We identified a strain of betacoronavirus BtKY72/Rhinolophus sp./Kenya/2007 (here BtKY72) from rectal swab samples in Kenyan bats. This paper reports the complete genomic sequence of BtKY72, which is closely related to BtCoV/BM48-31/Bulgaria/2008, a severe acute respiratory syndrome (SARS)-related virus from Rhinolophus bats in Europe.
ABSTRACT
We identified a strain of betacoronavirus BtKY72/Rhinolophus sp./Kenya/2007 (here BtKY72) from rectal swab samples in Kenyan bats. This paper reports the complete genomic sequence of BtKY72, which is closely related to BtCoV/BM48-31/Bulgaria/2008, a severe acute respiratory syndrome (SARS)-related virus from Rhinolophus bats in Europe.
ANNOUNCEMENT
The 2002 and 2003 outbreak of severe acute respiratory syndrome coronavirus (SARS-CoV) infection was a significant public health threat at the beginning of the 21st century (1–6). Initial identification of SARS-CoV in civet cats and other wild animals in live animal markets suggests zoonosis (7). Later, Rhinolophus sp. bats were identified as harboring severe acute respiratory syndrome-related CoV at high frequencies and were believed to be a natural reservoir host for SARS-CoV (8, 9).
During a 5-year bat coronavirus (CoV) surveillance study (2006 to 2010) in Kenya, we identified five bat betacoronaviruses by pan-CoV reverse transcription-PCR (RT-PCR) from fecal samples of Chaerephon and Rhinolophus bats (10, 11). The Institutional Animal Care and Use Committee (IACUC) of the Centers for Disease Control and Prevention and Kenya Wildlife Services approved all protocols related to the animal experiments in this study. These bat betacoronaviruses shared >98% nucleotide identity with each other and were clustered with other known bat SARS-related CoVs identified from Rhinolophus bats in China and Europe (8, 9, 12–15) based on a short amplicon sequence of open reading frame 1b (ORF1b) (121 bp). We selected RNA from the BtKY72 bat, which was one of the five betacoronavirus-positive bats from a previous study (11), for full genome sequencing. To determine the full genome sequence, consensus degenerate primers were designed from conserved sequences based on all known SARS-related CoVs (Table 1). Several small islands of sequences scattered throughout the genome were first determined from a Kenyan Rhinolophus bat using sets of seminested or nested consensus RT-PCR primers by Sanger sequencing. Then, sets of sequence-specific primers were used to fill the gaps and generate the full genome sequence, named BtKY72/Rhinolophus sp./Kenya/2007 (Table 1). The 5′ and 3′ ends of genome sequences were determined using a 5′/3′ rapid amplification of cDNA ends (RACE) kit (Roche). Complete genome sequencing was not performed due to limited viral loads in fecal samples from the other four betacoronavirus-positive bats.
