Abstract
Background
Starch biosynthesis in endosperm is a key process influencing grain yield and quality in maize. Although a number of starch biosynthetic genes have been well characterized, the mechanisms by which the expression of these genes is regulated, especially in regard to microRNAs (miRNAs), remain largely unclear.
Results
Sequence data for small RNAs, degradome, and transcriptome of maize endosperm at 15 and 25 d after pollination (DAP) from inbred lines Mo17 and Ji419, which exhibit distinct starch content and starch granule structure, revealed the mediation of starch biosynthetic pathways by miRNAs. Transcriptome analysis of these two lines indicated that 33 of 40 starch biosynthetic genes were differentially expressed, of which 12 were up-regulated in Ji419 at 15 DAP, one was up-regulated in Ji419 at 25 DAP, 14 were up-regulated in Ji419 at both 15 and 25 DAP, one was down-regulated in Ji419 at 15 DAP, two were down-regulated in Ji419 at 25 DAP, and three were up-regulated in Ji419 at 15 DAP and down-regulated in Ji419 at 25 DAP, compared with Mo17. Through combined analyses of small RNA and degradome sequences, 22 differentially expressed miRNAs were identified, including 14 known and eight previously unknown miRNAs that could target 35 genes. Furthermore, a complex co-expression regulatory network was constructed, in which 19 miRNAs could modulate starch biosynthesis in endosperm by tuning the expression of 19 target genes. Moreover, the potential operation of four miRNA-mediated pathways involving transcription factors, miR169a-NF-YA1-GBSSI/SSIIIa and miR169o-GATA9-SSIIIa/SBEIIb, was validated via analyses of expression pattern, transient transformation assays, and transactivation assays.
Conclusion
Our results suggest that miRNAs play a critical role in starch biosynthesis in endosperm, and that miRNA-mediated networks could modulate starch biosynthesis in this tissue. These results have provided important insights into the molecular mechanism of starch biosynthesis in developing maize endosperm.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5945-1) contains supplementary material, which is available to authorized users.
Keywords: Maize, Endosperm, Starch biosynthesis, miRNA, Regulatory network
Background
Maize (Zea mays L.) is grown extensively as a staple food and feed crop around the world. Starch is the most important component of maize endosperm, accounting for 70% of dry endosperm weight. The accumulation of starch in endosperm is one of the major factors determining maize yield, and the physical and chemical properties of starch can influence maize quality [1].
Starch biosynthesis is catalyzed by multiple, well-characterized enzymes, including ADP-glucose pyrophosphorylase (AGP), granule-bound starch synthase (GBSS), soluble starch synthase (SS), starch branching enzyme (SBE), starch debranching enzyme (DBE), and starch phosphorylase (PHO) [2–4]. Thus far, many genes associated with endosperm development have been cloned in maize. Among these genes, shrunken2 (Sh2), brittle2 (Bt2), GBSSI, SSI, SSIIa, SSIIIa, SBEI, SBEIIa, SBEIIb, and isoamylase 1 (ISA1) are predominantly involved in starch biosynthesis [2, 5–7]. Although the functions of many genes involved in starch biosynthesis are well understood, knowledge of the regulation of the expression of these genes remains limited.
As one of the main regulators of gene expression, transcription factors activate or repress the transcription of downstream genes upon binding to cis-acting elements in promoter regions. Only a few studies of the direct regulation of starch biosynthesis by transcription factors in cereals have been published. In barley, a WRKY transcription factor SUSIBA2 (sugar signaling in barley) can bind to the promoter of ISA1 to regulate its expression [8]. In maize, several transcription factors regulate the expression of starch biosynthetic genes. For example, ZmaNAC36 can regulate the expression of AGP large subunit 2 (AGPL2), AGP small subunit 2 (AGPS2), SSI, GBSSIIb, and SBEI [9]. ZmbZIP91 can regulate the expression of Bt2, SSI, SSIIIa, and ISA1 [10]; and ZmEREB156 can regulate the expression of Sh2 and SSIIIa [11]. Further, opaque2 (O2) and prolamine-box binding factor (PBF) can regulate the expression of pyruvate orthophosphate dikinase 1 (PPDK1), PPDK2, and SSIII [12].
Many metabolic processes in plants can be regulated by phytohormones [13]. Auxin (IAA), abscisic acid (ABA), brassinosteroid (BR), and gibberellin (GA) play important roles in starch biosynthesis and grain development. Palacios et al. [14] found that the AGP activity and starch content in Chlorella sorokiniana strains with normal levels of IAA accumulation were higher than in those with abnormal levels of IAA accumulation. In Arabidopsis, the sucrose induced expression of AGPL3 and SBE2.2 could be enhanced by ABA [15]. Similarly, the expression of SSI could be induced by ABA in maize endosperm [16]. In rice, increased BR increases UDP-glucose pyrophosphorylase (UGP) activity and starch content in grain [17]. During rice grain filling, higher GA content can lead to lower activities of AGP and other starch biosynthetic enzymes in grain and inhibit the accumulation of starch [18].
MicroRNAs (miRNAs) are a class of short non-coding RNAs that can modulate gene expression post-transcriptionally through sequence complementarity [19]. At present, there are only a few reports that miRNAs participate in the regulation of starch biosynthesis. miR394 can target sugar metabolism-related genes to regulate starch biosynthesis in cassava [20]. Overexpression of miR156 has increased starch content in plants such as Arabidopsis, maize, and switchgrass [21]. Recently, miRNAs have been shown to play an important role in grain development and grain filling. In rice, miR167 can regulate grain development through the auxin-miR167-ARF8 (auxin response factor)-OsGH3.2 (GH3, an auxin-binding protein) pathway [22]. Moreover, this pathway, together with the miR160-, miR164-, and miR390-mediated auxin signaling pathways, was confirmed to regulate grain filling in rice [23]. In addition, the miRNAs identified during endosperm development [24–26], grain filling [27], and kernel development [28] could have roles in starch accumulation in maize. miRNAs might modulate starch biosynthesis, which is critical for endosperm and grain development, by tuning the expression of transcription factors or mediating the effects of plant hormones.
Co-expression analyses have shown that co-expressed genes, those with similar expression profiles, can be involved in similar biological functions, which provides a feasible means to discover new genes [29, 30]. A recent study has reported a newly discovered starch synthase gene SSV by co-expression analysis [31]. Co-expression analysis is also an effective approach to identify genes related to a specific metabolic pathway. For instance, Rice Starch Regulator 1 (RSR1) in rice [32], and NAC36 and bZIP91 in maize [9, 10] were identified to regulate starch biosynthesis by co-expression analysis. Additionally, gene co-expression network analysis can be conducted using high-throughput gene expression profile data. Using available RNA sequencing (RNA-Seq) data, Huang et al. [33] constructed and optimized a large scale gene co-expression network in maize, and predicted a cell wall biosynthesis pathway from the merged network. Co-expression analysis has great utility for gene identification and functional analysis, and is an important strategy for enhancing our understanding of regulatory pathways.
There are few reports of the use of integrated high-throughput sequence data to unravel miRNA-mediated regulatory networks in developing maize endosperm. The present study analyzed small RNA, degradome, and transcriptome sequence data for endosperm at 15 and 25 d after pollination (DAP) from the maize inbred line Mo17 and its improved line Ji419, which exhibit distinct endosperm starch content and starch granule structure. These analyses identified and characterized differentially expressed miRNAs and their targets, and further, allowed construction of a co-expression regulatory network for starch biosynthesis in maize endosperm. Moreover, four miRNA-mediated regulatory pathways were chosen to implement expression pattern analysis, transient overexpression analysis, and transactivation assays to validate the modulating network. Our results will provide valuable information for a better understanding of the molecular mechanism of starch biosynthesis in maize endosperm.
