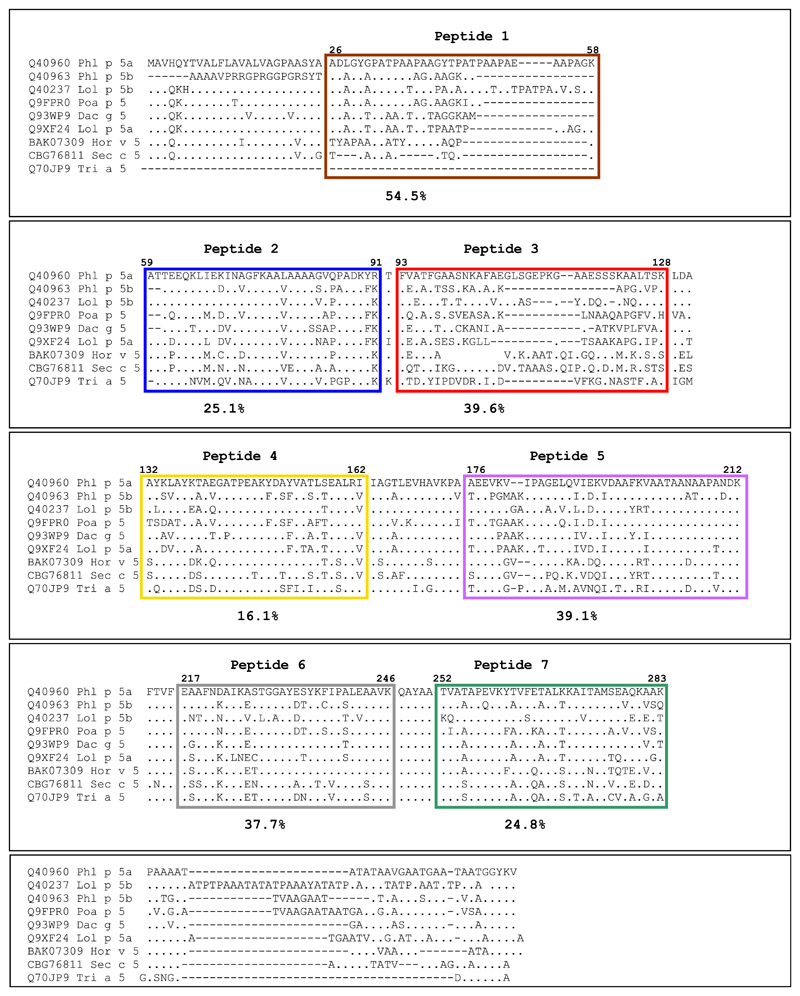
It was shown that allergen-derived peptides with a length of >25 amino acids coupled to a carrier gave robust peptide- and allergen-specific antibody responses on immunization.32,35 Solvent accessibility prediction indicated that the regions of Phl p 5a with predicted high hydrophilicity and surface accessibility can be covered with 7 peptides (P1-P7), ranging from 31 (P1, 3068 Dalton) to 38 (P5, 3853 Dalton) amino acids in length. Table E1 (in the Online Repository at www.jacionline.org) summarizes the position, length, and biochemical properties of the Phl p 5a-derived synthetic peptides. None of the peptides showed any residual secondary structure as determined by circular dichroism (CD) experiments (see Fig E1 in this article’s Online Repository at www.jacionline.org), whereas the CD spectrum of rPhl p 5a is characterized by 2 broad minima at 207 and 221 nm, typical for folded α helical proteins. P2, P3, and P4 represent surface-exposed areas on the N-terminal domain, whereas P5, P6, and P7 are part of the C-terminal domain (Fig 1). P1 represents a flexible region at the N-terminus of the first domain. In the given view of the model, P1, P2, P3, P5, and P6 appear on the front side, whereas P4 and P7 are located rather on the backside of the modeled structure (Fig 1).
Fig 1.
Mapping of the peptides onto a model of the 3-dimensional structure of Phl p 5. Peptides were highlighted in different colors (P1 aa 26-58, brown; P2 aa 59-91, blue; P3 aa 93-128, red; P4 aa 132-162, yellow; P5 aa 176-212, magenta; P6 aa 217-246: gray; P7 aa 252-283, green) in a ribbon (upper part) and surface representation of Phl p 5 (lower part).
Fig E2 (in the Online Repository at www.jacionline.org) shows a multiple sequence alignment of group 5 allergens from different grasses in which the peptides were highlighted to visualize their sequence homology with the corresponding regions in related grass pollen allergens from other species. The sequences for Phl p 5a, Phl p 5b, Lol p 5b, Poa p 5, Dac g 5, Lol p 5a, Hor v 5, and Sec c 5 are complete sequences, whereas for Tri a 5 only a partial sequence was available. Group 5 allergens possess an N-terminal hydrophobic leader peptide (eg, Phl p 5a: aa 1-25) that is absent in the mature allergens; therefore, it was not considered for epitope mapping. The peptides corresponding to P1 and P3 and the C-terminal portions in the different group 5 allergens have a considerable variability in length and a relatively low degree of sequence homology, whereas P2-, P4-, P5-, P6-, and P7-homologous peptides represented well-conserved regions in the group 5 allergens from the different grasses (Fig E2).
Phl p 5-derived peptides lack IgE reactivity and allergenic activity
First, we compared the IgE reactivity of the 7 Phl p 5 peptides with that of the complete Phl p 5 allergen by ELISA. For this purpose sera from 51 patients with grass pollen allergy (see Table E2 in this article’s Online Repository at www.jacionline.org) were tested for IgE reactivity to Phl p 5 and the purified peptides. Because coupling of multiple peptides to KLH may affect IgE reactivity, we also analyzed KLH-coupled versions of the peptides. Each of the 51 patients with grass pollen allergy experienced at least grass pollen-induced rhinitis and 11 had asthma (Table E2). They had varying levels of timothy grass pollen-specific IgE (3.2 to >100 kUA/L; mean, 49.53 kUA/L) and varying total IgE levels (18-4625 kU/L; mean, 583 kU/L). rPhl p 5-specific IgE levels ranged between OD 0.112 and >2.500 (max OD, 3.69; mean OD, 1.45). We found that none of the 36 patients showed any detectable IgE reactivity to the isolated peptides, to KLH-conjugated peptides, or to equimolar mixes thereof (ie, OD values ≤0.08; Table E2). In addition, recombinant Phl p 5 fragments comprising aa 26 to 176 and aa 176 to 258 showed no relevant IgE reactivity (data not shown). Serum from a person without allergy showed no IgE reactivity to grass pollen extract, rPhl p 5, or to any of the peptides. With the use of rabbit antipeptide antibodies, the coating of the peptides to the plates was confirmed (data not shown). The lack of IgE reactivity of the peptides was also found by dot blot experiments (data not shown).
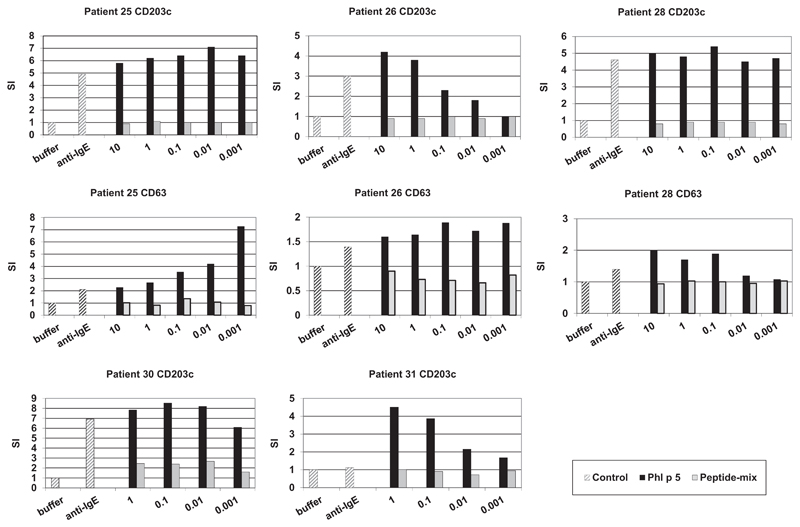
Fig 2 shows that Phl p 5 is an extremely potent allergen which induced in certain patients (patients 25 and 28) a full upregulation of CD203c expression at a concentration of 0.001 μg/mL. In the other 3 patients (patients 26, 30, and 31) a dose-dependent (10−4 to 10 μg/mL) increase in expression of CD203c was observed with Phl p 5. Neither the mix of isolated nor the mix of KLH-coupled peptides induced any relevant basophil activation even at the highest concentrations applied (1 or 10 μg/mL) in 4 of the 5 patients. Similar data were obtained for CD63 upregulation (Fig 2). A weak upregulation of CD203c expression by peptides was observed for concentrations starting with 0.01 μg/mL in patient 30 (Fig 2).
Fig 2.
Allergenic activity of rPhl p 5 and a peptide mix as measured by upregulation of CD203c and CD63 expression on patients’ basophils. Blood samples were incubated with serial dilutions of rPhl p 5 (0.001-1 or 10 μg/mL; black bars), equimolar mixes of unconjugated (patients 25, 26, and 28) or KLH-coupled peptides (patients 30 and 31; gray bars), anti-IgE, or buffer (x-axes). The stimulation indices (SI) are displayed on the y-axes.