TABLE 1.
Genomic PCR primers used in this study
| PCR or primer no. | First-round PCR primer |
Nested-round PCR primer |
||||
|---|---|---|---|---|---|---|
| Name | Sequence (5′→3′) | Nucleotide positiona | Name | Sequence (5′→3′) | Nucleotide positiona | |
| Consensus degenerate PCR primers | ||||||
| 1 | F20_Fwd | TACCCAGGAAAAGCCAACCAACC | 15–37 | F20_Fwd | TACCCAGGAAAAGCCAACCAACC | 15–37 |
| R328new_Rev | TGTAAAACAGGTAAACTGAGTTGGACGTG | 296–324 | R300_Rev | TGAAACCAGGGACAAGGCTCTCC | 254–284 | |
| 2 | F180_Fwd | AGACTGCAGACTGCTTACGGTTTCG | 174–198 | F220_Fwd | CATCAGCATACCTAGGTTTCGTCCG | 216–240 |
| R700_Rev | CACCTAACTCATAAGACTTTAGATCGATGCC | 668–698 | R490_Rev | CATCAGATCGTTTAATGAACACATAGGGC | 457–485 | |
| 3 | F1440_Fwd | ATTGAAACTCGACTCCGCAAGGG | 1436–1458 | F1470_Fwd | GGTAGGACTARATGTTTTGGRGGYTGTG | 1460–1487 |
| R2090_Rev | TACAAGACCACCWGTIACATAYGCCATRA | 2050–2079 | R2090_Rev | TACAAGACCACCWGTIACATAYGCCATRA | 2050–2079 | |
| 4 | F5810_Fwd | CAGAATATAAAGGACCAGTGACTGATGTTTTC | 5691–5722 | F5810_Fwd | CAGAATATAAAGGACCAGTGACTGATGTTTTC | 5691–5722 |
| R6580_Rev | GCTCGTTAGGTTTCTTAATGGTAATGCTTG | 6429–6458 | R6580_Rev | GCTCGTTAGGTTTCTTAATGGTAATGCTTG | 6429–6458 | |
| 5 | F8330_Fwd | ATGCCCAAGTAGCAARAAGYCACAATG | 8220–8246 | F8330_Fwd | ATGCCCAAGTAGCAARAAGYCACAATG | 8220–8246 |
| R9580_Rev | TGGTGAAATAGAATGTCAAGTACAAGTAAAAGA | 9441–9473 | R9470_Rev | TAGCAGCAACTACATGGTTGTACTCACC | 9345–9372 | |
| 6 | F10290_Fwd | GGCTTAAAGTTGATACYTCTAAYCCTAAGACACC | 10183–10216 | F10290_Fwd | GGCTTAAAGTTGATACYTCTAAYCCTAAGACACC | 10183–10216 |
| R11440_Rev | GCCCACATGGAAATAGCTTGATCTAARG | 11308–11335 | R11480_Rev | AACGACACCAGAATAGTTAGAGGTTACAGAA | 11345–11375 | |
| 7 | F11190_Fwd | TCTACATGCCTGCTAGYTGGGTGATG | 11079–11104 | F11220_Fwd | CGTATTATGACATGGCTYGAATTGGC | 11105–11130 |
| R12390_Rev | CGTGCATTGTTGATAATGTTGTTAAGTGC | 12252–12280 | R12390_Rev | CGTGCATTGTTGATAATGTTGTTAAGTGC | 12252–12280 | |
| 8 | F15280_Fwd | ACAGGRCTATGCCTAACATGCTTAG | 15170–15198 | F15300_Fwd | ATTATGGCTTCTCTTGTCCTTGCTCG | 15200–15225 |
| R15980_Rev | TTTCAATCATRAGTGTACCATCTGTTTTGAC | 15849–15879 | R15980_Rev | TTTCAATCATRAGTGTACCATCTGTTTTGAC | 15849–15879 | |
| 9 | F15830_Fwd | GACCTCAYGAATTTTGCTCWCAGC | 15729–15752 | F15850_Fwd | TCTCAGCAYACRAATGCTAGTTAAACAAGG | 15746–15775 |
| R16850_Rev | GTAGTACCTCTGTACACAACAGCATCWCC | 16718–16746 | R16840_Rev | GTACACAACAGCATCACCATAGTCACC | 16709–16735 | |
