Abstract
Although the human genome provides the blueprint for life, most of the proteins it encodes remain poorly studied. This perspective describes how one group of scientists, in seeking new targets for drug discovery, used open science through unrestricted sharing of small molecules to shed light on dark matter of the genome. Starting initially with a single pharmaceutical company before expanding to multiple companies, a precedent was established for sharing published kinase inhibitors as chemical tools. The integration of open science and kinase chemogenomics has supported the study of many new potential drug targets by the scientific community.
Extreme Open Science
Modern science is increasingly multidisciplinary and expensive, and the solutions to many of the grand challenges will require resources, skill sets and capital distributed among multiple academics and industrial institutions. Open science seeks to remove the barriers to tackling these problems through sharing research output, such as reagents and data, with the aim to increase the efficiency and quality of research by reducing duplication and improving reproducibility. Many academic institutions embrace the principle of open science, supporting the philosophy that their research endeavors are for the public good and should be accessible at no cost. For commercial organizations where the return on investment is paramount, the value proposition for engaging in open science has been less obvious.
The understanding of human biology and disease is one of the problems of equal relevance to the public and private sectors. Fifteen years after the sequencing of the human genome, and despite massive investments by the public and private sectors, the translation of genetic information into new drug therapies remains slow and ineffective. As a result, the pharmaceutical industry has become willing to explore new and more open models to support this research, including the selection of drug targets and the generation of research tools. The Structural Genomics Consortium (SGC) is a charitable research organization, supported by the pharmaceutical industry and public funders, that is pushing the boundaries of openness in biomedical research. The SGC mission is to catalyze research in new areas of human biology and drug discovery by focusing explicitly on less-studied areas of the human genome. The SGC laboratories practice a philosophy of extreme open science — all research output is made openly available to the scientific community with no strings attached, SGC scientists are encouraged to disclose their research findings in real time through the opennotebooks initiative,1 and all publications are deposited in open preprint sites such as bioRxiv prior to submission to a journal.
The Dark Kinases
In 2011, three scientists, Stefan Knapp, Susanne Müller, and Oleg Fedorov at the SGC Oxford laboratory, published a landmark paper in the journal Nature Chemical Biology titled “The (Un)targeted Cancer Kinome.”2 The paper described a counterintuitive situation. Protein kinases had emerged as one of the most successful sources of cancer drugs, yet scientists were focusing on only a few of the several hundred kinases found in the genome and ignoring the rest.
Sequencing of the human genome had uncovered 500 kinase enzymes that catalyze the transfer of phosphate from ATP to other proteins. Phosphorylation alters the charge and shape of a protein, leading to changes in its activity that affect cellular processes such as protein synthesis, cell division, signal transduction, and cell growth. Kinases modify as many as a quarter of all proteins and the importance of protein phosphorylation is such that it is often found to be dysregulated in cancer, inflammation, and neurodegeneration. Yet, as Stefan and his colleagues at the SGC noted, it appeared that the research community was focusing its effort on only a small subset of these enzymes, despite clear genetic evidence that many of the lesser-studied ‘dark’ kinases were implicated in cancer and many other diseases. This surprising behavior, which applies more broadly across the entire genome and to many protein families, continues to be the subject of intense discussion in the academic literature.3, 4 It has been proposed that the pragmatism of reagent availability drives this herd-like behavior (there are no small molecule inhibitors of the dark proteins) and the conservative nature of research funding constrains it (the less that is known about a protein, the harder it is to secure grant funding). The situation is frustrating. After all, in an ideal world, what scientist would want to study a well-trodden path of research rather than explore new avenues of discovery?
At the time, we were working in the Department of Chemical Biology at the pharmaceutical company GlaxoSmithKline (GSK). Our laboratory was part of a multidisciplinary effort to select the best new target proteins for drug discovery. The primary hurdle for starting any new drug discovery project is target validation; the evidence that inhibition of the target protein will translate into clinical efficacy in a human disease. Our view was that genetic evidence alone is not enough validation to invest in a new project. It needs to be backed up by a demonstration that inhibition of the target protein is therapeutic in a clinically relevant disease model.5 The dearth of academic publications on the dark kinases had a direct impact on our research. As chemists, we considered kinases to be highly druggable given the company’s in-house know-how and track record in kinase drug discovery.† However, kinases, in general, had fallen out of favor at GSK and at many other pharmaceutical companies. There was a view that vaccines, antibodies and other biopharmaceuticals were more profitable than small molecule drugs, in part because competitors were not as adept at bringing generic versions to market. In an effort to turn attention back towards the kinases, due to their proven tractability as a source of new medicines, we conceived a bold open science experiment to shine light on the dark kinases of the genome using small molecule inhibitors as chemical tools. We called the approach ‘kinase chemogenomics’ as others had coined this term to describe prior efforts in chemical genomics.6, 7
Kinase Chemogenomics
Our idea was to exploit a quirky feature of nearly all kinase inhibitors, namely their ability to inhibit more than one kinase, and to combine that with the power of open science to crowd-source the biological study of the dark kinases. Most small molecule kinase inhibitors are designed to displace ATP from the catalytic site of the enzyme. However, because the ATP binding sites of all kinases are quite similar in size and shape a single small molecule nearly always inhibits more than one kinase. Pharmaceutical companies had already exploited this cross-reactivity (some call it ‘promiscuity’) to treat new diseases. For example, the Abl kinase inhibitor Gleevec (imatinib) was developed as a drug to treat leukemia but also inhibits another kinase called Kit. This discovery led to its subsequent development as a drug to treat Kit-dependent gastrointestinal tumors.8 Given the inherent promiscuity of kinases inhibitors, we reasoned that some members of our internal library, though developed for specific kinases, would also inhibit many of the dark kinases as collateral targets. There was support for this concept from our internal kinase cross-profiling experiments as well as in publications from Bristol-Myers Squibb.9, 10 We reasoned that if we were to identify a set of inhibitors that targeted the dark kinases and make them freely-available, we could reach beyond the confines of GSK and enable the external scientific community to connect the dark kinases to human diseases.
