Abstract
Chromatin-modifying enzymes are frequently overexpressed in cancer cells, and their enzymatic activities play important roles in changing the epigenetic landscape responsible for tumorigenesis. However, many of these proteins also execute noncatalytic functions, which are poorly understood. In fission yeast, overexpression of Epe1, a histone demethylase homolog, causes heterochromatin defects. Interestingly, in our recent work, we discovered that overexpressed Epe1 recruits SAGA, a histone acetyltransferase complex important for transcriptional regulation, to disrupt heterochromatin, independent of its demethylase activity. Our findings suggest that overexpressed chromatin-modifying enzymes can alter the epigenetic landscape through changing their proteomic environments, an area that needs to be further explored in dissecting disease etiology associated with overexpression of chromatin regulators.
Keywords: Epe1, heterochromatin, H3K9 methylation, SAGA, transcription
Introduction
Changes in chromatin environments, especially the histone posttranslational modification profiles, play major roles in regulating gene expression and specifying cell identity. Mutations or aberrant expression of chromatin-modifying enzymes is frequently associated with human diseases.1 Among histone modifications, histone acetylation is generally associated with transcriptionally active regions, whereas histone methylation has distinct effects depending on the residue that is methylated and the degree of methylation. For example, trimethylation of H3K9 is frequently associated with constitutive heterochromatin, trimethylation of H3K27 is commonly found at repressed genes, and trimethylation of H3K4 often marks active gene promoters. These residue- and degree-specific methylations are deposited by histone methyltransferases and removed by histone demethylases.
Most histone demethylases contain a catalytic JmjC domain, which uses Fe2+and 2-oxoglutarate as cofactors to catalyze lysine demethylation.2 Interestingly, a number of JmjC domain–containing histone demethylases are overexpressed in distinct types of cancer cells.3 The cancer-promoting mechanism of these overexpressed proteins is often attributed to increased demethylase activities, leading to epigenetic changes more conducive to tumorigenesis. However, what remains often unexplored is whether changes in the levels of chromatin-modifying factors cause additional effects unrelated to their catalytic activities.
Overexpression of a JmjC Domain Protein Epe1 Disrupts Heterochromatin Through Its Noncatalytic Function
In fission yeast, similar to higher eukaryotes, H3K9 methylation is critical for the recruitment of HP1 family proteins to form constitutive heterochromatin at repetitive DNA elements to repress the transcription of and recombination between DNA repeats.4 Paradoxically, heterochromatin assembly requires transcription from the underlying DNA repeats, with these transcripts serving as a source for the production of double-stranded RNAs and small interfering RNAs (siRNAs) through the RNA interference pathway. The siRNAs are loaded into the RNA-induced transcriptional silencing (RITS) complex and guide RITS to the nascent transcripts still attached to repeat regions. RITS then recruits the H3K9 methyltransferase Clr4 to initiate H3K9 methylation at these regions.
A number of mechanisms exist that promote transcription within heterochromatin. For example, the HP1 homolog in flies, Rhino, recruits a transcriptional factor Moonshiner to initiate Pol II–mediated transcription within heterochromatin.5 In fission yeast, the transcription of repeat elements is elevated during the S phase of the cell cycle,6,7 suggesting that the DNA replication machinery may disassemble heterochromatin to provide an accessible chromatin environment for Pol II. During other stages of the cell cycle, transcription within heterochromatin is dependent on Epe1, although the mechanism by which Epe1 promotes transcription within heterochromatin is unknown.8
Epe1 is a resident heterochromatin protein that contains a JmjC domain found in many histone demethylases. H297A, a point mutation that disrupts the conserved iron-binding residue in the JmjC domain, phenocopies epe1∆: the expansion of H3K9me domains and the maintenance of ectopic H3K9me islands,8-13 suggesting that Epe1 is an active H3K9 demethylase. However, it is difficult to detect demethylase activity of Epe1 in vitro. Interestingly, overexpression of either the wild-type or the H297A mutant of Epe1 results in the loss of H3K9me within heterochromatin, suggesting that Epe1 has catalysis-independent functions for the removal of H3K9me.8,9
To further study the function of Epe1, we conducted a genetic screen with the fission yeast deletion library for mutants that can alleviate heterochromatin defects associated with Epe1 overexpression and identified mutations in several components of the SAGA complex.14 SAGA is a conserved multisubunit complex that serves as a transcriptional coactivator and has histone acetylation and deubiquitylation activities.15 We demonstrated that the histone acetyltransferase activity of SAGA is important for Epe1 function as a catalytic-dead point mutation of the acetyltransferase subunit Gcn5 rescues Epe1 overexpression. Through mass spectrometry analysis of proteins associated with overexpressed Epe1, we discovered that SAGA interacts with Epe1. Interestingly, one mutation outside of SAGA histone acetyltransferase module, tra1∆, was also identified from the genetic screen that rescues Epe1 overexpression. Tra1 is responsible for the recruitment of SAGA togene promoters by interacting with transcription factors, and tra1∆ does not affect SAGA integrity or itshistone acetyltransferase activity in fission yeast.15,16 Co-immunoprecipitation analysis showed that Tra1 mediates the interaction between Epe1 and SAGA. Chromatin immunoprecipitation analysis showed that overexpressed Epe1 recruits SAGA to heterochromatin, resulting in high histone acetylation and transcription, which contribute indirectly to the loss of H3K9me through transcription-mediated histone turnover. Further epistasis analysis showed that both the demethylase activity and interaction with SAGA contributed to heterochromatin defects when Epe1 is overexpressed.