| 10 | F16455_Fwd | TTGTGTGCTAATGGTCAGGTTTTTGG | 16347–16372 | F16455_Fwd | TTGTGTGCTAATGGTCAGGTTTTTGG | 16347–16372 |
| R17560_Rev | GTGTCRACAATTTCRGCAGGACAACG | 17427–17452 | R17510_Rev | ATGTCWGGACCTATTGTTTTCATRAGTCTGC | 17377–17407 | |
| 11 | F17990_Fwd | CGMAATGTGGCTACKTTACARGCAGAA | 17874–17903 | F17990_Fwd | CGMAATGTGGCTACKTTACARGCAGAA | 17874–17903 |
| R19170_Rev | TTACAATTCCAAAACAARCARACACCATC | 19038–19066 | R19195_Rev | CATTGGCYGGRTAACGATCAACG | 19069–19091 | |
| 12 | F18870_Fwd | CGCGTTGATTGGTCTGTTGAATAYC | 18768–18792 | F18870_Fwd | CGCGTTGATTGGTCTGTTGAATAYC | 18768–18792 |
| R20100_Rev | ATGTGACTCCATTGACRCTWGCTTG | 19959–19983 | R20110_Rev | TTTTACTGATTCTCCAATTAATGTGACTCC | 19974–20004 | |
| 13 | F19880_Fwd | TTTCTACAATAGGTRTCTGYACAATGACTG | 19773–19802 | F19900_Fwd | TGACTGACATTGMCAAGAAACCTACTG | 19797–19823 |
| R20730_Rev | GCGTTTCACCATAATTCTGAAGGTC | 20600–20625 | R20730_Rev | GCGTTTCACCATAATTCTGAAGGTC | 20600–20625 | |
| 14 | F20580_Fwd | GGTGTAAGGATGGACATGYTGAAACC | 20479–20504 | F20580_Fwd | GGTGTAAGGATGGACATGYTGAAACC | 20479–20504 |
| R21200_Rev | CCACCATGAGAAATRKCCCATAAGC | 21070–21096 | R21210_Rev | TTGTAACAAARGCTGTCCACCATGAG | 21083–21107 | |
| 15 | F24200_Fwd | TGGCATATAGGTTYAATGGCATTGGAG | 24089–24033 | F24220_Fwd | GGCATTGGAGTTRCYCAAAATGTTCTC | 24109–24126 |
| R25345_Rev | CTCATAACAAATCCATTAAGTTCGTTTATGTG | 25197–25229 | R25345_Rev | CTCATAACAAATCCATTAAGTTCGTTTATGTG | 25197–25229 | |
| 16 | F24970_Fwd | CAAAAATCATACATCACCWGATGTTGATC | 24854–24882 | F25005_Fwd | TTTCAGGCATTAAYGCTTCWGTCG | 24894–24918 |
| R26290_Rev | CGCAGTAAGGATGGCTAGTGTGACTA | 26127–26152 | R26235_Rev | AAAGAAGTACGCTATTAACTATTAACGTACCTG | 26070–26102 | |
| 17 | F26065_Fwd | ACACAATCGACGGCTCTTCAGGAG | 25945–25968 | F26120_Fwd | TGAGCCGACGACGACTACTAGCGT | 25988–26011 |
| R26890_Rev | GATCACAGCNCCAATGACAAGTTCAC | 26726–26751 | R26870_Rev | CAAGTTCACTTTCCARGAGCGGTCTG | 26709–26734 | |
| Specific PCR primers | ||||||
| 1 | contig10F1_Fwd | GGTAAGATGGAGAGCCTTGTCCCTG | 254–278 | contig10F2_Fwd | AACGAGAAAACTCACGTCCAACTCAG | 284–309 |
| contig10R1_Rev | CTGACATAGAAGCAAGAATAATTACTACTTCCTC | 1670–1703 | contig10R1_Rev | CTGACATAGAAGCAAGAATAATTACTACTTCCTC | 1670–1703 | |
| 2 | contig9-F1_Fwd | CACAAGCTGCTTGCGTGGTTAGG | 1872–1894 | contig9-F1_Fwd | CACAAGCTGCTTGCGTGGTTAGG | 1872–1894 |
| contig9-R1_Rev | AGAGTTTCCATTCCTTGTGCGTCATC | 6212–6237 | contig9-R2_Rev | GACAACGCAAACACCACATATTGGG | 6134–6158 | |
| 3 | contig11F1_Fwd | AGTCAAACACTTGTCTCTGAAGAAGTAGTGG | 6248–6278 | contig11F2_Fwd | GAAGTAGTGGAAACTCCTACCATACAGAAGG | 6269–6299 |
| contig8-R1_Rev | GCATGATAATGTAAAACAGACTAGCAACTAATACC | 8462–8495 | contig8-R2_Rev | CATGTGTTATTCAATTTACCACCCTTAAGTG | 8397–8427 | |
