Summary
It is well established that ferroptosis is primarily controlled by glutathione peroxidase 4 (GPX4). Surprisingly, we observed that p53 activation modulates ferroptotic responses without apparent effects on GPX4 function. Instead, ALOX12 inactivation diminishes p53-mediated ferroptosis induced by ROS stress and abrogates p53-dependent inhibition of tumor growth in xenograft models, suggesting that ALOX12 is critical for p53-mediated ferroptosis. The ALOX12 gene resides on human chromosome 17p13.1, a hot spot of monoallelic deletion in human cancers. Loss of one ALOX12 allele is sufficient to accelerate tumorigenesis in Eμ-Myc lymphoma models. Moreover, ALOX12 missense mutations from human cancers abrogate its ability to oxygenate polyunsaturated fatty acids and to induce p53-mediated ferroptosis. Notably, ALOX12 is dispensable for ferroptosis induced by erastin or GPX4 inhibitors; conversely, ACSL4 is required for ferroptosis upon GPX4 inhibition but dispensable for p53-mediated ferroptosis. Thus, our study identifies an ALOX12-mediated, ACSL4-independent ferroptosis pathway that is critical for p53-dependent tumor suppression.
Keywords: p53, ferroptosis, GPX4, ROS, ALOX12, oxidative stress, tumor suppressor
Although p53-mediated cell-cycle arrest, senescence and apoptosis serve as critical barriers to cancer development, accumulating evidence reveals that p53-mediated metabolic regulation also promotes tumor suppression1. We and others recently found that p53 plays an important role in modulating ferroptotic responses through its metabolic targets2-6. Nevertheless, the molecular factors that mediate p53-dependent ferroptosis have not been delineated and the mechanism by which this ferroptosis pathway is regulated remains unclear. Ferroptosis is a regulated form of cell death driven by excess accumulation of lipid peroxides7-10. Lipid peroxides are normally eliminated by glutathione peroxidase 4 (GPX4) and its co-factor glutathione (GSH), which convert lipid hydroperoxides to non-toxic lipid alcohols8-10. It is well established that ferroptosis is primarily controlled by glutathione peroxidase 4 (GPX4) 11-19. Although inactivation of p53 expression can partially reduce erastin-initiated cell death in certain cell types2-6, the ferroptotic responses induced by erastin or GPX4 inhibitors are not dependent on p53 status4. Thus, it remains unclear whether p53-dependent ferroptosis acts through modulation of GPX4 function.
Results
Identification of ALOX12 as an essential factor of p53-dependent ferroptosis.
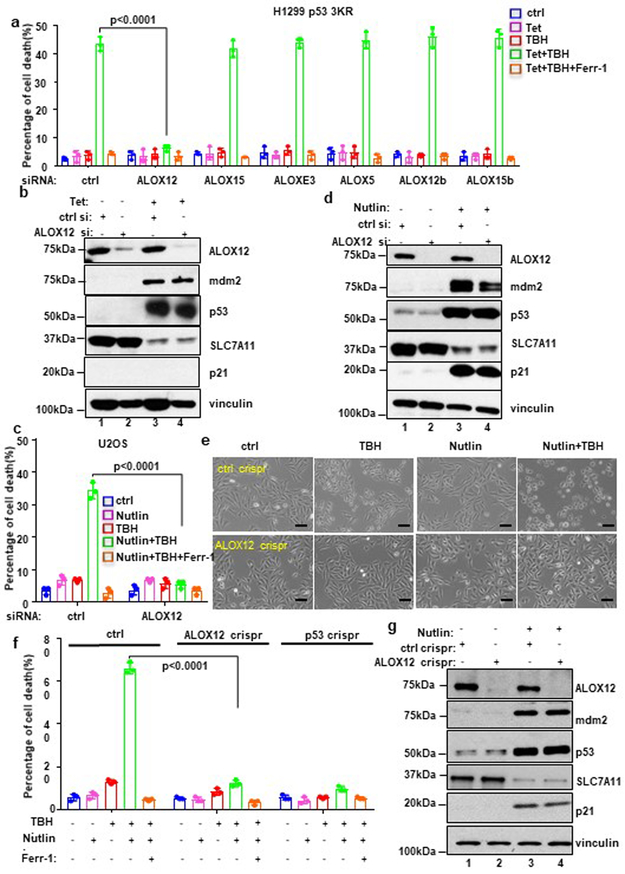
In our previous study, we established a ferroptosis assay that requires both p53 activation and reactive oxygen species (ROS)-induced stress (see methods)3. Indeed, the cell death induced by low levels of tert-Butyl hydroperoxide (TBH), a common ROS generator, is apparently p53-dependent and can be specifically inhibited by the ferroptosis inhibitor Ferr-1, but not by the inhibitors of other cell death pathways such as apoptosis, autophagy or necroptosis3. Since ferroptotic cell death is tightly regulated in response to oxidative stress in vivo8, we reasoned that this type of ferroptosis would better reflect p53 function during oxidative stress response. In addition to GPX4-mediated neutralization of lipid peroxidation11-18, the levels of cellular lipid peroxides can be induced enzymatically by the lipoxygenases. The mammalian lipoxygenase family consists of six isoforms (ALOXE3, ALOX5, ALOX12, ALOX12B, ALOX15, and ALOX15B) (Supplementary Fig. 1a) with differing substrate specificities8,10,19. To ascertain whether any of these lipoxygenases are required for p53-mediated ferroptosis, we performed an RNAi-mediated loss-of-function screen to test whether depletion of individual isoforms affects p53-dependent ferroptosis. qPCR analyses confirmed that expression of each of the six lipoxygenase isoforms was individually abrogated by RNAi-mediated depletion (Supplementary Fig. 1b). As expected, high levels of p53-mediated ferroptosis upon ROS-induced stress were detected in TBH-treated p53-3KR H1299 cells transfected with control siRNAs. However, ferroptosis was markedly and specifically blocked by RNAi-mediated depletion of ALOX12, but not by any of the other five lipoxygenases (Fig. 1a). Western analysis revealed that ALOX12 depletion had no effect on either p53 levels or p53-mediated downregulation of SLC7A11 (Fig. 1b).
Figure 1. ALOX12 is essential for p53-mediated ferroptosis upon ROS stress.
a) H1299 Tet-on p533KR cells transfected with control siRNA (ctrl) or a pool of ALOX family specific siRNAs pre-incubated with doxycycline (0.5μg/ml; Tet) for 12h, then treated with doxycycline (0.5ug/ml; Tet) and TBH (40uM) as indicated for 8h. Error bars are mean ± s.d., n=3 independent experiments.
b) Western blot analysis of H1299 Tet-on p533KR cells transfected with control or ALOX12 siRNA and then treated with doxycycline (0.5μg/ml) as indicated for 24h. The experiments were repeated twice, independently, with similar results.
c) U2OS cells transfected with control or ALOX12 siRNA were pre-incubated with Nutlin (10uM) for 12h, then the cells were treated with Nutlin (10uM) and TBH (300uM) as indicated for 8h. Error bars are mean ± s.d., n=3 independent experiments.
d) Western blot analysis of U2OS cells transfected with control or ALOX12 siRNA and then treated with Nutlin (10uM) as indicated for 48h. The experiments were repeated twice, independently, with similar results.
e) Representative phase-contrast images of U2OS ctrl crispr and ALOX12 crispr cells pre-incubated with Nutlin (10uM) for 12h were treated with TBH (300uM) and Nutlin (10uM) as indicated for 8h. Scale bars, 100μm. The experiments were repeated twice, independently, with similar results.
f) U2OS control, ALOX12, and p53 crispr cells pre-incubated with Nutlin (10uM) for 12h were treated with TBH (300uM), Nutlin (10uM), and Ferr-1 (2uM) as indicated for 8h. Error bars are mean ± s.d., n=3 independent experiments.
g) Western blot analysis of U2OS control and ALOX12 crispr cells treated with Nutlin (10uM) as indicated for 48h. The experiments were repeated twice, independently, with similar results. All P values (a,c,f) were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. Raw data are provided in Supplementary Table 1.
Activation of p53 can be readily induced by Nutlin, a small molecular inhibitor of Mdm2. While in most human cancer cells, Nutlin-mediated p53 activation induces reversible cell cycle arrest but not cell death, we previously showed that U2OS cells undergo p53-dependent, ROS-stress induced ferroptosis upon combined treatment with both Nutlin and TBH3. Again, although ALOX12 depletion had no obvious effect on p53 levels or expression of its transcriptional targets (e.g., SLC7A11, Mdm2, and p21) (Fig. 1d), p53-mediated ferroptosis was largely abrogated (Fig. 1c). To further validate the role of ALOX12 in p53-mediated ferroptosis, we used CRISPR/Cas9 technology to generate ALOX12 knockout subclones of U2OS. Indeed, ferroptosis was suppressed in ALOX12-knockout U2OS cells (Fig. 1e. 1f; Supplementary Fig. 1c, 1d and 1e). Taken together, these data indicate that ALOX12 is required for p53-dependent ferroptosis.
