Abstract
Background:
The molecular mechanisms underlying cervical cancer require elucidation to identify novel therapeutic targets. Apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1) is a multifunctional apurinic/apyrimidinic (AP) endonuclease that influences the transcription of many cancer-related genes via microRNome regulation. Herein, we examine the role of miR-92b-3p (hereinafter miR-92b), whose processing may be regulated by APE1, in cervical cancer progression.
Methods:
APE1’s processing of miR-92b from its pri-miR form was measured by a quantitative reverse transcription polymerase chain reaction (qRT-PCR)-based ratio. APE1’s endonuclease activity was measured with AP-site incision assays. APE1-DROSHA interaction was studied with immunofluorescence, confocal and proximity ligation analyses. The miR-92b’s targeting of low-density lipoprotein receptor (LDLR) was investigated with luciferase reporter assays. The miR-92b mimics and shRNA-based miR-92b silencing, as well as LDLR overexpression and short interfering RNA (siRNA)-based LDLR silencing, were employed in CaSki and SiHa cervical cancer cells. Cell proliferation and chemosensitivity to paclitaxel and cisplatin were assayed. Cell-cycle progression and apoptosis were assessed by flow cytometry. Tumor growth was studied in a murine xenograft model.
Results:
APE1’s endonuclease activity, via association with the DROSHA-processing complex, is necessary for processing mature miR-92b, thereby regulating expression of miR-92b’s direct target LDLR. The miR-92b promotes cell proliferation in vitro and in vivo, promotes cell-cycle progression, and reduces apoptosis and chemosensitivity. LDLR silencing recapitulated miR-92b’s transformative effects, while LDLR overexpression rescued these effects.
Conclusions:
APE1 enhances miR-92b processing, thereby suppressing LDLR expression and enhancing cervical carcinoma progression. Our identification of the novel APE1-miR-92b-LDLR axis improves our understanding of the complex pathogenesis of cervical carcinoma and reveals a novel therapeutic strategy for combating this disease.
Keywords: APE1, cervical cancer, LDLR, microRNA, miR-92b
Introduction
RNA damage induced by missing bases [e.g. by generation of apurinic/apyrimidinic (AP) sites], or by modification of bases (e.g. by oxidative damage), can alter RNA function and is increasingly found to participate in carcinogenesis.1 For instance, mRNA damaged by oxidation or AP-site formation can interfere with translational accuracy.1 In the case of microRNAs (miRNAs), oxidation can alter their regulatory activity, thus modifying expression of target genes.2 Consequently, degradation of damaged miRNAs in genotoxic environments can have significant repercussions in oncogenesis and for the development of chemoresistance in existing tumors.
Cells regulate the response to DNA damage via various DNA repair pathways.3 Damaged bases are fixed by base excision repair (BER), which first generates an AP site that is then cleaved by AP endonucleases followed by repair.4 AP endodeoxyribonuclease 1 (APE1) is a multifunctional AP endonuclease and BER protein that functions as a master regulator of cell fate under genotoxic stress.5 APE1 also has a role in chemoresistance by controlling expression of the tumor suppression PTEN (phosphatase and tensin homolog)6,7 and of MDR1 (multidrug resistance protein 1).8,9 The participation of APE1 in RNA processing and gene transcription has also been established.10–12 Numerous lines of evidence support a role for APE1 in RNA metabolism, including (a) binding of its N-terminal domain to structured RNA oligonucleotides in vitro,13 (b) endonuclease cleavage of AP sites on single-stranded RNAs, leading to RNA degradation,14 and (c) via exoribonuclease and RNA phosphatase activity at the 3’ end.14 APE1 is the sole enzyme found to date capable of AP site and oxidatively damaged RNA base removal or of 3’-RNA phosphatase activity.15
Knockdown (KD) of APE1 has been shown to influence the transcription of many genes related to malignant proliferation, invasiveness, and chemoresistance via miRNome regulation.15–17 In a recent study on cervical cancer cells, an exhaustive list of miRNAs directly regulated by APE1 during the cellular response to genotoxic treatment and that may specifically mediate resistance to chemotherapy has been recently detailed.15 Herein, we consider one candidate miRNA, miR-92b-3p (hereinafter miR-92b), whose processing may be regulated by APE1.15 We demonstrate that during genotoxic exposure, the APE1–miR-92b–low-density lipoprotein receptor (LDLR) axis mediates cervical cancer cell progression and that targeting this pathway may constitute a novel therapeutic approach to cervical cancer.
Methods
Ethics statement
This research was approved by the Ethics Committee of the First People’s Hospital of Yunnan Province (Kunming, China; KHYY20180814). The approval covered both the sample collection and the animal study. Human tissue specimens collected for this study required written informed consent from the donor. The animal procedures were conducted in accordance with the standards set forth in the Guide for the Care and Use of Laboratory Animals [eighth edition, National Institutes of Health (NIH)].
Cell cultures
SiHa, CaSki, and HeLa cervical carcinoma cells were procured from the American Type Culture Collection. Cultures were maintained in DMEM (Dulbecco’s modified Eagle’s medium; Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific) and antibiotics [penicillin (100 U/ml); streptomycin (10 μg/m1)]. HeLa clones overexpressing FLAG-tagged APE1 were maintained in the same media [DMEM, 10% FBS, penicillin (100 U/ml); streptomycin (10 μg/ml)], which additionally contained the selection antibiotics Zeocin™ (100 μg/ml), blasticidin (3 μg/ml), and geneticin (400 μg/ml; Invitrogen, Thermo Fisher Scientific) to preserve clones with APE1 overexpression. Short hairpin RNA (shRNA)-mediated KD of APE1 was induced by the addition of doxycycline (1 μg/ml; Sigma-Aldrich, St Louis, MO, USA) to the growth media and cultures were maintained for 10 days according to known protocols.18,19 Cultures were grown under an atmosphere of 5% CO2 and a temperature of 37°C, and were routinely assessed for contaminating mycoplasma.
Transient transfection of HeLa cells with short interfering RNA-resistant APE1 mutants and APE1-short interfering RNA
HeLa cells with stable APE1 KD (3 × 106) were plated and transiently transfected the following day with plasmid DNA (6 µg) employing Lipofectamine® 2000 transfection reagent (Invitrogen). The FLAG-bearing plasmids encoded either wild type (WT) or mutant APE1, whose mRNA contains mutations that render them resistant to cleavage by APE1-siRNA (short interfering RNA), but which translate into the intended protein sequence.15 The expressed APE1 were: (1) APE1WT, (2) APE1E96A, (3) APE1C65S, (4) APE1NΔ33, plus, as a control, (5) empty vector. Cells were harvested 24 hours following the transient transfection.
Transfection of cells for studies of siRNA-mediated KD of APE1 were performed using 100 pmol of siRNA oligonucleotides and DharmaFECT™ Transfection Reagent (Dharmacon, Lafayette, CO, USA): (1) APE1-targeted 5’-UAC UCC AGU CGU ACC AGA CCU-3’, or (2) scrambled negative control 5’-CCA UGA GGU CAU GGU CUG dTdT-3’.15 Cells were harvested 72 hours after transient transfection following by purification of RNA.
Inhibitor treatment of HeLa cells
HeLa cultures were incubated over a specified amount of time with a number of APE1 inhibitors: (1) fiduxosin (40 µM), which inhibits APE1’s endonuclease activity,20 (2) inhibitor #3 (20 µM), also an inhibitor of APE1’s endonuclease action,21 and (3) E3330 (100 µM), an inhibitor of the redox activity of APE1.22,23
Immunoprecipitation of APE1-bound RNA
HeLa cells (1 × 107 per dish) overexpressing FLAG-tagged APE1WT were plated into two 150-mm culture dishes for RNA immunoprecipitation (RIP) experiments, which were performed according to established protocols.24,25 Prior to the RIP assay, HeLa cells were rinsed with phosphate-buffered saline (PBS) buffer (two times), trypsinized, detached gently, and collected by centrifugation at low speed (250 × g) for a short duration (5 minutes) at 4°C. The supernatant was siphoned off and the pelleted HeLa cells were gently suspended in buffer (10 ml PBS). Cross-linking was started by formaldehyde (1% v/v, final concentration) at room temperature (RT). After 10 minutes, cross-linking was halted by glycine (125 mM, final concentration) and the HeLa cells were pelleted again. Pellets were rinsed two times with PBS supplemented with protease inhibitors [both a cocktail (1X, Sigma-Aldrich) and the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF; 0.5 mM)] and sodium fluoride (NaF; 1 mM), sodium orthovanadate (Na3VO4; 1 mM), and RNaseOUT™ (0.5 U/ml, Invitrogen). Cells were then pelleted followed by lysis in a 300 μl aliquot of RIP lysis buffer (50 mM Tris-HCl buffered at pH 7.4, with 150 mM NaCl, 1 mM EDTA, 1% w/v Triton X-100, supplemented with PMSF and protease cocktail inhibitors). Nucleic acids in HeLa cell lysates were sheared by sonication (three bursts lasting 30 seconds each) using a cell disrupter (Bioruptor®, Diagenode, Denville, NJ, USA). Insoluble debris was removed from lysates by high-speed ultracentrifugation for 10 minutes at 4°C. Cleared lysates with diluted with a 10-fold addition of RIP dilution buffer (16.7 mM Tris-HCl buffered at pH 8.1, with 167 mM NaCl, 1.2 mM ethylenediaminetetraacetic acid (EDTA), 1.1% w/v Triton X-100, 0.01% w/v sodium dodecyl sulfate (SDS), supplemented with PMSF and protease cocktail inhibitors and (0.5 U/ml RNaseOUT™).