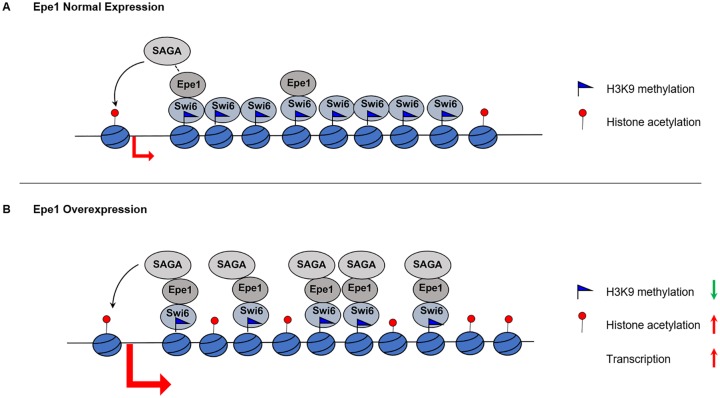
It is interesting to note that when Epe1 is expressed at normal levels, it only weakly interacts with SAGA, and such interaction recruits a small amount of SAGA to heterochromatin. Epe1 uses these weak interactions to facilitate the recruitment of Pol II to heterochromatin and transcription of the underlying repetitive DNA elements. These transcripts then serve both as a source for the production of siRNAs and as a platform for the recruitment of histone H3K9 methyltransferase to initiate heterochromatin formation. Therefore, at normal Epe1 expression levels, the weak interaction between Epe1 and SAGA is sufficient for transcription, but the transcription is not strong enough to disrupt the heterochromatin. However, when Epe1 is overexpressed, it recruits an excess amount of SAGA to the heterochromatin, acetylating more histones and triggering transcription strong enough to disrupt the heterochromatin (Figure 1). Thus, our findings suggest that overexpressed JmjC domain proteins can enhance relatively weak interactions, leading to the recruitment of excess epigenetic modifiers to alter the epigenetic landscape independent of JmjC domain’s enzymatic activity.
Figure 1.
Model for the role of Epe1-SAGA interaction in regulating heterochromatin stability. (A) At its normal expression levels, Epe1 weakly interacts with SAGA, resulting in the recruitment of small amount of SAGA to promote transcription for heterochromatin assembly. (B) When Epe1 is overexpressed, it recruits excess SAGA to promote histone acetylation and transcription strong enough to disrupt heterochromatin by transcription-mediated histone turnover.