Results
Transcriptome analysis of developing maize endosperm
Transcriptome analysis of developing endosperm revealed a total of 9241 differentially expressed genes (DEGs) between Mo17 and Ji419 at 15 DAP, of which 4590 genes were up-regulated and 4651 genes were down-regulated in Ji419 compared with Mo17. At 25 DAP, 1149 genes were differentially expressed, of which 552 genes were up-regulated and 597 genes were down-regulated in Ji419 compared with Mo17.
Gene Ontology (GO) term enrichment analysis suggested that the DEGs between Mo17 and Ji419 at 15 DAP were significantly enriched in biological processes including ‘glycogen biosynthesis’, cellular components such as ‘chloroplast’, and molecular functions including ‘catalytic activity’ (Additional file 1: Figure S1a, b). At 25 DAP, the DEGs were significantly enriched in biological processes including ‘auxin transport’, cellular components such as ‘phosphopyruvate hydratase’, and molecular functions including ‘phosphoenolpyruvate carboxylase activity’.
Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that the DEGs between Mo17 and Ji419 at 15 DAP were significantly enriched in biological pathways including ‘carbon metabolism’, ‘pyruvate metabolism’, and ‘glycolysis/gluconeogenesis’ (Additional file 1: Figure S1c, d). At 25 DAP, the DEGs were significantly enriched in biological pathways such as ‘carbon fixation in photosynthetic organisms’, ‘pyruvate metabolism’, ‘brassinosteroid biosynthesis’, and ‘other glycan degradation’.
Expression profiles of starch biosynthetic genes in developing endosperm
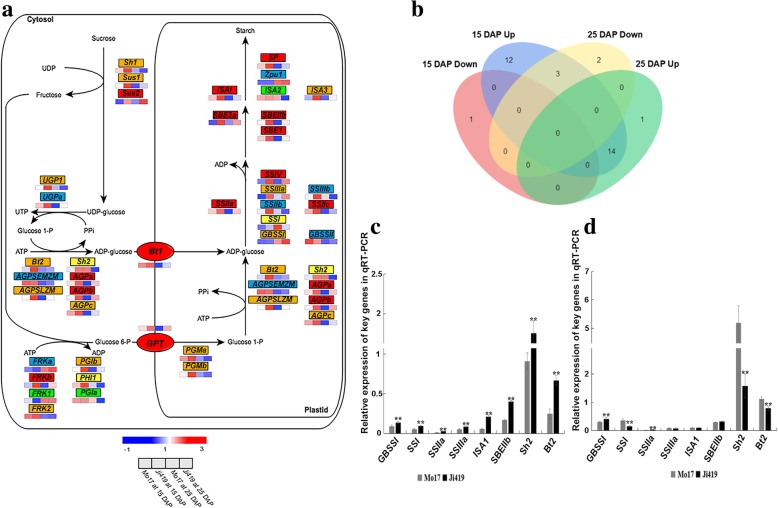
RNA-Seq analysis showed that 40 known starch biosynthetic genes were enriched in functional categories including ‘starch biosynthesis’, ‘glycogen biosynthesis’, ‘carbon metabolism’, and ‘glycolysis/gluconeogenesis’ (Additional file 2: Tables S1 and S2). The functions of these 40 genes, and the starch biosynthetic pathways in which they could be involved, are shown in Fig. 1a.
Fig. 1.
Expression profiles of starch biosynthetic genes. a The expression of 40 known starch biosynthetic genes and the pathways in which they could be involved in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP. The heat map represents the expression of each gene in a specific line. Red boxes represent up-regulated genes in Ji419 compared with Mo17 at both 15 and 25 DAP; orange boxes represent genes up-regulated in Ji419 at only one stage; yellow boxes represent genes up-regulated in Ji419 at one stage, but down-regulated at the other stage; green boxes represent genes down-regulated in Ji419 at only one stage; blue boxes represent genes with no differential expression between Mo17 and Ji419 at both 15 and 25 DAP. b Venn diagram of starch biosynthetic genes that are differentially expressed in the endosperm of Mo17 and Ji419 at 15 and 25 DAP. Red represents genes down-regulated in Ji419 compared with Mo17 at 15 DAP; blue represents genes up-regulated in Ji419 at 15 DAP; yellow represents genes down-regulated in Ji419 at 25 DAP; green represents genes up-regulated in Ji419 at 25 DAP. c-d Expression levels of eight key starch biosynthetic genes in the endosperm of Mo17 and Ji419 at 15 DAP (c) and 25 DAP (d). Black bars represent the expression of genes in Ji419; grey bars represent the expression of genes in Mo17. The data in c and d are means ± SD (n = 3). ** significant at p ≤ 0.01 by the Student’s t test
Among these 40 genes, 33 genes were differentially expressed, 12 genes were up-regulated only in Ji419 at 15 DAP, one gene was up-regulated only in Ji419 at 25 DAP, one gene was down-regulated only in Ji419 at 15 DAP, and two genes were down-regulated only in Ji419 at 25 DAP, compared to Mo17 (Fig. 1b). At both 15 and 25 DAP, 14 genes, including six starch biosynthetic genes SSIIa, SSIV, ISA1, SBEI, SBEIIa, and SBEIIb, were up-regulated in Ji419. In addition, three genes including Sh2, SSI, and phosphohexose isomerase1 (PHI1) were up-regulated in Ji419 at 15 DAP and down-regulated in Ji419 at 25 DAP (Fig. 1a, b).
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis showed that in Ji419 at 15 DAP, all eight key starch biosynthetic genes (GBSSI, SSI, SSIIa, SSIIIa, ISA1, SBEIIb, Sh2, and Bt2) were significantly up-regulated, and that in Ji419 at 25 DAP, GBSSI and SSIIa were significantly up-regulated, while SSI, Sh2 and Bt2 were significantly down-regulated compared with Mo17 (Fig. 1c, d), concordantly with the expression profiles obtained from RNA-Seq data (Additional file 2: Table S1).
Additionally, similar expression patterns were observed in these eight genes at six endosperm developmental stages: the transcript levels of these genes decreased from 15 to 20 DAP, increased from 20 to 25 DAP, and continuously decreased from 25 to 40 DAP (Fig. 2). The expression peak for the majority of these genes occurred at 15 or 25 DAP, indicating that starch metabolism in the endosperm of Ji419 and Mo17 might be more dynamic at these two stages. These eight genes could be further classified into two groups based on their expression patterns. One group includes SSI, SSIIIa, Sh2, and Bt2, which showed similar or higher expression in Ji419 compared with Mo17 at five developmental stages excluding 25 DAP. The other group includes GBSSI, SSIIa, ISA1, and SBEIIb, which exhibited higher expression in Ji419 than in Mo17 in all six developmental stages, which was consistent with the significantly higher starch content in the mature endosperm of Ji419 than in that of Mo17 (Additional file 3: Figure S2).
Fig. 2.
Analysis of the expression patterns of two miRNAs and their target genes, and eight key starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15, 20, 25, 30, 35, and 40 DAP. Red boxes represent miRNAs, yellow boxes represent target genes, and green boxes represent starch biosynthetic genes
Differential expression of miRNAs in developing endosperm
Sequencing of small RNAs generated 16,968,986 raw reads representing 5,065,474 unique reads from the endosperm of Mo17 at 15 DAP, and 14,965,571 raw reads representing 2,470,691 unique reads from the endosperm of Ji419. A further 15,215,476 raw reads representing 3,885,746 unique reads were generated from Mo17 endosperm at 25 DAP, and 16,315,425 raw reads representing 414,202 unique reads were generated from Ji419 endosperm at 25 DAP. In total, 314 miRNAs were detected, including 180 known miRNAs and 134 previously unknown ones. Most of these miRNAs ranged from 20 to 24 nt in length (Additional file 4: Figure S3a).
