Abstract
Background
Neuroblastoma is the most common pediatric extracranial solid tumor. Within conventional risk groups, there is considerable heterogeneity in outcomes, indicating the need for improved risk stratification.
Methods
In this study we analyzed the somatic mutational burden of 515 primary, untreated neuroblastoma tumors from three independent cohorts. Mutations in coding regions were determined by whole-exome/genome sequencing of tumor samples compared to matched blood leukocytes. Survival data for 459 patients were available for analysis of 5-year overall survival using the Kaplan–Meier method and log-rank test. All statistical tests were two-sided.
Results
Despite a low overall somatic mutational burden (mean = 3, range = 0–56), 107 patients were considered to have high mutational burden (>3 mutations). Unfavorable histology and age 18 months and older were associated with high mutational burden. Patients with high mutational burden had inferior 5-year overall survival (29.0%, 95% confidence interval [CI] = 17.2 to 41.8%) vs those with three or fewer somatic mutations (76.2%, 95% CI = 71.5 to 80.3%) (log-rank P < .001) and this association persisted when limiting the analysis to genes included on a 447-gene panel commonly used in clinical practice. On multivariable analysis, mutational burden remained prognostic independent of age, stage, histology and MYCN status.
Conclusions
This study demonstrates that mutational burden of primary neuroblastoma may be useful in combination with conventional risk factors to optimize risk stratification and guide treatment decisions, pending prospective validation.
Neuroblastoma is the most common extracranial solid tumor in children and accounts for approximately 15% of childhood cancer-related mortality (1). Clinical outcomes in neuroblastoma are highly variable, with some patients having an excellent prognosis even with limited to no therapy, while others have poor outcomes despite aggressive treatment (2). The current risk stratification paradigm is based on the International Neuroblastoma Risk Group (INRG) classification system that uses clinical characteristics (age, stage), histopathology, and biological features (MYCN status, segmental chromosomal aberrations, ploidy) to divide patients into four groups: very low, low, intermediate, and high risk (3). However, patients within individual risk groups have heterogeneous outcomes, leaving substantial room for improvement.
Prior studies have identified several gene alterations (eg, mutations/single-nucleotide variants, amplifications, and deletions) associated with specific disease characteristics or outcomes including ALK, PTPN11, ATRX, MYCN, NRAS, ARID1A/B, and TERT (4–10). However, like many other pediatric malignancies, neuroblastoma is associated with a low somatic mutation burden with a median of approximately 0.60 exonic mutations per Mb (4). Indeed, in one of the largest genetic landscaping studies in neuroblastoma published to date, ALK was the only frequently mutated gene to be statistically significantly associated with clinical outcome (4).
Since neuroblastoma is characterized by a heterogeneous mutation spectrum with relatively few recurrently mutated genes, it is unclear whether its biological behavior is driven more by direct genetic mutations vs expression-level modulation conferred through copy number alterations and epigenetic mechanisms. Indeed, there are discrepant findings in the literature regarding any association between somatic mutation burden and clinical characteristics/outcomes (4,11). An important limitation of these previous investigations was the focus on specific gene mutations or overall mutational burden in isolation. To address this issue, we undertook an integrated analysis of the prognostic value of mutational burden in the context of conventional risk factors.
Methods
Patients
Individual data from 1219 patients with untreated, primary neuroblastoma from three independent cohorts were assessed for eligibility: TARGET (n = 1089) (4), Amsterdam (n = 87) (11), and Germany (n = 56) (10). Since this study was based on publicly-available data, it was considered Institutional Review Board-exempt.