GPX4 inhibition is not required for p53-mediated ferroptosis upon ROS stress
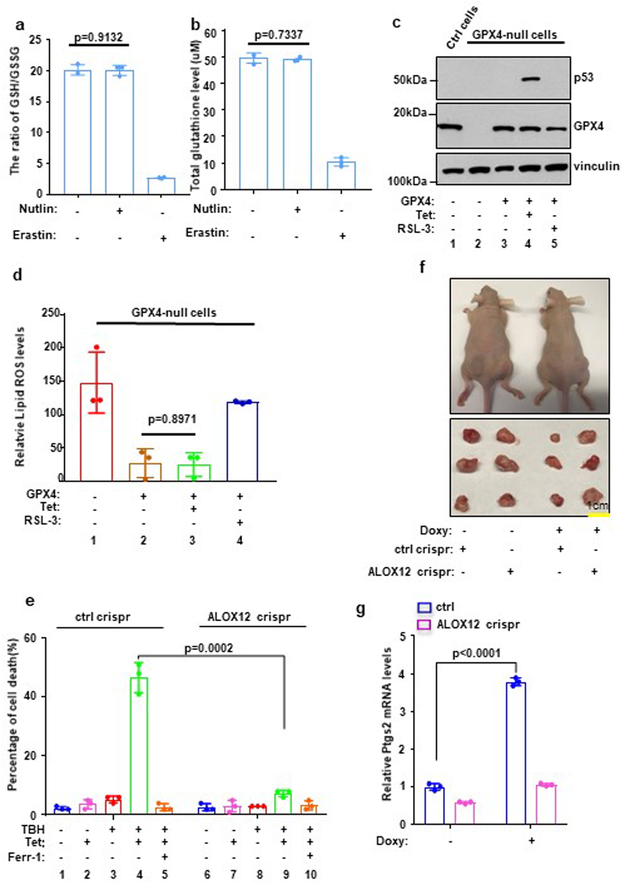
Interestingly, a systematic genome-wide screen failed to identify any lipoxygenase enzyme(s) required for ferroptosis induced by GPX4 inhibitors or erastin20. Thus, our finding that ALOX12 is specifically required for p53-mediated ferroptosis suppression is surprising and suggests that p53-mediated ferroptosis may act through a different mechanism. Lipid peroxides are normally eliminated by glutathione peroxidase 4 (GPX4) and its co-factor glutathione (GSH) and ferroptosis can be induced by pharmacological agents including GPX4 enzymatic inhibitors and erastin, which induces glutathione depletion and consequent GPX4 inactivation. To this end, we first examined the ratio of GSH/GSSG and levels of glutathione upon p53 activation. As shown in Fig. 2a and 2b, the ratio of reductive GSH to oxidative GSSG and the levels of glutathione were dramatically reduced upon erastin treatment; conversely, upon p53 activation by the treatment with Nutlin, no obvious effect was detected. Consistent with these observations, activation of p53 by nutlin treatment failed to suppress glutathione peroxidase activity (Supplementary Fig. 2a) and had no obvious effect on endogenous lipid peroxidation (Supplementary Fig. 2b and 2c). Nevertheless, increasing lipid peroxidation levels were detected upon combined treatment with both Nutlin and TBH (Supplementary Fig. 2b and 2c).
Figure 2. ALOX12 is critical for p53-mediated tumor growth suppression.
a) The ratio of reductive GSH to oxidative GSH was measured by GSH/GSSG quantification kit in U2OS cells with or without Nutlin (10uM) or Erastin (10uM) treatment. Error bars are mean ± s.d., n=3 independent experiments.
b) Total glutathione levels were measured by GSSG/GSH quantification kit in U2OS cells with or without Nutlin (10uM) or Erastin (10uM) treatment.
c) Western blot analysis of H1299 Tet-on p533KR GPX4 crispr cells transfected with GPX4 expression vectors. The experiments were repeated twice, independently, with similar results.
d) Quantification of lipid peroxidation levels from Fig. S2d is shown in H1299 Tet-on p533KR control or ALOX12 crispr clones pre-incubated with doxycycline (0.5μg/ml) were treated with doxycycline (0.5μg/ml), and RSL-3 (1uM) as indicated for 8h. Error bars are mean ± s.d., n=3 independent experiments.
e) H1299 Tet-on p533KR control or ALOX12 crispr clones pre-incubated with doxycycline (0.5ug/ml) were treated with doxycycline (0.5ug/ml) and TBH (60uM) as indicated for 8h. Error bars are mean ± s.d., n=3 independent experiments.
f) Xenograft tumors from H1299 Tet-on p533KR control and ALOX12 crispr cells fed with regular or doxycycline-containing chow (625mg kg−1) as indicated (see methods). The experiments were repeated twice, independently, with similar results.
g) Q-PCR of PTGS2 from tumors harvested in f; Error bars are mean ± s.d., n=3 independent experiments. All P values (a,d,e,g) were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. Raw data are provided in Supplementary Table 1.
To further support this notion, we examined whether p53 activation has any effect on GPX4-mediated activity of endogenous lipid peroxidation levels. To this end, we first established GPX4-null p53-tet-on H1299 cells by Crispr-mediated knockout (Fig. 2c) and then analyzed the levels of endogenous lipid peroxidation by flow cytometry with C11-BODIPY staining. As shown in Fig. 2d, high levels of endogenous lipid peroxidation were detected in the GPX4 null cells whereas upon GPX4 expression ectopically, the levels of lipid peroxidation were reduced dramatically (also see Supplementary Fig. 2d and 2e). Thus, the drastic reduction of lipid peroxidation levels represents the activity of GPX4 on endogenous lipid peroxidation. As expected, the treatment of a known GPX4 inhibitor RSL-3, largely abrogated the effects on lipid peroxidation reduction induced by GPX4 in those cells; however, activation of p53 failed to induce any obvious effect (Fig. 2d). Taken together, these data suggest that p53-dependent activation of ferroptosis may act through a distinct pathway, independent of GPX4 modulation.
Inactivation of ALOX12 abrogates p53-mediated tumor growth suppression.
Using xenograft tumor models in mice, we previously showed that the tumor suppression activity of p53-3KR, which is defective for the canonical p53 tumor suppression functions (cell cycle arrest, apoptosis, and senescence), is instead dependent on p53-mediated ferroptosis3. To ascertain whether ALOX12 is also required for p53-mediated tumor suppression in this setting, we first established isogenic lines of tet-inducible p53-3KR cells in which the ALOX12 gene has or has not been knocked out by CRISPR/Cas9 technology (Supplementary Fig. 3a). Indeed, the levels of ferroptosis were dramatically diminished in ALOX12-knockout cells (Fig. 2e, Supplementary Fig. 3b and 3c). To examine whether ALOX12 contributes to the tumor suppression activity of p53, we tested whether loss of ALOX12 expression affects tumor cell growth suppression by p533KR in xenograft tumor models. As expected, tetracycline-induced expression of p533KR markedly reduced tumor cell growth in this assay (Fig. 2f, Supplementary Fig. 3d and 3e); however, the tumor suppression effects of p533KR were ablated in ALOX12-knockout cells. Notably, up-regulation of Ptgs2, a marker of ferroptosis18, was also abrogated in ALOX12-knockout cells (Fig. 2g). These data demonstrate that ALOX12 is crucial for the tumor cell growth suppression activity of p53 in the absence of p53-mediated cell cycle arrest, apoptosis, and senescence.
Loss of one ALOX12 allele is sufficient to abrogate p53-mediated ferroptosis and accelerate tumorigenesis.
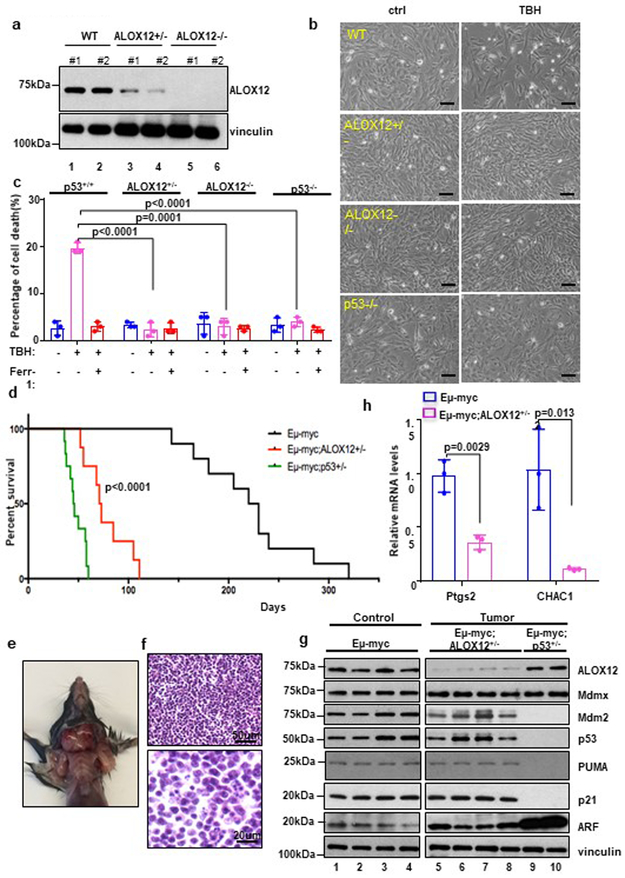
To further validate the role of ALOX12 in p53-mediated ferroptotic responses, we compared the sensitivity of isogenic ALOX12 mutant MEFs to p53-mediated ferroptosis. As shown in Figure 3a, ALOX12 protein levels were undetectable in ALOX12−/− MEFs and significantly reduced in ALOX12+/− MEFs. As expected, ferroptosis was readily induced by TBH in wild type, but not in p53-null, MEFs under the same conditions (Fig. 3b and 3c), indicating that TBH-induced ferroptosis is largely p53-dependent under these conditions. Strikingly, however, cell death was not observed in either homozygous (ALOX12−/−) or heterozygous (ALOX12+/−) MEFs. These data indicate that ALOX12 is haploinsufficient with respect to p53-mediated ferroptosis.
Figure 3. Loss of one ALOX12 allele is sufficient to accelerate tumorigenesis in Eμ-Myc lymphoma models.
a) Western blot analysis of wild-type (WT), ALOX12+/−, and ALOX12−/− MEFs. Two representative MEF cell lines for each genotype shown. The experiments were repeated twice, independently, with similar results.
b) Representative phase-contrast images of WT, ALOX12+/−, ALOX12−/−, and p53−/− MEFs treated with TBH (80uM) as indicated for 12h. Scale bars, 100μm. The experiments were repeated three times, independently, with similar results.
c) WT, ALOX12+/−, ALOX12−/−, and p53−/− MEFs treated with TBH (80uM) as shown in b. Error bars are mean ± s.d., n=3 independent experiments.
d) Kaplan-Meier survival curves of Eμ-myc (n=10 independent mice), Eμ-myc; ALOX12+/− (n=8 independent mice), and Eμ-myc; p53+/− (n=12 independent mice). P value (Eμ-myc versus Eμ-myc; ALOX12+/− background) was calculated using log-rank Mantel-Cox test.
e) Representative image of an early onset lymphoma in a 45 day-old Eμ-myc; ALOX12+/− mouse. The experiments were repeated three times, independently, with similar results.
f) Representative H&E of a high grade diffuse B-cell lymphoma developed from Eμ-myc; ALOX12+/− mouse (Upper panel scale bars, 50μm; Lower panel scale bars, 20μm). The experiments were repeated three times, independently, with similar results.
g) Western blot analysis of Eμ-myc control samples and tumor samples from Eμ-myc; ALOX12+/− and Eμ-myc; p53+/− mice. The experiments were repeated twice, independently, with similar results.
h) Q-PCR of PTGS2 from tumors harvested from of Eμ-myc control samples (n=3 independent samples) and tumor samples (n=3 independent samples) from Eμ-myc; ALOX12+/− mice. Error bars are mean ± s.d., n=3 independent experiments and P values(c,h) were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. Raw data are provided in Supplementary Table 1.