Each RIP assay contained: (1) 40 μl of resin, binds to FLAG-tagged APE1 (ANTI-FLAG® M2 Affinity Gel), (2) 0.4 μg salmon sperm DNA, and (3) 1 μg of bovine serum albumin (BSA), all from Sigma-Aldrich. RIP samples were rotated gently overnight in the cold room and then pelleted the following morning by centrifugation (8000 × g, 1 minute, 4°C). Supernatants were siphoned off, and the pelleted resin was rinsed one time using 1 ml of the following serial solutions: (1) Low-salt wash buffer (20 mM Tris-HCl buffered at pH 8.1, with 150 mM NaCl, 2 mM EDTA, 1% w/v Triton X-100, 0.1% w/v SDS); (2) High-salt wash buffer (20 mM Tris-HCl buffered at pH 8.1, with 500 mM NaCl, 2 mM EDTA, 1% w/v Triton X-100, 0.1% w/v SDS); and (3) LiCl wash buffer (10 mM Tris-HCl buffered at pH 8.1, with 0.25 M LiCl, 1 mM EDTA, 1% w/v deoxycholate, 1% w/v NP40), and two times using TE buffer (10 mM Tris-HCl buffered at pH 8.0, with 1 mM EDTA). All rinses were rotated for 5 minutes at RT and pelleted by centrifugation (8000 × g, 1 minute, RT) in between the various rinsing steps.
Immunoprecipitated APE1 and bound RNA were released from the resin by rotating for 15 minutes with two washes of RIP Elution Buffer (250 µl aliquot of 0.1 M NaHCO3, 1% w/v SDS, containing 0.5 U/ml RNaseOUT™). Cross-linking was reversed by adjusting NaCl levels of the eluate to 200 mM and heating at a temperature of 65°C for 2 hours. Protein material was eliminated from the freed RNA by digestion with proteinase K (Sigma-Aldrich) at a temperature of 42°C for 45 minutes in digestion buffer (20 µl aliquot of 1 M Tris-HCl buffered at pH 6.5, and 10 µl aliquot of 0.5 M EDTA). RNA was purified by QIAzol Lysis Reagent (Qiagen, Hilden, Germany) by following the supplier’s directions and a DNase on-column treatment step was added to remove any contaminating DNA.
Cervical tumor biopsy analysis of APE1 and LDLR by immunohistochemistry
Cervical tumor biopsies were obtained from 50 female patients whose tumors were surgically resected, but who did not receive chemotherapy or radiotherapy. Histopathological diagnosis of cervical cancer was confirmed by pathologists. Biopsy samples were formalin-fixed and then paraffin-embedded (FFPE), sectioned, and then treated in the cold room with primary antibodies for: (1) APE1 (1:5000, mouse, monoclonal, clone 13B8E5C2, catalog no. NB100-116, Novus Biologicals, Littleton, CO, USA), or (2) LDLR, 1:100, mouse, monoclonal, catalog no. sc-18823, Santa Cruz Biotechnology, Dallas, TX, USA). The following day, tissue sections were washed with PBS, treated with a peroxidase-conjugated secondary antibody (goat anti-mouse), washed in PBS again, and treated serially, first with 3,3’-diaminobenzidine (DAB), and then with Harris hematoxylin as a counterstain.
Semi-quantification of APE1 and LDLR proteins were performed by scoring staining levels according to a stratification scheme with four scores (0, 1, 2, or 3): (1) score of 0: no protein expression in cancer cells, (2) score of 1: faint protein expression in less than 10% of cancer cells, (3) score of 2: faint to intermediate expression in over 10% of cancer cells, and (4) score of 3: strong expression in over 10% of cancer cells. Scoring was performed by two independent, blinded researchers, and statistical assessment was accomplished by a Spearman’s rank correlation test.
mRNA purification and quantitative reverse transcription polymerase chain reaction analysis of LDLR in cell lines
Analysis of mRNA transcript levels in cell lines was achieved by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Total RNA content was isolated using an SV Total RNA Isolation System kit (Promega, Fitchburg, WI, USA). A 1-µg aliquot of RNA was used to synthesize cDNA by reverse transcription employing an iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). The cDNA was then used as a template for qRT-PCR employing iQ™ SYBR® Green Supermix (Bio-Rad) in 15 μl reactions that contained forward and reverse primers (10 μM each) setup within wells of 96-well plates. The PCR program used performed a denaturing step at 95°C for 10 seconds, followed by 40 repetitions of annealing and extending at 60°C for 30 seconds, using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). The primer pairs used in qRT-PCR assay were: (1) forward LDLR 5’-CAA TGT CTC ACC AAG CTC TGY-3’, (2) reverse LDLR 5’- TCT GTC TCG AGG GGT AGC TG −3’, (3) forward GAPDH 5’-CCT TCA TTG ACC TCA ACT ACA TG-3’, and (4) reverse GAPDH 5’-TGG GAT TTC CAT TGA TGA CAA GC-3’. Triplicates of every sample were included, as well as negative controls that contained no template. Melting curve analyses were conducted to validate the amplicons.
qRT-PCR analysis of pri-miRNA and miRNA in cell lines and cervical tumor biopsy FFPE samples
pri-miRNAs and miRNAs were purified for qRT-PCR analysis from cell lines and cervical tumor biopsy FFPE samples employing miRNeasy and miRNeasy FFPE kits (Qiagen), respectively. cDNA was prepared from total RNA employing either iScript™ cDNA Synthesis Kit (Bio-Rad) or TaqMan™ MicroRNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific) to analyze pri-miRNAs and miRNAs, respectively. For cDNA synthesis of miRNAs, a 10-ng aliquot of total RNA was used as template together with miRNA-targeting primers and RNase inhibitors. Confirmation of cDNA synthesis was performed with TaqMan™ Universal PCR Mix (no AmpErase™ UNG; Applied Biosystems) using miRNA-targeting primers and TaqMan™ probes. Quantification was accomplished on LightCycler® 480 (Applied Life Science, Penzberg, Germany). qRT-PCR analysis was performed by the ΔΔct (delta-delta threshold cycle) process and expression levels were normalized to GAPDH and RNU44 for pri-miRNA and miRNA, respectively.
Quantification of RNA containing AP sites
Oxidatively depurinated/depyrimidinated RNA was ARP-labeled and isolated with streptavidin magnetic beads as previously described.26 Briefly, the Xenopus elongation factor 1α (xEF-1α) control mRNA was added to total isolated RNA to achieve a total of 30 µg RNA. To quantify AP sites, RNA was reacted with aldehyde-reactive probes (ARPs; 2 mM, Invitrogen) in 50 mM sodium acetate buffered at pH 5.2 for 40 minutes at a temperature of 37°C. RNA was rendered insoluble by the addition of ethanol, pelleted, and then resolubilized in water. A portion was reserved to quantify total RNA transcripts, and the remainder was affinity purified using streptavidin-conjugated magnetic beads for 20 minutes at a temperature of 60°C. Beads were rinsed in consecutive buffers with urea, and oxidized RNA (i.e. RNA that had contained AP sites) was released from the magnetic beads upon treatment with biotin (2.5 mM) for 5 minutes at a temperature of 90°C. Ethanol was added to the released oxidized RNA to induce precipitation and then resolubilized in water. cDNA was prepared from the total and oxidized RNAs using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific) in the presence of DNase to eliminate contaminating DNA. The cDNA was then utilized for qRT-PCR using GTXpress™ Master Mix and the TaqMan™ probe for mature miR-92b (hereafter miR-92b-3p, both from Thermo Fisher Scientific) on a LightCycler® 480 qRT-PCR instrument. Quantification of oxidized miRNAs was achieved by calculating the difference (ΔCq) in Cq (quantification cycle) values between oxidized and total RNA. qRT-PCR was validated using: (1) negative controls lacking cDNA, (2) melting curve analyses to validate amplicons, and (3) serial dilution to construct calibration curve.
Western blot analysis and protein quantification
Cell cultures were rinsed with PBS buffer (two times), trypsinized, detached gently, and collected by centrifugation at low speed (250 × g) for a short duration (5 minutes) in the cold (4°C). The supernatant was siphoned off and the pelleted cells were gently rinsed in cold PBS, pelleted again by centrifugation, and lysed in buffer on ice for 30 minutes (lysis buffer was 50 mM Tris-HCl buffered at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1 mM Na3VO4, 1% w/v Triton X-100, containing protease inhibitors (1X cocktail, 0.5 mM PMSF). Debris was eliminated from lysates by high-speed centrifugation (12,000 × g) for 30 minutes at 4°C. Cleared lysates were assessed for protein content by a commercial Bradford assay (Bio-Rad Protein Assay) and 20 µg of total protein was loaded per well of a denaturing gel [12% SDS polyacrylamide gel electrophoresis (PAGE)]. Proteins were separated by electrophoresis and then electroblotted to a nitrocellulose membrane (Schleicher and Schuell BioScience, Keene, NH, USA).
Membranes were then soaked with antibodies for: (1) α-tubulin (1:2000, mouse, monoclonal, clone DM1A, catalog no. T9026, Sigma-Aldrich), (2) FLAG (1:1000, mouse, monoclonal, clone M2, catalog no. F1804, Sigma-Aldrich), (3) APE1 (1:1000, mouse, monoclonal, clone 13B8E5C2, catalog no. NB100-116, Novus Biologicals), (4) LDLR (1:1000, mouse, monoclonal, clone C7, catalog no. sc-18823, Santa Cruz Biotechnology), (5) Akt (1:1000, rabbit, polyclonal, catalog no. sc-1618, Santa Cruz Biotechnolog,), (6) Akt phosphorylated at serine 473 (phospho-Akt-Ser473; 1:1000, rabbit, polyclonal, catalog no. sc-7985-R, Santa Cruz Biotechnology), and (7) PTEN (1:1000, mouse, monoclonal, clone A2b1, catalog no. sc-7974, Santa Cruz Biotechnology). Membranes were then washed and incubated with the appropriate secondary antibody conjugated to an infrared dye (IRDye®): (1) anti-mouse immunoglobulin (Ig)G with IRDye® 800, or (2) anti-rabbit IgG with IRDye® 680. Membranes were then captured by a near-IR (NIR) fluorescence imaging system (Odyssey® CLx, Li-Cor, Lincoln, NE, USA) across two channels. Quantification of bands was achieved by an Odyssey® program package (Image Studio 5.0) and normalized to the α-tubulin loading control.