Somatic Mutations
Somatic mutations found in coding regions were determined by whole-exome/genome sequencing of tumor samples compared to matched blood leukocytes using validated mutation detection algorithms such as muTect (12), SNVMix2 (13), and MutSig (14). The detailed methods for somatic mutation calling can be found in the primary articles on each of the cohorts included in our study (4,10,11). For the analyses in this study, clinical and somatic mutation data from these cohorts were obtained from the cBioPortal for Cancer Genomics (15,16). The somatic mutation calls provided by each study (4,10,11) were compiled by cBioPortal and reannotated such that cBioPortal cancer genes (http://www.cbioportal.org/cancer_gene_list.jsp) with one or more mutations or any gene with two or more mutations were included. Furthermore, cBioPortal only reports somatic, nonsynonymous mutations (missense, nonsense, nonstart, nonstop, frameshift, inframe, splice site, truncation) that are predicted to result in a protein alteration.
Statistical Analysis
The primary endpoint of interest was overall survival (OS) measured from the date of diagnosis. The association of somatic mutation count with OS was analyzed by the Kaplan–Meier method, log-rank test, and Cox proportional hazards regression analysis (univariate and multivariable) using the following covariates: age, sex, INSS stage at diagnosis, histology, and MYCN status. We confirmed that the proportional hazards assumption was not violated by Schoenfeld residuals, log-log plot, and comparing the Kaplan–Meier observed survival curves with the Cox predicted survival curves. Multiple imputation by chained equations was used to incorporate patients with incomplete clinical data (histology, n = 101; MYCN, n = 3) into multivariable analysis (17). Fisher exact test was used to compare categorical proportions. All statistical tests were two-sided and a P value of less than .05 was considered statistically significant.
Results
Patients without somatic mutation data (n = 704) were excluded, leaving 515 patients, of whom 459 had survival data available (Figure 1). Baseline patient characteristics for the 515 patients with mutational data are provided in Supplementary Table 1 (available online). The median and mean somatic mutation counts were 1 and 3, respectively (range = 0 to 56). Cancer-associated genes that were mutated in one or more patients in this cohort are listed in Supplementary Table 2 (available online). We categorized patients at or below the group mean (3 or fewer mutations; n = 408) as having low mutational burden and patients above the group mean (more than 3 mutations; n = 107) as having high mutational burden. Patients with unfavorable histology (Fisher exact P = .04) or older than 18 months of age (Fisher exact P < .001) were more likely to have high mutational burden (Supplementary Table 1, available online).
Figure 1.
Flow diagram of patient selection for this study.
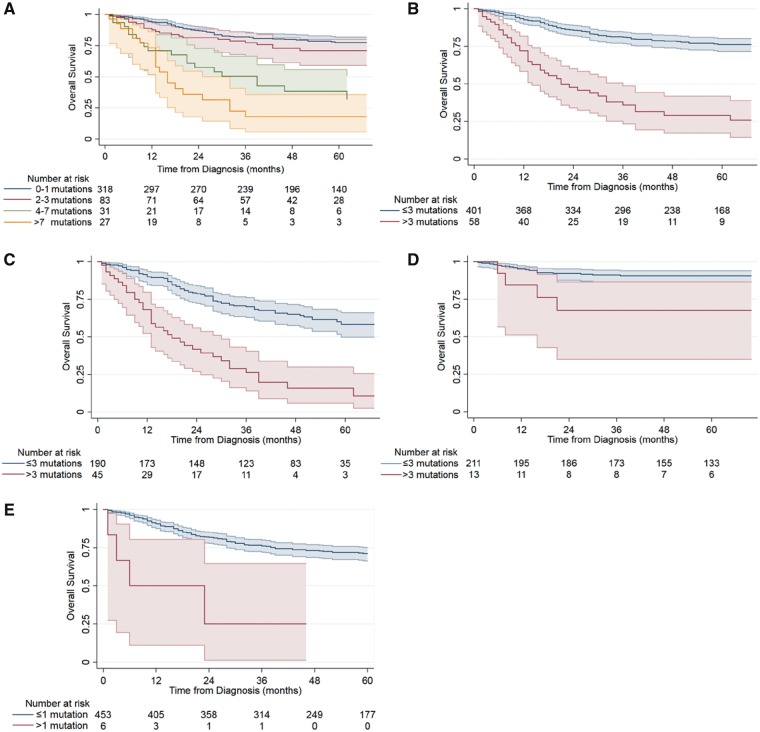
Despite the low overall mutational burden in neuroblastoma, increased mutation frequency was highly associated with worse OS among the 459 patients with survival data (Figure 2A,B; log-rank P < .001). The estimated 5-year OS was 76.2% (95% confidence interval [CI] = 71.5 to 80.3%) in patients with three or fewer somatic mutations (n = 401) compared to 29.0% (95% CI = 17.2 to 41.8%) in those with more than three mutations (n = 58). The statistical significance of this association persisted when three of the most frequently mutated genes in neuroblastoma, ALK, MYCN, ATRX, which may act as oncogenic drivers, were excluded (log-rank P = .005; data not shown).