To examine whether ALOX12 also affects tumor suppression in a haploinsufficient manner, we tested whether loss of one ALOX12 allele impacts Myc-induced tumorigenicity in the classic Eμ-Myc lymphoma model. Previous studies have shown that Eμ-Myc mice develop late-onset B-cell lymphomas between 6 and 12 months of age21-23, and that tumor formation is markedly accelerated upon loss of one p53 allele (in Eμ-Myc/p53+/− mice)21. Remarkably, we observed that the latency of tumor formation in Eμ-Myc mice (median tumor-free survival ~ 220 days) is also dramatically reduced by the absence of one ALOX12 allele (Eμ-Myc/ALOX12+/− mice; median survival ~70 days) (Fig. 3d). Of note, Eμ-Myc/ALOX12+/− mice presented with severe lymphadenopathy (Fig. 3e) and developed high-grade diffuse B-cell lymphomas (Fig. 3f) with a latency similar to that of Eμ-Myc/p53+/− mice (median tumor-free survival ~50 days), indicating that ALOX12-dependent ferroptosis contributes significantly to p53-mediated tumor suppression in the Eμ-Myc lymphoma model.
Although the lymphomas that arise in Eμ-Myc mice often exhibit defects in the ARF-p53 tumor suppression pathway21-23, neither the ARF nor p53 gene was deleted or mutated in tumors derived from Eμ-Myc/ALOX12+/− mice (Fig. 3g), which was further validated by sequencing analysis (Supplementary Fig. 4a). Likewise, the major p53 feedback inhibitors, Mdm2 and Mdmx, were not overexpressed in these tumors (Fig. 3g). Loss of functional p53 through the two-hit mechanism is among the most frequent events in human cancers and a ‘loss of heterozygosity’ (LOH) deletion is often associated with tumor development24. Indeed, LOH deletions of the p53 gene were confirmed in tumors derived from Eμ-Myc/p53+/− mice (Fig. 3g; Supplementary Fig. 4a). In contrast, however, LOH deletions of ALOX12 gene were not observed in tumors derived from Eμ-Myc/ALOX+/− mice (Fig. 3g; Supplementary Fig. 4b). Notably, the levels of Ptgs2 and CHAC1, two known markers of ferroptosis, were significantly downregulated in the tumors of Eμ-Myc/ALOX12+/− mice relative to the tumors of Eμ-Myc control mice (Fig. 3h); in contrast, the levels of p21 and PUMA, two well-known markers of p53-mediated cell cycle arrest and apoptosis, remained the same (Fig. 3g), suggesting that loss of p53-dependent ferroptosis is the key event during tumor development. Indeed, inactivation of ALOX12 had no obvious effect on p53-mediated cell cycle arrest and apoptosis (Supplementary Fig. 4c, 4d, 4e). Taken together, these findings demonstrate that loss of one ALOX12 allele is sufficient to accelerate Myc-induced tumorigenesis in a haploinsufficient manner even when the canonical functions of the p53 pathway in cell cycle arrest and apoptosis remain intact.
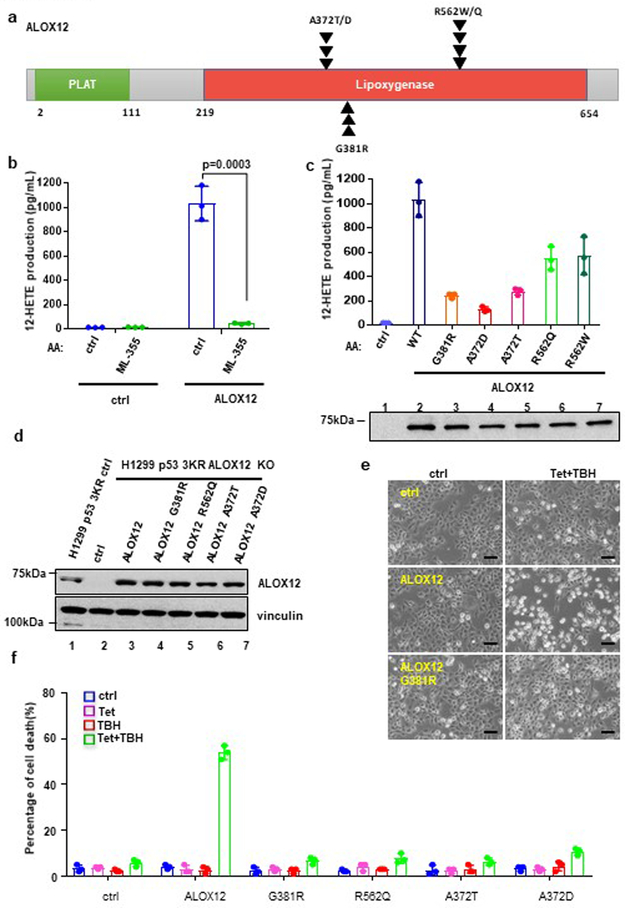
ALOX12 missense mutations from human cancers abrogate ALOX12 enzymatic activity and p53-mediated ferroptosis.
Since monoallelic loss of ALOX12 impairs both p53-mediated ferroptosis and p53-mediated tumor suppression, the above data suggest that ALOX12 itself functions as a haploinsufficient tumor suppressor. Consistent with this notion, examination of the Oncomine data base reveals that ALOX12 expression is down-regulated in a variety of human cancers, including cervical squamous cell carcinoma, head and neck squamous cell carcinoma, esophageal squamous cell carcinoma, and acute myeloid leukemia (Supplementary Fig. 5a and 5b). In addition, several ALOX12 missense mutations are present in the COSMIC data base of human tumor specimens, including R562Q/W (R→Q/or W mutation at amino acid 562), A372T/D (A→T or D mutation at amino acid 372) and G381R (G→R mutation at amino acid 381) (Fig. 4a, Supplementary Table 2). Since the affected residues are evolutionally conserved and localized within the lipoxygenase domain (Fig. 4a; Supplementary Fig. 5d), these mutations may have functional consequence. To this end, we first examined the effects of these mutations on lipoxygenase activity. As shown in Figure 4b, wildtype ALOX12 protein readily oxidized arachidonic acid (AA) in vitro and, as expected, its lipoxygenase activity was inhibited by ML-355, a specific ALOX12 inhibitor25. Moreover, p53-mediated ferroptosis is blocked by ML-355 (Supplementary Fig. 5e and 5f). Notably, this lipoxygenase activity was significantly impaired by each of the tumor-derived missense mutations tested (G381R, R562Q, R562W, A372T, and A372D) (Fig. 4c). Moreover, p53-mediated ferroptosis in ALOX12-null U2OS cells was restored by ectopic expression of wildtype ALOX12 but not by the tumor-derived ALOX12-G381R mutant (Fig. 4d and Supplementary 5g). Similar data was also obtained in ALOX12-null H1299 cells and further analysis showed that none of these tumor-derived mutants was able to fully induce cell death under the same conditions (Fig. 4e, 4f). Taken together, these data demonstrate that the ALOX12 gene is downregulated and mutated in human cancers, and that tumor-derived ALOX12 missense mutations are defective in both lipoxygenase activity and the ability to induce p53-mediated ferroptosis.
Figure 4. ALOX12 missense mutations from human cancers abrogate ALOX12 enzymatic activity and p53-mediated ferroptosis.
a) Schematic diagram of ALOX12 with frequent tumor mutations identified in the Lipoxygenase domain identified from the COSMIC database.
b) An in vitro catalytic activity assay of ALOX12 was measured by detecting 12-HETE levels (lipoxygenase activity) by ELISA. Highly purified ALOX12 was incubated with arachidonic acid and ML-355 (ALOX12 inhibitor) as indicated (see methods). Error bars are mean ± s.d., n=3 independent experiments.
c) An in vitro catalytic activity assay of ALOX12, as described in b, using indicated ALOX12 tumor mutants as shown in a. Western blot analysis of highly purified ALOX12 proteins (lower panel). The Western blot experiments were repeated twice, independently, with similar results. Error bars are mean ± s.d., n=3 independent experiments.
d) Western blot analysis of H1299 Tet-on p533KR ALOX12 crispr cells transfected with control and ALOX12 WT or mutant vectors as indicated. The experiments were repeated twice, independently, with similar results.
e) Representative phase-contrast images of H1299 Tet-on p533KR ALOX12 crispr cells transfected with control, ALOX12, or mutant ALOX12 G381R vectors. Cells were pre-incubated with doxycycline (0.5μg/ml) for 12h, then treated with doxycycline (0.5μg/ml) and TBH (60uM) as indicated for 8h. Scale bars, 100μm. The experiments were repeated twice, independently, with similar results.
f) The cell death of H1299 Tet-on p533KR ALOX12 crispr cells transfected with control or ALOX12 vectors as indicated, from e is shown. Error bars are mean ± s.d., n=3 independent experiments. All P values (b,c,f) were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. Raw data are provided in Supplementary Table 1.
Mechanistic insight into p53-mediated activation of ALOX12.