Complete Phl p 5 induces significantly higher lymphocyte proliferation and cytokine responses in PBMCs of patients with grass pollen allergy than the Phl p 5 peptides
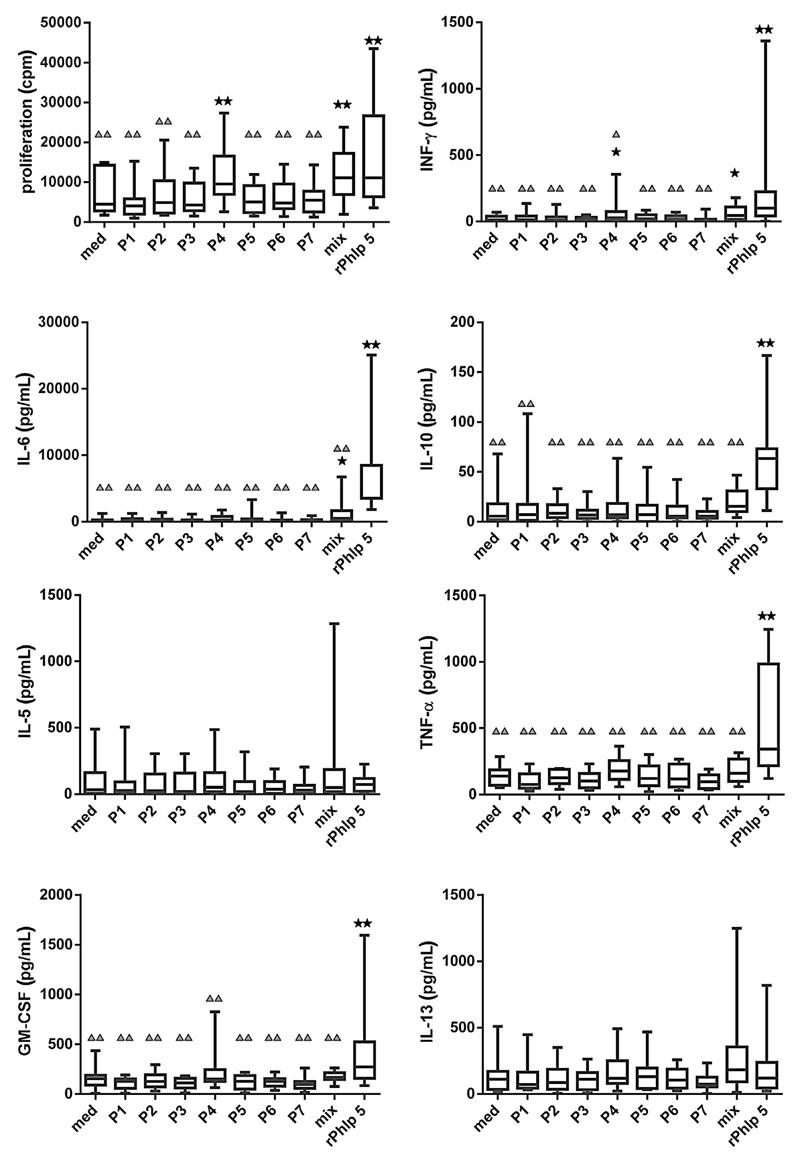
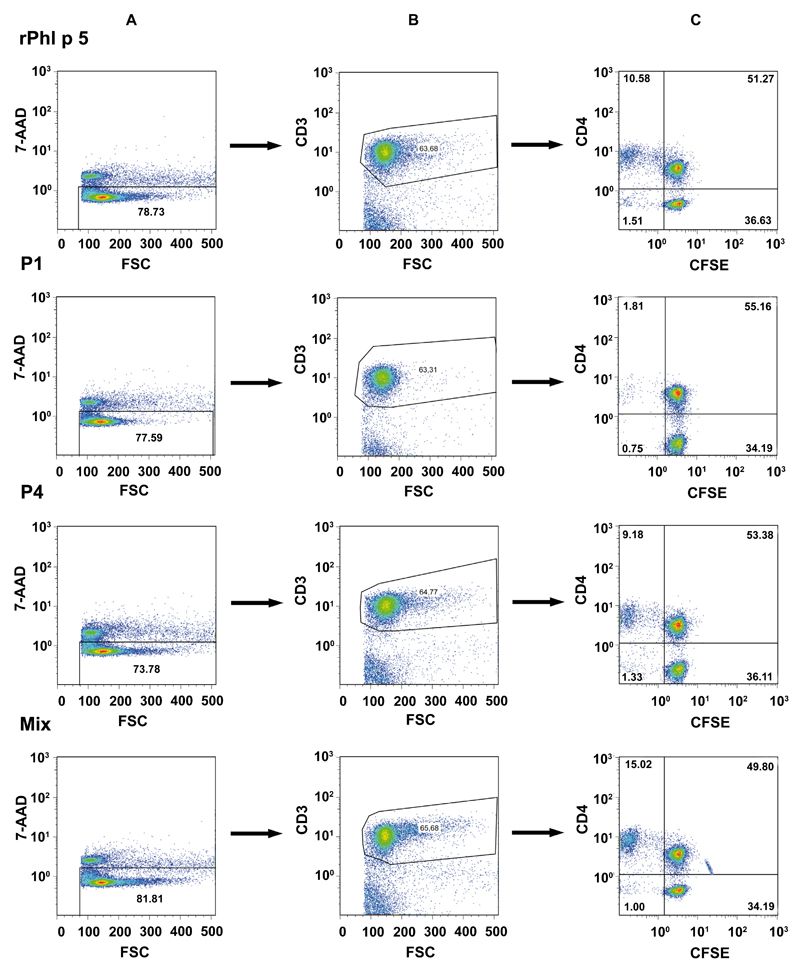
Next, we compared Phl p 5 peptides, the mix of all 7 peptides with complete Phl p 5, for the induction of lymphocyte proliferation and cytokine responses in PBMCs of patients with grass pollen allergy (Fig 3). Fig 3 shows that, compared with medium Phl p 5, the mix of all peptides and P4 alone induced significantly higher proliferation. A similar result was obtained for the induction of IFN-γ, a cytokine associated with chronic inflammation. In addition, Phl p 5 induced higher levels of other proinflammatory cytokines (eg, IL-6, TNF-α, and GM-CSF) and of the tolerogenic cytokine IL-10 than the individual peptides, whereas no significant differences were noted for TH2 cytokines (ie, IL-5, IL-13). IL-2, IL-4, and IL-12 levels have also been measured but were below the detection limit. To assure that the obtained proliferative responses were derived from specific T cells we performed CFSE staining and gated on CD4+ T cells (Table I; see also Fig E3 in this article’s Online Repository at www.jacionline.org). The CFSE analysis of samples from 10 patients with grass pollen allergy confirmed the results obtained in the proliferation experiments. The complete rPhl p 5 allergens induced the strongest CD4+ T-cell activation (mean, 9.21% CD4+ T cells), and, among the peptides, P4 induced the strongest expansion of specific CD4+ cells (mean, 5.61% CD4+ T cells). A similar CD4+ T-cell stimulation was only obtained with the mix of all peptides (mean, 5.33% CD4+ T cells).
Fig 3.
Phl p 5- and peptide-specific lymphocyte proliferation and cytokine responses. PBMCs from 13 patients with grass pollen allergy were stimulated with equimolar concentrations of rPhl p 5, the individual peptides (P1-P7), the peptide mix (mix), or medium alone (med) (x-axes). Box plots (horizontal bars, medians ± SDs) of thymidine incorporations (cpm values) and cytokine levels (pg/mL) are shown on the y-axes. Statistically significant differences between medium and stimuli (**P < .01; *P < .05) and between Phl p 5 and peptides (▲▲P < .01; ▲P < .05) are indicated.
Table I. Percentages of proliferating CD4+ T cells in PBMC cultures from patients with grass pollen allergy after stimulation with rPhl p 5 and Phl p 5-derived peptides.
| Patient |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 22 | 24 | 25 | 27 | 29 | 52 | 53 | 54 | 55 | Mean | |
| rPhl p 5 | 9.01 | 7.60 | 6.06 | 27.39 | 2.70 | 8.37 | 5.76 | 1.53 | 4.45 | 19.18 | 9.21 |
| P1 | 1.73 | 0.55 | 0.51 | 2.84 | 1.47 | 2.84 | 0.82 | 0.73 | 0.79 | 1.45 | 1.37 |
| P2 | 1.06 | 1.16 | 1.12 | 12.47 | 1.82 | 1.22 | 0.71 | 0.60 | 0.63 | 1.23 | 2.20 |
| P3 | 1.07 | 0.54 | 1.18 | 26.04 | 1.38 | 2.89 | 0.65 | 0.78 | 0.79 | 1.32 | 3.66 |
| P4 | 3.62 | 3.57 | 1.95 | 33.19 | 1.90 | 5.05 | 0.66 | 0.79 | 0.60 | 4.77 | 5.61 |
| P5 | 2.16 | 1.36 | 0.84 | 1.07 | 1.88 | 2.71 | 0.68 | 0.78 | 0.61 | 2.05 | 1.41 |
| P6 | 1.64 | 1.46 | 0.66 | 9.04 | 1.30 | 0.42 | 0.51 | 0.70 | 0.39 | 1.40 | 1.75 |
| P7 | 2.09 | 2.91 | 0.58 | 13.10 | 1.51 | 0.89 | 0.50 | 1.01 | 0.37 | 3.02 | 2.60 |
| Mix | 3.56 | 5.71 | 0.69 | 26.04 | 1.95 | 3.06 | 1.05 | 1.55 | 0.83 | 8.90 | 5.33 |
Peptide-specific antibodies bind to Phl p 5 and cross-react with group 5 allergens from different grasses to a varying degree
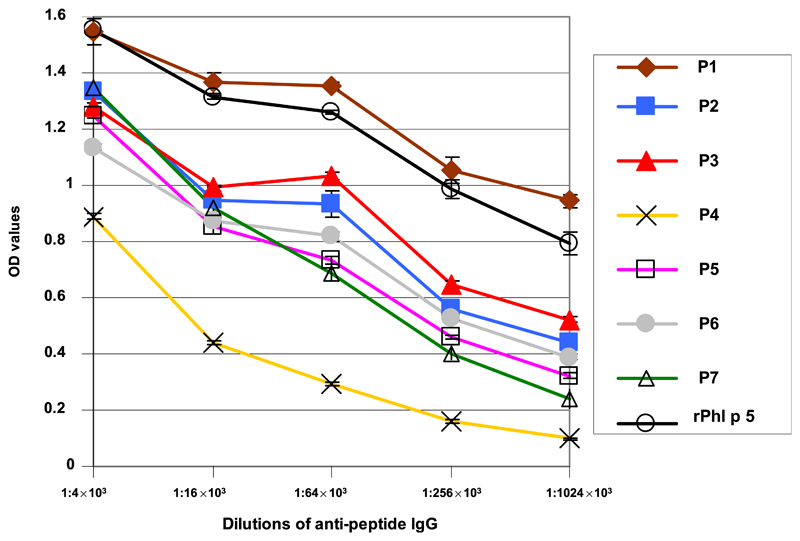
Interestingly, the highest anti-Phl p 5 IgG titers were obtained by immunization with KLH-coupled P1 which were even higher than those obtained by immunization with complete Phl p 5 (see Fig E4 in this article’s Online Repository at www.jacionline.org). The Phl p 5-specific IgG titers induced with KLH-coupled P2, P3, P5, P6, and P7 were comparable, albeit lower than those induced with Phl p 5. The lowest Phl p 5-specific IgG titers were achieved by immunization with P4, although the antiserum showed reactivity even at a dilution of 1:100,000 (Fig E3). The OD levels of the corresponding preimmune sera at a dilution of 1:4000 were between 0.003 and 0.1.