We were, however, cognizant of the obstacles to being able to conduct this truly open experiment from within a for-profit company. GSK, like all pharmaceutical companies, vigorously protects the chemical identity of its proprietary compounds. Disclosure of a chemical structure beyond the protective walls of the organization required the approval of senior management and a company lawyer following assessment of the risk and benefit. At the time, it was unprecedented for any company to unveil the chemical structures of hundreds of inhibitors, which would be required for a public chemogenomic experiment. Even though many potentially valuable kinase inhibitors were no longer being actively studied within the company, GSK was not interested in revealing these molecules to external investigators. How could we break this deadlock? One option was to share the compounds without disclosing the chemical structures, in effect blinded to the external scientist. However, we felt that concealing the structures ran against the spirit of an open science experiment. It would prevent academic scientists from being able to meet the publication standards of any good journal and would also fail to engage the minds of other scientists willing to interpret the relationship between chemical structure and biological activity that was at the core of the experiment. Karen Lackey, the chemist who had led the discovery of the drug lapatinib (an inhibitor of EGFR kinase), provided an elegant solution. Karen proposed that we build the chemogenomic set exclusively from kinase inhibitors that GSK chemists had already disclosed in the scientific literature. The company had encouraged its scientists to publish papers at the conclusion of a research project to promote their career development and to maintain the scientific rigor of the work. It is widely accepted that open publication of research has benefits to the scientific community and to the individual researcher.11 An upshot was that several thousand kinase inhibitors had been vetted through internal review and approved for publication. The concept was that by limiting the open science experiment to a pool of compounds that GSK chemists had previously published, we would benefit from the fact that the internal decision to disclose the chemical identity of each one had already been made.
Selection of the kinase inhibitors for the public set required several steps. First, we had to compile an accurate list of the kinase inhibitors that had been published by GSK. Second, we had to search each of the three internal compound stores to find physical samples of the inhibitors and remove compounds from the list that were no longer available. Third, we had to organize the pool of remaining inhibitors into their core chemical templates, creating groups of molecules based on the synthetic chemistry used to make them. Finally, we had to manually select inhibitors to balance chemical and biological diversity, such that each of 31 templates had multiple exemplars with their known activities spread across the largest number of kinases. Each step was time consuming, but none were insurmountable to an experienced medicinal chemist with access to the GSK inventory and a good knowledge of kinase inhibitor design. The combined collection of 367 compounds was named the GSK Published Kinase Inhibitor Set, or PKIS for short.
Although all of the compounds in PKIS had been published as kinase inhibitors, most of them had only been tested on a handful of kinases. Their full activity profile across every human kinase (known as the kinome) was not known. Fortunately, due to recent advances in screening technology, several commercial vendors had developed the capacity to run large panels of kinase screening assays. GSK was using some of these vendors for deep profiling of kinase inhibitors that were nominated for progression to clinical studies. The prevailing view was that kinase inhibitors with high selectivity for their primary target kinase would show fewer side effects due to off-target inhibition of nuisance kinases. However, translation of any selectivity profile into a prediction of drug safety was incredibly difficult, since the biology of hundreds of dark kinases was still unknown. For our chemogenomic experiment, deep annotation of off-target activity was the key to enabling the community to study the role of dark kinases in either disease biology or toxicology. Thus, profiling of PKIS on as many of the human kinases as possible was our next goal.
Commercial kinome-wide profiling assays were available in a variety of formats, ranging from conventional enzyme inhibition assays to affinity capture methods to mass spectrometry-based proteomics. Each technology has strengths and limitations, but the primary determinant of which vendor to select for our chemogenomic experiment was the price. Nanosyn, a company using microfluidics as its screening technology, generously offered to test PKIS across 224 kinases for the cost of only the assay reagents. It would allow them to showcase their kinase screening platform and increase visibility of the company through publication of the data. Nanosyn agreed to test all 367 compounds at two concentrations for inhibition of enzyme activity on 224 human kinases (367 × 224 × 2 = 164,416 assays). To further increase coverage of the darkest kinases for which no enzyme assay had been developed, Stefan Knapp at the SGC agreed to profile PKIS against 68 kinases by measuring their effect on thermal stability of the protein. In total, each molecule in PKIS was annotated for activity on 260 human kinases. The Nanosyn data showed that PKIS contained potent inhibitors for 2/3 of the kinases tested. There were only 18 kinases for which no inhibitor was found. Similar success was seen in the thermal stability screen where active compounds were identified for all but 4 of the kinases. Overall, PKIS contained small molecules with activity on more than 200 human kinases. In keeping with our open science objectives, the full dataset with all compound structures was published in the journal Nature Biotechnology and deposited at the web-accessible ChEMBL database.12, 13
A Path to True Open Science
We had to clear several internal hurdles before being allowed to distribute physical samples of PKIS as a public resource. Any exchange of compounds between a company and an external investigator requires a Material Transfer Agreement (MTA), signed by representatives of both the company and the investigator’s institution. The MTA usually identifies the company as the owner of the compound, assigns rights to the company to any new discoveries made while using the compound, and places restrictions on how the discoveries can be published. We believed that these reach through provisions and publication restrictions would limit significantly the uptake of the compound set and our ability to uncover unexpected therapeutic applications. Working with two GSK lawyers, John Lemanowicz and Barbara Carter, we created a simplified agreement to facilitate open sharing the compound set; the new abbreviated MTA placed no restriction on research performed with the compounds, gave no claim to GSK for any intellectual property arising from the research, but it did require that the resulting scientific data be made publicly available. Although the timing and scope of this disclosure was left undefined, the MTA made it clear that open publication of results was expected. GSK would still own the compounds, but external investigators would be free to use them and publish without fear that the company would claim rights to any of the data.