Duolink® proximity ligation assay and fluorescence confocal microscopy
Duolink® proximity ligation assay (PLA) was used to determine whether APE1 and DROSHA interact in situ in HeLa cells (Duolink® PLA Technology, Sigma-Aldrich). Duolink® PLA allows the microscopic detection and quantification of target protein-protein interactions (<40 nm) as individual fluorescent signals.27 The assay, and subsequent imaging by confocal microscopy, was performed according to the supplier’s directions and established protocols.25,28 Briefly, HeLa cells were incubated with hydrogen peroxide (H2O2, 1 mM) for a duration of 15, 30, and 60 min on coverslips. Then, cells were permeabilized for 10 min with PBS + Triton X-100 0.4%. After two PBS washes, cells were incubated for 30 min with blocking solution (37°C). Then, HeLa cells were serially treated with primary antibodies against: (1) APE1 (1:22, mouse, monoclonal, clone 13B8E5C2, catalog no. NB100-116, Novus Biologicals, Littleton, CO, USA) for 3 hours (37°C), followed by an antibody against (2) DROSHA (1:200, rabbit, polyclonal, catalog no. ab85027, Abcam, Cambridge, UK) overnight (37°C). The cover slips were washed two times with buffer A. The slips were then incubated with PLA probes in antibody diluent for 1 hour (37°C). After two washes with buffer A, ligation was executed with ligase diluted in ligation stock for 30 min (37°C), wherein the two oligonucleotides in the PLA probes are hybridized to the circularization oligonucleotides. Cells were twice washed with buffer A prior to 100 min incubation with amplification stock solution (37°C), which contains polymerase for rolling circle amplification and Cy3-labeled oligonucleotides. After two washes of with buffer B, cells were incubated with APE1-FITC (catalog no. LS‑C317203, LifeSpan BioSciences, Seattle, WA, USA) as a fluorescent staining reference for cell nuclei. The slips were PBS-washed and mounted with the Duolink® in situ mounting medium.
HeLa cells were imaged using confocal microscopy (Leica TCS SP5, Leica Microsystems, Wetzlar, Germany), with the microscope settings kept constant to enable comparison between images. Duolink® PLA signal counts were measured with the BlobFinder program (Centre for Image Analysis, Uppsala University, Uppsala, Sweden). Live cells with noncondensed nuclei were manually counted, and the Duolink® PLA signal count was normalized to the cell count to formulate the Duolink® PLA signals/cell metric. A total of 30 cells were chosen at random per experimental condition. We confirmed that negative control samples without the primary antibody treatment against DROSHA were devoid of PLA readout.
APE1 endonuclease activity by abasic site incision assay
The endonuclease action of APE1 was measured in HeLa cell lysates by tracking abasic site incision according to established protocols.18,20 The assay relies on a substrate, which is a double-stranded DNA 26-mer, that contains at position 14 one tetrahydrofuranyl unit that simulates an abasic site.20 Endonuclease action by APE results in substrate cleavage and the generation of a 14-mer fragment, which is detected on a gel. HeLa cell lysates were prepared to a volume of 10 µl containing 12.5 ng of protein in buffer [50 mM Tris-HCl buffer at pH 7.5, with 10 mM MgCl2, 50 mM KCl, 1 μg/ml BSA, and 1 mM dithiothreitol (DTT)]. A 26-mer substrate (100 nM) was added to the lysates, which were incubated for 15 minutes at a temperate of 37°C. Following the enzymatic reaction, products were annealed with: (1) a dye-conjugated probe 5’- DY-782-AAT TCA CCG GTA CCF TCT AGA ATT CG-3’ (F represents the tetrahydrofuran unit) to track cleaved products, and (2) a complementary oligonucleotide without a dye label 5’-CGA ATT CTA GAG GGT ACC GGT GAA TT-3’. Quenching was accomplished by a formamide buffer [96% formamide, 10 mM EDTA, and 6X gel-loading buffer (Fermentas, Thermo Fisher Scientific)] and DNA molecules were resolved on a PAGE gel (20%) under denaturing conditions. Bands were visualized by a NIR fluorescence imaging system (Odyssey® CLx, Li-Cor) and quantified as the fraction of substrate conversion to cleaved fragments employing an imaging program (Image Studio 5.0, Li-Cor).
Generation and stable infection of LDLR overexpression plasmid
The plasmid to induce overexpression of LDLR was generated within a lentiviral vector (pEZ-Lv105, GeneCopoeia, Rockville, MD, USA). The complete LDLR cDNA sequence was cloned into pEZ-Lv105 to generate the LDLR overexpression plasmid. The plasmid sequence was confirmed by DNA sequencing. pEZ-Lv105 lentiviral vectors were packaged into lentiviruses, which were employed to infect cell cultures to create stable lines according to established protocols.29
Generation and stable infection of anti-miR-92b small hairpin RNA plasmids
Plasmids carrying anti-miR-92b small hairpin RNA (shRNA; sh-miR-92b) or scrambled negative control shRNA were generated within a lentiviral vector (pGreenPuro, System Biosciences, Palo Alto, CA, USA).30 Plasmids were confirmed by DNA sequencing. The pGreenPuro lentiviral vectors were packaged into lentiviruses, which were employed to infect cell cultures to create stable lines according to established protocols.29
Transient transfection with siRNAs or mimics
The siRNAs against LDLR (si-LDLR) or scrambled negative control siRNA were generated by GenePharma. Cells were transiently transfected with siRNAs (20 nM) utilizing RNAi-mate Transfection Reagent (GenePharma, Shanghai, China). In addition, miR-92b mimics or scrambled negative control mimics were generated by GenePharma. Cells were transiently transfected with mimics (20 nM) utilizing Lipofectamine® 2000 transfection reagent (Invitrogen).
Luminescence reporter assay of miR-92b binding to LDLR 3’-UTR
Plasmids to report binding of miR-92b-3p to LDLR 3’-UTR were generated within luciferase reporter plasmids (pMIR-REPORT, Invitrogen). The sequence of either WT or mutant 3’-UTR of LDLR was cloned into pMIR-REPORT at the 3’-end of the firefly luciferase gene to generate WT or mutant 3’-UTR LDLR reporter plasmids, respectively, as follows: WT 3’-UTR of LDLR (containing the WT miR-92b binding sites; WT binding site A (3967-3986 bp): AGTGGCTC … ACGCCTGTAATC, WT binding site B (4100-4119 bp): GGTGGC … GGGCACC … TGTAGTC) or a mutant 3’-UTR of LDLR (containing the complementary sequence of the WT miR-92b binding sites (underlined); mutant binding site A: AGTCCGTC … TGCCCACTTTAC, mutant binding site B: GGTCCG … CCCCTGC … ACTTGAC). The use of the WT miRNA binding site’s complementary sequence is an acceptable method of creating a mutated 3’UTR reporter vector.31,32
The reporter assay of miR-92b binding to LDLR 3’-UTR was conducted in HEK 293T cultures in 96-well plates. Transfection of reporter plasmids (0.01 μg Renilla reporter and 0.1 μg pMIR-REPORT firefly reporter) was performed on cells that had reached a confluence of 70% using Lipofectamine® 2000 transfection reagent (Invitrogen). After 48 hours, the HEK 293T cells were lysed, and luminescence signal from the cell extracts was quantified with the Dual-Luciferase® Reporter Assay System. Results were expressed as average values of firefly luciferase signal relative to the Renilla luciferase signal.
Cellular viability and chemosensitivity assays for SiHa and CaSki cells
SiHa and CaSki cells were deposited to 96-well plates at 2000 cells/well (viability assays), or 5000 cells/well (chemosensitivity assays). The following day, they either underwent transient transfection with si-LDLRs, miR-92b mimics, or scrambled negative controls, or they underwent stable transfection with lentiviral sh-miR-92b.
For the viability assays, SiHa and CaSki cultures were grown and tested for viability at 0, 24, 48, 72, and 96 hour-time points using a Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Gaithersburg, MD, USA). The CCK-8 kit utilizes WST-8, which is processed by viable cells to produce a colorful product that is quantifiable on a microplate reader at 450 nm. For the chemosensitivity assays, SiHa and CaSki cultures were incubated with paclitaxel (ranging from 0 to 160 nM) and cisplatin (ranging from 0 to 40 μM; both Sigma-Aldrich) over 48 hours at which point cellular viability was quantified using CCK-8.
Flow cytometric analysis of apoptosis in SiHa and CaSki cells
Plated SiHa and CaSki cultures underwent transfection (vide supra). After 24 hours, the cells were harvested and incubated with propidium iodide (PI; 50 μg/ml) and fluorescein isothiocyanate (FITC)-conjugated Annexin V according to the manufacturer’s instructions (Annexin V-FITC/PI Apoptosis Detection Kit, Kaiji, Nanjing, China). Data analysis was performed on a flow cytometer with the ModFit program (BD Biosciences, Franklin Lakes, NJ, USA).
Flow cytometric analysis of cell-cycle progression in SiHa and CaSki cells
Plated SiHa and CaSki cultures underwent transfection (vide supra) After 24 hours, cells were incubated for 16 to 20 hours in media supplemented with nocodazole (100 ng/ml).30 SiHa and CaSki cells were harvested, fixed in ethanol (70%; −20°C, 24 hours), and incubated with PI (50 μg/ml, Kaiji). Data analysis was performed on a flow cytometer with the ModFit program (BD Biosciences).
Analysis of caspase-3/7 activation in SiHa and CaSki cells
SiHa and CaSki cells were deposited to 96-well plates at 4000 cells/well for overnight incubation. The growth media was exchanged for serum-free media, and cells were incubated for 24 hours. Caspase-3/-7 activation was quantified using Promega’s Caspase-Glo® assay kit and expressed as the change in readout upon treatment (relative to vehicle) over the average luminescence readout of control wells.