Figure 2.
Association of mutational burden with overall survival in neuroblastoma patients. Kaplan–Meier survival curves stratified by mutation count as a (A) continuous variable grouped for clarity of presentation (n = 459; P < .001), (B) dichotomous variable split at the group mean of three mutations (n = 459; P < .001), (C) dichotomous variable in high-risk patients defined as 18 months of age or older with International Neuroblastoma Staging System stage 4 disease (n = 235; P < .001), (D) dichotomous variable in non-high-risk patients (n = 224; P = .006), (E) dichotomous variable only using data from 447 genes included in OncoPanel (n = 459; P < .001). 95% confidence intervals are shown and risk tables are below each plot. A two-sided log-rank test was used to calculate the P values.
Next, we restricted the analysis to patients universally recognized as having high-risk disease (18 months of age or older, INSS stage IV; n = 235). In this subgroup, mutational burden remained a statistically significant predictor of increased mortality (Figure 2C; log-rank P < .001). The estimated 5-year OS was 58.4% (95% CI = 49.7 to 66.1%) in high-risk patients with three or fewer somatic mutations (n = 190) compared to 15.8% (95% CI = 5.9 to 30.0%) in those with more than three somatic mutations (n = 45). Similarly, we found that mutational burden was statistically significantly associated with survival in non-high-risk patients (Figure 2D; log-rank P = .006). We then limited the analysis to data from only those genes included in a validated 447-gene panel (OncoPanel) used in routine clinical practice at our institution (18,19). Overall survival was worse for patients with more than one somatic mutation in these genes compared to those with one or fewer mutations (Figure 2E; log-rank P < .001). The number of recurrently altered genes as determined by whole exome/genome sequencing in neuroblastoma is very few, with only 13 genes having a frequency of five or more nonsynonymous mutations in our pooled cohort. Of these 13 genes, only six (46.2%) are represented in the OncoPanel gene set, which is not surprising since the OncoPanel was not customized for neuroblastoma.
To determine the prognostic value of mutational burden in the context of established risk factors, we performed Cox regression analysis. We confirmed that the proportional hazards assumption was not violated by Schoenfeld residuals, log-log plot, and comparing the Kaplan–Meier observed survival curves with the Cox predicted survival curves. On univariate analysis, age, INSS stage, histology, MYCN amplification, and mutation count were each statistically significantly associated with OS (all P < .001) (Table 1). On multivariable analysis, stage, mutation count, and MYCN were independently associated with OS, while age and histology were not (Table 1).
Table 1.