| 4 | contig5-F1_Fwd | TTCTACCACGTGTGTTTAGTGCTGTTG | 8772–8798 | contig5-F1_Fwd | TTCTACCACGTGTGTTTAGTGCTGTTG | 8772–8798 |
| R10475_Rev | GTTAAAACCAACACTACCACATGANCCATT | 10334–10363 | R10410_Rev | ATTAGGTCTCATGGCACACTGRTAAACWC | 10281–10309 | |
| 5 | Contig7-F1_Fwd | AAAATGGCAGATCAGGCTATGACCC | 12129–12153 | Contig7-F2_Fwd | ACAGGCTAGGTCTGAAGACAAGAGGG | 12164–12189 |
| contig14R1_Rev | TTGTAGATTGCGGACATACTTGTCGG | 15444–15469 | contig14R2_Rev | CCATCAGTAGATAAGAGTGCATTCACATTAGC | 15401–15432 | |
| 6 | 500-c1-F1_Fwd | TCGATGGCCACTAATTATGACCTGAG | 17229–17254 | 500-c1-F1_Fwd | TCGATGGCCACTAATTATGACCTGAG | 17229–17254 |
| 500-c2-R1_Rev | AGCCCAAAGGACAAACACGACTC | 18369–18392 | 500-c2-R2_Rev | ACGCACTATGTTCCAAGGCAGACC | 18442–18464 | |
| 7 | 500-c3-F1_Fwd | AAGTTGGCATTAGGTGGTTCTGTGG | 21000–21024 | contig3-F2_Fwd | GCCATAAAGATTACAGAGCATTCGTGG | 21024–21050 |
| 500-R22790_Rev | CAGGTCCGATAGGTATATCACACTCATAGG | 23378–23406 | 500-R22740_Rev | TGGCTCCTAGAAGACAACCAGCTTG | 23338–23362 | |
| 8 | F23200_Fwd | CCGTGCTCTTTTGGTGGTGTKAGTG | 23161–23185 | F23200_Fwd | CCGTGCTCTTTTGGTGGTGTKAGTG | 23161–23185 |
| 500-c4-R1_Rev | CTGACATTTTAGTAGCAGCAAGATTAGCAG | 24334–24361 | 500-c4-R2_Rev | TCTGGACTTCAGCCTCAACTTTATCAAG | 24446–24475 | |
| 9 | 500C4F1_Fwd | GCTTAGCTACTTTGTTGCATCATTCAGG | 26593–26620 | 500C4F2_Fwd | ATTGGTGCTCATGATCATTCGTGGTT | 26735–26760 |
| oligodT anchor_Rev | GTTTCCCAGTCACGATATTTTTTTTTTTTTTTTV | 29273–29289 | oligodT anchor_Rev | GTTTCCCAGTCACGATATTTTTTTTTTTTTTTTV | 29273–29289 | |
Positions relative to the genome of BtKY72/Rhinolophus sp./Kenya/2007 (GenBank accession no. KY352407).
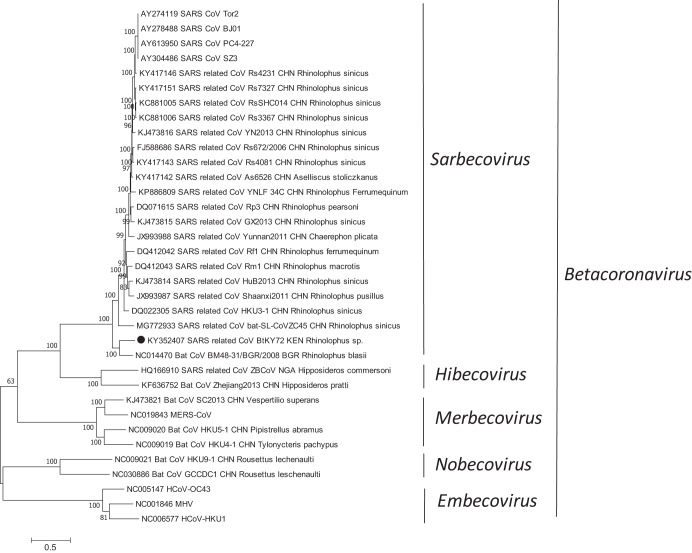
The genome of BtKY72 was 29,259 nucleotides long, including the poly(A) tail, with 39% G+C content. Sequence alignment and a BLAST search analysis of the full-length genome sequences showed that the BtKY72 genome shared an 81% overall nucleotide identity to its nearest relative, BtCoV/BM48-3, which was identified from a Rhinolophus bat in Europe (15), and that it has 93 to 94% amino acid identity in the seven concatenated, conserved replicase domains (ADP-ribose-1″-phosphatase [ADRP], nonstructural protein 5 [nsp5], and nsp12 to nsp16) to BtCoV/BM48-31 (Fig. 1). Phylogenetic analysis suggested that BtKY72 belongs to the subgenus Sarbecovirus of the genus Betacoronavirus (Fig. 1). The genome organization contained the following gene order: 5′ UTR-ORF1ab-S-ORF3a-E-M-ORF6-ORF7a-ORF7b-N-3′ UTR. Unlike SARS-CoV and other known SARS-CoV-related bat viruses, both ORF3b and ORF8 were absent in BtKY72. ORF8 was also missing in its closest neighbor, BtCoV/BM48-31 (15).
FIG 1.