Tumor xenograft grown in vivo in nude mice
Nude mice, aged 5 to 6 weeks, female, were procured from the National Rodent Laboratory Animal Resources (Shanghai, China). The mice were given four days to adapt to their new surroundings. At this point, CaSki cells (4.0 × 106 cells) with stable shRNA-mediated KD of miR-92b or scrambled negative controls (n = 9 per cohort) were implanted into the groin (right side) of each animal. The increase in tumor mass was registered in grams. Animals were sacrificed 28 days after implantation.
Statistical analyses
All reported values are represented as means ± standard errors of the mean (SEMs) of three biological replicates comprised of at least three technical replicates each (unless otherwise specified). Statistical comparisons were performed by using the Student’s t test or one-way analysis of variance with Bonferroni post hoc testing where appropriate. The application of other statistical procedures has been noted where necessary. A p value <0.05 was deemed statistically significant.
Results
APE1 binds precursor miR-92b forms
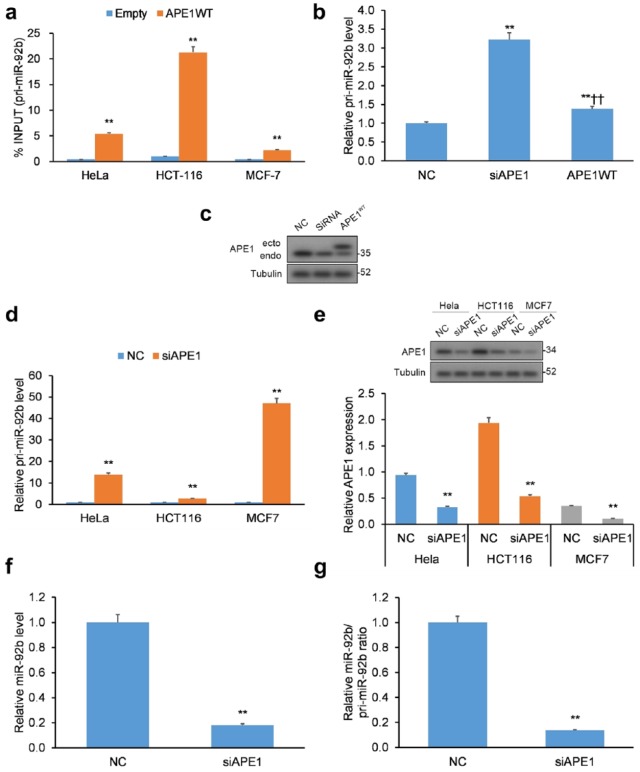
Our study sought to uncover the mechanism of miRNA regulation by APE1, centering our efforts on miR-92b that has been shown to be upregulated upon genotoxic stress but downregulated following APE1 KD.15 APE1 is known to directly interact with structured RNA oligonucleotides,10,11 so we surmised it could interact with the double-stranded, primary transcript pri-miR-17~92 (hereinafter pri-miRNA-92b).33 To assess this hypothesis, we conducted RIP on a human cervical (HeLa) cancer cell line transiently transfected with WT APE1 with a FLAG tag. By qRT-PCR, the levels of pri-miR-92b that immunoprecipitated with APE1 were significantly increased for all three cancer cell lines (i.e. HeLa, HCT116, and MCF7) examined [Figure 1(a)].
Figure 1.
APE1 binds to pri-miR-92b. (a) Validation of APE1-pri-miR-92b binding in the HeLa, HCT-116, and MCF-7 cancer cell lines. qRT-PCR of APE1-bound pri-miR-92b in cell lines transfected with APE1 WT FLAG-tag protein vector or empty vector. Data reported as proportion of immunoprecipitated pri-miR-92b relative to pri-miR-92b in total inputted RNA. (b) Pri-miR-92b levels evaluated in APE1-silenced HeLa cells. (c) Western blotting validation of APE1 silencing in HeLa cells with tubulin normalization. (d) Pri-miR-92b levels in APE1-silenced HeLa, HCT-116, and MCF-7 cell lines. (e) Western blotting validation of APE1 silencing in HeLa, HCT-116, and MCF-7 cells with tubulin normalization. (f) Mature miR-92b levels and (g) mature miR-92b-to-pri-miR ratios in APE1-silenced HeLa cells. Mature miR-92b levels were normalized to RNU44, while pri-miR-92b levels were normalized to GAPDH.
Data are represented as means ± SEMs. *p < 0.05, **p < 0.01.
APE1, apurinic/apyrimidinic endodeoxyribonuclease 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SEM, standard error of the mean; WT, wild type.
Due to APE1’s ability to cleave structured RNA,10,11 we hypothesized a possible function of APE1 in the processing of miR-92b. HeLa clones were transiently transfected with small interfering RNA (siRNA)-mediated KD of APE1 [Figure 1(b, c)] and assessed for pri-miR-92b levels. In addition, HeLa, HCT116, and MCF7 cells were transiently transfected with APE1-siRNA [Figure 1(d, e)] and assessed for pri-miR-92b levels. Under both transfection conditions, APE1 KD induced elevated expression of pri-miR-92b in HeLa cells. Similar results were observed in the two other cancer cell lines HCT116 and MCF7 [Figure 1(d, e)].
Elevated pri-miR-92b levels in cells with APE1 KD imply a build-up of primary transcripts induced by aberrant processing of pri-miR-92b. To test the possibility, we measured the levels of mature miR-92b in HeLa cells, which were lowered with a concurrent elevation of pri-miR-92b [Figure 1(f)] and resulted in a decreased miR/pri-miR ratio [Figure 1(g)] in response to APE1 KD. Mouse cells (CH12F3) with APE1 knockout (KO) were employed to confirm the role of APE1 on miR to pri-miR ratios (Supplementary Figure 1). Cumulatively, these findings imply that miR-92b levels and processing of pri-miR-92b are altered in cells with APE1 KD.
Mature miR-92b levels decrease upon inhibition of APE1
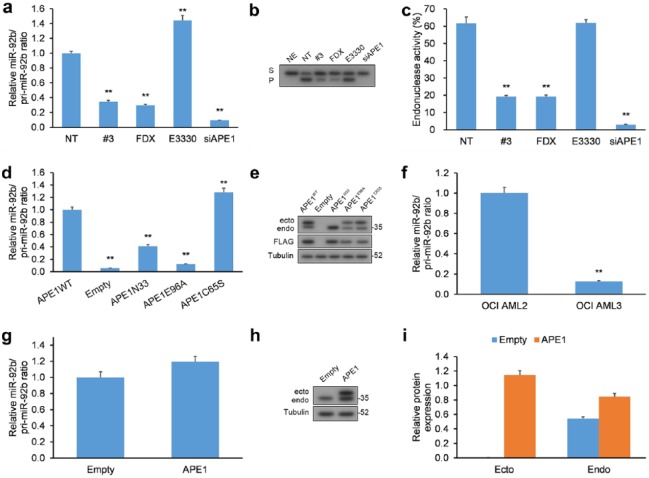
Since APE1 is a multifunctional enzyme with both endonuclease and redox activities,15 we set out to determine which was responsible for processing pri-miR-92b. HeLa cells were treated with distinct inhibitors of APE1’s activities: (a) fiduxosin, an inhibitor of the interaction of APE1 with nucleophosmin (NPM1),34 which localizes APE1 to the nucleus and activates BER,35 (b) compound #3, an inhibitor of the endonuclease activity of APE1 through a mechanism distinct to fiduxosin,34, 36 and (c) E3330, an inhibitor of the redox activity of APE1.37,38 HeLa cells were exposed to each APE1 inhibitor for 24 h, and both precursor (pri-miR-92b) and mature miR-92b forms were measured by qRT-PCR [Figure 2(a)]. Dosage and duration of treatment with inhibitors were selected according to their influence on the viability of cancer cells.34,39
Figure 2.
Inhibiting the endonuclease action of APE1 negatively impacts the processing of miR-92b. (a) HeLa cells incubated with fiduxosin (FDX; 40 µM), compound #3 (20 µM), or E3330 (100 µM) for 24 h or HeLa cells with siRNA-mediated APE1 KD had their levels of mature miR-92b and pri-miR-92b quantified by qRT-PCR. Mature miR and pri-miR levels were normalized to RNU44 and GAPDH, respectively. (b, c) Abasic site incision activity in HeLa extracts prepared from cells exposed to the same inhibitors or from cells with APE1 KD. (b) Typical image of polyacrylamide gel under denaturing conditions of the enzymatic transformation of substrate (S) containing an abasic site to incised product (P). (c) Histogram representation of the kinetics of cell-extract endonuclease activity denoted as transformation of S-to-P expressed as a percentage. (d) HeLa cells with APE1 KD were transiently transfected with FLAG-containing vectors encoding siRNA-resistant WT APE1, APE1E96A, APE1C65S, and APE1N-33 had their levels of mature miR-92b and miR-92b quantified by qRT-PCR. Mature miR and pri-miR levels were normalized to RNU44 and GAPDH, respectively. (e) WB of extracts from HeLa cells with APE1 KD (endo) and co-expressing siRNA-resistant FLAG-tagged WT and mutant APE1 (ecto). (f) OCI/AML3 (NPMc+) versus control OCI/AML2 (WT NPM1) cell lines had their levels of mature miR-92b and miR-92b quantified by qRT-PCR. Mature miR and pri-miR levels were normalized to RNU44 and GAPDH, respectively. (g) HeLa cells were transiently transfected with FLAG-containing vector encoding WT APE1 had their levels of mature miR-92b and miR-92b quantified by qRT-PCR. Mature miR and pri-miR levels were normalized to RNU44 and GAPDH, respectively. (h, i) Western blotting analysis of extracts from HeLa cells with overexpression of FLAG-tagged WT APE1.
Data are represented as means ± SEMs. *p < 0.05, **p < 0.01.
APE1, apurinic/apyrimidinic endodeoxyribonuclease 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KD, knockdown; NE, no cell extract, NT, nontreated cells; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SEM, standard error of the mean; WB, western blot; WT, wild type.