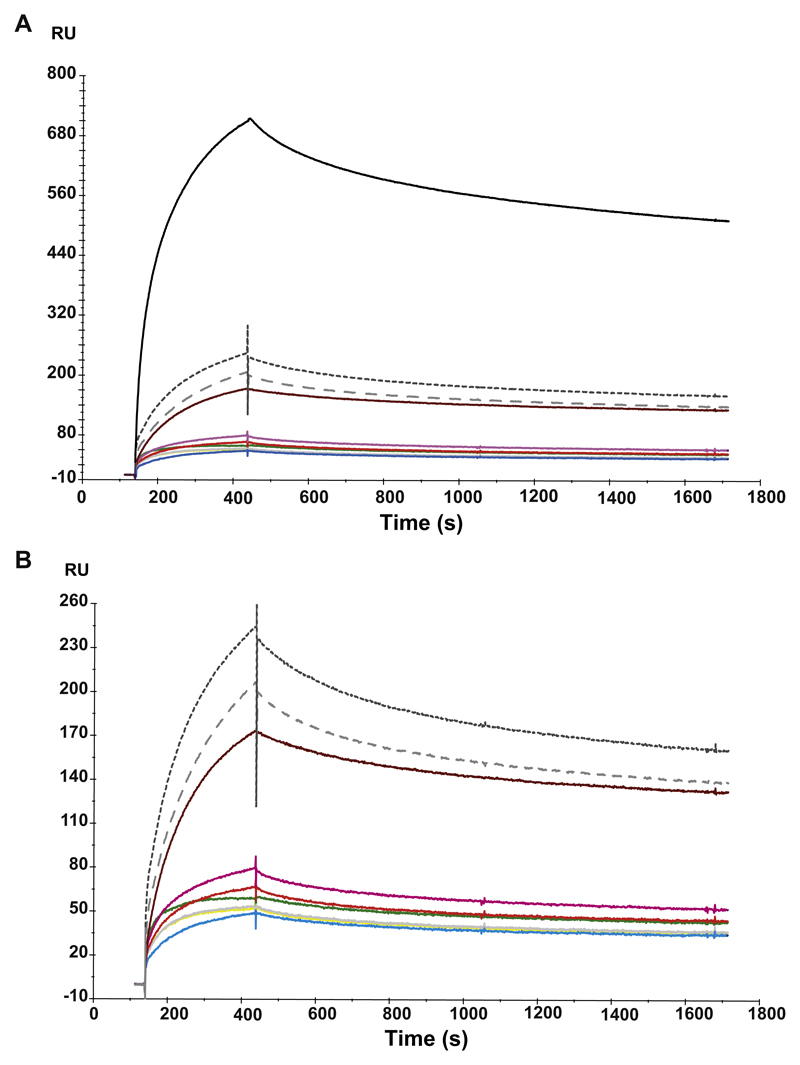
To further characterize the binding behavior of the allergen-specific and peptide-specific antisera, Biacore experiments were performed (see Fig E5 in this article’s Online Repository at www.jacionline.org). We found that the association and dissociation of peptide-specific antisera were comparable with that of an antiserum raised against the Phl p 5 allergen when comparing the association and dissociation curves (Fig E5, A and B), which suggests comparable avidities of the antisera. Interestingly, under the conditions tested, binding levels as measured by the response unit on the y-axis were not associated with the titers as determined by ELISA (Fig E4; Fig E5, B). The response was highest for Phl p 5 and lower for the individual peptide antisera even when a mixture of the antisera was tested. These results indicate that the peptide antisera target fewer areas on Phl p 5 than the anti-Phl p 5 antibodies.
Next, we investigated the cross-reactivity of Phl p 5 peptide-specific antibodies with group 5 allergens in different grass species (Table II). With the use of IgG antibodies raised against complete Phl p 5 we found that temperate grasses (eg, Lolium perenne, Poa pratensis, Dactylis glomerata, Secale cereale, Triticum aestivum, Avena sativa, Anthoxanthum odoratum, and Hordeum vulgare) contain allergens that cross-react with anti-Phl p 5 antibodies, whereas subtropical grasses (Cynodon dactylon, Phragmites communis) and Zea mays lack cross-reactive group 5 allergens (Table II). Because of the weakest sequence homology among group 5 allergens in the P3 region anti-P3 antibodies showed a rather selective reactivity with Phl p 5a, only a weak reactivity with Poa pratensis and Triticum aestivum, but no reactivity with the other grass species. With the exception of anti-P7 antibodies, which showed little reactivity to 3 common temperate grasses (ie, Lolium perenne, Dactylis glomerata, Secale cereale), the remaining antipeptide antibodies (ie, anti-P1, -P2, -P4, -P5, -P6) showed extensive cross-reactivity with each of the tested temperate grasses (Table II). The extensive cross-reactivity of antibodies obtained by immunization with P2, P4, P5, and P6 was in agreement with a high degree of sequence homology of these peptides among the different grasses. However, the extensive cross-reactivity of anti-P1 antibodies with the other temperate grasses could not be explained by sequence homology because the corresponding regions showed substantial variations regarding length and amino acid composition (Fig E2).
Table II. Heat map of the reactivity of antipeptide antisera with rPhl p 5 and natural group 5 allergens in different grasses species.
| anti-P1 | anti-P2 | anti-P3 | anti-P4 | anti-P5 | anti-P6 | anti-P7 | anti-rPhl p 5 | |
|---|---|---|---|---|---|---|---|---|
| rPhl p 5 | 2.218* | 1.690* | 1.537* | 1.336† | 2.142* | 1.600* | 1.540* | 3.530‡ |
| Phleum pratense | 1.155† | 2.500* | 0.187§ | 0.955† | 1.000† | 0.703† | 1.138† | 2.127† |
| Lolium perenne | 1.320† | 1.968* | 0.056§ | 0.834† | 2.495* | 0.238† | 0.163§ | 2.042† |
| Poa pratensis | 1.491† | 2.018* | 0.239† | 1.045† | 2.500* | 1.880* | 2.200* | 2.598‡ |
| Dactylis glomerata | 0.390† | 2.108* | 0.066§ | 0.728† | 0.657† | 0.689† | 0.154§ | 2.090* |
| Secale cereale | 0.292† | 1.519* | 0.062§ | 0.777† | 0.843† | 0.676† | 0.162§ | 1.720* |
| Triticum aestivum | 1.076† | 1.205† | 0.650† | 0.734† | 0.703† | 0.404† | 0.570† | 1.479† |
| Avena sativa | 0.790† | 2.438* | 0.063§ | 1.029† | 1.494† | 0.551† | 0.224† | 2.184* |
| Anthoxanthum odoratum | 1.209† | 0.854† | 0.058§ | 1.531* | 1.115† | 0.827† | 1.114† | 1.287† |
| Hordeum vulgare | 1.972* | 2.520‡ | 0.062§ | 1.150† | 1.513* | 1.184† | 0.602† | 2.501‡ |
| Cynodon dactylon | 0.140§ | 0.053§ | 0.030§ | 0.111§ | 0.044§ | 0.053§ | 0.000§ | 0.179§ |
| Phragmites communis | 0.064§ | 0.062§ | 0.061§ | 0.103§ | 0.066§ | 0.100§ | 0.000§ | 0.138§ |
| Zea mays | 0.180§ | 0.085§ | 0.064§ | 0.086§ | 0.043§ | 0.151§ | 0.004§ | 0.127§ |
Shown are IgG reactivities of antipeptide antisera (anti-P1 to anti-P7) and of an anti-rPhl p 5 antiserum to pollen extracts from grasses with (Phleum pratense, Lolium perenne, Poa pratensis, Dactylis glomerata, Secale cereale, Triticum aestivum, Avena sativa, Anthoxanthum odoratum, Hordeum vulgare) or without group 5 allergens (Cynodon dactylon, Phragmites communis, and Zea mays) as well as to Phl p 5.
OD, >1.5 to −2.5.
OD, >0.2 to 1.5.
OD, >2.5 to 3.5
OD, 0-0.2.
To better localize the portions of P1, which gave rise to the induction of cross-reactive antibodies, we synthesized 2 shorter peptides (P1a, aa 26-53; P1b, aa 31-58) and raised peptide-specific antisera. With the use of anti-P1a and anti–P1b-specific antisera we found that anti-P1a antibodies showed stronger cross-reactivity than anti-P1b antibodies with extracts from different grass pollen, indicating that the N-terminal amino acids of P1 are mainly involved in cross-reactivity (data not shown). Anti-P1 antibodies reacted exclusively with a recombinant fragment of Phl p 5, representing its N-terminal but not with a fragment representing its C-terminal half, which rules out the presence of a “duplicated” P1-like epitope within the Phl p 5 molecule (data not shown).
Mapping of conformational Phl p 5 IgE epitopes with peptide-specific antibodies
The lack of any relevant IgE reactivity of the 7 Phl p 5-derived peptides, P1 to P7 (Table E2), suggested that the major IgE epitopes of Phl p 5 belong to the discontinuous and/or conformational type. According to Arnon and Van Regenmoertel,31 it is possible to map such epitopes with the use of peptide-specific antibodies that can recognize the folded complete antigen by competitive binding experiments.