A total of 129 differentially expressed miRNAs were identified between Mo17 and Ji419 at 15 DAP, including 52 known miRNAs belonging to 21 miRNA families, and 77 newly discovered miRNAs (Additional file 2: Table S3). Among the 52 known miRNAs, 38 were down-regulated in Ji419 compared with Mo17, 22 of which were down-regulated by at least 3-fold. For example, miR528a-3p_L + 1 was down-regulated by 20-fold. Another 14 miRNAs were up-regulated in Ji419, of which seven were up-regulated by at least 3-fold, such as miR319b-5p, which was up-regulated by 17-fold. Among the 77 previously unknown miRNAs, 64 were down-regulated in Ji419, 31 of these were down-regulated by at least 3-fold, and PC-3p-104553_20 was down-regulated by 20-fold. Another 13 miRNAs were up-regulated in Ji419, while PC-3p-1338_720 was up-regulated by 253-fold.
A total of 131 differentially expressed miRNAs were identified between the two inbred lines at 25 DAP, including 52 known miRNAs belonging to 20 miRNA families and 79 newly discovered miRNAs (Additional file 2: Table S4). Among the 52 known miRNAs, 38 were down-regulated in Ji419 compared with Mo17, 29 of which were down-regulated by at least 3-fold. Among these, miR528a-3p_L + 1 and miR2118b were both down-regulated by 10-fold. In addition, 14 other miRNAs were up-regulated in Ji419, 10 of which were up-regulated by at least 3-fold, and one, miR159c-5p_L-1R-1, was up-regulated by 33-fold. Among the 79 newly discovered miRNAs, 68 were down-regulated in Ji419, 60 of which were down-regulated by at least 3-fold. Among these, PC-3p-383_2667, PC-5p-383_2667, and PC-5p-28889_59 were down-regulated by 10-fold. A further 11 miRNAs were up-regulated in Ji419, with PC-3p-1338_720 up-regulated by 170-fold.
Among these differentially expressed miRNAs, 35 and 11 were down- and up-regulated only in Ji419 at 15 DAP, respectively, while 39 and nine were down- and up-regulated only in Ji419 at 25 DAP, respectively (Additional file 4: Figure S3b). Meanwhile, 64 miRNAs were down-regulated and 13 were up-regulated in Ji419 at both 15 and 25 DAP. In addition, three miRNAs were up-regulated in Ji419 at 15 DAP and down-regulated in Ji419 at 25 DAP, while another three were down-regulated in Ji419 at 15 DAP and up-regulated in Ji419 at 25 DAP.
Targets of differentially expressed miRNAs in developing endosperm
Through degradome sequencing, 2.0 × 107 raw reads representing 2.9 × 106 unique reads were generated from the mixed degradome pools. In total, 55 target genes with 97 transcripts modulated by 56 miRNAs were identified.
Target genes of these differentially expressed miRNAs were further screened. As a result, 26 target genes with 47 transcripts regulated by 14 known miRNAs were detected (Figs. 3 and 4; Additional file 2: Tables S5 and S6). Of these miRNAs, both miR156a-5p and miR156j-5p_R-1 could target GRMZM5G806833 (unknown function), GRMZM2G061734 (squamosa promoter-binding protein transcription factor 5, SBP5), and GRMZM2G156621 (SBP31); miR159c-5p_L-1R-1 could target GRMZM2G015384 (actin related protein 4); miR167a-5p_R-2 could target GRMZM2G035405 (ARF18), GRMZM2G475882 (ARF30), and GRMZM2G078274 (ARF6); miR169a-5p could target GRMZM2G000686 (nuclear transcription factor Y subunit A1, NF-YA1), GRMZM2G037630 (CCAAT-HAP2-transcription factor 23, CA2P3), and GRMZM2G040349 (CCAAT-HAP2-transcription factor 24, CA2P4); miR169o-3p_L-1R + 1 could target GRMZM2G009530 (C2C2-GATA-transcription factor 9, GATA9) and GRMZM2G009808 (aconitase3); miR171c_R + 1 could target three GRAS (GIBBERELLIN-ACID INSENSITIVE (GAI), REPRESSOR OF GAI (RGA), SCARECROW (SCR)) family transcription factors (GRMZM2G110579, GRMZM5G825321, and GRMZM2G037792); miR393a-5p_L + 1R-2 could target GRMZM5G848945 (auxin signaling F-box, AFB); miR394a-5p could target GRMZM2G064954 (F-box protein 6); and miR396a-5p, miR396c_L-1, or miR396e-5p_1ss21TA could target six growth-regulating factors (GRMZM2G045977, GRMZM2G033612, GRMZM2G034876, GRMZM2G099862, GRMZM2G129147, and GRMZM2G041223). Also, miR396a-5p could regulate GRMZM2G024293 (encoding a nucleoside triphosphate hydrolase). In addition, miR397a-5p and miR827-5p_L + 1 could modulate GRMZM2G094699 (coatomer protein complex subunit beta 1, COPB1) and GRMZM2G044788 (unknown function), respectively.
Fig. 3.
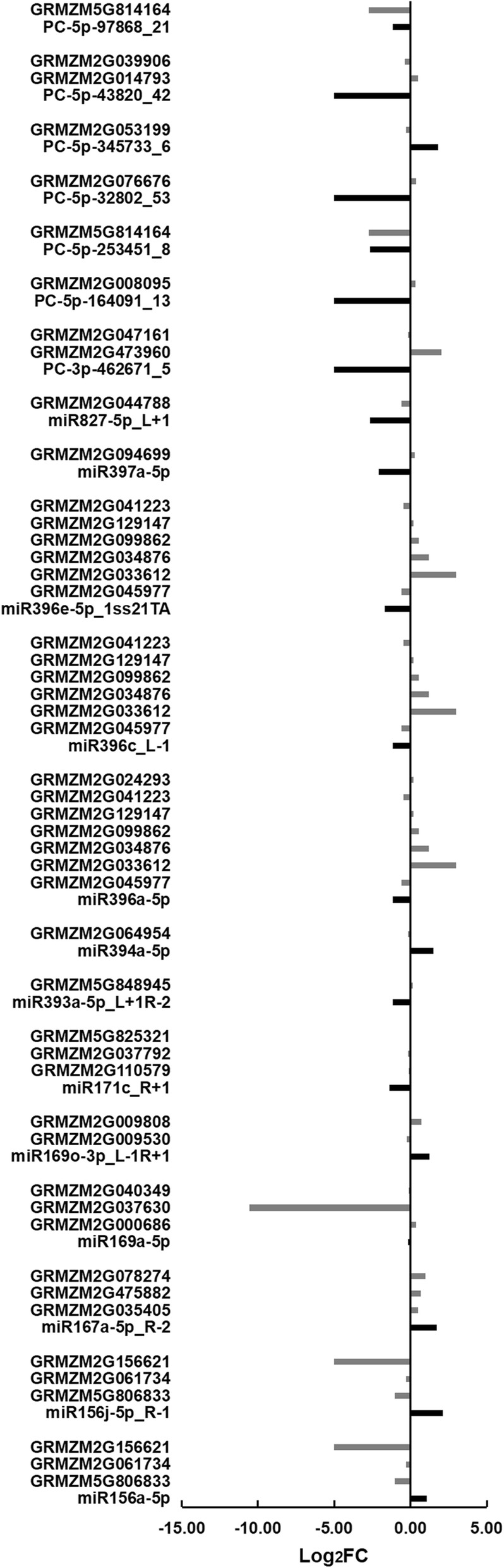
Differential expression of miRNAs and their target genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 DAP. FC, fold-change of the expression of the miRNAs/target genes in Ji419 relative to that in Mo17 at 15 DAP based on the sequencing data. Black bars represent expression of miRNAs; grey bars represent expression of target genes
Fig. 4.