Inhibiting the endonuclease action of APE1 by fiduxosin and compound #3, which was assayed by cleavage of a substrate with an abasic site in vitro, induced an increase in pri-miR-92b [Figure 2(a–c) and Supplementary Figure 2(a)]. On the other hand, E3330, the inhibitor of redox activity, only exhibited a marginal effect on the levels of mature miR-92b. Defective processing of miRNA induced by treatment with inhibitors of endonuclease activity did not affect net APE1 protein levels [Supplementary Figure 2(b)].
To corroborate our findings from the experiments employing fiduxosin, compound #3, and E3330 APE1 inhibitors, we transfected HeLa cells with APE1 KD of mutant versions of APE1: (a) APE1 with mutation to E96A (APE1E96A) devoid of nuclease activity,40 (b) APE1 with mutation to C65S (APE1C65S) devoid of redox activity,41 and (c) APE1 without the 33 N-terminal amino acids (APE1N-33) devoid of binding to NMP.5,12,42 All mutant APE1 levels were similar in these cells whose native, WT APE1 levels were very low [Figure 2(d, e)].
The experiments in HeLa cells transfected with APE1-bearing mutations [Figure 2(d, e)] confirm the findings from the experiments with APE1 inhibitors [Figure 2(a–c)], and attribute the endonuclease activity and N-terminal domain of APE1 as necessary features to correctly process pri-miR-92b. On the other hand, expression of APE1C65S, which is devoid of redox activity, induced a marginal elevation in mature miR-92b levels, attributable to secondary effects from overexpressing it in cells, as reported in the literature.43 Furthermore, due to the dual function of the 33 residue N-terminal domain of APE1 that provides both localization cues and interacts with proteins other than NPM1,16 we cannot eliminate the possibility that the effect of APE1N-33 on pri-miR-92b processing derives from loss of either of these functions.
OCI/AML3 cells have stable expression of a mutant NPM1 that localizes to the cytoplasm (NPMc+) with concurrent abnormal cytoplasmic localization of APE1, resulting in impaired BER in the nucleus.35 Additionally, these OCI/AML3 cells exhibit elevated levels of pri-miR-92b versus cells expressing WT NPM1 [Figure 2(f)]. Our findings in OCI/AML3 cells also mirror the results of fiduxosin inhibition34 and suggest that NPM1 positively influences APE1 in its role in the processing of pri-miRNAs.
Finally, since we observed that APE1 KD decreased pri-miR-92b processing, we determined whether overexpression (OE) of APE1 could exert an adverse result [Figure 2(g–i)]. Consequently, a vector of APE1 with a FLAG tag was transfected into HeLa cells, but resulted in no significant change to the miR/pri-miR ratio. The lack of any discernible effect could arise from other proteins involved in pri-miR-92b processing, which could limit the processing rate. Cumulatively, our findings demonstrate that the endoribonuclease property of APE1 may be essential in the early stages of miR-92b processing; however, contributions from other proteins may factor in.
Genotoxic stress augments the interaction between APE1 and DROSHA
Since APE1 KD induces a greater extent of RNA oxidation,12 we hypothesized that the dependency in the processing of pri-miR-92b on APE1 could arise from the influence of the DROSHA microprocessor complex on APE1. Indeed, previous research has demonstrated that APE1, through interacting with the DROSHA microprocessor complex, can promote miR maturity.15
As oxidative stress promotes the APE1/DROSHA interaction,15 an interaction between DROSHA and APE1 was induced upon addition of hydrogen peroxide (H2O2, 1 mM), which maximized after 15 minutes [Supplementary Figure 3(a, b)], and suggested an involvement by APE1 in the degradation of oxidatively damaged RNA.44 Hydrogen peroxide induced elevation in the level of pri-miR-92b within 15 minutes compared to untreated cells [NT; Supplementary Figure 3(c, d)], but did not affect the level of the mature miR-92b strand as evidenced by the kinetic studies of the ratio of miR-92b-to-pri-miR-92b. This situation could arise due to inhibition of pri-miR-92b maturation from H2O2-induced oxidative stress [Supplementary Figure 3(c, d)].
To gain further evidence, we employed a probe with an aldehyde-reactive group (ARP) to quantify the loss of oxidatively damaged bases from pri-miRNAs upon expression of APE1.26 APE1 KD induced a hike in the extent of oxidative damage on pri-miR-92b, which was reversed upon co-expression of WT APE1 [Supplementary Figure 3(e)]. These results validate APE1’s role in pri-miR-92b degradation, thereby impacting maturation of miR-92b in a genotoxic environment.
miR-92b directly suppresses LDLR expression in human cervical carcinoma cells
miRNAs function by binding to and regulating the transcription of their mRNA targets.45 Consequently, to clarify the role of miR-92b in cervical cancer, we set out to uncover its potential target genes. We queried the miRanda (microrna.org and miRbase), TargetScan (www.targetscan.org/), and PicTar (http://pictar.mdcberlin.de/) databases and identified LDLR as a possible miR-92b target [Supplementary Figure 4(a)].
In order to validate that miR-92b targets LDLR in cervical cancer cells, miR-92b mimics were transfected into CaSki and SiHa cells, which attenuated protein expression of LDLR [Supplementary Figure 4(b, c)]. We then created a reporter assay of LDLR gene expression by linking WT or mutant 3’-UTR of LDLR to the gene for firefly luciferase. HEK 293T cells were then co-transfected with either the WT or mutant 3’-UTR LDLR reporter plasmid with vectors expressing miR-92b mimics or negative control (NC). miR-92b mimics decreased the luminescence signal in HEK 293T cells transfected with the WT 3’-UTR LDLR reporter plasmid, but not with the mutant 3’-UTR LDLR reporter plasmid [Supplementary Figure 4(d)]. This results implicate the presence of at least one miR-92b binding site on the 3’-UTR of LDLR mRNA. As additional support, we measured the levels of miR-92b and LDLR in 50 human cervical cancer tumor biopsy samples and determined an inverse relationship between them [R2 = 0.2972, p < 0.01; Supplementary Figure 4(e)]. Overall, these results imply miR-92b’s targeting of LDLR in cervical cancer cells.
APE1 impacts the LDLR–PTEN pathway and corresponds to miR-92b levels
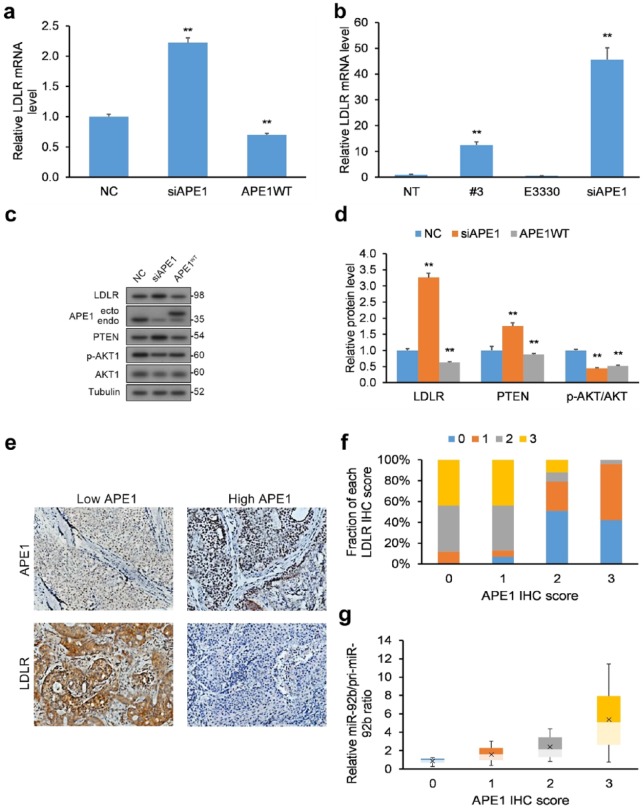
We next evaluated the functional applicability of our APE1/miR-92b-based experimental results by measuring the downstream effect of APE1 KD on miR-92b’s target LDLR. We quantified mRNA transcript and protein levels of LDLR in HeLa cells in response to APE1 KD [Figure 3(a, c, d)] and treatment with inhibitors [Figure 3(b)], which were both increased with a simultaneous decrease in the ratio of miR-92b-to-pri-miR-92b. We also assessed the levels of PTEN and Akt phosphorylation, since LDLR negatively impacts the Akt signaling pathway via PTEN upregulation.46 As anticipated, HeLa cells with APE1 KD displayed raised LDLR, elevated PTEN, and lowered Akt phosphorylation [Figure 3(c, d)], while APE1 overexpression (OE) rescued these effects.
Figure 3.
LDLR levels are influenced by miR-92b processing by APE1. (a) HeLa cells with WT APE1 and APE1 KD had their total RNA purified and their levels of LDLR mRNA transcripts quantified by qRT-PCR following reverse transcription. LDLR mRNA transcripts were normalized to GAPDH. (b) HeLa cells incubated with compound #3 (20 µM) or E3330 (100 µM) for 24 h or HeLa cells with siRNA-mediated APE1 KD had their levels of LDLR mRNA quantified by qRT-PCR following reverse transcription. LDLR mRNA transcripts were normalized to GAPDH. (c, d) HeLa cells with APE1 KD had their total levels of LDLR, PTEN, and p-Akt/Akt quantified by WB, and normalized to tubulin. Data are represented as means ± SEMs. *p < 0.05, **p < 0.01. (e) IHC analysis of LDLR and APE1 protein levels in human cervical cancer biopsy samples (n = 50), exemplified by typical IHC images. LDLR protein has higher in tumor biopsies with lower APE1 protein levels, and conversely lower in biopsies with greater APE1 levels. (f) Semi-quantification of LDLR scores (0, 1, 2, or 3) expressed as a percentage within each category of APE1 expression (0, 1, 2, or 3) represented as bar chart. Scoring was assessed by the following criteria: (i) score of 0: no protein expression in cancer cells, (ii) score of 1: faint protein expression in <10% of cancer cells, (iii) score of 2: faint to intermediate expression in >10% of cancer cells, and (iv) score of 3: strong expression in >10% of cancer cells. (g) miR-to-pri-miR ratios for miR-92b for each APE1 score level.