Table E3 (in the Online Repository at www.jacionline.org) shows the degree of inhibition of patients’ IgE that bind to Phl p 5 obtained by preincubation of the allergen with the individual peptide-specific antibodies and antibodies raised against the complete folded allergen for 29 patients with grass allergy. The strongest inhibition of patients’ IgE binding to Phl p 5 was obtained with anti-P1 antibodies, which caused a mean inhibition of IgE binding of 54.5% and thus came quite close to the inhibition obtained with antibodies raised against the complete Phl p 5 allergen (80.6% mean inhibition). A mean inhibition of IgE binding of >35% was obtained with antibodies specific for P3, P5, and P6, whereas inhibitions obtained with anti-P2 and anti-P7 were lower (mean inhibition of 25%). A low mean inhibition of IgE binding of 16% was obtained with anti-P4 antibodies. These results indicate that IgE epitopes are present on the N-terminal and C-terminal Phl p 5 domains (Fig E2) and cluster in the regions defined by P1, P2, and P3 on the N-terminal domain and by P5, P6, and P7 on the C-terminal domain, whereas the P4-defined area seemed to be less important for IgE recognition. Because the levels of the anti-P4 antibodies were lower than those of the other antipeptide antisera (Fig E3), we have performed the IgE competition experiments also with 3-fold higher concentrated anti-P4 antibodies, but the percentages of IgE competition could not be increased (data not shown).
When we used a mix of the 7 antipeptide antibodies, the mean inhibition of IgE binding was only 48.3%, which may be a result of steric hindrance and was comparable with the inhibition obtained with anti-P1 antibodies (data not shown). The best inhibition results that came close to those obtained with anti-Phl p 5 antibodies (mean inhibition 90.8) were obtained with a mix of anti-P1, -P2, -P5, and -P6 antibodies, giving mean inhibition rates of approximately 73% (Table III).
Table III. Rabbit anti-Phl p 5 peptide antisera inhibit allergic patient’s IgE binding to Phl p 5.
| Inhibition, % |
|||
|---|---|---|---|
| Patient | OD Phl p 5 | Anti-Phl p 5 | Anti-P1 + P2 + P5 + P6 |
| 30 | 3.33 | 99.1 | 79.3 |
| 31 | 1.82 | 96.4 | 78.5 |
| 33 | 2.25 | 98.7 | 64.9 |
| 36 | 2.61 | 91.8 | 77.9 |
| 37 | 2.09 | 96.6 | 80.9 |
| 38 | 3.23 | 98.7 | 81.2 |
| 39 | 1.74 | 96.3 | 71.4 |
| 40 | 1.136 | 69.4 | 66.1 |
| 41 | 2.82 | 96.2 | 77.9 |
| 42 | 3.11 | 98.1 | 78.4 |
| 43 | 0.255 | 80.7 | 65.6 |
| 44 | 2.27 | 97.2 | 75.1 |
| 45 | 3.59 | 98.6 | 90.3 |
| 46 | 3.55 | 98.0 | 81.5 |
| 47 | 3.69 | 98.2 | 86.1 |
| 48 | 3.32 | 96.1 | 45.6 |
| 49 | 0.79 | 77.8 | 47.4 |
| 50 | 0.216 | 72.8 | 75.4 |
| 51 | 0.66 | 63.6 | 57.7 |
| Mean | 2.2 | 90.8 | 72.7 |
Shown are OD values that correspond to Phl p 5-specific IgE levels after preincubation with normal rabbit serum and the percentages of inhibition of IgE binding (sera diluted 1:10) to Phl p 5 after preincubation of Phl p 5 with rPhl p 5-specific or peptide antisera (mix of anti-P1 + P2 + P5 + P6; diluted 1:250).
Inhibition of Phl p 5-induced basophil activation by a mix of antibodies directed to peptides P1, P2, P5, and P6
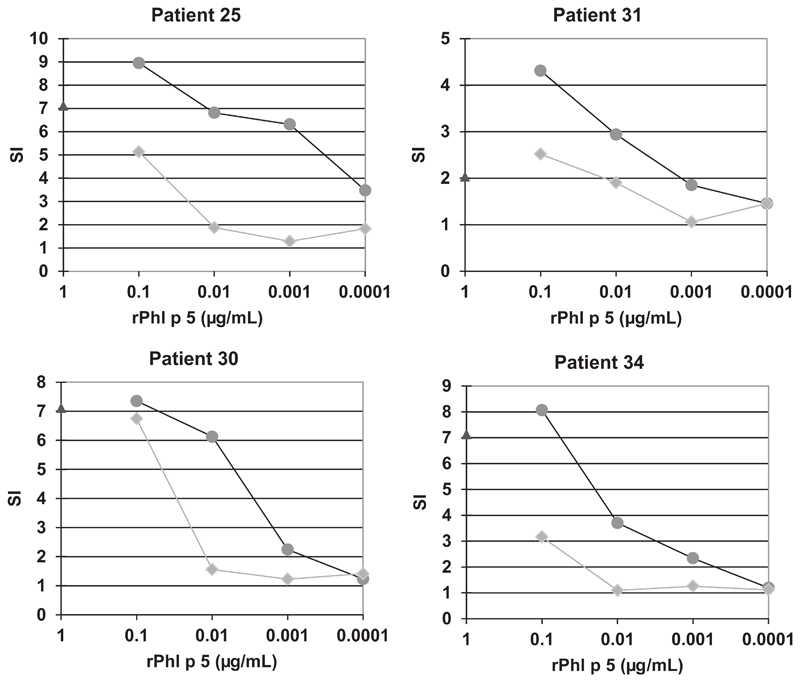
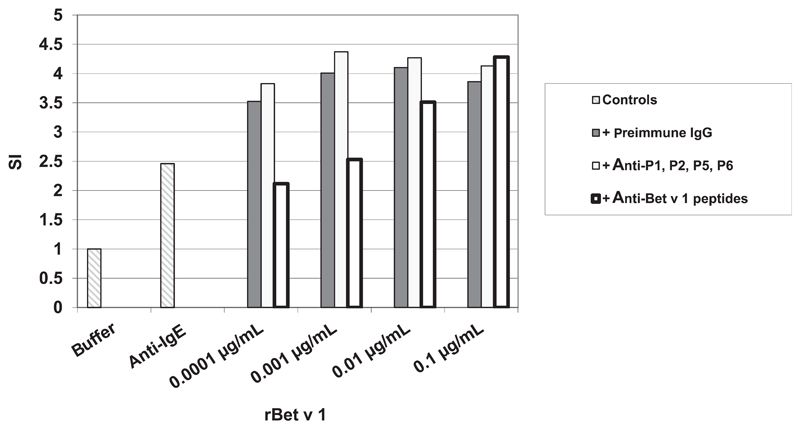
We found that after incubation of rPhl p 5 with the antipeptide antibodies, 10-fold (patients 30, 31, and 34) to 100-fold (for patient 25) more allergen was required to achieve similar levels of basophil activation (Fig 4) compared with incubation with preimmune antibodies. No inhibition was observed when the anti-Phl p 5 peptide IgG was used to inhibit basophil activation induced with a Phl p 5-unrelated allergen (eg, major birch pollen allergen, Bet v 1) in patients with birch pollen allergy (see Fig E6 in this article’s Online Repository at www.jacionline.org).
Fig 4.
Peptide-specific IgG antibodies inhibit allergen-induced basophil activation. Basophils from 4 patients with grass pollen allergy (patients 25, 30, 31, and 34) were exposed to different concentrations of Phl p 5 (1-0.0001 μg/mL) preincubated with peptide-specific rabbit IgG (squares, mix of anti-P1, -P2, -P5, and -P6) or preimmune IgG (dots). Response to anti-IgE is shown for 1 concentration (triangle). Allergen-induced upregulation of CD203c was calculated from mean fluorescence intensities obtained with stimulated (MFIstim) and unstimulated (MFIcontrol) cells, and is expressed as stimulation index (MFIstim : MFIcontrol) (SI) (y-axes).
Discussion
The major timothy grass pollen allergen Phl p 5 and homologous allergens present in most temperate grass species represent one of the most frequently recognized and potent respiratory allergens.3 It is therefore an essential component for specific preventive and therapeutic approaches for grass pollen allergy. New molecular vaccination and tolerance induction strategies require a detailed knowledge of IgE and T-cell epitopes.36 Here, we studied for a representative number of patients with grass pollen allergy the IgE and T-cell recognition of Phl p 5. For this purpose we used 7 synthetic peptides with a length between 31 and 38 amino acids that cover the Phl p 5 molecule. Although several studies provided evidence for the occurrence of sequential IgE epitopes in group 5 allergens, none of the tested patients showed detectable IgE reactivity to the peptides. Furthermore, the peptides exhibited no allergenic activity with patients’ basophils even when coupled in repetitive form to a carrier molecule. This finding can be explained by the fact that the peptides lacked fold and secondary structure as demonstrated by CD analysis, and it suggests that most of the Phl p 5-specific IgE epitopes are conformational. This assumption was also supported by the finding that recombinant unfolded Phl p 5 fragments comprising aa 26 to 176 and aa 176 to 258 showed no relevant IgE reactivity. Results obtained for Phl p 5 are thus in agreement with data obtained for most respiratory allergens that also contain mainly conformational IgE epitopes. These epitopes can be destroyed by fragmentation, truncation, or reassembly of the molecules,32,35,37–39 and represent a strategy to engineer recombinant hypoallergenic allergen molecules for immunotherapy.7,40
The mapping of conformational IgE epitopes is a laborious and time-consuming task, particularly when performed by structural biology methods such as the co-crystallization of allergen-IgE antibody complexes.41,42 We have recently shown that antibodies raised against allergen-derived peptides can be used for inhibiting IgE binding to allergens in patients with allergy.30 Peptide-specific antibodies may therefore be useful for the fast mapping of conformational IgE epitopes on allergens. The latter may be explained by the ability of peptide-specific antibodies to recognize portions within the conformational epitopes and thus to compete directly with the binding of IgE or by causing steric hindrance. We found that each of the 7 peptide-specific IgG antibodies recognized the complete Phl p 5 allergen, indicating that they can bind to the folded protein. Six of them also showed extensive cross-reactivity with natural group 5 allergens from 8 temperate grass species known to contain group 5 allergens. Results of IgE inhibition studies showed that Phl p 5 contains several IgE binding sites that could be visualized on a model of the Phl p 5 structure. The IgE binding sites could be located on the N-terminal as well as C-terminal domain of Phl p 5, occupying one side of the molecule, whereas the backside areas defined by P4 and partly by P7 appeared to be less important for IgE binding. In fact the majority of immune-dominant IgE epitopes appeared to be defined by P1, P2, P5, and P6 because antibodies against these 4 peptides inhibited IgE binding to Phl p 5 in patients with allergy almost as good (72% inhibition) as antibodies induced by immunization with the complete Phl p 5 allergen (90% inhibition). The fact that inhibition of patients’ IgE binding could not be increased by using a mix of all 7 antipeptide antisera instead of the 4 may be explained by steric hindrance when too many specificities are present in the mix. Accordingly, a mix of antibodies specific for P1, P2, P5, and P6 inhibited Phl p 5-induced basophil activation, indicating that the IgE binding sites defined by these antibodies on the 2 Phl p 5 domains are a target for an oligoclonal Phl p 5-specific allergenic IgE response. In fact, each of the 2 Phl p 5 domains seems to contain binding sites for several IgE species and thus have allergenic activity as it has been described earlier for the isolated N-terminal domain that was recognized by >70% of patients with Phl p 5 allergy and strongly induced basophil activation.23
When we tested the peptides for the induction of lymphocyte proliferation and cytokine responses with the use of PBMCs from patients with grass pollen allergy, we noted an interesting dissociation of IgE and T-cell recognition of Phl p 5. In fact, P4 that induced the strongest T-cell proliferation and cytokine responses was not a major site involved in IgE recognition because anti-P4 antibodies only weakly inhibited IgE binding to Phl p 5. Because we could not obtain inhibitions of IgE binding even when the anti-P4 antiserum was used in higher concentrations, the lack of inhibition of IgE binding was not because of the fact that the anti-Phl p 5-specific IgG titers of the anti-P4 antiserum were lower than those of the other peptide antisera. Moreover, Biacore experiments indicated that the association and dissociation of the peptide-specific antisera were similar to that of the antiserum raised against the complete Phl p 5 allergen, suggesting that no major differences were found in the avidity of the antisera.