Differential expression of miRNAs and their target genes in the endosperm of maize inbred lines Mo17 and Ji419 at 25 DAP. FC, fold-change of the expression of the miRNAs/target genes in Ji419 relative to that in Mo17 at 25 DAP based on the sequencing data. Black bars represent expression of miRNAs; grey bars represent expression of target genes
Nine target genes with 47 transcripts were detected, which were regulated by eight newly discovered miRNAs including PC-3p-24264_68 and PC-5p-164091_13 (Figs. 3 and 4; Additional file 2: Tables S5 and S6). Of these miRNAs, both PC-5p-32802_53 and PC-5p-43820_42 were specifically expressed in Mo17 at 15 and 25 DAP, with PC-5p-32802_53 targeting GRMZM2G076676 (encoding a glycine-rich protein), and PC-5p-43820_42 targeting both GRMZM2G014793 (encoding a glycosyl transferase) and GRMZM2G039906 (encoding a cell cycle control protein). Both PC-3p-462671_5 and PC-5p-164091_13 were specifically expressed in Mo17 at 15 DAP, with PC-3p-462671_5 targeting both GRMZM2G473960 (encoding a seed maturation protein) and GRMZM2G047161 (unknown function), and PC-5p-164091_13 targeting GRMZM2G008095 (encoding a vesicle transport protein). Both PC-5p-97868_21 and PC-3p-24264_68 were specifically expressed in Mo17 at 25 DAP, with PC-5p-97868_21 targeting GRMZM5G814164 (peroxin3), and PC-3p-24264_68 targeting GRMZM2G035985 (encoding a transduction protein). Additionally, PC-5p-253451_8 and PC-5p-345733_6 could regulate GRMZM5G814164 and GRMZM2G053199 (encoding a protein phosphatase), respectively. Notably, GRMZM5G814164 could be regulated by both PC-5p-97868_21 and PC-5p-253451_8.
Among all the miRNA-target pairs, 29 pairs including miR393a-5p_L + 1R-2-GRMZM5G848945, miR169o-3p_L-1R + 1-GRMZM2G009530, and miR169a-5p-GRMZM2G000686, and eight pairs including PC-5p-43820_42-GRMZM2G014793, miR396a-5p-GRMZM2G034876, and miR169a-5p-GRMZM2G037630, showed reverse expression patterns at 15 and 25 DAP, respectively (Figs. 3 and 4).
None of the starch biosynthetic genes that were directly regulated by miRNAs were detected through degradome sequencing. However, bioinformatic prediction showed that the starch synthase genes SSIIIa and SSIIb-2 might be the targets of two newly discovered miRNAs, PC-3P-336668_6 and PC-3p-169926_13, respectively (Additional file 2: Table S7).
Co-expression network of genes involved in starch biosynthesis in developing endosperm
A set of 35 target genes was identified via combined analysis of the degradome and differentially expressed miRNAs. Co-expression analysis of these 35 genes and eight key genes involved in maize endosperm starch biosynthesis suggested that 34 target genes were significantly co-expressed with at least one of the eight starch biosynthetic genes (Additional file 5: Figure S4; Additional file 2: Table S8). Co-expression analysis revealed a complex network modulating starch biosynthesis in developing maize endosperm, in which 19 miRNAs, including 11 known and eight newly discovered miRNAs, could regulate 19 target genes associated with starch biosynthesis (Fig. 5). For example, miR169a and miR169o could directly target NF-YA1 and GATA9, respectively, to control starch biosynthetic genes.
Fig. 5.
Co-expression network of miRNA-target-starch biosynthesis in developing maize endosperm. Blue boxes represent miRNAs, yellow boxes represent target genes, and green boxes represent starch biosynthetic genes. PCC, Pearson’s correlation coefficient
Validation of the regulatory pathways for starch biosynthesis mediated by miR169a-NF-YA1 and miR169o-GATA9
To verify the regulatory pathways involved in starch biosynthesis, the expression patterns of the miRNA-target pairs miR169a-NF-YA1 and miR169o-GATA9 were analyzed at six endosperm developmental stages by qRT-PCR (Fig. 2). The results revealed a reverse expression pattern for miR169a-NF-YA1 during endosperm development. The expression of miR169o was relatively high, whereas that of GATA9 was extremely low, implying that miR169o might repress the expression of GATA9.
Furthermore, transient expression assays of miR169a, miR169o, NF-YA1, and GATA9 in maize endosperm showed that NF-YA1 was significantly down-regulated in miR169a-overexpressing endosperm, suggesting that miR169a could negatively regulate the expression of NF-YA1. GATA9 was significantly up-regulated in miR169o-overexpressing endosperm, indicating that miR169o might positively regulate the expression of GATA9 (Fig. 6). Meanwhile, GBSSI was significantly down-regulated, whereas SSIIIa was significantly up-regulated in NF-YA1-overexpressing endosperm; both SSIIIa and SBEIIb were significantly up-regulated in GATA9-overexpressing endosperm (Fig. 6). Moreover, transactivation assays of the promoters of GBSSI/SSIIIa and SSIIIa/SBEIIb by NF-YA1 and GATA9, respectively, indicated that NF-YA1 could enhance the promoter activity of SSIIIa and decrease the promoter activity of GBSSI; GATA9 could enhance the promoter activities of SSIIIa and GBSSI (Fig. 7). These results demonstrated that miR169a-NF-YA1 and miR169o-GATA9 could be involved in the regulatory pathways for starch biosynthesis in maize endosperm. Overall, the regulatory interactions between miR169a-NF-YA1-GBSSI/SSIIIa and miR169o-GATA9-SSIIIa/SBEIIb that could affect target expression were confirmed in the present study.
Fig. 6.
Transient expression analysis in maize endosperm. a The expression of miR169a in miR169a-overexpressing endosperm (pUbi:miR169a) and the control (pUbi:Gus); b The expression of miR169o in miR169o-overexpressing endosperm (pUbi:miR169o) and the control (pUbi:Gus); c The expression of NF-YA1 in miR169a/NF-YA1-overexpressing endosperm (pUbi:miR169a and pUbi:NF-YA1) and the control (pUbi:Gus); d The expression of GATA9 in miR169o/GATA9-overexpressing endosperm (pUbi:miR169o and pUbi:GATA9) and the control (pUbi:Gus); e The expression of GBSSI in NF-YA1-overexpressing endosperm (pUbi:NF-YA1) and the control (pUbi:Gus); f The expression of SSIIIa in NF-YA1/GATA9-overexpressing endosperm (pUbi:NF-YA1 and pUbi:GATA9) and the control (pUbi:Gus); g The expression of SBEIIb in GATA9-overexpressing endosperm (pUbi:GATA9) and the control (pUbi:Gus). All data are means ± SD (n = 3). *, ** significant at p ≤ 0.05 and p ≤ 0.01 by the Student’s t test
Fig. 7.