Even with the abbreviated MTA drafted, we still needed authorization from GSK management to allow PKIS to be freely distributed. At the time, no company had been bold enough to pursue a truly open experiment in which hundreds of compound structures, biological data, and physical samples were made available to investigators without restrictions. Some other pharmaceutical companies had explored sharing their proprietary compounds through mechanisms that were branded ‘open innovation.’ Examples included the Lilly Target Drug Discovery Initiative and the Pfizer Phenotypic Screening Library.14 These efforts, which sought to balance open access with return on investment, shared compounds that were blinded to the academic scientist and the chemical structures were only revealed after data was reported back to the company. The data review and the blinded structures provided a degree of control over the external experiments. How could we convince GSK to take a more radical approach; to allow unrestricted access to compounds without any guarantee of something in return?
Many of our colleagues in GSK embraced the innovation of the PKIS experiment. There was near consensus that GSK needed more new targets with strong scientific validation to fuel its drug discovery. The support for using open science was founded on the premise that, as a large pharmaceutical company, GSK would benefit from access to more publically validated targets, since value (and exclusivity) could be created from the final drug molecule even without a competitive advantage at the outset of the drug discovery project. Not surprisingly, there were pockets of resistance too. Reasonable issues were raised about the time and money that it would take to assemble and distribute the compounds as well as the administrative burden required to execute the MTA. Others voiced the concern “what if we share these compounds and someone identifies a drug?” Our counterargument was that the published inhibitors would create more value for GSK in the biology that they would uncover. That was the real treasure for drug discovery. We likened the situation to the model of ‘eaters and bakers’‡ that had been popularized by Guy Kawasaki.15 We used the analogy that open availability of PKIS in the scientific community would be akin to baking a bigger pie; the more the set was used in diverse biological assays, the more public data would be available to GSK to support the selection of new drug discovery targets. We were also able to address a specific concern of the ‘eaters’ who feared that the size of the kinase drug discovery pie was finite. We implemented an internal operating agreement that (paradoxically) limited our ability to collaborate openly with the recipients of PKIS. Specifically, we agreed to refrain from sharing additional unpublished analogs of the compounds in PKIS with external investigators, since there was still a very small chance that any new inhibitor could be a novel and patentable drug. By agreeing to this restriction, we were given permission to ship PKIS to external investigators. Our final hurdle was to make the scientific community aware of PKIS and how to request a copy. We reached out to several prominent scientists in the field of kinase research to gauge their interest in using PKIS and to help spread the word of its availability. We also made multiple presentations at scientific conferences and published a commentary describing PKIS as a tool for kinase research in the journal Current Topics in Medicinal Chemistry.16
Initial uptake of the set was brisk. While there were also commercially available kinase inhibitor sets (e.g. from Selleck Chemicals, Cayman Chemical, and Tocris Bioscience) for investigators to choose from, they lacked the deep kinase activity annotation of PKIS. By the end of 2013, GSK had shipped copies of PKIS to several hundred laboratories. However, it quickly became clear that the restriction on being able to make and share new compounds was hindering the ability of external investigators to confirm their preliminary results and strengthen the validation of dark kinases as drug targets. For example, many investigators, upon generating an exciting result in a cell assay would approach us with a request for an analog that was optimized for activity in an animal model. We were not able to engage in these collaborations under our open science operating agreement. In an attempt to remove the restriction on synthesis of new analogs, we decided to seek support for the PKIS experiment from the GSK CEO, Sir Andrew Witty, who was scheduled to visit our laboratory in January 2014. Sir Andrew was a known advocate for increased pharmaceutical company research into diseases of the developing world and had supported open and collaborative approaches to drug development for neglected tropical diseases, where the human burden outweighed the focus on business profits.17 We were able to secure a short meeting to describe our open science experiment, and pitched the concept of sharing published GSK kinase inhibitors without restrictions as a mechanism to identify new kinase targets for drug discovery. Although prepared to be peppered with questions about intellectual property, ownership, and return on investment, we were surprised by Sir Andrew’s response. Within a few minutes, he understood the concept of our experiment. He recognized that open distribution of PKIS could potentially give GSK access to scientific experts in many fields of biology and medicine and stated: “I want my scientists to be exposed to as many great and diverse ideas as possible. What can I do to help?”
The Next Chemogenomic Set
Having secured support for our open science experiment from the CEO, we turned our attention to improving the utility of PKIS as a tool to study dark kinases. As the first reports of its use emerged in the scientific literature we became aware of some of the technical limitations of PKIS as a chemogenomic set. Analysis of the activity profile of PKIS showed that many of the dark kinases were only inhibited by the most promiscuous compounds in the set. So, although while we had established that these dark kinases could be inhibited by small molecules, there were no inhibitors in PKIS with sufficient selectivity to easily connect them to a disease. To address this flaw, we decided to build a follow-up set (named PKIS2) using inhibitors from different chemical templates with different kinase inhibition profiles. Our initial thought was to add inhibitors from sources outside of GSK, since other companies and academics would likely have access to additional chemical diversity.18 We reached out to scientists at other pharmaceutical companies, but disappointingly their responses ranged from “Good idea but we’ll never be able to do that here” to “No thanks. We would rather tackle the problem by ourselves.” It was clear that many companies were not ready to work cooperatively with GSK to shed a common light on the dark kinases. Our discussions with Paul Gillespie from Roche were more positive. He assembled a set of 253 kinase inhibitors that had been published by Roche scientists and used our success with sharing PKIS to convince his management to allow a similar experiment to proceed. Ultimately, however, the Roche PKIS was shared under a much more restrictive MTA that included intellectual property stipulations and required the vetting of data before public disclosure.