Data are represented as means ± SEMs. *p < 0.05, **p < 0.01.
APE1, apurinic/apyrimidinic endodeoxyribonuclease 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IHC, immunohistochemistry; KD, knockdown; LDLR, low-density lipoprotein receptor; NE, no cell extract, NT, nontreated cells; PTEN, phosphatase and tensin homolog; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SEM, standard error of the mean; WB, western blot; WT, wild type.
We next assessed the comparative expression of APE1 mRNA, miR-92b, and LDLR mRNA levels in human cervical cancer tumor biopsy samples (n = 50) relative to matching healthy cervical tissue (n = 50). We found significantly higher levels of miR-92b in cervical tumors relative to matched healthy cervical tissue (Supplementary Figure 5). However, we did not discover significant differences in APE1 mRNA expression or LDLR mRNA expression (Supplementary Figure 5).
We next assessed the interrelationship between APE1 and miR-92b processing to LDLR protein levels in human cervical cancer tumor biopsy samples (n = 50). Immunohistochemistry (IHC) was used to determine LDLR and APE1 protein levels, which were semi-quantitatively scored [0, 1, 2, or 3; Figure 3(e)]. An inverse correlation emerged between high LDLR protein levels in cervical cancer biopsy samples to low APE1 protein levels [Figure 3(f)]. qRT-PCR was employed to quantify mature and primary miR-92b levels and their ratios. Another relationship emerged between greater miR-to-pri-miR ratios (more miR-92b processing) and higher APE1 protein levels [Figure 3(g)]. The results in cervical cancer biopsies corroborate the findings in HeLa cells in vitro, namely that APE1’s miR-92b processing negatively correlates with LDLR protein levels.
miR-92b increases the proliferation of human cervical carcinoma cells in vitro and in xenograft tumors in vivo
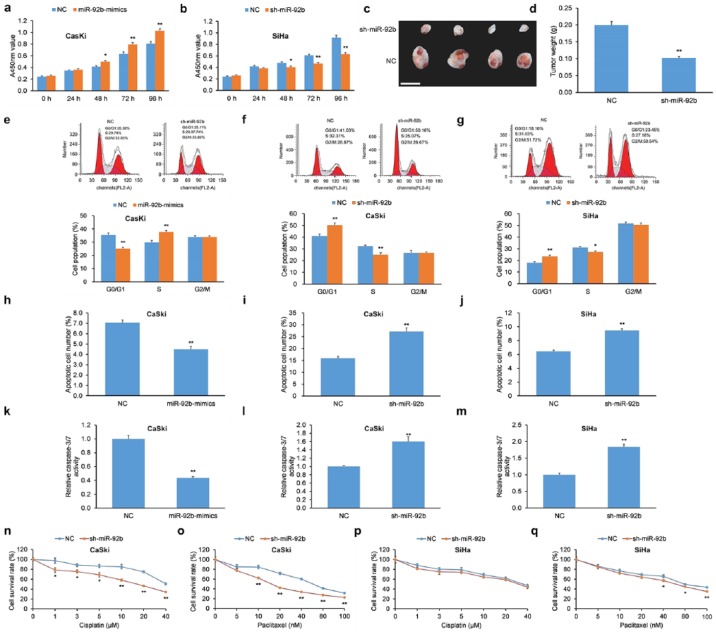
We hypothesized that miR-92b could impact cellular proliferation of cervical cancer cells since its expression levels have an inverse relationship with the tumor-suppressive LDLR–PTEN pathway.46 We employed two human cervical carcinoma lines CaSki and SiHa for these experiments. CaSki cells were transiently transfected with miR-92b mimics, while SiHa cells were infected with a shRNA against miR-92b (sh-miR-92b) to silence miR-92b expression. Transfection of miR-92b mimics in CaSki cells increased cellular proliferation compared with negative controls [NCs; Figure 4(a)], while infection of sh-miR-92b in SiHa cells decreased cellular proliferation compared with NCs [Figure 4(b)].
Figure 4.
miR-92b increases proliferation, promotes cell-cycle progression, decreases apoptosis, and reduces chemosensitivity of cervical carcinoma cells. (a) The proliferation of CaSki cells with miR-92b mimics was greater versus cells with scrambled NC as determined by a CCK-8 assay. (b) The proliferation of SiHa cells with sh-miR-92b was lowered versus cells with scrambled NC as determined by a CCK-8 assay. (c) Morphological features of xenograft tumors in nude mice implanted with CaSki cells with sh-miR-92b or scrambled NC. Scale bar = 1 cm. (d) Xenograft tumor masses from nude mice implanted with sh-miR-92b CaSki cells were lower versus implanted CaSki cells with scrambled NC. (e) CaSki cells with miR-92b mimics exhibited lower proportion of cells in G0/G1 phase and greater proportion in S phase upon treatment with nocodazole. (f, g) CaSki and SiHa cells with sh-miR-92b elicited G0/G1 phase arrest upon treatment with nocodazole. (h–j) Apoptosis levels of (h) CaSki cells with miR-92b mimics and (i, j) CaSki and SiHa cells with sh-miR-92b were assessed by Annexin V-FITC/PI-based flow cytometry. (k–m) Caspase-3/7 activity was assessed in (k) CaSki cells with miR-92b mimics and (l, m) CaSki and SiHa cells with sh-miR-92b. (n–q) Chemosensitivity to cisplatin and paclitaxel was assessed in CaSki and SiHa cells with sh-miR-92b or scrambled NC.
Data are represented as means ± SEMs. *p < 0.05, **p < 0.01.
CCK-8, Cell Counting Kit-8; FITC, fluorescein isothiocyanate; NC, negative control; PI, propidium iodide; SEM, standard error of the mean.
To assess the influence of miR-92b on the proliferation of cervical carcinoma cells in vivo, CaSki cells with shRNA-mediated miR-92b KD or NC counterparts were implanted into nude mice using a subcutaneous injection. Animals were sacrificed after 28 days and their tumors were excised and weighed. The final masses of tumor xenografts derived from implanted CaSki cells with miR-92b KD were less than xenografts implanted with NC cells [Figure 4(c, d)]. Like the in vitro findings, these data indicate that miR-92b promotes proliferation of human cervical cancer cells in vivo.
miR-92b elicits cell-cycle progression of human cervical carcinoma cells
Since miR-92b increased cellular proliferation of human cervical cancer cells, we then determined whether it also had an influence of cell-cycle progression in these cells. CaSki cells transfected with miR-92b mimics were incubated with nocodazole, and cell-cycle analysis revealed a smaller fraction in the G0/G1 phase and a greater proportion in the S phase [Figure 4(e)] compared with NCs. In agreement, shRNA-mediated miR-92b KD elicited an opposite effect in CaSki and SiHa cells, which resulted in enhanced G0/G1 phase arrest [Figure 4(f, g)].
miR-92b decreases apoptosis and chemosensitivity of human cervical carcinoma cells
Due to its influence in cellular proliferation and cell-cycle arrest in human cervical cancer, we next evaluated whether miR-92b influenced apoptosis and chemosensitivity. CaSki cells with miR-92b mimics had lower apoptosis [Figure 4(h)], while CaSki and SiHa cells with shRNA-mediated miR-92b KD displayed higher apoptosis [Figure 4(i, j)] as measured by Annexin V/PI flow cytometry. To corroborate those findings, CaSki cells with miR-92b mimics had lower caspase-3/7 activity [Figure 4(k)] while CaSki and SiHa cells with shRNA-mediated miR-92b KD displayed higher caspase-3/7 activity [Figure 4(l, m)].
Next, the role of miR-92b on the susceptibility of human cervical carcinoma cells to chemotherapy was tested using cisplatin and paclitaxel. CaSki cells with shRNA-mediated miR-92b KD displayed greater chemosensitivity to cisplatin versus NCs [Figure 4(n)]. Moreover, CaSki and SiHa cells with shRNA-mediated miR-92b KD displayed greater chemosensitivity to paclitaxel than NCs [Figure 4(o, q)]. However, SiHa cells with shRNA-mediated miR-92b KD did not achieve statistical significance with respect to cisplatin chemosensitivity [Figure 4(p)].
KD of LDLR mimics the influence of miR-92b in human cervical carcinoma cells
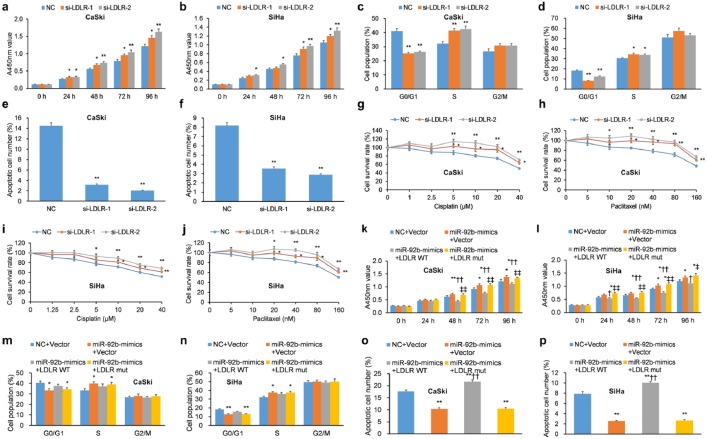
LDLR influences a number of cellular pathways important in carcinogenesis,46,47 but its impact in cervical cancer is presently unknown. To analyze its role in cervical cancer, three LDLR-targeting siRNAs were designed and their efficacy, particularly for si-LDLR-1 and si-LDLR-2, was demonstrated in CaSki and SiHa cells (Supplementary Figure 6). si-LDLR-1 and si-LDLR-2 promoted cellular proliferation of CaSki and SiHa cells [Figure 5(a, b)] and promoted G1/S phase progression upon addition of nocodazole [Figure 5(c, d)]. Furthermore, si-LDLR-1 and si-LDLR-2 decreased apoptosis in CaSki and SiHa cells [Figure 5(e, f)] and reduced their chemosensitivity to cisplatin and paclitaxel [Figure 5(g–j)]. Overall, LDLR KD enhanced cellular proliferation, promoted cell-cycle progression, reduced apoptosis, and reduced chemosensitivity, features common to the influence of miR-92b mimics in human cervical cancer cells.