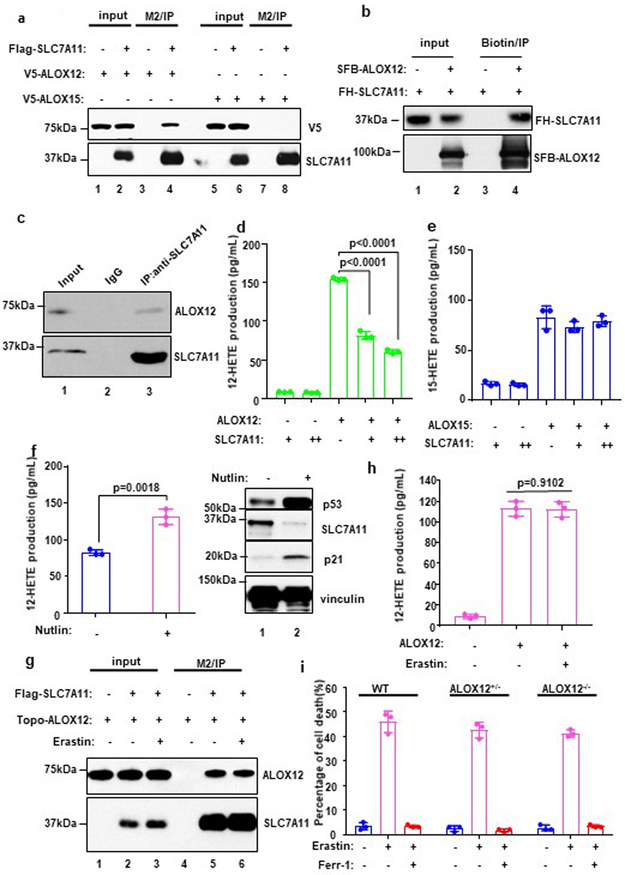
The specific requirement for ALOX12 in p53-dependent ferroptosis raised the possibility that ALOX12 activity may be regulated by p53. Since no obvious effect on ALOX12 levels was detected upon p53 activation (Fig. 1d and Fig. 1g), we examined whether ALOX12 function is modulated by known p53 targets involved in ferroptosis such as SLC7A11. To this end, we first tested whether SLC7A11 interacts with ALOX12. As shown in Figure 5a, ALOX12 (lane 4), but not ALOX15 (lane 8), readily co-immunoprecipitated with Flag-HA tagged SLC7A11 (FH-SLC7A11). Indeed, none of other ALOX family members (ALOXE3, ALOX5, ALOX12B, and ALOX15B) were detectable in the Flag-SLC7A11 immunoprecipitates (Supplementary Fig. 6a, b, c, d), indicating that SLC7A11 interacts specifically with ALOX12. Purified SLC7A11 bound a recombinant ALOX12 in an in vitro pull-down assay (Fig. 5b), indicating that ALOX12 and SLC7A11 interact directly. Moreover, co-immunoprecipitation analysis confirmed that the endogenous ALOX12 and SLC7A11 polypeptides also interact in vivo (Fig. 5c). Together, these data demonstrate that ALOX12 is a bona fide binding partner of SLC7A11.
Figure 5. Mechanistic insight into p53-mediated activation of ALOX12.
a) Western blot analysis of the interaction among SLC7A11, ALOX12 and ALOX15. 293T cells were co-transfected with indicated constructs, and the extract was analyzed by Co-IP assays. The experiments were repeated twice, independently, with similar results.
b) In vitro binding assay between purified SLC7A11 and ALOX12. The experiments were repeated twice, independently, with similar results.
c) Western blot analysis of endogenous interaction between SLC7A11 and ALOX12 in H1299 cells. The experiments were repeated twice, independently, with similar results.
d) An in vivo catalytic activity assay of ALOX12 was measured by detecting 12-HETE levels (lipoxygenase activity) by ELISA. U2OS cells were co-transfected with SLC7A11 and ALOX12. Error bars are mean ± s.d., n=3 independent experiments.
e) An in vivo catalytic activity assay of ALOX15 was measured by detecting 12-HETE levels (lipoxygenase activity) by ELISA. U2OS cells were co-transfected with SLC7A11 and ALOX15. Error bars are mean ± s.d., n=3 independent experiments.
f) An in vivo catalytic activity assay of ALOX12 was measured by detecting 12-HETE levels (lipoxygenase activity) by ELISA. U2OS cells were transfected with ALOX12 upon Nutlin (10uM) treatment or not. Western blot analysis of U2OS cells with or without Nutlin treatment. Error bars are mean ± s.d., n=3 independent experiments. The Western blot experiments were repeated twice, independently, with similar results.
g) Western blot analysis of the interaction between SLC7A11 and ALOX12 with or without Erastin (5uM) treatment. The experiments were repeated twice, independently, with similar results.
h) An in vivo catalytic activity assay of ALOX12 was measured by detecting 12-HETE levels (lipoxygenase activity) by ELISA. U2OS cells were transfected with ALOX12 with or without Erastin (5uM) treatment. Error bars are mean ± s.d., n=3 independent experiments.
i) WT, ALOX12+/−, ALOX12−/− MEFs were treated with Erastin (10uM) and Ferr-1 (2uM) for 12h. Error bars are mean ± s.d., n=3 independent experiments. All P values (d,e,f,h,i) were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. Raw data are provided in Supplementary Table 1.
To elucidate the functional consequence of this interaction, we tested whether SLC7A11 affects ALOX12 lipoxygenase activity. As shown in Figure 5d and 5e, the lipoxygenase activity of ALOX12, but not ALOX15, was suppressed in a dosage-dependent manner by SLC7A11 expression (Supplementary Fig. 6e, 6f). Although the protein structure of SLC7A11 has not been solved yet, it is predicated as a 12-pass transmembrane protein with several domains located inside the cell26. Further analyses demonstrate that the intracellular loop of SLC7A11(a.a:98-131) plays a key role in binding and repressing ALOX12 lipoxygenase activity (Supplementary Fig. 6h, i, and j). Since SLC7A11 levels are downregulated by p53, this observation suggested that p53 can promote ALOX12 function indirectly by downregulating SLC7A11. Indeed, as shown in Figure 5f, SLC7A11 levels were reduced (right panel) and ALOX12 activities elevated (left panel) in human U2OS cells upon Nutlin-induced p53 activation. Together, these data demonstrate 1) that SLC7A11 specifically binds, and inhibits the enzymatic activity of, ALOX12, and 2) that p53 induces ALOX12 lipoxygenase activity by repressing SLC7A11 expression.
ALOX12 is dispensable for ferroptosis induced by erastin or GPX4 inhibitors
To elucidate differences between p53-mediated and erastin-induced ferroptosis, we first examined the impact of erastin on ALOX12 function. Erastin treatment did not affect the interaction between SLC7A11 and ALOX12 (Fig. 5g) or the levels of ALOX12 activity (Fig. 5h). In contrast to p53-mediated ferroptosis, erastin-induced ferroptosis was not significantly affected in isogenic ALOX12+/− and ALOX12−/− MEFs (Fig. 5i, Supplementary Fig. 7a) or ALOX12-null U2OS cells (Supplementary Fig. 7b and 7c). Together, these data demonstrate that ALOX12 is not required for erastin-induced ferroptosis.
Thus, although p53 and erastin both induce ferroptosis by targeting the same factor (SLC7A11), the mechanisms are strikingly different, as exemplified by the fact that the former is ALOX12 dependent while the latter is ALOX12 independent. On one hand, p53 promotes ferroptosis through transcriptional repression of SLC7A11, which in turn releases the lipoxygenase activity of ALOX12 from SLC7A11 inhibition. On the other hand, although erastin suppresses the antiporter activity of SLC7A1127, it fails to abrogate SLC7A11 inhibition of ALOX12. Nevertheless, erastin treatment ultimately induces ferroptosis by blocking cystine import, glutathione biosynthesis, and GPX4 activity7-8. Indeed, most of described ferroptotic responses, including that induced by erastin, entail inhibition of GPX48-10,19. In this regard, we also found that loss of ALOX12 expression has no obvious effect on ferroptosis induced by RSL-3, a specific GPX4 inhibitor (Supplementary Fig. 7d). Thus, the mechanism of p53-dependent ferroptosis is distinct from that of ferroptotic responses induced by erastin or GPX4 inhibitors.
ACSL4 is required for ferroptosis induced by erastin or GPX4 inhibitors but dispensable for p53-mediated ferroptosis.
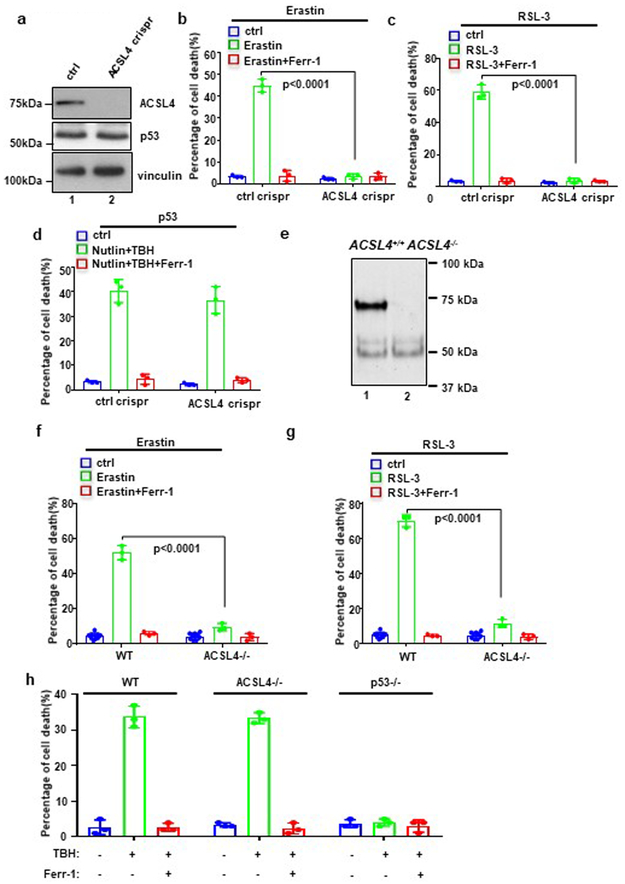
Recent studies show that the ferroptotic responses induced by either erastin or GPX4 inhibitors are dependent on acyl-CoA synthetase long-chain family member 4 (ACSL4), an enzyme that promotes biosynthesis of unsaturated phospholipids, the main substrates for lipid peroxidation28,29. To further elucidate the mechanism of p53-mediated ferroptosis, we generated ACSL4 knockout cell lines of U2OS (Fig. 6a). As expected, ACSL4-null cells are resistant to ferroptosis induced by either erastin (Fig. 6b) or the GPX4 inhibitor RSL-3 (Fig. 6c). In contrast, however, ACSL4-null cells are fully susceptible to p53-mediated ferroptosis (Fig. 6d). Moreover, we generated Acsl4 mutant mice (Supplementary Fig. 8a, 8b and 8c) and obtained Acsl4−/− MEFs for ferroptotic analysis. As expected, ACSL4 protein was not detectable in Acsl4−/− MEFs (Fig. 6e) and the levels of ferroptosis induced by erastin (Fig. 6f) or RSL-3 (Fig. 6g) were largely suppressed. Again, however, p53-dependent ferroptosis was fully retained in Acsl4−/− MEFs (Fig. 6h). Together, these data demonstrate that p53-mediated ferroptosis is ALOX12-dependent but ACSL4-independent.
