Pioneer transcription factors are necessary, but not sufficient, for the transcription of many target genes during development. To carry out their function, these sequence-specific transcription factors usually recruit chromatin-modifying factors to specific enhancers and promoters, which regulate histone post-translational modifications (PTMs) and nucleosome position to facilitate RNA Polymerase II occupancy and transcription of a specific battery of genes. This collaboration between transcription factors and chromatin-modifying factors is essential for numerous cell-lineage commitments. For example, the cardiogenic transcription factors TBX5, NKX2-5, and GATA4 work in concert with SWI/SNF chromatin-remodeling complexes to regulate gene expression during cardiomyocyte specification and differentiation (1,2). And functionally, transfection of two cardiogenic transcription factors plus one SWI/SNF subunit can convert naïve, non-cardiac mesoderm into beating cardiomyocytes (CM) (3). Understanding these types of regulatory networks has translational potential, including the prospect of inducing cardiomyocyte regeneration in patients who have suffered heart attacks and/or heart failure.
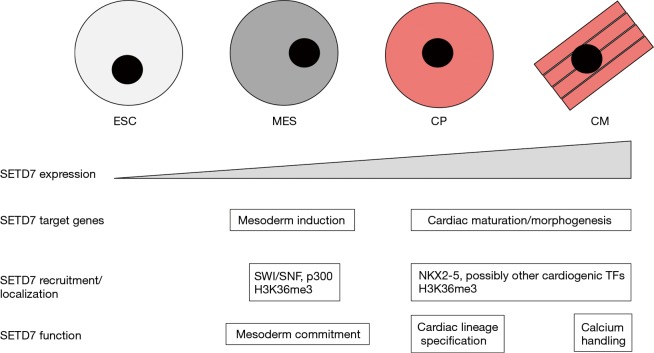
Our knowledge of chromatin-modifying factors in cardiac development has been growing (2) but is still quite limited Chang, 2012 #16. Therefore, a recent study by Lee et al. identifying a role for SETD7 in multiple steps of cardiomyocyte development is a welcome addition (4). SETD7 is expressed in cardiac as well as other tissues where it functions as a lysine methyltransferase that catalyzes histone 3 lysine 4 monomethylation (H3K4me1) (5). Similar to other “histone methyltransferases”, SETD7 also methylates non-histone proteins (5), and it is an ongoing challenge to understand the relative importance of histone versus non-histone targets. To interrogate SETD7 function, Lee et al. took advantage of an experimental platform where human embryonic stem cells (ESC) are sequentially differentiated into mesoderm (MES), cardiac progenitors (CP), and terminally-differentiated CM. Their first observation was that SETD7 expression is correlated with differentiation status with the highest levels of SETD7 mRNA and protein occurring in CP and CM (Figure 1). ChIP-seq experiments demonstrated that SETD7 occupancy was surprisingly dynamic at promoters and gene bodies of target loci. For example, at the MES stage, SETD7 occupancy was enriched at genes involved in mesoderm induction (e.g., T, EVX1, EOMES, Nodal) plus the Notch and Wnt signaling pathways, but this occupancy was transient and fell off in CP and CM (Figure 1). Interestingly, there was a shift in occupancy at the CP and CM stages with SETD7 binding enriched at cardiac maturation/morphogenesis genes (e.g., TNNT1, TNNT2, ACTC1, MYH6) (Figure 1).
Figure 1.
SETD7 expression, recruitment/occupancy, and function during cardiomyocyte development. Depicted at the top are human embryonic stem cells (ESC) as they are progressively differentiated into mesoderm (MES), cardiac progenitors (CP), and terminally-differentiated cardiomyocytes (CM). Shown below are SETD7 relative expression levels, SETD7 target genes (based on SETD7 ChIP occupancy profiles combined with RNA-seq data identifying misexpressed genes in SETD7 knockdown cells as compared to controls), factors believed to recruit/localize SETD7 to promoters and gene bodies, and SETD7 function at different stages (based on SETD7 knockdown phenotypes).