Phylogenetic analysis of whole-genome sequences of betacoronaviruses. The phylogenetic tree is inferred using the maximum likelihood (ML) method available in PhyML version 3.0 (16), assuming a general time-reversible (GTR) model with a discrete gamma-distributed rate variation among sites (Γ4) and a subtree pruning and regrafting (SPR) tree-swapping algorithm. The sequences are labeled with accession number, strain name, geographic (three-letter country code), and host (species) information. BtKY72/Rhinolophus sp./Kenya/2007, sequenced in this study, is highlighted with a solid circle. The genus taxonomy information is shown to the right side of the phylogeny. The maximum likelihood bootstrap is indicated next to the nodes. The scale bar indicates the estimated number of nucleotide substitutions per site. KEN, Kenya; CHN, China; BGR, Bulgaria; NGA, Nigeria; MERS-CoV, Middle East respiratory syndrome coronavirus; HCoV, human coronavirus; MHV, mouse hepatitis virus; ZBCoV, Zaria bat coronavirus.
In conclusion, our study demonstrates that the SARS-related CoVs that were identified from Rhinolophus bats in China and Europe were also present in Kenyan Rhinolophus bats (Fig. 1). The discovery of SARS-related CoVs in Kenyan bats adds to the diversity and geographic range of CoVs in Rhinolophus bats. The genome data for BtKY72 will facilitate understanding of the molecular evolutionary characteristics of bat SARS-related CoV.
Data availability.
The complete genome sequence of BtKY72 is available in GenBank under the accession number KY352407.
ACKNOWLEDGMENTS
We thank Ivan Kuzmin and Michael Niezgoda from the Poxvirus and Rabies Branch, Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention (CDC), Atlanta, GA, and Bernard Agwanda from the National Museum, Kenya Wildlife Service, Nairobi, Kenya, for excellent technical and logistical assistance and field study.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Cherry JD, Krogstad P. 2004. SARS: the first pandemic of the 21st century. Pediatr Res 56:1–5. doi: 10.1203/01.PDR.0000129184.87042.FC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF, Lui SF, Szeto CC, Chung S, Sung JJ. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY, members of the SARS Study Group . 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong N, Ding Y, Mao Y, Wang Q, Wang G, Wang D, Cong Y, Li Q, Liu Y, Ruan L, Chen B, Du X, Yang Y, Zhang Z, Zhang X, Lin J, Zheng J, Zhu Q, Ni D, Xi X, Zeng G, Ma D, Wang C, Wang W, Wang B, Wang J, Liu D, Li X, Liu X, Chen J, Chen R, Min F, Yang P, Zhang Y, Luo H, Lang Z, Hu Y, Ni A, Cao W, Lei J, Wang S, Wang Y, Tong X, Liu W, Zhu M, Zhang Y, Zhang Z, Zhang X, Li X, Chen W, Xhen X, Lin L, Luo Y, Zhong J, Weng W, Peng S, Pan Z, Wang Y, Wang R, Zuo J, Liu B, Zhang N, Zhang J, Zhang B, Zhang Z, Wang W, Chen L, Zhou P, Luo Y, Jiang L, Chao E, Guo L, Tan X, Pan J, Chinese Medical Association , China Association of Chinese Medicine. 2003. Consensus for the management of severe acute respiratory syndrome. Chin Med J (Engl) 116:1603–1635. [PubMed] [Google Scholar]
- 5.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, Group SW. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 8.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A 102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 10.Tong S, Conrardy C, Ruone S, Kuzmin IV, Guo X, Tao Y, Niezgoda M, Haynes L, Agwanda B, Breiman RF, Anderson LJ, Rupprecht CE. 2009. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg Infect Dis 15:482–485. doi: 10.3201/eid1503.081013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S. 2017. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol 91:e01953-16. doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y, Xia L, Zhou J, Zhen W, Feng Y, Guo H, Zhang H, Tu C. 2014. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J Virol 88:7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, Cheung CL, Xu KM, Song WJ, Vijaykrishna D, Poon LL, Peiris JS, Smith GJ, Chen H, Guan Y. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol 80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan J, Hon CC, Li Y, Wang D, Xu G, Zhang H, Zhou P, Poon LL, Lam TT, Leung FC, Shi Z. 2010. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J Gen Virol 91:1058–1062. doi: 10.1099/vir.0.016378-0. [DOI] [PubMed] [Google Scholar]
- 15.Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, Seebens A, Niedrig M, Pfefferle S, Yordanov S, Zhelyazkov L, Hermanns U, Vallo P, Lukashev A, Muller MA, Deng H, Herrler G, Drosten C. 2010. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol 84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of BtKY72 is available in GenBank under the accession number KY352407.