A more constructive solution to building PKIS2 emerged when a large collection of new compounds was disclosed by GSK scientists. For many years, GSK had been committed to research on malaria to develop treatments for this life-threatening disease of the developing world. The company had established a laboratory at the Tres Cantos site outside of Madrid to screen for compounds to inhibit the Plasmodium parasite that causes malaria. With the support of Sir Andrew Witty, the Spanish scientists had recently embraced an open science strategy and released the structures and screening data of the Tres Cantos Antimalarial Set (TCAMS).19 It comprised over 13,500 compounds that had been tested as inhibitors of the parasite. Their goal was to provide the scientific community with new chemical starting points for the development of antimalarial drugs. At the same time, it greatly increased the pool of publicly-disclosed GSK compounds. We recognized that many of the compounds in TCAMS had been synthesized originally as kinase inhibitors for long-since defunct projects. By choosing molecules from TCAMS as well as kinase inhibitors published since the PKIS had been selected, we assembled 600 new GSK kinase inhibitors as PKIS2. The set contained molecules from 55 different chemical templates, the vast majority of which were not represented in the original PKIS.
Opportunity from Adversity
In late 2014, around the time that we finished assembling PKIS2 and were preparing to address the limitation on working with only published compounds, our open experiment hit an unexpected roadblock. GSK was entering a financial crisis. Despite having succeeded in developing more new medicines than many of its competitors over the prior decade, sales had been disappointing and the company stock price had fallen by 20% in the past year. There were concerns that GSK, itself the product of multiple mergers, would be bought by a rival company and stripped of its assets. With income falling as sales of older blockbuster drugs were eroded by generic versions, the decision was made to cut expenses in research and development. But who or what was on the chopping block? Senior management made a disappointing decision to close the former Glaxo research site in North Carolina, the one where our laboratory was located. We were given the option of moving the laboratory to the GSK research site at Upper Providence, about 30 miles northwest of Philadelphia. However, faced with the uncertainties of a new research environment and the risk that that open science would no longer be supported if the CEO was ousted, we opted to explore moving our laboratory to an academic institute. Dean Robert Blouin of the UNC Eshelman School of Pharmacy and Al Edwards, CEO of the SGC, were very responsive to our predicament. They saw an opportunity to bring a team of scientists with pharmaceutical drug discovery experience to UNC that would complement ongoing academic research in oncology and neurosciences. With their support and generous grants from the Eshelman Institute for Innovation and the UNC Lineberger Cancer Research Fund, we moved the laboratory to the University of North Carolina at Chapel Hill in July 2015 to create the first SGC site in the USA. GSK, an original founding member of the SGC, was also generous in allowing us to bring samples of PKIS and PKIS2 to UNC and donating used equipment to the laboratory. The SGC-UNC laboratory operates under the consortium principles of extreme open science to accelerate drug discovery on the dark kinases.
Moving the laboratory to UNC had an immediate positive impact on the kinase chemogenomic experiment. We were no longer restricted to working with only published GSK kinase inhibitors and could make new analogs that would fill the gaps in the set. Also, now that we were working at an academic institution, many additional pharmaceutical companies expressed interest in contributing. The SGC is currently supported by eight pharmaceutical companies, and each of them signaled their willingness to donate additional inhibitors to the public set. Pfizer and Takeda quickly identified published kinase inhibitors from their internal compound collections. Working with Dan Treiber and his colleagues at DiscoverX, we struck a deal that would allow both PKIS2 and these newly donated inhibitors to be profiled against 400 human kinases using their affinity capture KINOMEscan technology. For DiscoverX it provided an opportunity to cement KINOMEscan as the gold standard for measuring kinome-wide selectivity. Thus, by the Fall of 2016, we had on hand a large dataset of 1000 inhibitors screened across hundreds of human kinases to provide a platform on which to build an improved kinase chemogenomic set. We also had garnered feedback from the PKIS user community that provided additional insight into how the set had been used to explore the biology of dark kinases.
Chemogenomics in Practice – User Impact and Examples
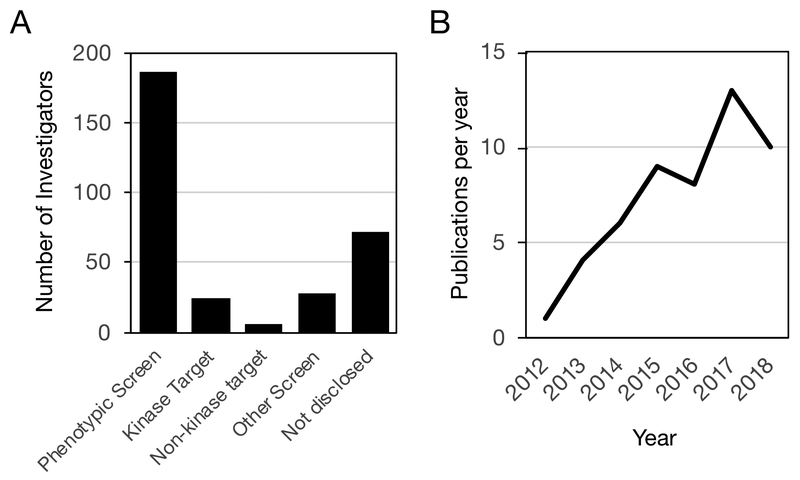
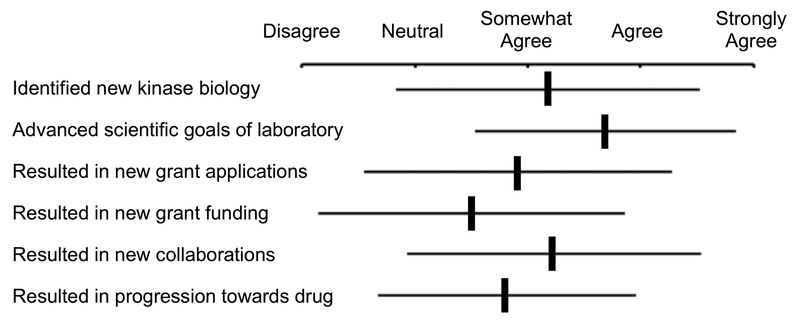
PKIS and PKIS2 were distributed by GSK and the SGC-UNC from 2011 to 2017 to over 300 laboratories. When completing the MTA, researchers were asked to briefly describe their planned use of the set (Figure 1A). Most of the laboratories indicated that the set would be used to identify kinases that affected the biology of a disease-relevant cell assay (known as a phenotypic screen). The simplest measure of impact was a steady increase in scientific publications over this period (Figure 1B). However, a recent survey of scientists that received PKIS or PKIS2 has also captured the positive impact of the chemogenomic sets on the progression of the research in their laboratories (Figure 2). The majority of respondents reported that use of the sets led to the discovery of new kinase biology, advanced their scientific goals, and was supporting new collaborations. Evidence of new grant applications and funding were also reported. Importantly, many of the recipients noted that they had made progress towards a new drug therapy. A few selected highlights of the research performed with PKIS and PKIS2 are described below:
Figure 1.