Figure 6. p53-mediated ferroptosis is ALOX12-dependent but ACSL4-independent.
a) Western blot analysis of U2OS ctrl crispr and ACSL4 crispr cells. The experiments were repeated twice, independently, with similar results.
b) U2OS control, ACSL4 crispr cells treated with Erastin (20uM) and Ferr-1 (2uM) as indicated for 12h. Error bars are mean ± s.d., n=3 independent experiments.
c) U2OS control, ACSL4 crispr cells treated with RSL (4uM) and Ferr-1 (2uM) as indicated for 12h. Error bars are mean ± s.d., n=3 independent experiments.
d) U2OS control, ACSL4 crispr cells pre-incubated with Nutlin (10uM) for 12h were treated with TBH (300uM), Nutlin (10uM) and Ferr-1 (2uM) as indicated for 12h. Error bars are mean ± s.d., n=3 independent experiments.
e) Western blot analysis of WT and ACSL4−/− MEFs. The experiments were repeated twice, independently, with similar results.
f) WT, ACSL4−/− MEFs treated with Erastin (10uM) and Ferr-1 (2uM) as indicated for 9h. Quantification of cell death is shown; Error bars are mean ± s.d., n=3 independent experiments except ctrl n=9.
g) WT, ACSL4−/− MEFs treated with RSL-3 (1uM) and Ferr-1 (2uM) as indicated for 9h. Quantification of cell death is shown; Error bars are mean ± s.d., n=3 independent experiments except ctrl n=9.
h) WT, ACSL4−/− MEFs pre-incubated with Nutlin (10uM) for 12h were treated with TBH (80uM) and Ferr-1 (2uM) as indicated for 9h. Quantification of cell death is shown; Error bars are mean ± s.d., n=3 independent experiments. All P values (b,c,d,f,g,h) were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. Raw data are provided in Supplementary Table 1.
p53-mediated ferroptosis is deregulated in human cancer cell lines expressing wildtype p53.
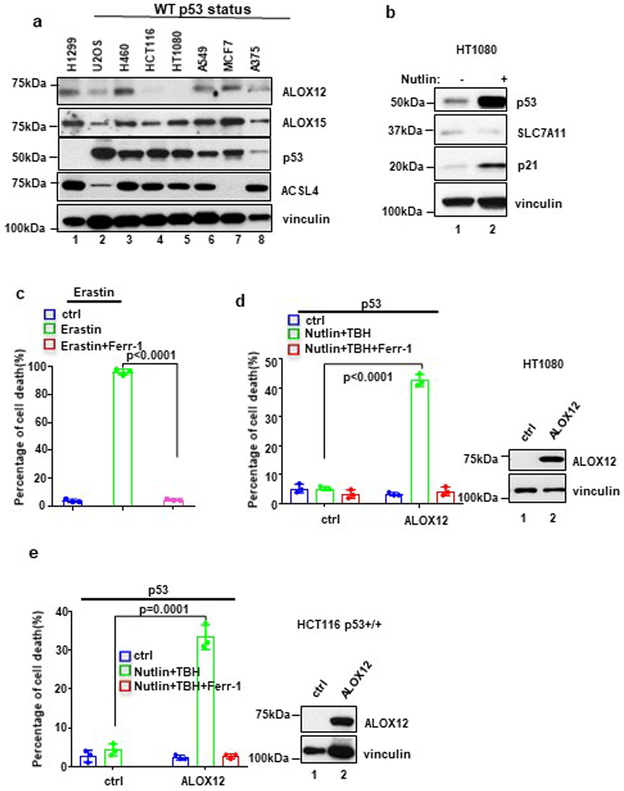
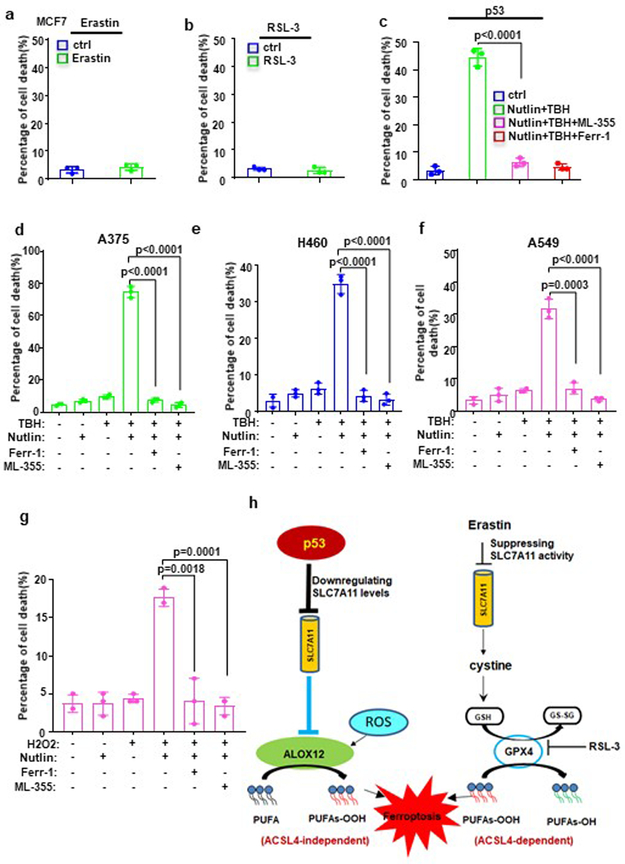
To examine whether p53-mediated ferroptosis is deregulated in human cancers, we examined ALOX12 protein levels in a number of human cancer cell lines that retain wild type p53. As shown Figure 7a, ALOX12 was undetectable in human fibrosarcoma cell line HT1080, the original cell line characterized for erastin-induced ferroptosis7. As expected, ferroptosis was readily induced in the cells by erastin (Fig. 7c). In contrast, HT1080 cells were not susceptible to p53-mediated ferroptosis (left panel, Fig. 7d), despite the fact that SLC7A11 levels were downregulated upon p53 activation (Fig. 7b). However, these cells were rendered susceptible to p53-mediated ferroptosis by ectopic expression of ALOX12 (Fig. 7d). Similar results were also obtained in the human colorectal cancer cell line HCT116 (left panel, Fig. 7e). Of note, it was recently reported that p53-induced ferroptosis was not observed in HCT116 and HT1080 cell lines; instead, p53 stabilization led to repression of erastin-induced ferroptosis in these cells30-31, further validating the specific role of ALOX12 in p53-mediated ferroptosis. Notably, ACSL4 is not expressed in the human breast cancer cell line MCF-7 (lane 7, Fig. 7a), and indeed these cells are resistant to both erastin (Fig. 8a) and RSL-3 (Fig. 8b) mediated ferroptosis. Nonetheless MCF-7 cells are clearly susceptible to p53-mediated ferroptosis induced by ROS stress (Fig. 8c). Moreover, p53-mediated ferroptosis was also readily detected in other cell lines in which both p53 and ALOX12 are expressed, including human lung carcinoma lines A549 and H460 and human melanoma line A375 (Fig. 8d, 8e and 8f). As expected, p53-mediated ferroptosis in these cells was completely suppressed by the ALOX inhibitor ML355 (Fig. 8d, 8e and 8f), further validating the specific requirement for ALOX12 in p53-mediated ferroptosis.
Figure 7. Mechanistic insight into p53-mediated ferroptosis.
a) Western blot analysis of a panel of cancer cell lines. The experiments were repeated twice, independently, with similar results.
b) Western blot analysis of HT1080 cells with or without Nutlin (10uM) treatment. The experiments were repeated twice, independently, with similar results.
c) HT1080 cells treated with Erastin (10uM) and Ferr-1 (2uM) treatment for 8h. Quantification of cell death is shown. Error bars are mean ± s.d., n=3 independent experiments.
d) HT1080 cells transfected with ALOX12 were pre-incubated with Nutlin (10uM) for 12h, then treated with TBH (200uM), Nutlin (10uM) and Ferr-1 (2uM) for 8h. Quantification of cell death is shown. Western blot analysis of HT1080 cells transfected with ALOX12. Error bars are mean ± s.d., n=3 independent experiments. The Western blot experiments were repeated twice, independently, with similar results.
e) HCT116 p53+/+ cells were pre-incubated with Nutlin (10uM) for 12h, then treated with Nutlin (10uM), TBH (200uM), and Ferr-1 (2uM) as indicated for 12h. Quantification of cell death is shown. Western blot analysis of HCT116 p53+/+ cells transfected with ALOX12. Error bars are mean ± s.d., n=3 independent experiments. All P values (c,d,e) were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. The Western blot experiments were repeated twice, independently, with similar results. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. Raw data are provided in Supplementary Table 1.
Figure 8. ALOX12 in regulating p53-mediated ferroptosis in human cancer lines.
a) MCF7 cells treated with Erastin (20uM) and Ferr-1 (2uM) as indicated for 12h. Error bars are mean ± s.d., n=3 independent experiments.
b) MCF7 cells treated with RSL (4uM) and Ferr-1 (2uM) as indicated for 12h. Error bars are mean ± s.d., n=3 independent experiments.
c) MCF7 cells pre-incubated with Nutlin (10uM) for 12h were treated with TBH (100uM), Nutlin (10uM) and Ferr-1 (2uM) as indicated for 12h. Error bars are mean ± s.d., n=3 independent experiments.
d) A375 cells pre-incubated with Nutlin (10uM) for 12h were treated with TBH (150uM), Nutlin (10uM), ML-355 (4uM) and Ferr-1 (2uM) as indicated for 8h. Quantification of cell death is shown. Error bars are mean ± s.d., n=3 independent experiments.
e) H460 cells pre-incubated with Nutlin (10uM) for 12h were treated with TBH (300uM), Nutlin (10uM), ML-355 (4uM) and Ferr-1 (2uM) as indicated for 8h. Quantification of cell death is shown. Error bars are mean ± s.d., n=3 independent experiments.
f) A549 cells pre-incubated with Nutlin (10uM) for 12h were treated with TBH (150uM), Nutlin (10uM), ML-355 (4uM) and Ferr-1 (2uM) as indicated for 8h. Quantification of cell death is shown. Error bars are mean ± s.d., n=3 independent experiments.
g) U2OS cells pre-incubated with Nutlin (10uM) for 12h were treated with Nutlin (10uM), H2O2 (100uM), Ferr-1 (2uM), and ML-355 (4uM) as indicated for 48h. The percentage of cell death is shown. Error bars are mean ± s.d., n=3 independent experiments.
h) A model of p53-mediated ferroptosis compared to Erastin-induced ferroptosis. All P values (a,b,c,d,e,f,g)were calculated using two-tailed unpaired Student’s t-test. Detailed statistical tests are described in the Methods. Raw data are provided in Supplementary Table 1.