Transactivation assays of the promoters of GBSSI/SSIIIa and SSIIIa/SBEIIb by NF-YA1 and GATA9, respectively. a Diagram of the effector, reporter, and internal constructs; b Regulatory interaction between NF-YA1 and the promoter of GBSSI; c Regulatory interactions between NF-YA1/GATA9 and the promoter of SSIIIa; d Regulatory interaction between GATA9 and the promoter of SBEIIb. All data are means ± SD (n = 3). ** significant at p ≤ 0.01 by the Student’s t test
Discussion
DEGs related to starch biosynthesis in developing maize endosperm
During endosperm development, active expression of most genes associated with primary metabolism can be observed [34, 35]. In the present study, transcriptome analysis of developing endosperm showed that most genes expressed differentially between Mo17 and Ji419 at 15 and 25 DAP participate in biological processes such as carbohydrate metabolism (Additional file 1: Figure S1a-d). As indicated in previous studies [26, 31, 36], these DEGs may directly or indirectly influence starch biosynthesis, resulting in divergent endosperm starch content and starch granule structure.
RNA-Seq data indicated that of the 33 differentially expressed starch biosynthetic genes, up-regulated genes outnumbered down-regulated genes in Ji419 (Fig. 1b), indicating that endosperm starch biosynthesis was more dynamic in Ji419 than in Mo17 at both 15 and 25 DAP, and possibly explaining why the endosperm starch content of Ji419 is higher than that of Mo17. In addition, some genes were up-regulated at one stage but down-regulated at the other. This might have contributed less to the difference in endosperm starch content between the two lines if the negative effect on endosperm starch content of the down-regulation of some genes at a specific stage had been offset by the up-regulation of other genes [36–38]. Here, Sh2 was down-regulated, while AGPL embryo (AGPLEMZM) and AGPb were up-regulated in Ji419 at 25 DAP (Fig. 1a). Both AGPLEMZM and AGPb, which are homologous to Sh2, encode large subunits of AGP [37], and their up-regulation might counteract any negative effect of down-regulation of Sh2.
In the current study, transcript abundances of the eight key starch biosynthetic genes determined by qRT-PCR conformed to those determined from RNA-Seq data (Fig. 1c, d; Additional file 2: Table S1). Meanwhile, the expression patterns of these eight genes at 15, 20, 25, 30, 35, and 40 DAP (Fig. 2) were in accordance with results of previous research [39]. Among these genes, Sh2 and Bt2 determine the direct substrate for starch biosynthesis; GBSSI mainly plays a role in amylose biosynthesis, whereas SSI, SSIIa, SSIIIa, SBEIIb, and ISA1 affect amylopectin biosynthesis [2, 5–7]. Mutations in these genes would result in reduced starch content or altered starch structure. For instance, the starch content of grains from sh2 and bt2 mutants is significantly lower than that from wild-type maize [40]. Mutations in GBSSI and SBEIIb cause alteration in the structure of starch [41, 42]. Therefore, differential expression of these starch biosynthetic genes might be at least one reason for the distinct endosperm starch content and starch granule structure of Mo17 and Ji419 (Additional file 3: Figure S2; Additional file 6: Figure S5). As regulation by miRNAs could be a major factor leading to the differential expression of these genes, the regulation by miRNAs of starch biosynthesis was further analyzed.
miRNAs and corresponding targets associated with starch biosynthesis in developing endosperm
Although few miRNAs have been reported to regulate starch biosynthesis, miR156 can target SPL (SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE)/SBP family transcription factors [43], and its overexpression in Arabidopsis, maize, and switchgrass results in significantly suppressed expression of SPL/SBP transcription factors and increased starch content [21]. SPL/SBP transcription factors can be involved in the initiation of lateral primordia, and can also influence the accumulation of carbohydrates [44, 45]. Thus, miR156-SPL/SBP transcription factors might participate in the regulation of starch biosynthesis. In the present study, two miR156 family members, miR156a-5p and miR156j-5p_R-1, might regulate starch biosynthesis by targeting GRMZM5G806833 (unknown function), GRMZM2G061734 (SBP5), and GRMZM2G156621 (SBP31) (Additional file 2: Tables S5 and S6). In rice, miR167 can regulate grain development through the auxin-miR167-ARF8-GH3.2 pathway [22]. miR393 can also target the auxin receptor genes OsTIR1 and OsAFB2 to modulate the development of roots and grains [46, 47]. Therefore, auxin-mediated miR167 and miR393 might participate in starch biosynthesis in endosperm. In the present study, miR167a-5p_R-2 might target GRMZM2G035405 (ARF18), GRMZM2G475882 (ARF30), and GRMZM2G078274 (ARF6), and miR393a-5p_L + 1R-2 could target GRMZM5G848945 (AFB) to modulate endosperm starch biosynthesis (Additional file 2: Tables S5 and S6). Growth regulators, a class of specific transcription factors in plants [48], play critical roles in the development of stems, leaves [49–51], roots [52–55], and grains [56, 57]. Notably, miR396 can modulate the expression of such growth regulators [48]. In rice, miR396c can regulate grain development and yield through the miR396c-GRF4 (growth regulating factor)-GIF1 (grain incomplete filling) pathway, which might be associated with starch biosynthesis [58]. In the present study, miR396a-5p, miR396c_L-1, and miR396e-5p_1ss21TA could target six growth regulator genes (GRMZM2G045977, GRMZM2G033612, GRMZM2G034876, GRMZM2G099862, GRMZM2G129147, and GRMZM2G041223) that might participate in the regulation of endosperm starch biosynthesis (Additional file 2: Tables S5 and S6).
Additionally, the newly identified miRNAs PC-5p-32802_53 and PC-5p-43820_42 were specifically expressed in the endosperm of Mo17 at 15 and 25 DAP, PC-3p-462671_5 and PC-5p-164091_13 were specifically expressed in Mo17 at 15 DAP, and PC-5p-97868_21 and PC-3p-24264_68 were specifically expressed in Mo17 at 25 DAP (Additional file 2: Tables S3 and S4). The target genes of these miRNAs, including GRMZM2G014793 (encoding a glycosyl transferase), GRMZM2G039906 (encoding a cell cycle control protein), and GRMZM2G473960 (encoding a seed maturation protein) (Additional file 2: Tables S5 and S6), may be closely associated with carbohydrate metabolism and grain development. Thus, these newly discovered miRNAs might regulate the differential expression of their target genes in the endosperm of Mo17 and Ji419. This could further differentially modulate biological processes such as starch biosynthesis and grain development, leading to divergent endosperm starch content and starch granule structure in these two lines (Additional file 3: Figure S2; Additional file 6: Figure S5).
Regulatory network of starch biosynthesis in developing endosperm
Thus far, no research has been reported on an miRNA-mediated regulatory network for starch biosynthesis in maize. Our study of the co-expression of miRNA targets and known starch biosynthetic genes has established the existence of a complex miRNA-target regulatory network for starch biosynthesis in endosperm (Fig. 5). In this network, 19 miRNAs could target 19 genes to regulate endosperm starch biosynthesis, which reveals an miRNA-mediated mechanism modulating starch biosynthesis in developing maize endosperm.