Results obtained in our study should be useful for the development of immunotherapy strategies that selectively target the T-cell and IgE response against Phl p 5. For example, it should be possible to use the P4-defined area of Phl p 5 for the development of tolerance induction strategies that target Phl p 5-specific T cells by peptide vaccines as described for the major cat allergen, Fel d 1.43,44 However, P1, P2, P5, and P6 can be linked with carrier molecules to develop vaccines that induce allergen-specific IgG toward the IgE epitopes of Phl p 5 as described for several major allergens.45,46 The dissection of IgE and T-cell epitopes as exemplified for the major grass pollen allergen Phl p 5 should thus allow the development of new molecular forms of allergen-specific immunotherapy that target different mechanisms underlying allergy.
Methods
Natural allergen extracts and recombinant allergens
Pollen from several grasses (Phleum pratense, Lolium perenne, Poa pratensis, Dactylis glomerata, Secale cereale, Triticum aestivum, Avena sativa, Hordeum vulgare, Anthoxanthum odoratum, Phragmites communis, Cynodon dactylon, and Zea mays) was purchased from Allergon Thermo Fisher Scientific (Ängelholm, Sweden) and aqueous pollen extracts were prepared as described.E1 Purified recombinant Phl p 5a that had been expressed in Escherichia coliE2 was obtained from Biomay (Vienna, Austria).
Characterization of patients with allergy and patients’ sera
Patients with grass pollen allergy (n = 66) were characterized by a case history indicative of grass pollen allergy (ie, symptoms of allergic rhinoconjunctivitis and/or asthma during the grass pollen season), positive skin prick test reactions to grass pollen extract, and/or timothy grass pollen-specific IgE antibodies as determined by CAP-FEIA (Phadia, Thermo Fisher Scientific, Uppsala, Sweden) testing. Anonymized residual serum samples from these patients and from persons without grass pollen allergy were used for the serologic experiments (IgE-binding and competition experiments) with approval of the ethics committee of the Medical University of Vienna.
Total IgE levels in the sera were determined by CAP measurements (Phadia). IgE antibodies specific for rPhl p 5 were determined by ELISA to identify 51 patients positive for Phl p 5. Heparinized blood samples for T-cell proliferation, cytokine assays, and CD203c assays were obtained from 12 of these patients with grass pollen allergy after informed consent was obtained.
Peptide synthesis and coupling to KLH
According to multiple sequence alignment, secondary structure and solvent accessibility prediction of group 5 and group 6 grass pollen allergens performed with the program by Rost and Sander,E3,E4 regions in Phl p 5a that were rich in exposed amino acids were identified. Peptides with a length between 31 and 38 amino acids that covered these regions were synthesized on the Applied Biosystems peptide synthesizer Model 433A (Foster City, Calif) by using the Fmoc-approach and HBTU activation (0.1 mmol small-scale cycles).E5 An additional cysteine residue was added to each of the synthetic peptides to facilitate coupling to carriers. Whether the cysteine was added to the N-terminus (peptides 2, 3, 5, and 6) or C-terminus (peptides 1, 4, and 7) of the peptide was dictated by aspects of the synthetic peptide chemistry such as yields and purity of the peptides.
Peptides were purified by HPLC and characterized by mass spectrometry (piChem, Graz, Austria).E5 Furthermore, they were analyzed for residual fold by CD spectroscopy with the use of a Jasco J-810 spectropolarimeter.E4
The far-UV spectra were recorded at 20°C at wavelengths from 185 nm to 260 nm as an average of 5 scans. Protein and peptide solutions of 0.3 mg/mL in MilliQ water were measured with a 0.05-cm path length cylindrical cell by using a scan speed of 100 nm/min and recording intervals of 0.2 nm.
Peptides were coupled to KLH (Pierce, Rockford, Ill) and were purified with a Conjugation Kit according to the manufacturer’s advice (Sigma, St Louis, Mo).
IgE reactivity of complete Phl p 5 and Phl p 5-derived peptides
ELISA plates (Nunc Maxisorp, Roskilde, Denmark) were coated with each of the Phl p 5-derived peptides (5 μg/mL), each of the peptide-KLH conjugates containing 5 μg/mL of the coupled peptide, or rPhl p 5 (5 μg/mL); washed and blocked.E5 Subsequently, plates were incubated with 1:10 diluted sera from patients with grass pollen allergy or from a nonatopic person overnight at 4°C. Bound IgE antibodies were detected with a 1:1000 diluted alkaline-phosphatase-coupled mouse monoclonal anti-human IgE antibody (Pharmingen, San Diego, Calif). The presence of the peptides on the ELISA plates was confirmed with rabbit antipeptide antisera and the corresponding preimmune sera as negative control.E5 The lack of IgE reactivity of the peptides was also shown with nitrocellulose-dotted peptides as described.E4 In addition, in the dot blot experiments, the presence of the peptides on the nitrocellulose was confirmed with rabbit antipeptide antibodies.
Multiple sequence alignment and modeling of the Phl p 5a structure
Sequences of group 5 grass pollen allergens were retrieved from GenBANK by comparing the Phl p 5a sequence with the deposited sequences with the use of the BLASTp program. Sequences were selected to represent most temperate grasses, and a multiple sequence alignment was made with ClustalW. The alignment was edited by hand for maximal fit; identical amino acids were shown as dots and gaps by dashes. A model of the putative Phl p 5a structure was built, based on the coordinates of Phl p 6 deposited in the protein data bank (www.rcsb.org/pdb/home/home.do; PDB 1nlx),E6 which shows 58% sequence identity with the Phl p 5a N-terminal portion and on the x-ray crystal structure of the isolated Phl p 5b C-terminal portion (PDB 1l3p),E7 which has 78% sequence identity with the Phl p 5a C-terminus. The model was made by using the program SWISS-MODEL.E8 The 2 domains comprising aa 59 to 172 and aa 182 to 283 of Phl p 5a could be modeled, whereas the N-terminal residues, the residues comprising the interdomain linker as well as the positions of the N-terminal and C-terminal domains to each other, could not be exactly predicted.
Biacore experiments
The interaction of Phl p 5- and of peptide-specific antisera with rPhl p 5 was measured by surface plasmon resonance with the use of a BIAcore 2000 instrument (GE Healthcare, BIAcore, Uppsala, Sweden) at 25°C. The sensor chip C1 (BIAcore) surface was activated by injection of a 1:1 mixture of 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride and N-hydroxysuccinimide at a flow rate of 5 μL/min for 7 minutes. Purified rPhl p 5 (10 μg/mL) diluted in 10 mmol/L sodium acetate (pH 4.5) was then covalently coupled to the activated surface by amine coupling by injecting the protein at 5 μL/min into the flow cell. The unreacted active ester groups were blocked with 1 mol/L ethanolamine (pH 8.5) at a flow rate of 5 μL/min for 7 minutes. The final immobilization level was 68.4 response units (flow cell 2). In the reference cell, the unrelated birch pollen allergen rBet v 1 was covalently coupled as described above, reaching a final immobilization level was 47.6 response units. Binding experiments were performed by injecting peptide-specific antisera diluted 1:160 in HBS-EP buffer (0.01 mol/L HEPES, 0.15 mol/L NaCl, 3 mmol/L EDTA, 0.005% [vol/vol] surfactant P20 [pH 7.4]; BIAcore). During the association phase, peptide-specific antibodies were passed over the buffer-equilibrated chip surface at a rate of 30 μL/min for 5 minutes. An identical dilution of 1:160 was chosen for each antiserum and within mixes of antisera the dilution of each antiserum was also 1:160. Dissociation was investigated by injection of HBS-EP buffer at 30 μL/min for a period of 30 minutes. Regeneration of the sensor chip surface after each injection cycle was performed by 2 brief pulses (30 seconds, 20 μL/min) of 10 mmol/L glycine-HCl (pH 1.5).
Binding of the peptide-specific antisera to the immobilized rPhl p 5 was analyzed with BIAEvaluation 3.0 software (BIAcore).