Discussion
Although the precise mechanisms by which p53 induces ferroptosis still require further elucidation, our study emphasizes the importance of ROS in this process. In addition to treatment with TBH, p53-mediated ferroptosis can also be induced by other forms of ROS, such as low levels of H2O2 and paraquat (Fig. 8g and Supplementary Fig. 8d). Thus, p53 promotes tumor suppression at least, in part by sensitizing cancer cells to ferroptotic death upon oxidative stress. Since high levels of cell proliferation are generally accompanied by increased ROS production, cancer cells are likely to select for various strategies, such as SLC7A11 overexpression or ALOX12 inactivation, to protect themselves from ROS-induced ferroptosis.
The ALOX12 gene resides on human chromosome 17p13.1 at a position very close to the p53 locus (Supplementary Fig. 8e). Since loss of one copy of chromosome 17p13.1 is very common in human cancer, it is likely that many human tumors have lost one allele of ALOX1224,32-34. Interestingly, recent studies indicate that a substantial fraction (31-36%) of human tumors containing chromosome 17p deletion retain a wildtype TP53 allele24. In light of our data that ALOX12 haploinsufficiency inhibits p53-mediated ferroptosis and accelerates Myc-induced tumorigenesis without obvious alterations in the ARF-p53 pathway, it is conceivable that monoallelic loss of ALOX12 is an etiological factor in this subset of patients. Notably, in contrast to the severe phenotypes associated with GPX4 mutant mice12-17,35, deletion of ALOX12 gene does not elicit major developmental defects in mice36. Thus, ALOX12, in a manner reminiscent of p53, is largely dispensable for most developmental processes that entail ferroptosis. Since ALOX12 is deleted, downregulated and mutated in human cancers, the p53-ALOX12 axis may act as a potential barrier to cancer development.
Although numerous studies implicate GPX4 inhibition as a central node for the induction of ferroptosis, our data identify a distinct mechanism through which p53 induces the ferroptotic response. In particular, we found that p53 is able to activate ALOX12 function indirectly by transcriptional repression of SLC7A11, resulting in ALOX12-dependent ferroptosis upon ROS stress (Fig. 8h). Further investigations are needed to examine whether other metabolic targets of p53 also contribute to ALOX12 activation2-5. Notably, in contrast to the effects by erastin treatment, we observed that p53 activation alone by Nutlin treatment does not significantly induce downregulation of the GSH/GSSG ratio, glutathione levels and GPX4 activity in human cancer cells (Fig. 2a, 2b and 2d). Although downregulation of SLC7A11 induced by p53 is able to partially inhibit cystine uptake3, the overall effects on the GSH/GSSG ratio and glutathione levels are apparently neutralized, probably by activation of other p53 targets such as TIGAR and p21. For example, activation of TIGAR by p53 was found to reduce ROS levels and increase the GSH/GSSG ratio37,38; p53-mediated activation of p21 was reported to promote the conservation of glutathione30. Our data have further demonstrated that ALOX12-mediated ferroptosis is independent of ACSL4 (Fig. 8h). Since ACSL4 is required for ferroptosis induced by erastin or GPX4 inhibitors28,29, our study reveals a previously uncovered ferroptotic pathway, distinct from the current ferroptosis model mainly focusing on GPX4 modulation.
Methods
Cell culture and stable lines.
U2OS, H1299, A549, A375, MCF7, HCT116, HT1080, H460 and SJSA cancer lines were obtained from American Type Culture Collection (ATCC) and have been proven to be negative for mycoplasma contamination. No cell lines used in this work were listed in the ICLAC database. All cells were cultured in a 37 °C incubator with 5% CO2. All media used for cancer cells were supplemented in DMEM with 10% FBS, 100U/mL penicillin and 100 ug/mL streptomycin (all from Gibco). Mouse embryonic fibroblasts (MEFs) were generated from day 13.5 embryos and supplemented in DMEM with 10% heat-inactivated FBS, 100U/mL penicillin, 100 ug/mL streptomycin and 1% nonessential amino acids. H1299 Tet-on p533KR cells were previously described (Jiang et al 2015). To obtain ALOX12 CRISPR-cas9-knockout U2OS and H1299 Tet-on p533KR cells were generated by transfecting ALOX12 double nickase plasmid (sc-403010-NIC; Santa Cruz). The cells were selected with puromycin (1ug/mL) for 4-6 days. ALOX12 knockout single clones were then screened and acquired after continuing to culture 2-3 weeks without selective antibiotics. Identical strategies were used to generate ALOX15 knockout, p53 knockout, ACSL4 knockout cells (ALOX15, sc-401591-NIC; p53, sc-416469-NIC; ACSL4, sc-401649-NIC). To generate ALOX12 stable cell line, wild-type or mutant pcin4-Flag-HA-ALOX12 was transfected into U2OS cells, followed by selection and maintenance with 1mg/mL G418 (Sigma) in DMEM medium containing 10% FBS. Single clones were selected and screened by western blot.
Plasmids.
pcDNA-ALOX12 plasmid was generously gifted from Dr. Colin D. Funk (Queen’s University). For wild-type ALOX12, full length ALOX12 was subcloned into pcDNA3.1/v5-His-Topo vector (Invitrogen), and mutant ALOX12 Topo plasmids were constructed using QuickChange XL Site-Directed Mutagenesis Kit (Agilent) according to the standard protocol. For Flag-HA wild-type and mutant ALOX12, full length ALOX12 was subcloned into pcin4-Flag-HA vector. For Flag-HA-SLC7A11, full length SLC7A11 was subcloned into pcin4-Flag-HA vector. For V5-ALOXe3, ALOX5, ALOX12, ALOX12B, ALOX15, ALOX15B, full length ALOXs were subcloned into pcDNA3.1/v5-His-Topo vector.
Western Blotting and Antibodies.
Protein extracts were analyzed by Western Blotting according to standard protocols using primary antibodies specific for p53 (DO-1) (sc-126 ; Santa Cruz;1:1000 dilution), p53 (FL-393) (sc-6243 ; Santa Cruz;1:1000 dilution), MDM2 (Ab-5) Mouse mAb (4B2C1.11)(OP-145; Millipore; 1:100 dilution), p21 (SX118) (sc-53870; Santa Cruz; 1:1000 dilution), MDMX (A300-287A; Bethyl; 1:1000 dilution), p19-ARF antibody (5C-3) (ab-26696; Abcam; 1:1000 dilution), PUMA antibody(H-136) (sc-28226; Santa Cruz; 1:500 dilution), SLC7A11 antibody (D2M7A) (12691s; Cell Signaling; 1:500 dilution), ACSL4 antibody (A5) (sc-271800; Santa Cruz; 1:1000 dilution), V5 (R960-25; Invitrogen; 1:1000 dilution), HA (11867423001; Sigma; 1:1000 dilution), ALOX12 antibody (C-5) (sc-365194; Santa Cruz; 1:200 dilution), ALOX15 (ab-80221; Abcam; 1:1000 dilution) and vinculin (V9264; Sigma-Aldrich; 1:5000 dilution). HRP-conjugated anti-mouse and anti-rabbit secondary antibody (GE Healthcare) and anti-rat (Southern Biotech) were used. The more detailed information is provided in the Supplementary Table 3.
Ablation of endogenous ALOX family by RNAi.
Knock-down of ALOX family proteins was performed by transfection of U2OS and H1299 Tet-on p533KR cells with siRNA duplex oligoset (ON-TARGET plus SMARTpool: ALOXE3: L-009022-00-0005; ALOX12B: L-009025-00-0005; ALOX12: L-004558-00-0005; ALOX5: L-004530-00-0005; ALOX15B: L-009026-00-0005; ALOX15: L-003808-00-0005; Dharmacon) with Lipofectamine 2000 (Invitrogen) for 24h and then transfected again according to the manufacturer’s protocol.
RNA extraction and qRT-PCR and sequencing of tumor samples.
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s protocol. cDNA was reversed using SuperScript™ IV VILO™ Master Mix (Invitrogen). For the qRT-PCR analysis of human transcripts the following primers were used: PTGS2 forward 5’-CTTCACGCATCAGTTTTTCAAG-3’, PTGS2 reverse 5’ -TCACCGTAAATATGATTTAAGTCCAC-3’; ALOX15 forward 5’-AGCCTGATGGGAAACTCTTG-3’, ALOX15 reverse 5’-AGGTGGTGGGGATCCTGT-3’; ALOXe3 forward 5’- GTGGATTGAAGGCTACTGCAC-3’, ALOXe3 reverse 5’- GGGGAAGAGAGTCCTGACAA-3’; ALOX5 forward 5’- TTTCATCGTGGACTTTGAGC-3’, ALOX5 reverse 5’- ATCTTGTTGGCCAGGTTCTT-3’; ALOX12 forward 5’- GAGGAATTTTTGATAAGGCAGTG-3’, ALOX12 reverse 5’- CCCGACGGAGCAACTGTA-3’; ALOX12B forward 5’- GACACTTCCCGGACATTCAC-3’, ALOX12B reverse 5’- GGAACGCCTCTATGCTCCT-3’; ALOX15B forward 5’- GATCTTCAACTTCCGGAGGAC-3’; ALOX15B reverse 5’- ACTGGGAGGCGAAGAAGG-3’ and GAPDH forward 5’-ATCAATGGAAATCCCATCACCA-3’, GAPDH reverse 5’-GACTCCACGACGTACTCAGCG-3’.