Univariate and multivariable Cox regression analyses of factors associated with overall survival in neuroblastoma patients
| Variable | Univariate analysis |
Multivariable analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P * | HR (95% CI) | P * | |
| Age, months | ||||
| ≥18 | 2.54 (1.57 to 4.10) | <.001 | 1.16 (0.64 to 2.09) | .63 |
| <18 | 1.00 (Reference) | 1.00 (Reference) | ||
| Sex | ||||
| Male | 0.83 (0.59 to 1.18) | .30 | — | — |
| Female | 1.00 (Reference) | — | ||
| INSS stage at diagnosis | ||||
| 4 | 6.83 (3.97 to 11.76) | <.001 | 4.42 (2.48 to 7.86) | <.001 |
| Other | 1.00 (Reference) | 1.00 (Reference) | ||
| Histology | ||||
| Unfavorable | 5.08 (2.45 to 10.54) | <.001 | 1.66 (0.72 to 3.81) | .23 |
| Favorable | 1.00 (Reference) | 1.00 (Reference) | ||
| MYCN status | ||||
| Amplified | 2.34 (1.66 to 3.30) | <.001 | 1.59 (1.11 to 2.29) | .01 |
| Not amplified | 1.00 (Reference) | 1.00 (Reference) | ||
| Mutation count per 1 unit increase | 1.09 (1.06 to 1.11) | <.001 | — | — |
| Mutation count | ||||
| >3 | 4.77 (3.27 to 6.96) | <.001 | 3.43 (2.27 to 5.18) | <.001 |
| ≤3 | 1.00 (Reference) | 1.00 (Reference) | ||
All statistical tests were two-sided. CI = confidence interval; HR = hazard ratio; INSS = International Neuroblastoma Staging System.
We compared the baseline characteristics of the 515 patients with mutation data available with the 704 patients without mutation data and found that there was no statistically significant difference between groups in terms of age, sex, histology, or MYCN amplification (Supplementary Table 3, available online). However, there were statistically significantly more stage IV patients in the group without mutation data (78.1% vs 53.8%; P < .001).
Discussion
This study represents the largest pooled analysis of the clinical implications of mutational burden in neuroblastoma. Higher somatic mutational burden in coding regions of the genome is strongly associated with inferior survival in patients with neuroblastoma, including those with high-risk disease. This difference is not driven by a small number of critical driver mutations, as the disparity in outcomes remains after we removed ALK, MYCN, and ATRX mutations from the analysis. We demonstrate that the prognostic value of mutation count persists in the context of conventional risk factors including age, stage, MYCN amplification, and histology. Routine determination of somatic mutation counts through whole-exome sequencing is currently still clinically impractical. Hence, we also limited the analysis to the 447 genes represented in the OncoPanel used routinely at our own institution (18,19) and found that the prognostic value of mutational burden remained. The fact that limiting our analysis to the OncoPanel gene set still yielded a statistically significant survival difference based on mutational burden highlights the robustness of these findings and suggests that a gene panel tailored specifically for neuroblastoma may be even more impactful. In one of the previously published cohorts, higher mutational burden was associated with older age and advanced disease stage (11). While our pooled analysis confirmed the association between mutational burden and age, the association between disease stage and mutation frequency was not observed.
Limitations of this study include the potential heterogeneity introduced by pooling three patient cohorts and the lack of treatment information. The imbalance of missing histology data between the high and low mutational burden groups (Supplementary Table 1, available online) may introduce bias into the imputed data. Furthermore, there were 704 patients missing mutation data, which could serve as a source of bias. To address this concern, we compared the baseline characteristics of the 515 patients with mutation data available with the 704 patients without mutation data and found that the groups were comparable except for statistically significantly more stage IV patients in the group without mutation data. Since we found that mutational burden was associated with survival in both the high-risk group (18 months of age or older, stage IV) and nonhigh-risk group independently, it is unlikely that any bias resulting from this imbalance would qualitatively alter the results of this study.
While neuroblastoma overall has a low mutation frequency, approximately 1–2% of tumors are associated with alterations in DNA repair pathway genes such as MLH1, DDB1, POLE, and POLD1 resulting in greater mutational burden and in rare cases hypermutation (>10 mutations/Mb) (4,20,21). In a recent pan-cancer study focused on hypermutation, the only pediatric cancer types that involved cases of ultrahypermutation (>100 mutations/Mb) were malignant gliomas, colorectal cancers, and leukemias/lymphomas (21). Germline events in the mismatch repair pathway and other mechanisms of hypermutation are exceedingly rare in neuroblastoma and therefore unlikely to influence the results observed in this study.