PKIS and PKIS2 have been used in chemogenomic experiments in over 300 laboratories. A) Primary use of the sets as reported by the investigator in their abbreviated Material Transfer Agreement. B) Number of publications reporting data on PKIS or PKIS2 in each calendar year from 2012 to 2018 as captured in PubMed and SciFinder on November 12th, 2018. The full list of publications is available at www.sgc-unc.org/kcgs-and-pkis-publications/.
Figure 2.
PKIS user survey conducted by Maryann Feldman (UNC Business School) on August 28th, 2018. Recipients of PKIS or PKIS2 were asked how it had impacted their research. Vertical bars represent the average of the 94 responses with the standard deviation shown by the horizontal bars.
An algorithm for analysis of chemogenomic screens:
Kinase chemogenomics utilizes the ability of small molecule inhibitors to bind to multiple kinases. The inhibitors in PKIS show a range of promiscuity; some compounds inhibit a small number of kinases while others show activity on many kinases. The deconvolution of data from a biological assay in the context of these activity profiles is the key to being able to identify which kinase or combination of kinases causes the effect. Hassan Al-Ali in the Lemon-Bixby laboratory at the University of Miami created an algorithm that analyzed the full data set of kinase inhibition by the compounds in PKIS to help answer this question.20 Hassan applied the algorithm to the interpretation of data from the screening of PKIS on the growth of primary neurons isolated from rats. Kinases were identified that promoted or hindered neurite growth alone or in combination. Importantly, the algorithm can be generalized to any screen using PKIS and underscores the value making all PKIS activity data publicly available.
A new drug therapy for chordoma:
Chordoma is a rare bone cancer with a median survival of fewer than 7 years and no approved drug therapy. Researchers have developed cellular models of chordoma to better understand the disease and to provide a means to identify potential drug targets. Susanne Scheipl at University College in London screened PKIS and PKIS2 to identify compounds with the ability to stop the growth of chordoma cell lines.21 Susanne identified multiple compounds that were annotated as inhibitors of EGFR. The results were replicated with selective EGFR inhibitors that had been developed to treat breast cancer. In an exciting development, the EGFR inhibitor afatinib is now being tested in a clinical trial in chordoma patients. The chemogenomic experiment successfully defined a new kinase-disease connection that contributed to the repurposing of a known kinase inhibitor drug.
Identification chemical tools for a pseudokinase:
About 10% of human kinases have changes in their amino acids that render them unable to catalyze the transfer of phosphate to other proteins.22 These pseudokinases are still expressed in cells, but most of them are dark kinases that have not been well studied and lack chemical tools. Indeed, a critical question is whether pseudokinases are tractable at all to inhibition be small molecules. Patrick Eyers at the University of Liverpool was interested in the TRIB family pseudokinases and the role they play in the development of solid tumors and blood cancers. He screened PKIS and PKIS2 to identify compounds that either increased or decreased the stability of the TRIB2 protein.23 Identification of active compounds within a common chemotype laid the foundation for the development of a series of analogs that bind covalently to TRIB2 and trigger its rapid degradation in cells. The TRIB2 degrader molecules are being used as chemical tools to study the role of the pseudokinase in cancer biology.
Identification of a new antifungal drug target:
Kinases are ancient proteins that are found in all eukaryotic organisms including animals, plants, and microbiota. Although PKIS and PKIS2 were designed to explore the biology of human kinases, they have also been screened in assays from a variety of nonhuman organisms. Identification of inhibitors of fungal kinases is one exciting example. Systemic fungal infections in immunocompromised individuals are often deadly, with mortality rates near 50%. Leah Cowen at the University of Toronto screened PKIS against a strain of the fungal yeast Candida albicans that had resistance to caspofungin, a first line antifungal drug. Several compounds from the pyrazolopyridine chemotype showed antifungal activity that was further potentiated by caspofungin (Cowen, L. unpublished results). Genetic complementation experiments identified Yck2 as the fungal kinase targeted by these inhibitors. Development of selective Yck2 inhibitors may lead to a new approach to treat life-threatening yeast infections.