For RT-PCR: mouse full length p53 forward 5’- CCCTCCCACGTGCTCACCCTG-3’, p53 reverse 5’- CTCAGAGAGGGGGCTGAGCTAC-3’. For ALOX12 genotyping: mouse ALOX12 Wild type 5’- CCTCAGTGCAGGAACCTGTG-3’, ALOX12 Mutant 5’- TACTTCCATTTGTCACGTCCTG-3’, Common primer for Wild-type and Mutant ALOX12 5’- CTGCTACCCATGGCTATCCAG-3’.
Cell death assay.
For p53-dependent ferroptosis, we used the assay established in our previous study3, showing a ferroptotic response that requires both p53 activation and ROS stress, generated by low levels of tert-Butyl hydroperoxide (TBH). The ROS generator TBH, which is converted into peroxyl and alkoxyl radicals by cytochrome P45039, is more stable than traditional oxidative reagents, such as hydrogen peroxide (H2O2)40,41. Although high levels of TBH treatment may induce different types of cell death, no significant cell death was observed under low levels of TBH treatment3. Nevertheless, the combination of low levels of TBH treatment and p53 activation (e.g. nutlin treatment) is able to induce ferroptoiss. Typically, careful titration is performed to determine the concentration of TBH as well as the incubation time for p53-dependent ferroptosis on specific cell lines used. Similar assays with TBH treatment for ferroptosis were also reported by others6,9. Cells were trypsinized, collected and stained with trypan blue, and then counted with a hemocytometer using the cell number counter (Life technologies countess II). Living cells and dead cells were all counted according to the cell size, and cells stained blue were considered as dead cells. The cell death assays were also further confirmed by FACS with the similar results. To validate whether these are ferroptotic cell death, we treated the cells with Ferostatin-1 or other cell death inhibitors. If the cell death can be completely rescued by Ferostatin-1 but not by other cell death inhibitors, the cell death is ferroptotic.
Drugs and inhibitors.
For ROS generation, tert-butyl hydroxide solution (TBH, Sigma) was used at different doses depending on the experiment; see respective figure legends. Specific GPX4 inhibitor (B6095; APExBIO) was used. Specific cell death inhibitor and ALOX12 activity inhibitor were used in the experiments: Ferrostatin-1(ferroptosis inhibitor; Xcess Biosciences) and ML-355 (ALOX12 inhibitor; Cayman Chemical) was used at different doses depending on the experiment; see respective figure legends.
Expression, purification and activity assay of ALOX12 in vitro.
For purification of ALOX12 proteins from U2OS cells, U2OS cells were transfected with pCIN4-Flag-HA-ALOX12 and grown for 24h. Then the cells were harvested and lysed using BC100 buffer (20 mM Tris·Cl, pH 7.3, 100 mM Nacl, 10% glycerol, 0.2 mM EDTA, 1% NP-40, proteinase inhibitor, with a final concentration of 0.1 mM TPP (triphenylphosphine) on ice for 1 h. The cell lysate was centrifuged for 15 min at 15,000 rpm at 4°C. The supernatants were used for Flag/M2 immunoprecipitations by anti-Flag antibody-conjugated agarose (Sigma) and incubated overnight. The bound ALOX12 proteins were eluted with 10ug/mL Flag peptide (Sigma) in BC100 solution.
Considering arachidonic acid is a substrate of ALOX12, active ALOX12 will metabolize arachidonic acid into 12-HETE. Specifically, active ALOX12 reduces 12(S)-hydroperoxy tetraenoic eicosatetraenoic acid [12(S)-HpETE] into 12(S)-HETE (12-HETE). The amount of 12-HETE can be measured by ELISA. Therefore, for the ALOX12 catalytic activity assay, aliquots of the purified ALOX12 enzyme (500ng) were incubated with arachidonic acid (100uM) for 10 min in PBS containing 0.1mM ATP (New England Biolabs), and 0.4 mM Ca2+ with or without 4uM ML-355 (ALOX12 inhibitor; as in Figure 4b). Then the reaction solution was acidified with acetic acid to a pH of approximately 3-4 as determined by standard pH paper. The samples were extracted with equal volume ethyl acetate and then dried and dissolved in 20uL ethanol.
To detect 12-HETE levels we performed a 12-HETE ELISA. A 100uL of purified 12-HETE was used to generate a serial-dilution for the standard curve (1mg/mL; Detroit R&D). The samples from the purified ALOX12 reaction buffer were diluted to 500uL with 1x sample dilution buffer.
Then, 100uLs of each of the 12-HETE standards (from 1,000,000pg/mL-10pg/ml) and the reaction samples were loaded into a 96-well plate coated with anti-12-HETE antibody (Detroit R&D). Next, the 12-HETE-HRP conjugate was added to each the well and the plate was incubated at room temperature for 2 h. The plate was washed three times after incubation and 200 uL of TMB substrate was added to all the wells and incubated for 15-30 minutes, then the reaction was stopped with 50 uL 2N Sulfuric Acid and the plate was read at 450 nm.
Measurement of glutathione in vivo.
To measure total glutathione (GSH) and the ratio of reductive GSH to oxidative GSH, we used GSSG/GSH quantification kit (Donjindo Molecular Technologies). U2OS cells were collected and washed with PBS. The cells were lysed by freezing and thawing twice upon 10mM HCl addition. The cells were then added 5% 5-sulfosalicylic acid (5-SSA) and centrifuge at 8,000 xg for 10 min. The supernatants with final 0.5% 5-SSA are used for assays. To detect glutathione levels, GSH standard solution was used to generate a serial-dilution for the standard curve (200uM).
Then, 40uL of GSH and GSSG standard solution and samples were loaded into a 96-well plate. Next, 60uL of Buffer solution was added to each well and incubate the plate at 37°C for 1h. 60uL of substrate working solution was added to each well and subsequently with 60uL enzyme/coenzyme working solution and incubate at 37°C for 10 min and the plate was read at 405 nm.
Measurement of GPX4 activity in vivo.
To measure GPX4 activity, we used glutathione peroxidase assay kit (Abcam). U2OS cells transfected with exogenous GPX4 were harvested and washed with cold PBS. The cells were lysed with 200uL Assay buffer and centrifuge at 10,000 xg for 15 min at 4°C and transfer the supernatants to new tubes. NADPH standard solution was used to generate a serial-dilution for the standard curve (1mM).
Then, 100uL of NADPH standard solution and 40ul samples were loaded into a 96-well plate. Next, 40uL of reaction mix was added to sample wells, then mix well and incubate at room temperature for 15 min. 10uL cumene hydroperoxide solution was added to sample wells and measure output (A1) on a microplate reader at 340 nm. The plate was incubated at 25°C for 5 min and measure output (A2) at 340 nm.
Lipid ROS assay using flow cytometer.
Cells were incubated with DMEM containing 5uM of C-11 BODIPY dye and the cells were returned to the tissue culture incubator for 30 min. Cells were then harvested and washed twice with PBS followed by re-suspending in 500 ul of PBS. ROS levels were analyzed using a Becton Dickinson FACSCalibur machine through the FL1 channel, and the data were analyzed using CellQuest. 10,000 cells were analyzed in each sample.
GPX4 activity assay using flow cytometer.
To measure GPX4 activity on phospholipid hydroperoxides, we used Tet-on p53 3KR H1299 GPX4 crispr cells cultured with DMEM containing 1uM Ferr-1. Cells were transfected with exogenous GPX4. Ferr-1 was then withdrew 24h after transfection with or without RSL-3 (GPX4 inhibitor) treatment. GPX4 activity were measured by Lipid ROS levels using flow cytometer as above.
Mouse xenograft.
H1299 Tet-on p533KR ctrl crispr and ALOX12 crispr cells were pretreated with or without doxycycline (0.5ug/mL) for 48 h, then the cells were trypsinized and counted. 1.0× 106 cells were mixed with Matrigel (BD Biosciences) at 1:1 ratio (v/v) and injected subcutaneously into nude mice (NU/NU; Charles River). Mice were fed with either control food or food containing doxycycline hyclate (Harlan, 625mg/kg). After four weeks, the mice were killed and the tumors were weighed and recorded. The study is compliant with all relevant ethical regulations for animal experiments. All the experimental protocols were approved by the Institutional Animal Care and Use Committee of Columbia University.
Mice.
Eμ-Myc, Trp53+/− and ALOX12−/− mice were ordered from Jackson Laboratories. Eμ-Myc, Eμ-Myc; ALOX12+/− and Eμ-Myc; p53+/− mice were monitored weekly for disease state defined by enlarged solid lymph nodes and/or paralysis, and recorded the date of death. Then the mice were dissected and the harvest tumors were subjected to H&E staining (after being parafinized), Western Blotting, or RT-PCR. For ACSL4−/− mice, gene targeting of mouse ACSL4 gene was designed to flank the second coding exon (exon 2) with loxP sites (flox) to generate a conditional knockout allele of ACSL4. The cre mediated deletion of exon 2 causes translational reading frame shift and truncation of the rest of the ACSL4 protein. Briefly a gene targeting construct was generated containing ACSL4 exon 2 and flanking genomic DNA. The first loxP site was inserted 5’ to the exon 2 and the second loxp site along with neomycin selection cassette was inserted 3’ to exon 2 using recombineering. The ACSL4 targeting vector was electroporated into mouse embryonic stem cells (ES cells) and the neomycin resistant ES cell clones were selected using drug G418. The correctly targeted mouse ES cell clones were identified by southern blot using 5’ probe and BamHI digested ES cell genomic DNA, which displays wild type band at 14 kb and mutant band at 7 kb. Southern blot using a 3’ probe was also performed to confirm the targeted ES clones (WT band 14 kb and mutant band 7 kb using BamH1 digested genomic DNA). The confirmed targeted mouse ES cells were injected into blastocyst to generate chimeras, from which germline transmission of the exon 2-floxed allele of ACSL4 was achieved. The ACSL4 conditional knockout mice were maintained on a mixed background of 129Sv and C57BL/6J through breeding between littermates. Maintenance and handling protocols were approved by Institutional Animal Care and Use Committee (IACUC) of Columbia University. The ACSL4 FL/+ female mice were crossed with rosa26cre/cre mice to generate ACSL4 knockout MEFs. Genotyping of ACSL4 knockout MEFs were done by PCR using mixed primers of WT 5’ primer: CTTCTGAGCATTCACATCATGGGA; WT 3’ primer: TCGCGTGCACACACTAAGTGCTA; and KO 5’ primer ATCCGGTGGCGACGAATTCATAA
ALOX12 gene expression analysis.