Noncatalytic Functions of Other JmjC Histone Demethylases
Many other JmjC histone demethylases possess noncatalytic functions as well. For example, Kdm2b, an H3K36 demethylase, recruits the Polycomb Repressive Complex 1 (PRC1) to unmethylated CpG islands of developmentally regulated genes through Kdm2b’s zinc finger domain.17 The catalytic activity of Kdm2b does not play a role in this process as mutating the JmjC domain of Kdm2b has no effect on the recruitment of PRC1. In addition, JMJD1A, an H3K9 demethylase, is phosphorylated by protein kinase A in response to β-adrenergic receptor activation and forms a complex with SWI/SNF and peroxisome proliferator-activated receptor gamma (PPARγ) to mediate long-range enhancer-promoter interactions. Similar to Kdm2b, JMJD1A’s histone demethylase activity is dispensable for complex formation and for regulating gene expression. Furthermore, Drosophila dKDM4A, an H3K36 demethylase, affects position effect variegation independent of its demethylase activity.18 Finally, UTX, an H3K27 demethylase, modulates enhancer activation by recruiting the H3K4 methyltransferase MLL4, which cooperates with histone acetyltransferase p300 to activate gene expression. Again, the demethylase activity of UTX is dispensable for this interaction.19
Nonenzymatic Functions of Other Chromatin-Modifying Enzymes Overexpressed in Cancer
Besides histone demethylases, other chromatin-modifying enzymes overexpressed in cancer also possess catalytic-independent functions. For example, G9a, an H3K9 methyltransferase, is overexpressed in highly invasive lung cancer cells, and its overexpression is associated with poor prognosis.20 Interestingly, G9a induces DNA methylation independently of its methyltransferase activity in embryonic stem cells,21,22 although whether the noncatalytic function of G9a plays a role in lung cancer tumorigenesis remains to be explored. In addition, EZH2, which is the H3K27 methyltransferase in the PRC2 complex, is a known oncogene and is upregulated in several cancer types including prostate and breast cancers.23 Cancer cells with SWI/SNF mutations depend on EZH2 to proliferate, but only partially on the catalytic activity of EZH2.24 Moreover, EZH2 promotes transcription of androgen receptor independent of its H3K27 methyltransferase activity or PRC2 in prostate cancer cells.25 With these observations, it is clear that overexpressed chromatin-modifying enzymes, including JmjC demethylases, can promote cancer through their nonenzymatic functions.
Concluding Remarks
In summary, our article showed that a noncatalytic function of an overexpressed JmjC domain protein Epe1 contributes to heterochromatin defects. Similarly, many other JmjC histone demethylases and chromatin-modifying enzymes possess noncatalytic functions and are overexpressed in cancer cells. Therefore, studying whether and how these noncatalytic functions modulate cellular functions will elucidate how the dysregulation of chromatin-modifying enzymes contributes to disease.
Note added in proof: Since the acceptance of our article, Sorida et al, 2019, PLoS Genetics 15(6): e1008129 also show that Epe1 has JmjC-independent functions.
Acknowledgments
We thank Tasneem Ebrahim and Kendra Zhang for critical reading of the manuscript.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Work in the Jia lab is supported by National Institutes of Health grant R35-GM126910.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: KB wrote the paper with guidiance from SJ.
References
- 1. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006;439:811–816. [DOI] [PubMed] [Google Scholar]
- 3. Johansson C, Tumber A, Che K, et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics 2014;6:89–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet 2007;8:35–46. [DOI] [PubMed] [Google Scholar]
- 5. Andersen PR, Tirian L, Vunjak M, Brennecke J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 2017;549:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 2008;451:734–737. [DOI] [PubMed] [Google Scholar]
- 7. Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 2008;18:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zofall M, Grewal SIS. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Molec Cell 2006;22:681–692. [DOI] [PubMed] [Google Scholar]
- 9. Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J 2007;26:4670–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SIS. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science (New York, NY) 2012;335:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ragunathan K, Jih G, Moazed D. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science (New York, NY) 2014;348:1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Audergon PN, Catania S, Kagansky A, et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science (New York, NY) 2015;348:132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J, Reddy BD, Jia S. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. Elife 2015;2015:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bao K, Shan CM, Moresco J, Yates J, III, Jia S. Anti-silencing factor Epe1 associates with SAGA to regulate transcription within heterochromatin. Genes Dev 2019;33:116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koutelou E, Hirsch CL, Dent SYR. Multiple faces of the SAGA complex. Curr Opin Cell Biol 2010;22:374-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helmlinger D, Marguerat S, Villén J, et al. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J 2011;30:2843–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nature Cell Biol 2013;15:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Colmenares SU, Swenson JM, Langley SA, Kennedy C, Costes SV, Karpen GH. Drosophila histone demethylase KDM4A has enzymatic and non-enzymatic roles in controlling heterochromatin integrity. Dev Cell 2017;42:156.e5-169.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang SP, Tang Z, Chen CW, et al. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol Cell 2017;67:308.e6-321.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen MW, Hua KT, Kao HJ, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res 2010;70:7830–7840. [DOI] [PubMed] [Google Scholar]
- 21. Dong KB, Maksakova IA, Mohn F, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 2008;27:2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 2008;27:2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res 2011;17:2613–2618. [DOI] [PubMed] [Google Scholar]
- 24. Kim KH, Kim W, Howard TP, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015;21:1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim J, Lee Y, Lu X, et al. Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep 2018;25:2808.e4–2820.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]