Figure 5.
LDLR KD mimics miR-92b, while LDLR overexpression mitigates the influence of miR-92b, in human cervical carcinoma cells. (a, b) The proliferation of SiHa and CaSki cells with transient transfection of LDLR siRNAs was greater compared to cells transfected with scrambled vector as assessed by a CCK-8 assay. (c, d) siRNA-mediated KD of LDLR elicited G1 arrest in (c) CaSki and (d) SiHa cells incubated with nocodazole. (e, f) SiHa and CaSki cells with transient transfection of LDLR-targeted siRNAs exhibited decreased apoptosis as determined by incubation with 50 μg/ml PI and FITC-conjugated Annexin V, and evaluated on a flow cytometer. (g–j) The proliferation of SiHa and CaSki cells with transient transfection of LDLR-targeted siRNAs was quantified by CCK-8 after a 48-hour incubation with paclitaxel (0–160 nM) and cisplatin (0–40 μM) to assess chemosensitivity. Transiently transfected si-LDLR-1 and si-LDLR-2 attenuated the chemosensitivity of SiHa and CaSki cells to paclitaxel and cisplatin relative to NCs. (k, l) The proliferation of SiHa and CaSki cells with transient transfection of miR-92b mimics with LDLR-WT, LDLR-mut, or NC was assessed by CCK-8 assay. (m, n) The cell-cycle progression of SiHa and CaSki cells with transient transfection of miR-92b mimics with LDLR-WT, LDLR-mut, or NC was determined by flow cytometry upon incubation with nocodazole. (o, p) Apoptosis of SiHa and CaSki cells with transient transfection of miR-92b mimics with LDLR-WT, LDLR-mut, or NC was determined by incubation with 50 μg/ml PI and FITC-conjugated Annexin V, and evaluated on a flow cytometer.
Data are represented as means ± SEMs. *p < 0.05, **p < 0.01.
CCK-8, Cell Counting Kit-8; FITC, fluorescein isothiocyanate; KD, knockdown; LDLR, low-density lipoprotein receptor; NC, negative control; PI, propidium iodide; SEM, standard error of the mean; siRNA, short interfering RNA; WT, wild type.
OE of LDLR mitigates the influence of miR-92b on proliferation and apoptosis
We hypothesized that, as LDLR is a functional target of miR-92b, OE of LDLR would rescue the impact of miR-92b mimics on proliferation and apoptosis of cervical cancer cells. To shed light on the hypothesis, lentiviral constructs containing the LDLR sequence with WT or mutant LDLR 3’-UTRs (LDLR-WT and LDLR-mut, respectively) were infected into CaSki and SiHa cells. These cells were also transfected with miR-92b mimics. As anticipated, cells with miR-92b mimics and OE of LDLR-WT displayed decreased cervical cancer cell proliferation [Figure 5(k, l)], enhanced G1 phase arrest [Figure 5(m, n)], and promoted apoptosis [Figure 5(o, p)]. Notably, these effects were not observed in cells with miR-92b mimics and OE of LDLR-mut. Cumulatively, these experiments illustrate that OE of WT LDLR reverses the pro-malignant impact of miR-92b mimics on cellular proliferation, cell-cycle progression, and apoptosis in cervical cancer cells. These findings advocate LDLR as a functional intermediary of miR-92b.
Discussion
Here, we reveal that the BER endonuclease APE1, via interacting with the DROSHA microprocessor complex, enhances miR-92b maturity, thereby suppressing LDLR–PTEN pathway expression and upregulating Akt activation under baseline conditions. As oxidative stress enhances the APE1-DROSHA interaction and APE1 KD promotes oxidized pri-miR-92b levels, our findings also validate APE1’s role in regulating degradation of primary miRNA transcripts. Noting that APE1’s endoribonuclease activity affects many oxidized and abasic miRNA species,12 our findings may be generalized to other oxidative stress-regulated miRNAs which are also subject to oxidative, alkylative, or abasic damage.48 As APE1 appears to preferentially target the pri-miRNA duplexes of these oxidative stress-regulated miRNA species 17, APE1’s role in regulating responses to genotoxicity and in carcinogenesis should be closely investigated.
miR-92b is a product of the polycistronic miR-17~92 cluster situated on chromosome 13, whose aberrant overexpression is present in several cancers.49–53 miR-92b has been shown to target several anticancer genes, including the tumor and metastasis suppressor RECK,54 the tumor suppressor DKK3,55 and the epithelial–mesenchymal transition (EMT) suppressor DAB2IP.56 More importantly, miR-92b promotes cell proliferation, invasiveness, and chemoresistance in various cancer cell types via target gene suppression.54–57 Thus, rescuing anticancer gene expression via miR-92b suppression may be a promising therapeutic strategy against many malignancies. As APE1 promotes miR-92b maturation, the application of APE1 inhibitors may represent a novel adjuvant approach to bolster chemotherapeutic efficacy in certain chemoresistant tumors.
Herein, our results reveal that miR-92b suppresses human cervical carcinoma growth in vitro and in vivo via LDLR suppression. LDLR is best-known for its role in maintaining proper cholesterol homeostasis, thereby reducing proatherosclerotic foam cell development.58 Notably, LDLR also upregulates PTEN, a potent tumor suppressor that suppresses the prosurvival Akt pathway.46 Therefore, miR-92b-mediated LDLR–PTEN pathway downregulation produces Akt pathway activation, which has been shown to participate in the transformation and chemoresistance of cervical cells.59,60 These findings are consistent with previous work showing that APE1 KD upregulates PTEN expression via suppressing miR-221/22 maturation.15 On the basis of this combined evidence, we propose a regulatory model where APE1 negatively regulates the tumor-suppressive LDLR–PTEN pathway on two levels: via post-transcriptional regulation of LDLR expression by miR-92b and PTEN expression by miR-221/22. Further research is needed to see if this multi-tier regulatory model can be carried over to other cancer cell signaling pathways that are subject to APE1 regulation.
It should be noted that we identified LDLR as one miR-92b target that suppresses the proliferation and cell-cycle progression while promoting apoptosis and chemosensitivity in cervical carcinoma cells. However, based on our analysis of the miRNA databases, other tumor suppressor genes could also be targets of miR-92b. Further research is needed to identify other gene targets that may be involved in miR-92b-mediated cervical carcinoma progression.
We also observed increased chemosensitivity to cisplatin and paclitaxel in miR-92b-silenced CaSki cells and decreased chemosensitivity to cisplatin and paclitaxel in LDLR-silenced CaSki cells. Moreover, we also observed that miR-92b-silenced SiHa cells responded differently to cisplatin and paclitaxel, showing no significant chemosensitivity to cisplatin and a less profound chemosensitivity to paclitaxel. This evidence suggests that the APE1-miR-92b-LDLR axis may be involved in regulating chemosensitivity to cisplatin and paclitaxel in cervical carcinoma cells, albeit in varying degrees depending on the particular cell line. Interestingly, preliminary trans-omics data from epithelial ovarian carcinoma cells suggests that LDLR promotes cisplatin chemosensitivity via the LDLR- lysophosphotidylcholine (LPC)-family with sequence similarity 83-member B (FAM83B)- fibroblast growth factor receptors axis.61 However, we did not investigate the mechanism underlying this phenomenon in cervical carcinoma cells. Further research is needed to investigate the mechanism(s) that may be involved in APE1-miR-92b-LDLR- mediated chemosensitivity to cisplatin and paclitaxel in cervical carcinoma cells.
Conclusion
APE1 enhances miR-92b processing, thereby suppressing LDLR expression and enhancing cervical carcinoma progression. Our identification of the novel APE1-miR-92b-LDLR axis improves our understanding of the complex pathogenesis of cervical carcinoma and reveals a novel therapeutic strategy for combating this disease.
Supplemental Material
Supplemental material, TAM_Supplementary_Information for The endonuclease APE1 processes miR-92b formation, thereby regulating expression of the tumor suppressor LDLR in cervical cancer cells by Yi Sun, Yun Feng, Guiqian Zhang and Ya Xu in Therapeutic Advances in Medical Oncology
Footnotes
Author Contributions: The following author contributions are noted: Yi Sun conceived and designed the study. Yun Feng and Guiqian Zhang performed the experimental procedures. Guiqian Zhang analyzed the data. Ya Xu and Yi Sun drafted the manuscript.
Funding: This work was supported by the Yunnan Provincial Health and Family Planning Commission Medical Discipline Leaders Training Program (grant no. D-201670). The Martin Expert Workstation used in this study was supported by a separate grant (grant no. 2018IC106). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Yi Sun, Department of Clinical Laboratory Medicine, the First People’s Hospital of Yunnan Province, No. 157, Jinbi Road, Kunming, Yunnan Province, China.
Yun Feng, Department of Reproductive Gynecology, The First People’s Hospital of Yunnan Province, Kunming, Yunnan Province, China.
Guiqian Zhang, Department of Clinical Laboratory Medicine, The First People’s Hospital of Yunnan Province, Kunming, Yunnan Province, China.
Ya Xu, Department of Clinical Laboratory Medicine, The First People’s Hospital of Yunnan Province, Kunming, Yunnan Province, China.