The ALOX12 gene expression data for normal versus cancer human tissues were derived from Oncomine database(https://www.oncomine.org/resource/). Box plots derived from gene expression data in Oncomine comparing the expression of ALOX12 mRNA in different cancer types. For Cervical normal tissues n=10 independent samples, tumor tissues n=21 independent samples; Head and Neck normal tissues n=13 independent samples, tumor tissues n=41 independent samples; Esophageal normal tissues n=17 independent samples, tumor tissues n=17 independent samples; Acute Myeloid Leukemia, normal tissues n=8 independent samples, tumor tissues n=285 independent samples. Box plots with centre line at median, box limits at 25th/75th centiles. The association of ALOX12 with overall survival in patients with Pancreatic Adenocarcinoma were derived from publicly available clinical information provided by cBioportal for Cancer Genomics databases(http://www.cbioportal.org/). Kaplan-Meier survival curves were generated for overall survival (months) by stratifying patient samples with Pancreatic Adenocarcinoma (TCGA, Provisional) from cBioPortal (n = 186) based on ALOX12 expression levels. The patients were quartiled for ALOX12 expression [ALOX12 low (n = 45; black) and ALOX12 high (n =45; red)], Log-rank Mantel-Cox test was used (p=0.016).
Statistics and reproducibility.
Figs. 1b,d-e,g, 2c,f, 3a,g, 4c(bottom panel), d-e, 5a-c,f(right panel),g, 6a,e, 7a-b, d(right panel),e(right panel) and Supplementary Figs. 1c, 3a,e, 4a-c, 6a-f,h-i, 8b-c were independently repeated twice. All the other experiments were independently repeated at least three times. Statistical analysis was carried out using Microsoft Excel software and GraphPad Prism to assess the differences between experimental groups. Statistical significance was determined by using a two-tailed, unpaired Student’s t-test with a confidence interval (CI) of 95%. P ≤0.05 was denoted as statistically significant. Statistical analysis of all survival curves data was performed using log-Rank (Mantel-Cox) test.
Code availability.
No computational code was used in this study.
Data availability.
The ALOX12 gene expression data for normal versus cancer human tissues were derived from Oncomine database(https://www.oncomine.org/resource/). The association of ALOX12 with overall survival in patients with Pancreatic Adenocarcinoma were derived from publicly available clinical information provided by cBioportal for Cancer Genomics databases (http://www.cbioportal.org/). Source data for Figs. 1a,c,f, 2a-b, d-e,g, 3c-d,h, 4b-c,f, 5d-f,h-i, 6b-d,f-h, 7c-e and 8a-g and Supplementary Figs. 1b,d-e, 2b, 3b-d, 5c-d,6j, 7a-c and 8d have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Brent Stockwell, Dr. Richard Baer, Xujun Jiang, Dr. Mykola Bordyuh, and Dr. Raul Rabadan for critical suggestions in this study. We also appreciated Dr. C.D. Funk for critical reagents. This work was supported by the National Cancer Institute of the National Institutes of Health under Award 5R01CA216884, 5R01CA190477, 5R01CA085533 and 5RO1CA224272 to W.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Kastenhuber ER & Lowe SW Putting p53 in Context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jennis M et al. (2016). An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes & development 30, 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang L et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62, doi: 10.1038/nature14344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao M, Monian P, Quadri N, Ramasamy R & Jiang X Glutaminolysis and Transferrin Regulate Ferroptosis. Molecular cell 59, 298–308, doi: 10.1016/j.molcel.2015.06.011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang SJ et al. Acetylation Is Crucial for p53-Mediated Ferroptosis and Tumor Suppression. Cell Rep 17, 366–373, doi: 10.1016/j.celrep.2016.09.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saint-Germain E et al. SOCS1 regulates senescence and ferroptosis by modulating the expression of p53 target genes. Aging 9, 2137–2162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon SJ et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072, doi: 10.1016/j.cell.2012.03.042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stockwell BR et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 171, 273–285, doi: 10.1016/j.cell.2017.09.021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YL et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol 20, 1181–1192, doi: 10.1038/s41556-018-0178-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angeli JPF, Shah R, Pratt DA & Conrad M Ferroptosis Inhibition: Mechanisms and Opportunities. Trends Pharmacol Sci 38, 489–498, doi: 10.1016/j.tips.2017.02.005 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Viswanathan VS et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457, doi: 10.1038/nature23007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angeli JPF et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16, 1180–U1120, doi: 10.1038/ncb3064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsushita M et al. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J Exp Med 212, 555–568, doi: 10.1084/jem.20140857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai H et al. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem. Biophys. Res. Commun 305, 278–286 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Ingold I et al. Expression of a Catalytically Inactive Mutant Form of Glutathione Peroxidase 4 (Gpx4) Confers a Dominant-negative Effect in Male Fertility. J Biol Chem 290, 14668–14678, doi: 10.1074/jbc.M115.656363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wortmann M et al. Combined Deficiency in Glutathione Peroxidase 4 and Vitamin E Causes Multiorgan Thrombus Formation and Early Death in Mice. Circ Res 113, 408–417, doi: 10.1161/Circresaha.113.279984 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Schriever SC et al. Alterations in Neuronal Control of Body Weight and Anxiety Behavior by Glutathione Peroxidase 4 Deficiency. Neuroscience 357, 241–254, doi: 10.1016/j.neuroscience.2017.05.050 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Yang WS et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 156, 317–331, doi: 10.1016/j.cell.2013.12.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai H, Matsuoka M, Kumagai T, Sakamoto T & Koumura T Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis. Curr Top Microbiol 403, 143–170, doi: 10.1007/82_2016_508 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Dixon SJ et al. , Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem Biol. 10, 1604–9. doi: 10.1021/acschembio.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eischen CM, Weber JD, Roussel MF, Sherr CJ & Cleveland JL Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes & development 13, 2658–2669, doi:DOI 10.1101/gad.13.20.2658 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Post SM et al. p53-dependent senescence delays E mu-myc-induced B-cell lymphomagenesis. Oncogene 29, 1260–1269, doi: 10.1038/onc.2009.423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valente LJ, Grabow S, Vandenberg CJ, Strasser A & Janic A Combined loss of PUMA and p21 accelerates c-MYC-driven lymphoma development considerably less than loss of one allele of p53. Oncogene 35, 3866–3871, doi: 10.1038/onc.2015.457 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Liu Y et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531, 471–457, doi: 10.1038/nature17157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luci D et al. Discovery of ML355, a Potent and Selective Inhibitor of Human 12-Lipoxygenase. in Probe Reports from the NIH Molecular Libraries Program (2010–2013). [Google Scholar]
- 26.Gasol E, Jimenez-Vidal M, Chillaron J, Zorzano A & Palacin M Membrane topology of system xc- light subunit reveals a re-entrant loop with substrate-restricted accessibility. J Biol Chem 279, 31228–31236, doi: 10.1074/jbc.M402428200 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Dixon SJ et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 3, doi:ARTN e02523 10.7554/eLife.02523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doll S et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 13, 91–98, doi: 10.1038/Nchembio.2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagan VE et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol 13, 81–90, doi: 10.1038/Nchembio.2238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarangelo A et al. p53 Suppresses Metabolic Stress-Induced Ferroptosis in Cancer Cells. Cell Rep 22, 569–575, doi: 10.1016/j.celrep.2017.12.077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie YC et al. The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep 20, 1692–1704, doi: 10.1016/j.celrep.2017.07.055 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Ferretti E, De Smaele E, Di Marcotullio L, Screpanti I & Gulino A Hedgehog checkpoints in medulloblastoma: the chromosome 17p deletion paradigm. Trends Mol Med 11, 537–545, doi: 10.1016/j.molmed.2005.10.005 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Schnaiter A & Stilgenbauer S 17p Deletion in Chronic Lymphocytic Leukemia Risk Stratification and Therapeutic Approach. Hematol Oncol Clin N 27, 289–301, doi: 10.1016/j.hoc.2013.01.008 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Tam CS & Stilgenbauer S How best to manage patients with chronic lymphocytic leuekmia with 17p deletion and/or TP53 mutation? Leukemia Lymphoma 56, 587–593, doi: 10.3109/10428194.2015.1011641 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Ingold I, et al. , (2018) Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell. 172, 409–422. [DOI] [PubMed] [Google Scholar]
- 36.Johnson EN, Brass LF & Funk CD Increased platelet sensitivity to ADP in mice lacking platelet-type 12-lipoxygenase. Proceedings of the National Academy of Sciences of the United States of America 95, 3100–3105 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bensaad K et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120, doi: 10.1016/j.cell.2006.05.036 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Kruiswijk F, Labuschagne CF, Vousden KH p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 7, 393–405 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Davies MJ Detection of peroxyl and alkoxyl radicals produced by reaction of hydroperoxides with rat liver microsomal fractions. The Biochemical journal 257, 603–606 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes RH, Silva VA, Ahmed I, Shreiber DI & Morrison B 3rd Neuroprotection by genipin against reactive oxygen and reactive nitrogen species-mediated injury in organotypic hippocampal slice cultures. Brain research 1543, 308–314 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Jiang H, Chen S, Du F & Wang X The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ALOX12 gene expression data for normal versus cancer human tissues were derived from Oncomine database(https://www.oncomine.org/resource/). The association of ALOX12 with overall survival in patients with Pancreatic Adenocarcinoma were derived from publicly available clinical information provided by cBioportal for Cancer Genomics databases (http://www.cbioportal.org/). Source data for Figs. 1a,c,f, 2a-b, d-e,g, 3c-d,h, 4b-c,f, 5d-f,h-i, 6b-d,f-h, 7c-e and 8a-g and Supplementary Figs. 1b,d-e, 2b, 3b-d, 5c-d,6j, 7a-c and 8d have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.