Extended Data
Fig E1.
CD analysis shows a comparison of rPhl p 5a and the 7 peptides. X-axis shows the molecular ellipticity and the y-axis the wave length.
Fig E2.
Position of peptides in a sequence alignment of group 5 allergens from different grasses. Comparison of the amino acid sequences of Phl p 5a and homologous grass allergens by using ClustalW. Each allergen sequence is preceded by its database entry code (Q40960: Phl p 5a, timothy grass; Q40963: Phl p 5b, timothy grass; Q40237: Lol p 5b, ryegrass, Q9FPR0: Poa p 5, Kentucky bluegrass; Q93WP9 Dac g 5, Q9XF24: Lol p 5a, ryegrass; CBG76811: Sec c 5, rye; BAK07309: Hor v 5, barley; Q70JP9: Tri a 5, wheat). Dashes indicate gaps, points represent identical amino acids. Colored frames indicate the positions of peptides 1 to 7 and their borders. Mean percentages of inhibition of patients’ (n = 9) IgE binding to Phl p 5 after preincubation of Phl p 5 with antipeptide antisera are displayed for each peptide.
Fig E3.
Illustration of the fluorescence-activated cell sorting strategy to detect allergen- and peptide-specific CD4+ cells by CFSE staining. PBMCs from a patient with grass pollen allergy were stimulated with rPhl p 5, peptides 1 and 4, or the peptide mix. A, The 7-AAD dead cell exclusion is shown. The gating on CD3+ (B) and on CD4+ (C) T cells is shown. Percentages of positive cells are displayed. FSC, Forward scatter.
Fig E4.
Reactivity of antipeptide IgG and anti-rPhl p 5 IgG with Phl p 5 by ELISA titration. OD values ± SDs corresponding to bound IgG (y-axis) are shown for 5 different dilutions (x-axis: 1:4 × 103, 1:16 × 103, 1:64 × 103, 1:256 × 103, 1:1024 × 103).
Fig E5.
Analysis of the interaction between rPhl p 5 and specific antisera with the use of surface plasmon resonance (BIAcore) measurements. Binding is expressed in response units (RUs) on the y-axis over time. A, The association and dissociation of the antisera (anti-rPhl p 5, black line; mix of all peptide antisera [anti-P1 to anti-P7], dark gray broken line; mix of anti-P1, -P2, -P5, and -P6 antisera, light gray broken line; antisera against individual peptides: P1, brown line, P2, blue line; P3, red line; P4, yellow line; P5, magenta line; P6, gray line; P7, green line) is shown over time (in seconds; x-axis). B, Shown is a magnification of the reactivity of the antipeptide antisera without the anti-rPhl p 5 antiserum.
Fig E6.
Specific inhibition of allergen-induced basophil activation by peptide antisera. Upregulation of CD203c expression on basophils (SI, stimulation index; y-axis) of a patient with birch pollen allergy by different concentrations of rBet v 1 (x-axis) in the presence of anti-P1, -P2, -P5, and -P6 antisera; a mix of the corresponding preimmune sera; or anti-Bet v 1 peptide antisera by buffer alone or by anti-IgE.
Table E1. Biochemical characteristics of Phl p 5-derived synthetic peptides.
| Peptide | Position, aa | Sequence | No. of aa | Molecular weight | Isoelectric point |
|---|---|---|---|---|---|
| 1 | 26-58 | ADLGYGPATPAAPAAGYTPATPAAPAEAAPAGKC | 34 | 3068 | 4.37 |
| 2 | 59-91 | CATTEEQKLIEKINAGFKAALAAAAGVQPADKYR | 34 | 3578 | 8.14 |
| 3 | 93-128 | CFVATFGAASNKAFAEGLSGEPKGAAESSSKAALTSK | 37 | 3592 | 8.16 |
| 4 | 132-162 | AYKLAYKTAEGATPEAKYDAYVATLSEALRIC | 32 | 3482 | 6.29 |
| 5 | 176-212 | CAEEVKVIPAGELQVIEKVDAAFKVAATAANAAPANDK | 38 | 3853 | 4.66 |
| 6 | 217-246 | CEAAFNDAIKASTGGAYESYKFIPALEAAVK | 31 | 3236 | 4.87 |
| 7 | 252-283 | TVATAPEVKYTVFETALKKAITAMSEAQKAAKC | 33 | 3501 | 8.98 |
Cysteines added to facilitate coupling are boldface and underlined.
Table E2. Demographic, clinical, and serologic characterization of patients with grass pollen allergy.
| Patient | Sex | Age, y | Total IgE (kU/L) |
IgE timothy (kUA/L) |
IgE (OD) rPhl p 5 |
P1 P1 KLH |
P2 P2 KLH |
P3 P3 KLH |
P4 P4 KLH |
P5 P5 KHL |
P6 P6 KLH |
P7 P7 KLH |
Mix P1-P7 |
Symptoms | Treatment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 51 | 370 | 90.1 | 1.296 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,u,a | SIT |
| 2 | F | 30 | 1004 | 59.8 | 0.573 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | p |
| 3 | M | 27 | 760 | n.d | 1.574 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | - |
| 4 | F | 43 | n.d. | 85 | 1.000 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | - |
| 5 | M | 30 | 19.5 | 6.38 | 0.407 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 6 | M | 29 | 60.8 | 30.8 | 1.208 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | - |
| 7 | M | 30 | 219 | 76.7 | 1.405 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,a | SIT |
| 8 | F | 26 | 18 | 6.85 | 0.521 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 9 | M | 19 | 118 | 61 | 1.347 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | SIT |
| 10 | F | 24 | n.d | 27.4 | 0.209 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | - |
| 11 | F | 36 | 134 | 37 | 0.878 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 12 | M | 32 | 386 | 50.9 | 1.091 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | p |
| 13 | M | 57 | 57.9 | 14.5 | 0.682 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 14 | M | 31 | 115 | 15.5 | 1.221 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | p |
| 15 | M | 44 | 75.2 | 17.4 | 2.200 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | p |
| 16 | M | 43 | 86 | 17.6 | 1.160 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | SIT,p |
| 17 | F | 37 | 104 | 32.7 | 1.030 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 18 | F | 36 | 184 | 15.8 | 2.030 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 19 | M | 44 | 111 | 24.1 | 1.930 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 20 | M | 56 | 2000 | 100 | 2.5 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,d | p |
| 21 | F | 30 | 107 | 38 | 1.380 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | - |
| 22 | M | 24 | 117 | 3.2 | 0.112 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 23 | F | 27 | 4625 | 100 | 1.774 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 24 | M | 31 | 150 | 33.6 | 0.360 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | SIT |
| 25 | M | 44 | 128 | 20.7 | 0.725 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | - |
| 26 | F | 27 | 28.4 | 6.3 | 0.409 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | SIT |
| 27 | M | 22 | 80.6 | 5.93 | 0.401 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | - |
| 28 | M | 26 | 529 | 37.8 | 0.766 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | p |
| 29 | M | 23 | 3082 | 51.6 | 2.014 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | d | p |
| 30 | M | 28 | 45.7 | 26.3 | 1.285 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 31 | M | 29 | 3432 | 100 | 1.820 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 32 | F | 26 | 1005 | 6.08 | 0.310 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | SIT |
| 33 | F | 31 | 1683 | 100 | 2.431 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | p |
| 34 | M | 25 | 86.3 | 23.4 | 1.204 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | SIT,p |
| 35 | M | 45 | 262 | 45.3 | 0.481 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | SIT,p |
| 36 | M | 29 | 81.2 | 31.3 | 1.331 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | SIT,p |
| 37 | F | 13 | 147 | 74.4 | 2.090 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 38 | M | 10 | 370 | 97.1 | 3.230 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,a | p |
| 39 | F | 26 | 314 | 100 | 1.740 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 40 | F | 25 | 307 | 95.9 | 1.136 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 41 | M | 48 | 1234 | 100 | 2.820 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 42 | F | 13 | 178 | 97.6 | 3.110 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 43 | F | 35 | 463 | 71.2 | 0.255 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,a | p |
| 44 | M | 29 | 231 | 84 | 2.270 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 45 | M | 64 | 260 | 24.1 | 3.590 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 46 | M | 24 | 362 | 74.9 | 3.550 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 47 | F | 40 | 39 | 6.08 | 3.690 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 48 | F | 36 | 1347 | 100 | 3.320 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c,a | p |
| 49 | F | 17 | 840 | 100 | 0.790 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r,c | p |
| 50 | F | 30 | 811 | 100 | 0.216 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| 51 | F | 13 | 416 | 53.2 | 0.660 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | r | p |
| N | F | 40 | 5 | 0.35 | 0.049 | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | - | - |
Shown are sex, age, total serum IgE levels (kU/L), timothy grass pollen allergen-specific IgE (kUA/L), IgE antibodies specific for rPhl p 5, and the 7 isolated (P1-P7) and KLH-coupled (KLH-P1 to KLH-P7) peptides (ODs), symptoms during grass pollen season, and treatment. Dashes indicate a lack of detectable IgE reactivity.
a, Asthma; c, conjunctivitis; d, dermatitis; F, female; M, male; N, non-allergic person; OD, optical density; p, pharmacotherapy; r, rhinitis; SIT, specific immunotherapy; u, urticaria.