In order to verify the regulatory network, gene expression patterns in four pathways involving miR169a-NF-YA1-GBSSI/SSIIIa and miR169o-GATA9-SSIIIa/SBEIIb were analyzed, and further transient expression and transactivation assays were conducted (Figs. 2, 6, and 7). The analyses of gene expression pattern in our study implied that miR169a and miR169o might negatively modulate their target genes NF-YA1 and GATA9, respectively. The transient overexpression results indicated that miR169a could negatively regulate NF-YA1, whereas miR169o could positively regulate GATA9 (Figs. 2 and 6). Most previous studies have shown that miRNAs can negatively regulate the expression of their target genes at the post-transcriptional level [59–63]. Also, there have been reports of positive regulation of target genes by miRNAs in humans and animals [64–66]. Therefore, it is possible that miR169o could positively regulate the target gene GATA9; however, further studies are needed to elucidate this mechanism. Nevertheless, it seemed that the regulatory relationship between miR169o and GATA9 observed from expression pattern analysis was inconsistent with that from transient expression analysis. It might be because regulators other than miRNAs, such as transcription factors, hormones, or environmental signals could tune the expression of these target genes in normal maize, which would not be unexpected as the regulation of biological processes is often complex. In previous studies, NF-YA family transcription factors have been reported as the targets of miR169 [67–69]. Specifically, NF-YA1, NF-YA5, NF-YA6, and NF-YA9 play important roles in the development of male gametes, embryos, and seeds in Arabidopsis [70], and might be associated with starch biosynthesis. Two GATA family transcription factors, GNC and GNL/CGA1, can act as regulators of the gibberellin signaling pathway in Arabidopsis [71]. As gibberellin can regulate starch biosynthesis [18], GATA transcription factors might participate in starch biosynthesis through regulation of the gibberellin signaling pathway. In our study, the transient overexpression and transactivation results comprehensively confirmed that miR169a-NF-YA1-GBSSI/SSIIIa and miR169o-GATA9-SSIIIa/SBEIIb could interact and influence starch biosynthesis in maize endosperm (Figs. 6 and 7). In summary, the regulatory network described here will be of great significance for further in-depth research into the molecular mechanism of starch biosynthesis in developing maize endosperm. Moreover, although endosperm is the organ for synthesis and accumulation of starch, the evidence for maternal contribution to seed development (seed weight, seed size, etc.) in maize has revealed that starch content may be also controlled by maternal effect [72, 73]. Therefore, a comprehensive study on the coordinated roles of maternal effect and starch biosynthetic pathways will be further considered for a better understanding of the mechanism of starch biosynthesis in seed, which will facilitate the improvement of grain yield and quality in future maize breeding programs.
Methods
Plant materials
Two maize inbred lines, Mo17 and its improved line Ji419, were grown at the Changping Station at the Institute of Crop Science, Chinese Academy of Agricultural Sciences in 2015. Ji419 was derived from a cross between B68Ht and Mo17. These two lines differ significantly in endosperm starch content and starch granule structure (Additional file 3: Figure S2; Additional file 6: Figure S5). Further details of the kernel morphological characteristics of Mo17 and Ji419 have been detailed in our previous study [74]. The experimental field consisted of 40 rows that were 3 m long and 0.6 m wide, with spacing of 0.3 m between plants within rows. Common cultural practices for growing maize were used. Endosperm tissues were collected from Mo17 and Ji419 at 15, 20, 25, 30, 35, and 40 DAP. Three biological replicates were sampled, with each replicate sampled from a separate ear chosen from each line. The endosperm samples from kernels sampled from the central portion of the ear were frozen immediately in liquid nitrogen and stored at − 80 °C.
RNA extraction, library construction, and sequencing
Total RNA was extracted from each sample using Trizol reagent (Invitrogen, Carlsbad, USA). The quantity and purity of the total RNA were checked using NanoDrop ND-1000 (NanoDrop, Wilmington, USA) and Agilent 2100 with a minimum RNA Integrity Number (RIN) threshold value > 7.0. Approximately 15 μg of total RNA were used to prepare each transcriptome sequencing library, and a total of 12 libraries were constructed for each biological replicate of the Mo17 and Ji419 endosperm samples taken at 15 and 25 DAP. RNA-Seq was performed by OE-Biotech Co., Ltd., Shanghai, China.
Approximately 1 μg of total RNA was used to prepare a small RNA library according to the protocol of TruSeq Small RNA Sample Prep Kit (Illumina, San Diego, USA), and four libraries were constructed from equally mixed RNA quantities from the three biological replicates of the Mo17 and Ji419 endosperm samples taken at 15 and 25 DAP. Approximately 20 μg of total RNA were used to prepare a degradome library, and two libraries were constructed from equally mixed RNAs from the six endosperm samples of Mo17 and Ji419 at both 15 and 25 DAP. Both small RNA and degradome sequencing were performed on an Illumina HiSeq 2500 platform at the LC-Biotech Co., Ltd., Hangzhou, China, following the manufacturer’s recommended protocol.
Transcriptome analysis
Clean reads were obtained after filtering adaptor sequences, and empty, low-quality, and one-copy tags from the raw reads. Next, reads were aligned to the maize reference gene set based on the B73 genome (release 5b.60) using the SOAPaligner/SOAP2 program [75]. Gene expression was quantified as fragments per kb per million reads (FPKM) [76]. Genes with P-value less than 0.05 were considered to have altered expression and were designated as DEGs.
DEGs were subjected to GO functional enrichment and KEGG pathway analyses. GO annotation was conducted using Blast2GO software (v.2.5.0) [77], and the significance of enrichment of each functional term was computed using the hypergeometric distribution test. KEGG Automatic Annotation Server (KAAS) provides functional annotation of genes by blast against the manually managed KEGG database. The results contain KEGG Orthology (KO) assignments and automatically generated KEGG pathways.
Forty known starch biosynthetic genes [78] were chosen to construct pathways for starch biosynthesis, and the expression profiles of these genes were also determined from the RNA-Seq data. Further, the expression of eight key starch biosynthetic genes (GBSSI, SSI, SSIIa, SSIIIa, ISA1, SBEIIb, Sh2, and Bt2) in Mo17 and Ji419 at 15 and 25 DAP was validated using qRT-PCR. The first-strand cDNAs of these eight genes were synthesized using the Fast Quant RT Kit (TianGen, Beijing, China). Then qRT-PCR was carried out using the Bio-Rad iQ5 (Bio-Rad, USA) following the instructions for the SuperReal PreMix Plus (SYBR Green) (TianGen, Beijing, China). Three technical replicates of all reactions were performed, and 18S rRNA was used as the internal reference for expression. The primers used for qRT-PCR were listed in Additional file 2: Table S9.
miRNA analysis
Raw reads were initially processed by an in-house program, ACGT101-miR (LC Sciences, Houston, USA). Clean reads were obtained after filtering adapters, low-quality reads, common RNA families (rRNA, tRNA, snRNA, snoRNA), and repeats. The filtered unique sequences with length in 18 to 25 nucleotides were mapped to the precursors of specific species in miRBase 20.0 by blast to identify known miRNAs and previously unknown 5p- and 3p-derived miRNAs. Length variation at both 3′ and 5′ ends and one mismatch inside the sequences were allowed in the alignment. Unique sequences that match one of the arms in the stem-loops of mature miRNAs of specific species were identified as known miRNAs. Any unique sequences that mapped to the other arm of the stem-loops of precursors of known specific species opposite to the annotated mature miRNA-containing arm were considered to be candidates for previously unknown 5p- or 3p-derived miRNAs. The remaining sequences were mapped to the precursors of other selected species (with the exclusion of specific species) in miRBase 20.0 by blast, and the mapped pre-miRNAs were used as further blast queries against the specific species genomes to determine their genomic locations. The miRNAs in the above two categories were then defined as known miRNAs. The unmapped sequences were aligned against the specific genomes, and the hairpin RNA structures containing sequences were predicated from the flanking 120 nucleotides sequences using RNAfold software. miRNAs with at least twofold change in expression (Log2FC ≥ 1) and at least 10 reads in a dataset were identified here as differentially expressed miRNAs.