References
- 1. Antoniali G, Malfatti MC, Tell G. Unveiling the non-repair face of the Base Excision Repair pathway in RNA processing: a missing link between DNA repair and gene expression? DNA Repair 2017; 56: 65–74. [DOI] [PubMed] [Google Scholar]
- 2. Wang J-X, Gao J, Ding S-L, et al. Oxidative modification of miR-184 enables it to target Bcl-xL and Bcl-w. Mol Cell 2015; 59: 50–61. [DOI] [PubMed] [Google Scholar]
- 3. Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 2016; 16: 35. [DOI] [PubMed] [Google Scholar]
- 4. Friedberg EC. A history of the DNA repair and mutagenesis field: the discovery of base excision repair. DNA Repair 2016; 37: A35–A39. [DOI] [PubMed] [Google Scholar]
- 5. Li M, Wilson III DM. Human apurinic/apyrimidinic endonuclease 1. Antioxid Redox Signal 2014; 20: 678–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fantini D, Fantini D, Vascotto C, et al. APE1/Ref-1 regulates PTEN expression mediated by Egr–1. Free Radic Res 2008; 42: 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abbotts R, Jewell R, Nsengimana J, et al. Targeting human apurinic/apyrimidinic endonuclease 1 (APE1) in phosphatase and tensin homolog (PTEN) deficient melanoma cells for personalized therapy. Oncotarget 2014; 5: 3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chattopadhyay R, Das S, Maiti AK, et al. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol 2008; 28: 7066–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sengupta S, Mantha AK, Mitra S, et al. Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene 2011; 30: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berquist BR, McNeill DR, Wilson III DM. Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J Mol Biol 2008; 379: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barnes T, Kim W-C, Mantha AK, et al. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res 2009; 37: 3946–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vascotto C, Fantini D, Romanello M, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol 2009; 29: 1834–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poletto M, Vascotto C, Scognamiglio PL, et al. Role of the unstructured N-terminal domain of the hAPE1 (human apurinic/apyrimidinic endonuclease 1) in the modulation of its interaction with nucleic acids and NPM1 (nucleophosmin). Biochem J 2013; 452: 545–557. [DOI] [PubMed] [Google Scholar]
- 14. Chohan M, Mackedenski S, Li W-M, et al. Human apurinic/apyrimidinic endonuclease 1 (APE1) has 3′ RNA phosphatase and 3′ exoribonuclease activities. J Mol Biol 2015; 427: 298–311. [DOI] [PubMed] [Google Scholar]
- 15. Antoniali G, Serra F, Lirussi L, et al. Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nat Commun 2017; 8: 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vascotto C, Cesaratto L, Zeef LA, et al. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics 2009; 9: 1058–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai N, Zhong Z, Cun Y, et al. Alteration of the microRNA expression profile in human osteosarcoma cells transfected with APE1 siRNA. Neoplasma 2013; 60: 384–394. [DOI] [PubMed] [Google Scholar]
- 18. Vascotto C, Fantini D, Romanello M, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol 2009; 29: 1834–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vascotto C, Cesaratto L, Zeef LA, et al. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics 2009; 9: 1058–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poletto M, Malfatti MC, Dorjsuren D, et al. Inhibitors of the apurinic/apyrimidinic endonuclease 1 (APE1)/nucleophosmin (NPM1) interaction that display anti-tumor properties. Mol Carcinog 2016; 55: 688–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gangadhar GD, Tamas V, Michelle DW, et al. Synthesis, structure−activity relationships, and biological evaluation of fatty alcohol phosphates as lysophosphatidic acid receptor ligands, activators of PPARγ, and inhibitors of autotaxin. J Med Chem 2005; 48: 4919–4930. [DOI] [PubMed] [Google Scholar]
- 22. Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med 2007; 28: 375–395. [DOI] [PubMed] [Google Scholar]
- 23. Kelley MR, Luo M, Reed A, et al. Functional analysis of novel analogues of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref–1. Antioxid Redox Signal 2011; 14: 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antoniali G, Lirussi L, D’Ambrosio C, et al. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol Biol Cell 2014; 25: 532–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lisa L, Giulia A, Carlo V, et al. Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells. Mol Biol Cell 2012; 23: 4079–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka M, Han S, Küpfer PA, et al. Quantification of oxidized levels of specific RNA species using an aldehyde reactive probe. Anal Biochem 2011; 417: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alam MS. Proximity ligation assay (PLA). Curr Protoc Microbiol 2018; 123: e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carlo V, Elena B, Li M, et al. Knock-in reconstitution studies reveal an unexpected role of Cys-65 in regulating APE1/Ref-1 subcellular trafficking and function. Mol Biol Cell 2011; 22: 3887–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 2009; 4: 495–505. [DOI] [PubMed] [Google Scholar]
- 30. Scherr M, Venturini L, Battmer K, et al. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res 2007; 35: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X, Liu J, Zhang X, et al. MiR-520b promotes the progression of non-small cell lung cancer through activating Hedgehog pathway. J Cell Mol Med 2019; 23: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oever MV, Muldoon D, Mathews W, et al. miR-29 regulates type VII collagen in recessive dystrophic epidermolysis bullosa. J Invest Dermatol 2016; 136: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Concepcion CP, Bonetti C, Ventura A. The miR-17–92 family of microRNA clusters in development and disease. Cancer J 2012; 18: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poletto M, Malfatti MC, Dorjsuren D, et al. Inhibitors of the apurinic/apyrimidinic endonuclease 1 (APE1)/nucleophosmin (NPM1) interaction that display anti-tumor properties. Mol Carcinog 2016; 55: 688–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vascotto C, Lirussi L, Poletto M, et al. Functional regulation of the apurinic/apyrimidinic endonuclease 1 by nucleophosmin: impact on tumor biology. Oncogene 2014; 33: 2876. [DOI] [PubMed] [Google Scholar]
- 36. Rai G, Vyjayanti VN, Dorjsuren D, et al. Synthesis, biological evaluation, and structure–activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem 2012; 55: 3101–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med 2007; 28: 375–395. [DOI] [PubMed] [Google Scholar]
- 38. Zhang J, Luo M, Marasco D, et al. Inhibition of apurinic/apyrimidinic endonuclease I’s redox activity revisited. Biochemistry 2013; 52: 2955–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cesaratto L, Codarin E, Vascotto C, et al. Specific inhibition of the redox activity of ape1/ref-1 by e3330 blocks tnf-α-induced activation of IL-8 production in liver cancer cell lines. PLoS One 2013; 8: e70909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barzilay G, Walker LJ, Robson CN, et al. Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res 1995; 23: 1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walker L, Robson C, Black E, et al. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol 1993; 13: 5370–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fantini D, Vascotto C, Marasco D, et al. Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. Nucleic Acids Res 2010; 38: 8239–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vascotto C, Bisetto E, Li M, et al. Knock-in reconstitution studies reveal an unexpected role of Cys-65 in regulating APE1/Ref-1 subcellular trafficking and function. Mol Biol Cell 2011; 22: 3887–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tell G, Wilson DM, Lee CH. Intrusion of a DNA repair protein in the RNome world: is this the beginning of a new era? Mol Cell Biol 2010; 30: 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5: 522. [DOI] [PubMed] [Google Scholar]
- 46. Basu V. Role of low-density lipoprotein receptor as a potential suppressor of growth and survival of colorectal cancer cells. Electronic theses and dissertations. 5929. https://scholar.uwindsor.ca/etd/59292017.
- 47. Furuya Y, Sekine Y, Kato H, et al. Low-density lipoprotein receptors play an important role in the inhibition of prostate cancer cell proliferation by statins. Prostate Int 2016; 4: 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simms CL, Zaher HS. Quality control of chemically damaged RNA. Cell Mol Life Sci 2016; 73: 3639–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005; 65: 9628–9632. [DOI] [PubMed] [Google Scholar]
- 50. Takakura S, Mitsutake N, Nakashima M, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci 2008; 99: 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mu P, Han Y-C, Betel D, et al. Genetic dissection of the miR-17∼ 92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 2009; 23: 2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsuchida A, Ohno S, Wu W, et al. miR-92 is a key oncogenic component of the miR-17–92 cluster in colon cancer. Cancer Sci 2011; 102: 2264–2271. [DOI] [PubMed] [Google Scholar]
- 53. Tsz-fung FC, Mankaruos M, Scorilas A, et al. The miR-17–92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. J Urol 2010; 183: 743–751. [DOI] [PubMed] [Google Scholar]
- 54. Lei L, Huang Y, Gong W. Inhibition of miR-92b suppresses non-small cell lung cancer cells growth and motility by targeting RECK. Mol Cell Biochem 2014; 387: 171–176. [DOI] [PubMed] [Google Scholar]
- 55. Li Q, Shen K, Zhao Y, et al. MiR-92b inhibitor promoted glioma cell apoptosis via targeting DKK3 and blocking the Wnt/beta-catenin signaling pathway. J Transl Med 2013; 11: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang J, Wang B, Hui K, et al. miR-92b targets DAB2IP to promote EMT in bladder cancer migration and invasion. Oncol Rep 2016; 36: 1693–1701. [DOI] [PubMed] [Google Scholar]
- 57. Li Y, Li L, Guan Y, et al. MiR-92b regulates the cell growth, cisplatin chemosensitivity of A549 non-small cell lung cancer cell line and target PTEN. Biochem Biophys Res Commun 2013; 440: 604–610. [DOI] [PubMed] [Google Scholar]
- 58. Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol 2013; 13: 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma Y-Y, Wei S-J, Lin Y-C, et al. PIK3CA as an oncogene in cervical cancer. Oncogene 2000; 19: 2739. [DOI] [PubMed] [Google Scholar]
- 60. Li L, Wei X, Pan Y, et al. LAPTM4B: a novel cancer-associated gene motivates multidrug resistance through efflux and activating PI3K/AKT signaling. Oncogene 2010; 29: 5785. [DOI] [PubMed] [Google Scholar]
- 61. Chang W-C, Wang H-C, Cheng W-C, et al. Low-density lipoprotein receptor-mediated lipidome-transcriptome reprogramming impulses to cisplatin insensitivity. 2019: SSRN 3311843. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, TAM_Supplementary_Information for The endonuclease APE1 processes miR-92b formation, thereby regulating expression of the tumor suppressor LDLR in cervical cancer cells by Yi Sun, Yun Feng, Guiqian Zhang and Ya Xu in Therapeutic Advances in Medical Oncology