Temporally-induced knockdowns of SETD7 in these cells at different stages demonstrated that SETD7 is required for each step of differentiation: for mesoderm-lineage commitment at MES, cardiac-lineage specification at CP, and for normal calcium-handling properties in CM (Figure 1). RNA-seq identified misexpressed genes in knockdown cells at each stage, and integration of the RNA-seq data with the ChIP-seq data suggested that SETD7 was primarily acting as a transcriptional activator at the MES and CP-CM stages. Because most chromatin-modifying factors lack DNA-binding specificity, they usually are recruited to specific sites. To address SETD7 recruitment, candidates were identified based on SETD7 protein-protein interactions via co-immunoprecipitation (co-IP) assays and co-occupancy at promoters of select target genes via ChIP assays. At the MES stage, two SWI/SNF subunits (BRG1 and BAF60a) and the p300 histone acetyltransferase emerged as the strongest candidates for SETD7 recruitment (Figure 1). However, these are also chromatin-modifying factors that lack DNA-binding specificity and presumably are recruited by as-yet-unidentified transcription factors. At the CP-CM stages, the cardiogenic transcription factor NKX2-5 emerged as the strongest candidate for SETD7 recruitment (Figure 1). Other cardiogenic transcription factors could participate in this recruitment, and there seems to be a feedback loop where SETD7 binds to the promoters of several cardiogenic transcription factors and vice versa.
Lee et al. also identified a novel mechanism regulating SETD7 localization, which in this case was shared by the MES, CP, and CM. ChIP-seq data demonstrated that SETD7 occupancy was not restricted to the promoters of actively transcribed genes but extended to the corresponding gene bodies marked by H3K36me3 (Figure 1). When RNA Polymerase II transcribes genes, it is physically associated with SETD2, which catalyzes H3K36me3 of gene bodies (exons and introns) to prevent cryptic anti-sense transcripts that have the potential to be deleterious (6). Unlike SETD2, a SETD7 knockdown did not diminish H3K36me3 levels. This finding is consistent with SETD7 catalyzing H3K4me1 rather than H3K36me3. However, SETD2 knockdown cells not only exhibited diminished H3K36me3 in gene bodies but also had diminished SETD7 occupancy at these sites. Based on histone peptide arrays, SETD7 physically bound H3K36 in a PTM-dependent manner with the following relative affinities: H3K36me3 > H3K36ac > HeK36me2 > H3K36me1 > H3K36 unmodified. Lacking a domain known to bind methylated histones such as a chromodomain, PHD fingers, or a tudor domain, the part of SETD7 domain responsible for binding H3K36me3 was not reported and awaits identification. It could also be an indirect physical interaction. Also, considering that H3K36me3 labels gene bodies as they are transcribed, SETD7 binding to H3K36me3 likely represents a transcriptional-maintenance mechanism or a way to prevent inappropriate intragenic transcription rather than a transcriptional-activation mechanism as described for SETD7 promoter occupancy.
In addition to providing new mechanistic insights, the study by Lee et al. raises several questions that may lead to further studies that provide future insights. The first topic involves SETD7 function in cardiac development in vivo. On the one hand, the idea that SETD7 is required for cardiomyocyte development during ESC differentiation is compatible with a SETD7 requirement for skeletal muscle development in previous work utilizing C2C12 myoblast cultures (7,8). However, on the other hand, it is not clear how these in vitro results should be interpreted considering that SETD7 constitutive knockout mice do not exhibit any reported developmental defects and are viable and fertile (9). In addition, a MyoD-Cre-driven conditional mutation of SETD7 recently demonstrated that SETD7 is dispensable for skeletal muscle development but is required for adult skeletal muscle regeneration (10). Therefore, even if SETD7 is not required for cardiomyocyte development in vivo, there is a possibility that it could be manipulated to increase the regenerative potential of adult CM in response to damage. The SETD7 phenotype of knockout mice also does not preclude a role for SETD7 in terminally-differentiated CM, including calcium-handling properties, that are dispensable for viability under sedentary husbandry conditions. SETD7 might be required for optimal cardiac function, as revealed by stress tests, and an impairment could contribute to cardiac dysfunction including heart failure.
The second topic is that the experiments performed by Lee et al. did not distinguish whether SETD7 function in CM is mediated by histone or non-histone targets. This represent a general challenge for the field but is not an intractable problem. For example, a recent study was able to separate the functional importance of SETD2 methylation of histone (H3K36) and non-histone (tubulin) targets (11). Interestingly, this work demonstrated that tubulin methylation, not H3K36me3, was responsible for preventing mitotic defects and genomic instability that drive cancer. In this regard, it is noteworthy that SETD7 exhibits stronger H3K4me1 activity on free histones than nucleosomes (the true physiologic substrate), which has led some investigators to favor the relevance of SETD7 non-histone substrates (5).