Degradome analysis
Potentially cleaved targets of 20 to 21 nucleotides in the extracted sequence reads generated by degradome sequencing were identified using CleaveLand 3.0. The degradome reads were then mapped to the transcriptome data. We constructed t-plots using the transcriptome data for highly efficient analysis of potential miRNA targets. Finally, the targets of differentially expressed miRNAs were identified as candidate target genes.
Co-expression analysis of candidate targets and eight starch biosynthetic genes
Using our RNA-Seq data, as well as those from the study performed by Li et al. [35], co-expression analysis among the candidate target genes and eight key starch biosynthetic genes (Sh2, Bt2, GBSSI, SSI, SSIIa, SSIIIa, ISA1, and SBEIIb) was conducted in SPSS software (v.24.0). Candidate target genes were considered associated with endosperm starch biosynthesis only for Pearson’s correlation coefficient of ≥0.60 between their expression and that of starch biosynthetic gene(s). Based on the co-expression analysis, a model molecular regulatory network of miRNA-target-starch biosynthesis in developing maize endosperm was constructed using Cytoscape software (v.3.6.0).
Expression pattern analysis
The expression patterns of the pathways involving miR169a-NF-YA1-GBSSI/SSIIIa and miR169o-GATA9-SSIIIa/SBEIIb were analyzed by qRT-PCR. Total RNAs and miRNAs from Mo17 and Ji419 at 15, 20, 25, 30, 35, and 40 DAP were used for analysis of gene expression patterns by qRT-PCR to clarify any possible regulatory relationship between the chosen miRNA-target pairs in developing maize endosperm. Two target genes, NF-YA1 and GATA9, were analyzed using qRT-PCR as described above. The first-strand cDNAs of these miRNAs were synthesized using the miRcute miRNA First-Strand cDNA Synthesis Kit (TianGen, Beijing, China). qRT-PCR was then carried out to analyze expression of these miRNAs using the Bio-Rad iQ5 (Bio-Rad, USA) following the instructions for the miRcute miRNA qRT-PCR Detection Kit (SYBR Green) (TianGen, Beijing, China). Three technical replicates were also performed, and 5S rRNA was used as internal reference. The primers used for qRT-PCR were listed in Additional file 2: Table S9.
Transient expression analysis
The pre-miR169a/169o precursor and the coding sequences of NF-YA1 and GATA9 were cloned and inserted into PUbi:Gus by replacing the β-glucuronidase (Gus) reporter gene and driven by the maize ubiquitin promoter (pUbi:miR169a, pUbi:miR169o, pUbi:NF-YA1 and pUbi:GATA9). Transient overexpression of the above vectors in maize endosperm was mediated by particle bombardment, and the pUbi:Gus was used as a control. Three biological replicates were performed. The endosperm used for particle bombardment was isolated from immature maize kernels (10 DAP), which had been surface-sterilized with 75% ethanol and plated on MS medium (Murashige and Skoog salts containing 0.85% agar and 12% sucrose) for 4 h prior to bombardment. Particle bombardment was performed as previously described [16]. After particle bombardment, the medium plates containing bombarded endosperm were incubated at 28 °C in the dark for 16–24 h. Total RNA was extracted from the endosperm, and the expression of related miRNAs, target genes, and starch biosynthetic genes were analyzed using qRT-PCR as described above to characterize the starch biosynthetic pathways. The primers used for qRT-PCR were listed in Additional file 2: Table S9.
Transactivation assay
To determine the regulatory relationship between NF-YA1 and the promoters of GBSSI/SSIIIa, and that between GATA9 and the promoters of SSIIIa/SBEIIb, transactivation assays were conducted. pUbi:NF-YA1 and pUbi:GATA9 served as effector constructs. The promoters of GBSSI, SSIIIa, and SBEIIb were cloned into the reporter construct pPromoter:Luc (pGBSSI:Luc, pSSIIIa:Luc, and pSBEIIb:Luc). The pUbi:Gus was used as an internal construct. The effector construct, the reporter construct, and the internal construct were combined at a molar ratio of 2:2:1 and then co-bombarded into maize endosperm. Three biological replicates were performed. After the medium plates containing bombarded endosperm were incubated at 28 °C in the dark for 24 h, the activity of Gus and Luc were analyzed using a Luminoskan™ Ascent (Thermo, USA). The primers used for transactivation assays were listed in Additional file 2: Table S9.
Additional files
Figure S1. Analysis of GO term and KEGG pathway enrichment among genes differentially expressed in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP. a-b GO terms of differentially expressed genes in the endosperm of Mo17 and Ji419 at 15 DAP (a) and 25 DAP (b); c-d KEGG pathways of differentially expressed genes in the endosperm of Mo17 and Ji419 at 15 DAP (c) and 25 DAP(d). (TIF 1660 kb)
Table S1. The expression of 40 starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S2. GO term and KEGG pathway enrichment of 40 starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S3. Differentially expressed miRNAs in the endosperm of maize inbred lines Mo17 and Ji419 at 15 DAP; Table S4. Differentially expressed miRNAs in the endosperm of maize inbred lines Mo17 and Ji419 at 25 DAP; Table S5. Differentially expressed miRNAs and their target genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 DAP; Table S6. Differentially expressed miRNAs and their target genes in the endosperm of maize inbred lines Mo17 and Ji419 at 25 DAP; Table S7. Predicted target genes of miRNAs in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S8. Pearson’s correlation coefficients between the expression of 35 target genes and eight key starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S9. Primers used in this study. (XLSX 508 kb)
Figure S2. Starch content in the mature endosperm of maize inbred lines Mo17 and Ji419. All data are means ± SD (n = 3). *, ** significant at p ≤ 0.05 and p ≤ 0.01 by the student’s t test. (TIF 840 kb)
Figure S3. Size distribution of unique miRNAs (a) and Venn diagram of miRNAs differentially expressed (b) in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP. Red represents miRNAs down-regulated in Ji419 compared with Mo17 at 15 DAP; blue represents miRNAs up-regulated in Ji419 at 15 DAP; yellow represents miRNAs down-regulated in Ji419 at 25 DAP; green represents miRNAs up-regulated in Ji419 at 25 DAP. (TIF 358 kb)
Figure S4. Heat map depicting co-expression analysis based on Pearson’s correlation coefficients between the expression of 35 target genes and eight key starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP. (TIF 401 kb)
Figure S5. Scanning electron micrograph of starchy endosperm cells in seeds of maize inbred lines Mo17 and Ji419 at different developmental stages. a Endosperm of Mo17 at 15 DAP; b Endosperm of Mo17 at 25 DAP; c Endosperm of Mo17 at 35 DAP; d Mature endosperm of Mo17; e Endosperm of Ji419 at 15 DAP; f Endosperm of Ji419 at 25 DAP; g Endosperm of Ji419 at 35 DAP; h Mature endosperm of Ji419. Scale bar, 50 μm. (TIF 3589 kb)
Acknowledgments
We would like to express our sincere gratitude to Mary Ann Cushman from Alum Canyon Editing for our manuscript editing.