Building a Better Chemogenomics Set
Having successfully established the SGC-UNC laboratory with its primary focus to develop chemical tools for the dark kinases, we decided to challenge ourselves to build the ultimate public chemogenomic set with complete coverage of all human kinases.24 To formulate a robust scientific plan, we organized a scientific retreat and invited key collaborators and representatives from ten pharmaceutical and biotechnology companies to participate. We picked an isolated, yet appealing location — in mid-October the North Carolina beaches are uncrowded and often blessed with sunny and warm weather — we had a great turn out! The collected academic and industry scientists were motivated by the need for an improved chemogenomic set for the study of dark kinases. At the gathering, the lessons we had learned from the assembly and use of PKIS and PKIS2 were reviewed. The kinase selectivity profile of the ideal chemogenomic compound was debated. A list of the dark kinases that were not inhibited by PKIS or PKIS2 was defined. Most importantly, we agreed to coordinate efforts to build the optimal public kinase chemogenomic set. The key recommendations were to remove all promiscuous inhibitors from PKIS and PKIS2 and select the best of the remaining inhibitors, to increase the chemical diversity of the set by adding inhibitors from multiple companies and academic collaborators, and to include additional compounds from the scientific literature that met our new selectivity criteria.§ Each of the pharmaceutical companies agreed to search their internal databases for published compounds to donate to the set. We decided to use the DiscoverX KINOMEscan for broad screening of the inhibitors, since these assays covered the largest number of human kinases, and to use Promega NanoBRET assays to confirm activity in live cells. Finally, we decided to call the new set the Kinase Chemogenomics Set or KCGS.25
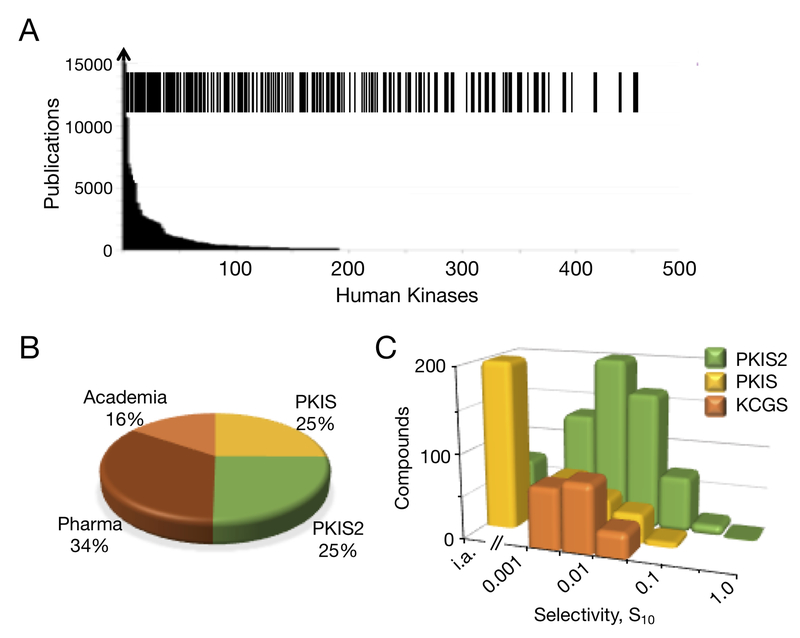
For inclusion in KCGS an inhibitor has to demonstrate activity on its target kinase at 100 nM or less, while also showing activity on less than 5% of the kinases surveyed. Statistical modeling shows that these narrow spectrum inhibitors are ideally suited to assign function to dark kinases; it maximizes coverage of the kinome, while over-sampling in the set (where more than one inhibitor covers each kinase) allows biology to be assigned to a specific kinase. Culling of PKIS and PKIS2 left just under 50 compounds from each set that met these potency and selectivity criteria. In addition to the compounds received from Pfizer and Takeda, kinase inhibitors were made available by AbbVie, AstraZeneca, Bayer, Boehringer Ingelheim, Merck & Co, and Vertex. We also acquired inhibitors from the laboratories of Nathanael Gray (Harvard), Richard Angell (University College London), Julian Blagg (Cancer Research UK), and new analogs synthesized in the SGC laboratories. The set will continue to grow over time, but as of November 2018, we have completed kinome-wide profiling of over several hundred new inhibitors for inclusion in KCGS and the first release is ready for distribution.** KCGS v1.0 contains 188 narrow spectrum inhibitors that cover 212 kinases, representing over 50% of the screenable human kinome as defined by the DiscoverX KINOMEscan (Figure 3A). Half of the new set contains the best inhibitors from PKIS and PKIS2, while the other half is composed of newly acquired or synthesized inhibitors (Figure 3B).
Figure 3.
The Kinase Chemogenomic Set, KCGS. A) Coverage of human kinases by KCGS. Each line in the barcode indicates a kinase that has at least one potent inhibitor in the set. Shown in the background is the number of publications on each kinase as reported by Knapp et al.2 B) Origin of the kinase inhibitors in KCGS. Half of the set was selected from PKIS and PKIS2. An additional third of the set was sourced from multiple pharmaceutical companies. The remaining inhibitors originated from academic laboratories. C) Comparison of the range of kinase selectivities of the inhibitors in PKIS, PKIS2, and KCGS. Compounds were binned by their calculated kinase selectivity index (S10 at 1 μM) from data collected in the Nanosyn, KINOMEscan,or other large kinase panels. KCGS contains only narrow spectrum inhibitors with a selectivity index S10 < 0.05. PKIS2 contains many promiscuous inhibitors with S10 > 0.05. i.a. indicates no kinases inhibited by 90% at 1 μM. PKIS contains many inhibitors with < 90% inhibition of kinase activity at 1 μM.
KCGS v1.0 is a much higher quality chemogenomic set than either PKIS or PKIS2. Nominally the sets cover a similar number of kinases, but KCGS does so with potent and selective inhibitors (Figure 3C). In PKIS and PKIS2, many of the dark kinases were only covered by promiscuous inhibitors, which made it difficult to assign a phenotype to an individual kinase. In addition, the inhibitors in PKIS were often only active at higher concentrations. KCGS contains many more useful inhibitors than either PKIS or PKIS2, which will greatly aid the deconvolution of screening data and the association of disease biology to specific kinases. Expansion of KCGS towards our goal of covering all human kinases is an ongoing project. Additional inhibitors donated by our collaborators to improve the set will be added in subsequent releases. We are also working with the NIH Illuminating the Druggable Genome program26 to identify inhibitors for 160 dark kinases that will fill many of the remaining gaps in the chemogenomic set.