For the third topic, assuming that SETD7 regulates cardiomyocyte development via H3K4 methylation, its occupancy and function should focus on enhancers. The underlying rationale is that SETD7 catalyzes H3K4me1, which is a stronger mark for poised/active enhancers than poised/active promoters. This analysis could be performed, in part, by comparing SETD7 occupancy to H3K4me1 and H3K27ac profiles. Although Lee et al. did not perform ChIP-seq for H3K4me1, they did report ChIP-seq profiles for H3K27ac (plus H3K4me3, H3K36me3, and H3K27me3). Considering that enhancer-promoter loops occur at many actively expressed genes, there might not be a large difference in SETD7 enhancer occupancy as compared to the SETD7 promoter occupancy that was already reported. However, one would expect to see higher enrichment of SETD7 at H3K4me1- and H3K27ac-marked enhancers than H3K4me3- and H3K9ac-marked promoters. To interrogate enhancer function, SETD7 knockdown cells could be analyzed for H3K4me1 (as a directly regulated mark), H3K27ac (as an indirect but independent mark of enhancers), enhancer RNAs (eRNAs) at select targets, and chromatin loop formation between enhancers and promoters at select targets (via 3C assays). Finally, it would be interesting to characterize bivalent domains, which have active marks (e.g., H3K4me3, H3K4me1) juxtaposed with inactive marks (e.g., H3K27me3) (12), in their experimental platform. One would expect bivalent domains to be most abundant in ESC because they are associated with developmental genes poised for expression, but the analysis of the MES, CP, and CM stages might also be revealing and would provide insight into the kinetics with which the bivalent domains in ESC are converted to an on/off state.
Regardless of how the unanswered questions and future studies play out, the findings by Lee et al. are an important step forward and have clinical implications. During the last 5–10 years, drugs targeting chromatin-modifying factors have been gaining traction, particularly in the oncology sector (13). This class of drugs may also prove to be an effective intervention for cardiovascular disease. The number of potential chromatin-related drug targets for cardiovascular disease is growing as exemplified by work demonstrating a role for the BRG1 catalytic subunit of SWI/SNF complexes in cardiac hypertrophy (14) and the prevention of arrhythmias associated with heart failure (15). SETD7 adds to this growing list, and it has an advantage of being “druggable”. A potent SETD7 inhibitor has been characterized and validated in preclinical models (10,16), suggesting that clinical-grade inhibitors or activators are plausible.
Acknowledgements
Funding: SJ Bultman is supported by funding from the National Institutes of Health (CA125237) and the USDA.
Footnotes
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- 1.Bruneau BG. Chromatin remodeling in heart development. Curr Opin Genet Dev 2010;20:505-11. 10.1016/j.gde.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 2.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol 2012;74:41-68. 10.1146/annurev-physiol-020911-153242 [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009;459:708-11. 10.1038/nature08039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J, Shao NY, Paik DT, et al. SETD7 Drives Cardiac Lineage Commitment through Stage-Specific Transcriptional Activation. Cell Stem Cell 2018;22:428-44.e5. 10.1016/j.stem.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keating ST, El-Osta A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics 2013;8:361-72. 10.4161/epi.24234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDaniel SL, Strahl BD. Shaping the cellular landscape with Set2/SETD2 methylation. Cell Mol Life Sci 2017;74:3317-34. 10.1007/s00018-017-2517-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao Y, Neppl RL, Huang ZP, et al. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol 2011;194:551-65. 10.1083/jcb.201010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum R, Vethantham V, Bowman C, et al. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev 2012;26:2763-79. 10.1101/gad.200113.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurash JK, Lei H, Shen Q, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell 2008;29:392-400. 10.1016/j.molcel.2007.12.025 [DOI] [PubMed] [Google Scholar]
- 10.Judson RN, Quarta M, Oudhoff MJ, et al. Inhibition of Methyltransferase Setd7 Allows the In Vitro Expansion of Myogenic Stem Cells with Improved Therapeutic Potential. Cell Stem Cell 2018;22:177-90.e7. 10.1016/j.stem.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park IY, Powell RT, Tripathi DN, et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 2016;166:950-62. 10.1016/j.cell.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125:315-26. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 13.Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature 2013;502:480-8. 10.1038/nature12751 [DOI] [PubMed] [Google Scholar]
- 14.Hang CT, Yang J, Han P, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010;466:62-7. 10.1038/nature09130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willis MS, Holley DW, Wang Z, et al. BRG1 and BRM function antagonistically with c-MYC in adult cardiomyocytes to regulate conduction and contractility. J Mol Cell Cardiol 2017;105:99-109. 10.1016/j.yjmcc.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barsyte-Lovejoy D, Li F, Oudhoff MJ, et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc Natl Acad Sci U S A 2014;111:12853-8. 10.1073/pnas.1407358111 [DOI] [PMC free article] [PubMed] [Google Scholar]