Tumor mutational burden has been associated with survival in other malignancies, primarily in adults (22,23). We hypothesize that a high mutational burden representative of a complex genetic profile may be a hallmark of more aggressive and treatment-refractory disease. With further validation, mutational burden could potentially be used as a key additional piece of information to help determine the disease trajectory of patients with neuroblastoma. Given the low overall frequency of recurrent mutations in neuroblastoma, further work is needed to identify other biological alterations that drive the development and progression of neuroblastoma. Ultimately, an optimized genomic classifier incorporating mutational burden information is likely to improve prognostication, risk stratification, and management.
Funding
This work was supported by a Pedals for Pediatrics research grant to WLH and NIH Medical Scientist Training Program grant (T32GM007753) to RLW.
Notes
Affiliations of authors: Harvard Radiation Oncology Program, Boston, MA (WLH); Harvard Medical School, Boston, MA (WLH, RLW, AN, KJM, SGDB, DHK); Department of Radiation Oncology, Massachusetts General Hospital, Boston, MA (AN); Department of Radiation Oncology, Dana-Farber Cancer Institute, Boston, MA (KJM, DHK); Department of Radiation Oncology, Brigham & Women’s Hospital, Boston, MA (KJM, DHK); Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Boston, MA (SGDB).
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the article; and the decision to submit the article for publication.
The authors have no conflicts of interest to disclose.
Supplementary Material
References
- 1. Maris JM, Hogarty MD, Bagatell R, Cohn SL.. Neuroblastoma. Lancet. 2007;3699579:2106–2120. [DOI] [PubMed] [Google Scholar]
- 2. Brodeur GM, Bagatell R.. Mechanisms of neuroblastoma regression. Nat Rev Clin Oncol. 2014;1112:704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohn SL, Pearson ADJ, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG task force report. J Clin Oncol. 2009;272:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;453:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. George RE, Sanda T, Hanna M, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;4557215:975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;4557215:971–974. [DOI] [PubMed] [Google Scholar]
- 7. Janoueix-Lerosey I, Lequin D, Brugières L, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;4557215:967–970. [DOI] [PubMed] [Google Scholar]
- 8. Sausen M, Leary RJ, Jones S, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;451:12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheung N-KV, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;30710:1062.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peifer M, Hertwig F, Roels F, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;5267575:700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Molenaar JJ, Koster J, Zwijnenburg DA, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;4837391:589–593. [DOI] [PubMed] [Google Scholar]
- 12. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;313:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goya R, Sun MGF, Morin RD, et al. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010;266:730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang J, Liu J, Sun J, Chen C, Foltz G, Lin B.. Identifying driver mutations from sequencing data of heterogeneous tumors in the era of personalized genome sequencing. Brief Bioinform. 2014;152:244–255. [DOI] [PubMed] [Google Scholar]
- 15. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6269:pl1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;25:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayati Rezvan P, Lee KJ, Simpson JA.. The rise of multiple imputation: a review of the reporting and implementation of the method in medical research. BMC Med Res Methodol. 2015;151:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;21:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;119:e87062.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma X, Liu Y, Liu Y, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;5557696:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;1715:1042–1056.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Owada-Ozaki Y, Muto S, Takagi H, et al. Prognostic impact of tumor mutation burden in patients with completely resected non–small cell lung cancer: brief report. J Thorac Oncol. 2018; doi:10.1016/J.JTHO.2018.04.003 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 23. Grassi E, Durante S, Astolfi A, et al. Mutational burden of resectable pancreatic cancer, as determined by whole transcriptome and whole exome sequencing, predicts a poor prognosis. Int J Oncol. 2018;526:1972–1980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.