Sustainability of the chemogenomic set remains a key challenge. When PKIS and PKIS2 were distributed directly from GSK, we could honor nearly all requests for compounds. As long as there was a reserve supply of each inhibitor in the GSK compound stores, requests could be filled for almost any quantity of the set free of charge. This led to some exploitation of the system; for example, a third of investigators reported that they had still not used the set within 3 months of receipt. At the SGC-UNC, we are more constrained. We received only a limited supply of PKIS and PKIS2 from GSK, and thus, the sets were distributed with liquid volumes limited to a few microliters to preserve their availability. Likewise, KCGS is available in limited quantity using the compound supply donated by our collaborators. To maintain KCGS as a sustainable resource for open science we have now embarked on a campaign to resynthesize each of the inhibitors in the set to ensure that ample supplies are available for all researchers.
Dark Matter of the Genome
The PKIS experiment set out to identify new kinase drug targets by making available a set of highly annotated chemical tools for the dark kinases. The creation, annotation, and distribution of a public kinase chemogenomic set by a pharmaceutical company broke new ground as a truly open science experiment. In the process, important precedents were established. The knowledge created through use of a public chemogenomic set far exceeded the perceived value of the original chemical matter. Full annotation of the previously published inhibitors was absolutely essential to their utility as chemical tools. Unrestricted screening of the chemogenomic set led to ideas being tested in a diverse range of assays that could never be conceived and carried out within a single organization. The set was also put into the hands of external scientists with technical expertise not always available internally. Important lessons were learned on the design of chemogenomic tools that have been applied to the creation of a new and improved kinase set, KCGS. How then will the value of this open science experiment be measured? Although, it has not yet led to a renaissance in kinase drug discovery at GSK, we would argue that the reputational value has been enormous. GSK is recognized as a leader in open drug discovery and has continued to pioneer efforts in open science through its support of Open Targets and the Tres Cantos Open Lab Foundation as two examples.27 More importantly, even if the kinase-disease connections uncovered by use of the chemogenomic set leads to only a single new drug being developed, it will have returned immeasurable value to patients. Ultimately, society will benefit from the sharing of knowledge, ideas, and research tools even if no single company reaps any direct financial benefit.
Can the chemogenomics concept be applied more broadly to proteins beyond kinases? Analysis of all major protein families shows that the majority of research occurs on only a small fraction of each family; kinases are not unique in this respect. Indeed, there are over 4000 proteins that belong to one of the families of chemically tractable proteins.28 Examples include G-protein coupled receptors, ion channels, solute carrier proteins, histone methyl transferases, acetyl lysine reader domains, and hydrolase enzymes. In each of these protein families, one or more small molecule drugs have already been discovered. Creation of public chemogenomic sets for these druggable protein families through a coordinated effort by academic institutions and pharmaceutical companies could deliver hundreds of new drug targets from the dark matter of the genome. Such an endeavor would yield a biomedical tool kit of immense value to society.
Acknowledgements
The open science kinase chemogenomics experiment would not have succeeded without a large community of supporters. The following individuals were critical to the conception, assembly, and distribution of PKIS and PKIS2: Francis Atkinson (ChEMBL), Paul Bamborough (GSK), Louisa Bellis (ChEMBL), Barbara Carter (GSK), Jon Elkins (SGC), Brian Hardy (GSK), Stefan Knapp (SGC), Karen Lackey (GSK), John Lemanowicz (GSK), Jowita Mikolajczyk (Nanosyn), John Overington (ChEMBL), Cathy Pieshoff (GSK), Daniel Price (GSK), Sergei Romanov (Nanosyn), and Sir Andrew Witty (GSK). Individuals who facilitated the move of our laboratory to the UNC Eshelman School of Pharmacy: Dean Robert Blouin (UNC), Al Edwards (SGC), Fred Eshelman (EII), and Stacey Jones (GSK). Critical contributions to the design, planning, annotation and assembly of KCGS: Hassan Al-Ali (U. Miami), David Andrews (AstraZeneca), Richard Angell (UCL), Alison Axtman (UNC), Mark Bunnage (Pfizer), Carolyn Buser-Doepner (GSK), Stephen Capuzzi (UNC), Rajiah Aldrin Denny (Pfizer), Peter Ettmayer (Boehringer Ingelheim), Maryann Feldman (UNC), Christian Fischer (Merck), Matthias Frederiksen (Novartis), Nathanael Gray (Harvard), Alice Hooper (UCL), Opher Gileadi (SGC), Ulrick Luecking (Bayer), Mike Michaelides (Abbvie), Susanne Müller-Knapp (SGC), Eugene Muratov (UNC), Dafydd Owen (Pfizer), Jackie Quay (UNC), Matt Robers (Promega), Kumar Singh Saikatendu (Takeda), Dan Treiber (DiscoverX), plus the students and staff at the SGC-UNC. For assistance in the preparation of Figure 3: Shawn Gomez (UNC). For critical review of the manuscript: Cheryl Arrowsmith (SGC), Al Edwards (SGC), Dave Morris (UNC), Caroline Slade (UNC), and Mya Willson. We apologize to anyone whom we have inadvertently overlooked. Funding for the SGC-UNC was provided by The Eshelman Institute for Innovation, UNC Lineberger Comprehensive Cancer Center, PharmAlliance, and National Institutes of Health (1R44TR001916–02, 1R01CA218442–01, and 1U24DK116204–01). The SGC is a registered charity that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (ULTRA-DD 115766), Wellcome Trust, Janssen, Merck Kga, Merck Sharp & Dohme, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, and Takeda.
Footnotes
GSK has brought four small molecule kinase inhibitors to market as cancer therapeutics: lapatinib, pazopanib, trametinib, and dabrafenib.
“There are two kinds of people and organizations in the world: eaters and bakers. Eaters want a bigger slice of an existing pie; bakers want to make a bigger pie. Eaters think that if they win, you lose, and if you win, they lose. Bakers think that everyone can win with a bigger pie.”