Table E3. Inhibition of patients’ IgE binding to Phl p 5 by anti-sera.
| Patient | Anti-P1 | Anti-P2 | Anti-P3 | Anti-P4 | Anti-P5 | Anti-P6 | Anti-P7 | Anti-Phl p 5 |
|---|---|---|---|---|---|---|---|---|
| 1 | 45 | 0 | 31 | 25 | 34 | 28 | 30 | 85 |
| 2 | 46 | 31 | 44 | 36 | 46 | 43 | 49 | 91 |
| 3 | 28 | 21 | 34 | 10 | 25 | 17 | 17 | 91 |
| 4 | 72 | 20 | 32 | 27 | 58 | 51 | 36 | 92 |
| 5 | 70 | 21 | 44 | 33 | 50 | 53 | 46 | 88 |
| 6 | 70 | 29 | 44 | 33 | 47 | 53 | 40 | 93 |
| 7 | 58 | 16 | 48 | 28 | 30 | 36 | 33 | 84 |
| 8 | 59 | 20 | 43 | 45 | 54 | 48 | 56 | 80 |
| 9 | 59 | 31 | 51 | 3 | 19 | 30 | 17 | 89 |
| 10 | 69 | 17 | 30 | 20 | 23 | 40 | 40 | 24 |
| 11 | 56 | 24 | 38 | 10 | 55 | 37 | 23 | 93 |
| 12 | 62 | 19 | 37 | 29 | 50 | 49 | 35 | 83 |
| 13 | 36 | 13 | 38 | 12 | 29 | 30 | 18 | 83 |
| 14 | 22 | 0 | 0 | 18 | 11 | 9 | 23 | 56 |
| 15 | 59 | 42 | 46 | 27 | 50 | 51 | 32 | 89 |
| 16 | 56 | 22 | 43 | 0 | 40 | 37 | 9 | 87 |
| 17 | 43 | 10 | 32 | 10 | 10 | 27 | 12 | 87 |
| 18 | 58 | 10 | 43 | 0 | 24 | 24 | 1 | 90 |
| 19 | 55 | 41 | 48 | 23 | 49 | 48 | 31 | 90 |
| 20 | 31 | 15 | 25 | 8 | 5 | 21 | 14 | 88 |
| 21 | 62 | 19 | 47 | 6 | 33 | 38 | 15 | 87 |
| 22 | 60 | 46 | 51 | 13 | 55 | 49 | 26 | 62 |
| 23 | 55 | 29 | 37 | 4 | 43 | 32 | 11 | 78 |
| 24 | 57 | 35 | 41 | 9 | 45 | 38 | 19 | 72 |
| 25 | 62 | 42 | 48 | 7 | 56 | 44 | 20 | 78 |
| 26 | 62 | 46 | 51 | 11 | 58 | 49 | 22 | 76 |
| 27 | 59 | 44 | 47 | 9 | 52 | 44 | 20 | 67 |
| 28 | 48 | 21 | 30 | 6 | 31 | 25 | 11 | 76 |
| 29 | 61 | 46 | 45 | 4 | 51 | 40 | 15 | 78 |
| Mean | 54.5 | 25.1 | 39.6 | 16.1 | 39.1 | 37.7 | 24.8 | 80.6 |
The percentage of inhibition of patients’ IgE binding to Phl p 5 after preincubation of Phl p 5 with the individual antipeptide antibodies (anti-P1 to anti-P7) or with anti-rPhl p 5 antibodies is shown.
Clinical implications.
The dissociation of the major IgE- and T-cell–reactive domains in Phl p 5 provides a basis for the development of novel forms of immunotherapy selectively targeting IgE or T-cell responses.
Acknowledgments
We thank Karoline Sonneck from the Department of Internal Medicine I, Division of Hematology and Hemostaseology, Medical University of Vienna, Austria, for CD203c analysis of peptides.
Supported by the Christian Doppler Research Association, by a research grant from Biomay AG, Vienna, Austria, and by projects F4604, F4605, F4611 of the Austrian Science Fund (FWF).
Abbreviations used
- 7-AAD
7-Amino-actinomycin D
- CD
Circular dichroism
- CFSE
Carboxyfluorescein diacetate succinimidyl ester
- KLH
Keyhole-limpet hemocyanin
- P
Peptide
- PBMC
Peripheral blood mononuclear cell
Footnotes
Disclosure of potential conflict of interest: R. Valenta has consultant arrangements with Biomay AG and Phadia and has received research support from the Austrian Science Fund (FWF), Biomay AG, and the Christian Doppler Research Association. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Kay AB. Allergy and Allergic Diseases. Oxford, United Kingdom: Blackwell Science; 2008. [Google Scholar]
- 2.Heinzerling L, Frew AJ, Bindslev-Jensen C, Bonini S, Bousquet J, Bresciani M, et al. Standard skin prick testing and sensitization to inhalant allergens across Europe–a survey from the GALEN network. Allergy. 2005;60:1287–300. doi: 10.1111/j.1398-9995.2005.00895.x. [DOI] [PubMed] [Google Scholar]
- 3.Suphioglu C, Singh MB, Taylor P, Bellomo R, Holmes P, Puy R, et al. Mechanism of grass-pollen-induced asthma. Lancet. 1992;339:569–72. doi: 10.1016/0140-6736(92)90864-y. [DOI] [PubMed] [Google Scholar]
- 4.Pollart SM, Reid MJ, Fling JA, Chapman MD, Platts-Mills TA. Epidemiology of emergency room asthma in northern California: association with IgE antibody to ryegrass pollen. J Allergy Clin Immunol. 1988;82:224–30. doi: 10.1016/0091-6749(88)91003-2. [DOI] [PubMed] [Google Scholar]
- 5.Blackley CH. Experimental Researches on the Causes and Nature of Catarrhus Aestivus (Hay-Fever or Hay-Asthma) Whitefish (MT): Kessinger Publishing; 1873. [Reprinted] [Google Scholar]
- 6.Noon L. Prophylactic inoculation against hay fever. Lancet. 1911;1:1572–3. [Google Scholar]
- 7.Calderon M, Cardona V, Demoly P. One hundred years of allergen immunotherapy European Academy of Allergy and Clinical Immunology celebration: review of unanswered questions. Allergy. 2012;67:462–76. doi: 10.1111/j.1398-9995.2012.02785.x. [DOI] [PubMed] [Google Scholar]
- 8.Möller C, Dreborg S, Ferdousi HA, Halken S, Host A, Jacobsen L, et al. Pollen immunotherapy reduces the development of asthma in children with seasonal rhinoconjunctivitis (the PAT-study) J Allergy Clin Immunol. 2002;109:251–6. doi: 10.1067/mai.2002.121317. [DOI] [PubMed] [Google Scholar]
- 9.Durham SR, Walker SM, Varga EM, Jacobson MR, O’Brien F, et al. Long-term clinical efficacy of grass-pollen immunotherapy. N Engl J Med. 1999;341:468–75. doi: 10.1056/NEJM199908123410702. [DOI] [PubMed] [Google Scholar]
- 10.Thomas WR. The advent of recombinant allergens and allergen cloning. J Allergy Clin Immunol. 2011;127:855–9. doi: 10.1016/j.jaci.2010.12.1084. [DOI] [PubMed] [Google Scholar]
- 11.Valenta R, Ferreira F, Focke-Tejkl M, Linhart B, Niederberger V, Swoboda I, et al. From allergen genes to allergy vaccines. Annu Rev Immunol. 2010;28:211–41. doi: 10.1146/annurev-immunol-030409-101218. [DOI] [PubMed] [Google Scholar]
- 12.Singh MB, Bhalla PL. Hypoallergenic derivatives of major grass pollen allergens for allergy vaccination. Immunol Cell Biol. 2003;81:86–91. doi: 10.1046/j.0818-9641.2002.01144.x. [DOI] [PubMed] [Google Scholar]
- 13.Valenta R. The future of antigen-specific immunotherapy of allergy. Nat Rev Immunol. 2002;2:446–53. doi: 10.1038/nri824. [DOI] [PubMed] [Google Scholar]
- 14.Larche M. T cell epitope-based allergy vaccines. Curr Top Microbiol Immunol. 2011;352:107–19. doi: 10.1007/82_2011_131. [DOI] [PubMed] [Google Scholar]
- 15.Focke M, Swoboda I, Marth K, Valenta R. Developments in allergen-specific immunotherapy: from allergen extracts to allergy vaccines bypassing allergen-specific immunoglobulin E and T cell reactivity. Clin Exp Allergy. 2010;40:385–97. doi: 10.1111/j.1365-2222.2009.03443.x. [DOI] [PubMed] [Google Scholar]
- 16.Focke-Tejkl M, Valenta R. Safety of engineered allergen-specific immunotherapy vaccines. Curr Opin Allergy Clin Immunol. 2012;12:555–63. doi: 10.1097/ACI.0b013e328357ca53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor PE, Flagan RC, Valenta R, Glovsky M. Release of allergens as respirable aerosols: a link between grass pollen and asthma. J Allergy Clin Immunol. 2002;109:51–6. doi: 10.1067/mai.2002.120759. [DOI] [PubMed] [Google Scholar]
- 18.Singh MB, Hough T, Theerakulpisut P, Avjioglu A, Davies S, Smith PM, et al. Isolation of cDNA encoding a newly identified major allergenic protein of rye-grass pollen: intracellular targeting to the amyloplast. Proc Natl Acad Sci U S A. 1991;88:1384–8. doi: 10.1073/pnas.88.4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvanovich A, Astwood J, Zhang L, Olsen E, Kisil F, Sehon A, et al. Nucleotide sequence analysis of three cDNAs coding for Poa p IX isoallergens of Kentucky bluegrass pollen. J Biol Chem. 1991;266:1204–10. [PubMed] [Google Scholar]
- 20.Vrtala S, Sperr WR, Reimitzer I, van Ree R, Laffer S, Müller WD, et al. cDNA cloning of a major allergen from timothy grass (Phleum pratense) pollen; characterization of the recombinant Phl pV allergen. J Immunol. 1993;151:4773–81. [PubMed] [Google Scholar]
- 21.Maglio O, Saldanha JW, Vrtala S, Spitzauer S, Valenta R, Pastore A. A major IgE epitope-containing grass pollen allergen domain from Phl p 5 folds as a four-helix bundle. Protein Eng. 2002;15:635–42. doi: 10.1093/protein/15.8.635. [DOI] [PubMed] [Google Scholar]
- 22.Bufe A, Betzel C, Schramm G, Petersen A, Becker WM, Schlaak M, et al. Crystallization and preliminary diffraction data of a major pollen allergen. Crystal growth separates a low molecular weight form with elevated biological activity. J Biol Chem. 1996;271:27193–6. doi: 10.1074/jbc.271.44.27193. [DOI] [PubMed] [Google Scholar]
- 23.Flicker S, Vrtala S, Steinberger P, Vangelista L, Bufe A, Petersen A, et al. A human monoclonal IgE antibody defines a highly allergenic fragment of the major timothy grass pollen allergen, Phl p 5: molecular, immunological, and structural characterization of the epitope domain. J Immunol. 2000;165:3849–59. doi: 10.4049/jimmunol.165.7.3849. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Olsen E, Kisil FT, Hill RD, Sehon AH, Mohapatra SS. Mapping of antibody binding epitopes of a recombinant Poa p IX allergen. Mol Immunol. 1992;29:1383–9. doi: 10.1016/0161-5890(92)90175-w. [DOI] [PubMed] [Google Scholar]
- 25.Suphioglu C, Blaher B, Rolland JM, McCluskey J, Schäppi G, Kenrick J, et al. Molecular basis of IgE-recognition of Lol p 5, a major allergen of rye-grass pollen. Mol Immunol. 1998;35:293–305. doi: 10.1016/s0161-5890(98)00050-9. [DOI] [PubMed] [Google Scholar]
- 26.Blaher B, Suphioglu C, Knox RB, Singh MB, McCluskey J, Rolland JM. Identification of T-cell epitopes of Lol p 9, a major allergen of ryegrass (Lolium perenne) pollen. J Allergy Clin Immunol. 1996;98:124–32. doi: 10.1016/s0091-6749(96)70234-8. [DOI] [PubMed] [Google Scholar]
- 27.Müller WD, Karamfilov T, Kahlert H, Stüwe HT, Fahlbusch B, Cromwell O, et al. Mapping of T-cell epitopes of Phl p 5: evidence for crossreacting and non-crossreacting T-cell epitopes within Phl p 5 isoallergens. Clin Exp Allergy. 1998;28:1538–48. doi: 10.1046/j.1365-2222.1998.00432.x. [DOI] [PubMed] [Google Scholar]
- 28.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, et al. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–55. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoecklinger A, Scheiblhofer S, Roesler E, Lang A, Fastner G, Sedlmayer F, et al. T cell epitopes of the timothy grass pollen allergen Phl p 5 of mice and men and the detection of allergen-specific T cells using class II ultimers. Int Arch Allergy Immunol. 2012;158:326–34. doi: 10.1159/000333551. [DOI] [PubMed] [Google Scholar]
- 30.Gieras A, Cejka P, Blatt K, Focke-Tejkl M, Linhart B, Flicker S, et al. Mapping of conformational IgE epitopes with peptide-specific monoclonal antibodies reveals simultaneous binding of different IgE antibodies to a surface patch on the major birch pollen allergen, Bet v 1. J Immunol. 2011;186:5333–44. doi: 10.4049/jimmunol.1000804. [DOI] [PubMed] [Google Scholar]
- 31.Arnon R, Van Regenmortel MH. Structural basis of antigenic specificity and design of new vaccines. FASEB J. 1992;6:3265–74. doi: 10.1096/fasebj.6.14.1385242. [DOI] [PubMed] [Google Scholar]
- 32.Focke M, Mahler V, Ball T, Sperr WR, Majlesi Y, Valent P, et al. Non-anaphylactic synthetic peptides derived from B cell epitopes of the major grass pollen allergen, Phl p 1, for allergy vaccination. FASEB J. 2001;15:42–4. doi: 10.1096/fj.01-0016fje. [DOI] [PubMed] [Google Scholar]
- 33.Hauswirth AW, Natter S, Ghannadan M, Majlesi Y, Schernthaner GH, Sperr WR, et al. Recombinant allergens promote expression of CD203c on basophils in sensitized individuals. J Allergy Clin Immunol. 2002;110:102–9. doi: 10.1067/mai.2002.125257. [DOI] [PubMed] [Google Scholar]
- 34.Jahn-Schmid B, Kelemen P, Himly M, Bohle B, Fischer G, Ferreira F, et al. The T cell response to Art v 1, the major mugwort pollen allergen, is dominated by one epitope. J Immunol. 2002;169:6005–11. doi: 10.4049/jimmunol.169.10.6005. [DOI] [PubMed] [Google Scholar]
- 35.Focke M, Linhart B, Hartl A, Wiedermann U, Sperr WR, Valent P, et al. Non-anaphylactic surface-exposed peptides of the major birch pollen allergen, Bet v 1, for preventive vaccination. Clin Exp Allergy. 2004;34:1525–33. doi: 10.1111/j.1365-2222.2004.02081.x. [DOI] [PubMed] [Google Scholar]
- 36.Larche M, Akdis CA, Valenta R. Immunological mechanisms of allergen-specific immunotherapy. Nat Rev Immunol. 2006;6:761–71. doi: 10.1038/nri1934. [DOI] [PubMed] [Google Scholar]
- 37.Vrtala S, Hirtenlehner K, Vangelista L, Pastore A, Eichler HG, Sperr WR, et al. Conversion of the major birch pollen allergen, Bet v 1, into two nonanaphylactic T cell epitope-containing fragments: candidates for a novel form of specific immunotherapy. J Clin Invest. 1997;99:1673–81. doi: 10.1172/JCI119330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mothes-Luksch N, Stumvoll S, Linhart B, Focke M, Krauth MT, Hauswirth A, et al. Disruption of allergenic activity of the major grass pollen allergen Phl p 2 by reassembly as a mosaic protein. J Immunol. 2008;181:4864–73. doi: 10.4049/jimmunol.181.7.4864. [DOI] [PubMed] [Google Scholar]
- 39.Westritschnig K, Focke M, Verdino P, Goessler W, Keller W, Twardosz A, et al. Generation of an allergy vaccine by disruption of the three-dimensional structure of the cross-reactive calcium-binding allergen, Phl p 7. J Immunol. 2004;172:5684–92. doi: 10.4049/jimmunol.172.9.5684. [DOI] [PubMed] [Google Scholar]
- 40.Niederberger V, Horak F, Vrtala S, Spitzauer S, Krauth MT, Valent P, et al. Vaccination with genetically engineered allergens prevents progression of allergic disease. Proc Natl Acad Sci U S A. 2004;101(suppl 2):14677–82. doi: 10.1073/pnas.0404735101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niemi M, Jänis J, Jylhä S, Kallio JM, Hakulinen N, Laukkanen ML, et al. Characterization and crystallization of a recombinant IgE Fab fragment in complex with the bovine beta-lactoglobulin allergen. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:25–8. doi: 10.1107/S174430910706160X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G, et al. High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J Immunol. 2009;182:2141–51. doi: 10.4049/jimmunol.0803018. [DOI] [PubMed] [Google Scholar]
- 43.Worm M, Lee HH, Kleine-Tebbe J, Hafner RP, Laidler P, Healey D, et al. Development and preliminary clinical evaluation of a peptide immunotherapy vaccine for cat allergy. J Allergy Clin Immunol. 2011;127:89–97. doi: 10.1016/j.jaci.2010.11.029. 97.e1–14. [DOI] [PubMed] [Google Scholar]
- 44.Oldfield WL, Kay AB, Larche M. Allergen-derived T cell peptide-induced late asthmatic reactions precede the induction of antigen-specific hyporesponsiveness in atopic allergic asthmatic subjects. J Immunol. 2001;167:1734–9. doi: 10.4049/jimmunol.167.3.1734. [DOI] [PubMed] [Google Scholar]
- 45.Edlmayr J, Niespodziana K, Linhart B, Focke-Tejkl M, Westritschnig K, Scheiblhofer S, et al. A combination vaccine for allergy and rhinovirus infections based on rhinovirus-derived surface protein VP1 and a nonallergenic peptide of the major timothy grass pollen allergen Phl p 1. J Immunol. 2009;182:6298–306. doi: 10.4049/jimmunol.0713622. [DOI] [PubMed] [Google Scholar]
- 46.Niespodziana K, Focke-Tejkl M, Linhart B, Civaj V, Blatt K, Valent P, et al. A hypoallergenic cat vaccine based on Fel d 1-derived peptides fused to hepatitis B PreS. J Allergy Clin Immunol. 2011;127:1562–70.e6. doi: 10.1016/j.jaci.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E1.Vrtala S, Grote M, Duchêne M, van Ree R, Kraft D, Scheiner O, et al. Properties of tree and grass pollen allergens: reinvestigation of the linkage between solubility and allergenicity. Int Arch Allergy Immunol. 1993;102:160–9. doi: 10.1159/000236567. [DOI] [PubMed] [Google Scholar]
- E2.Vrtala S, Susani M, Sperr WR, Valent P, Laffer S, Dolecek C, et al. Immunologic characterization of purified recombinant timothy grass pollen (Phleum pratense) allergens (Phl p 1, Phl p 2, Phl p 5) J Allergy Clin Immunol. 1996;97:781–7. doi: 10.1016/s0091-6749(96)80156-4. [DOI] [PubMed] [Google Scholar]
- E3.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–99. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- E4.Twaroch TE, Focke M, Civaj V, Weber M, Balic N, Mari A, et al. Carrier-bound, nonallergenic Ole e 1 peptides for vaccination against olive pollen allergy. J Allergy Clin Immunol. 2011;128:178–84.e7. doi: 10.1016/j.jaci.2011.03.011. [DOI] [PubMed] [Google Scholar]
- E5.Focke M, Mahler V, Ball T, Sperr WR, Majlesi Y, Valent P, et al. Non-anaphylactic synthetic peptides derived from B cell epitopes of the major grass pollen allergen, Phl p 1, for allergy vaccination. FASEB J. 2001;15:2042–4. doi: 10.1096/fj.01-0016fje. [DOI] [PubMed] [Google Scholar]
- E6.Vrtala S, Fischer S, Grote M, Vangelista L, Pastore A, Sperr WR, et al. Molecular, immunological, and structural characterization of Phl p 6, a major allergen and P-particle-associated protein from Timothy grass (Phleum pratense) pollen. J Immunol. 1999;163:5489–96. [PubMed] [Google Scholar]
- E7.Bufe A, Betzel C, Schramm G, Petersen A, Becker WM, Schlaak M, et al. Crystallization and preliminary diffraction data of a major pollen allergen. Crystal growth separates a low molecular weight form with elevated biological activity. J Biol Chem. 1996;271:27193–6. doi: 10.1074/jbc.271.44.27193. [DOI] [PubMed] [Google Scholar]
- E8.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modelling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]