Selectivity S10 at 1 μM < 0.05, where S10 is the number of kinases with 90% inhibition (or 10% activity remaining) divided by the number of kinases tested.
KCGS can be requested from the SGC at https://www.sgc-unc.org/request-kcgs/
References
- 1.The SGC Research Informatics . https://openlabnotebooks.org/ (accessed Nov 30, 2018).
- 2.Fedorov O; Müller S; Knapp S The (Un)targeted Cancer Kinome. Nat Chem Biol 2010, 6, 166–169. [DOI] [PubMed] [Google Scholar]
- 3.Edwards AM; Isserlin R; Bader GD; et al. Too Many Roads not Taken. Nature 2011, 470, 163–5. [DOI] [PubMed] [Google Scholar]
- 4.Stoeger T; Gerlach M; Morimoto RI; et al. Large-Scale Investigation of the Reasons why Potentially Important Genes are Ignored. PLoS Biol 2018, 16, e2006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunnage ME; Chekler EL; Jones LH Target Validation using Chemical Probes. Nat Chem Biol 2013, 9, 195–9. [DOI] [PubMed] [Google Scholar]
- 6.Caron PR; Mullican MD; Mashal RD; et al. Chemogenomic Approaches to Drug Discovery. Curr Opin Chem Biol 2001, 5, 464–70. [DOI] [PubMed] [Google Scholar]
- 7.Rognan D Chemogenomic Approaches to Rational Drug Design. Br J Pharmacol 2007, 152, 38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capdeville R; Buchdunger E; Zimmermann J; et al. Glivec (STI571, imatinib), a Rationally Developed, Targeted Anticancer Drug.Nat Rev Drug Discov 2002, 1, 493–502. [DOI] [PubMed] [Google Scholar]
- 9.Bamborough P; Drewry D; Harper G; et al. Assessment of Chemical Coverage of Kinome Space and its Implications for Kinase Drug Discovery. J Med Chem 2008, 51, 7898–914. [DOI] [PubMed] [Google Scholar]
- 10.Posy SL; Hermsmeier MA; Vaccaro W; et al. Trends in Kinase Selectivity: Insights for Target Class-focused Library Screening. J Med Chem 2011, 54, 54–66. [DOI] [PubMed] [Google Scholar]
- 11.McKiernan EC; Bourne PE; Brown CT; et al. How Open Science Helps Researchers Succeed.Elife 2016, 5, e16800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkins JM; Fedele V; Szklarz M; et al. Comprehensive Characterization of the Published Kinase Inhibitor Set. Nat Biotechnol 2016, 34, 95–103. [DOI] [PubMed] [Google Scholar]
- 13.European Bioinformatics Institute Accessed November 30th, 2018. https://www.ebi.ac.uk/chembldb/extra/PKIS/
- 14.Jones LH; Bunnage ME Applications of Chemogenomic Library Screening in Drug Discovery. Nat Rev Drug Discov 2017, 16, 285–296. [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki G Enchantment: the Art of Changing Hearts, Minds and Actions. Portfolio Penguin: London, 2011. [Google Scholar]
- 16.Drewry DH; Willson TM; Zuercher WJ Seeding Collaborations to Advance Kinase Science with the GSK Published Kinase Inhibitor Set (PKIS). Curr Top Med Chem 2014, 14, 340–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witty A New Strategies for Innovation in Global Health: a Pharmaceutical Industry Perspective. Health Aff (Millwood) 2011, 30, 118–26. [DOI] [PubMed] [Google Scholar]
- 18.Knapp S; Arruda P; Blagg J; et al. A Public-Private Partnership to Unlock the Untargeted Kinome.Nat Chem Biol 2013, 9, 3–6. [DOI] [PubMed] [Google Scholar]
- 19.Gamo FJ; Sanz LM; Vidal J; et al. Thousands of Chemical Starting Points for Antimalarial Lead Identification. Nature 2010, 465, 305–10. [DOI] [PubMed] [Google Scholar]
- 20.Al-Ali H; Lee DH; Danzi MC; et al. Rational Polypharmacology: Systematically Identifying and Engaging Multiple Drug Targets To Promote Axon Growth.ACS Chem Biol 2015, 10, 1939–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheipl S; Barnard M; Cottone L; et al. EGFR Inhibitors Identified as a Potential Treatment for Chordoma in a Focused Compound Screen. J Pathol 2016, 239, 320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiterer V; Eyers PA; Farhan H Day of the Dead: Pseudokinases and Pseudophosphatases in Physiology and Disease. Trends Cell Biol 2014, 24, 489–505. [DOI] [PubMed] [Google Scholar]
- 23.Foulkes DM; Byrne DP; Yeung W; et al. Covalent Inhibitors of EGFR Family Protein Kinases Induce Degradation of Human Tribbles 2 (TRIB2) Pseudokinase in Cancer Cells.Sci Signal 2018, 11, eaat7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Axtman AD; Counago R; Drewry DH; et al. Drugging the Kinome. In Kinase Drug Discovery: Modern Approaches; 1st ed; Ward RA; Goldberg FW , ; RSC Drug Discovery Series; Royal Society of Chemistry: London, 2019; Chapter 10, pp. 253–280. [Google Scholar]
- 25.Drewry DH; Wells CI; Andrews DM; et al. Progress Towards a Public Chemogenomic Set for Protein Kinases and a Call for Contributions.PLoS One 2017, 12, e0181585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illuminating the Druggable Genome . https://druggablegenome.net/ (accessed Nov 30, 2018). [Google Scholar]
- 27.Koscielny G; An P; Carvalho-Silva D; et al. Open Targets: a Platform for Therapeutic Target Identification and Validation. Nucleic Acids Res 2017, 45, D985–D994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finan C; Gaulton A; Kruger FA; et al. The Druggable Genome and Support for Target Identification and Validation in Drug Development.Sci Transl Med 2017, 9, eaag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]