Abbreviations
- ABA
Abscisic acid
- AFB
Auxin signaling F-box
- AGP
ADP-glucose pyrophosphorylase
- AGPL
AGP large subunit
- AGPLEMZM
AGPL embryo
- AGPS
AGP small subunit
- ARF
Auxin response factor
- BR
Brassinosteroid
- Bt2
Brittle2
- CA2P3
CCAAT-HAP2-transcription factor 23
- CA2P4
CCAAT-HAP2-transcription factor 24
- COPB1
Coatomer protein complex subunit beta 1
- DAP
D after pollination
- DBE
Starch debranching enzyme
- DEGs
Differentially expressed genes
- FPKM
Fragments per kb per million reads
- GA
Gibberellin
- GATA9
C2C2-GATA-transcription factor 9
- GBSS
Granule-bound starch synthase
- GIF
Grain incomplete filling
- GO
Gene Ontology
- GRAS
GIBBERELLIN-ACID INSENSITIVE (GAI), REPRESSOR OF GAI (RGA), SCARECROW (SCR)
- GRF
Growth regulating factor
- Gus
β-glucuronidase
- IAA
Auxin
- ISA1
Isoamylase 1
- KAAS
KEGG Automatic Annotation Server
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG Orthology
- miRNAs
microRNAs
- NF-YA1
Nuclear transcription factor Y subunit A1
- O2
opaque2
- PBF
Prolamine-box binding factor
- PHI1
Phosphohexose isomerase 1
- PHO
Starch phosphorylase
- PPDK1
Pyruvate orthophosphate dikinase 1
- qRT-PCR
quantitative real-time polymerase chain reaction
- RIN
RNA Integrity Number
- RNA-Seq
RNA sequencing
- RSR1
Rice Starch Regulator 1
- SBE
Starch branching enzyme
- SBP5
Squamosa promoter-binding protein transcription factor 5
- Sh2
Shrunken2
- SPL
Squamosa Promoter-binding Protein-Like
- SS
Soluble starch synthase
- SUSIBA
Sugar signaling in barley
- UGP
UDP-glucose pyrophosphorylase
Authors’ contributions
XZ and SX analyzed the data and wrote the paper. JH, YZ, CL, ZZ, FW, ZC, JZ, YH, ZH, ML, DZ, HY, and YH performed the experiments and revised the paper. JW and XL designed and supervised the experiments. All authors read and approved the final manuscript.
Funding
This work was jointly supported by the National Key Research and Development Program of China (2016YFD0100303), the National Natural Science Foundation of China (31471509), the China Postdoctoral Science Foundation (2018 M631642), and the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences (CAAS).
Availability of data and materials
Raw sequence data for small RNAs, degradome, and transcriptome in this study are available at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under accession number SRP099290.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaocong Zhang and Sidi Xie contributed equally to this work.
Contributor Information
Jianfeng Weng, Email: wengjianfeng@caas.cn.
Xinhai Li, Email: lixinhai@caas.cn.
References
- 1.Sonnewald U, Kossmann J. Starches—from current models to genetic engineering. Plant Biotechnol J. 2013;11:223–232. doi: 10.1111/pbi.12029. [DOI] [PubMed] [Google Scholar]
- 2.Tetlow IJ. Understanding storage starch biosynthesis in plants: a means to quality improvement. Can J Bot. 2006;84:1167–1185. doi: 10.1139/b06-089. [DOI] [Google Scholar]
- 3.Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang SK, et al. Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell. 2008;20:1833–1849. doi: 10.1105/tpc.107.054007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tetlow IJ. Starch biosynthesis in developing seeds. Seed Sci Res. 2011;21:5–32. doi: 10.1017/S0960258510000292. [DOI] [Google Scholar]
- 5.Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y. Starch biosynthesis in cereal endosperm. Plant Physiol Bioch. 2010;48:383. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Keeling PL, Myers AM. Biochemistry and genetics of starch synthesis. Annu Rev Food Sci Technol. 2010;1:271. doi: 10.1146/annurev.food.102308.124214. [DOI] [PubMed] [Google Scholar]
- 7.Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol. 2010;61:209. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 8.Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Chen J, Yi Q, Hu Y, Liu H, Liu Y, et al. Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol Biol. 2014;84:359. doi: 10.1007/s11103-013-0153-x. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Yi Q, Cao Y, Wei B, Zheng L, Xiao Q, et al. ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J Exp Bot. 2016;67:1327. doi: 10.1093/jxb/erv527. [DOI] [PubMed] [Google Scholar]
- 11.Huang H, Xie S, Xiao Q, Wei B, Zheng L, Wang Y, et al. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci Rep. 2016;6:27590. doi: 10.1038/srep27590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Zheng X, Yang J, Messing J, Wu Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. P Natl Acad Sci USA. 2016;113:10842. doi: 10.1073/pnas.1613721113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.León P, Sheen J. Sugar and hormone connections. Trends Plant Sci. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 14.Palacios OA, Choix FJ, Bashan Y, de Bashan LE. Influence of tryptophan and indole-3-acetic acid on starch accumulation in the synthetic mutualistic Chlorella sorokiniana-Azospirillum brasilense system under heterotrophic conditions. Res Microbiol. 2016;167:367–379. doi: 10.1016/j.resmic.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26:421–433. doi: 10.1046/j.1365-313X.2001.2641043.x. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Li Y, Zhang J, Liu H, Tian M, Huang Y. Binding of ABI4 to a CACCG motif mediates the ABA-induced expression of the ZmSSI gene in maize (Zea mays L.) endosperm. J Exp Bot. 2012;63:5979. doi: 10.1093/jxb/ers246. [DOI] [PubMed] [Google Scholar]
- 17.Wu C, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20:2130–2145. doi: 10.1105/tpc.107.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Zhang J, Wang Z, Zhu Q, Wang W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 2001;127:315–323. doi: 10.1104/pp.127.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Jia T, Chen X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017;216:1002–1017. doi: 10.1111/nph.14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Xia J, Xia Z, Zhang H, Zeng C, Lu C, et al. Potential functions of microRNAs in starch metabolism and development revealed by miRNA transcriptome profiling of cassava cultivars and their wild progenitor. BMC Plant Biol. 2015;15:33. doi: 10.1186/s12870-014-0355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, et al. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. P Natl Acad Sci USA. 2011;108:17550–17555. doi: 10.1073/pnas.1113971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue L, Zhang J, Xue H. Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 2009;37:916–930. doi: 10.1093/nar/gkn998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng T, Sun H, Du Y, Zhang J, Li J, Liu Y, et al. Characterization and expression patterns of microRNAs involved in rice grain filling. PLoS One. 2013;8:e54148. doi: 10.1371/journal.pone.0054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Y, Liu Y, Zhang J, Liu H, Hu Y, Du H, et al. Identification and characterization of microRNAs in the developing maize endosperm. Genomics. 2013;102:472. doi: 10.1016/j.ygeno.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Xin M, Yang G, Yao Y, Peng H, Hu Z, Sun Q, et al. Temporal small RNA transcriptome profiling unraveled partitioned miRNA expression in developing maize endosperms between reciprocal crosses. Front Plant Sci. 2015;6:744. doi: 10.3389/fpls.2015.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, Long J, Zheng L, Li Y, Hu Y, Yu G, et al. Identification and characterization of microRNAs in maize endosperm response to exogenous sucrose using small RNA sequencing. Genomics. 2016;108:216. doi: 10.1016/j.ygeno.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Jin X, Fu Z, Lv P, Peng Q, Ding D, Li W, et al. Identification and characterization of microRNAs during maize grain filling. PLoS One. 2015;10:e0125800. doi: 10.1371/journal.pone.0125800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Liu Z, Lei G, Wang L, Gao M, Jiao Z, et al. Genome-wide identification and characterization of microRNAs in developing grains of Zea mays L. PLoS One. 2016;11:e0153168. doi: 10.1371/journal.pone.0153168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics – ‘majority report by precogs’. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Higashi Y, Saito K. Network analysis for gene discovery in plant-specialized metabolism. Plant Cell Environ. 2013;36:1597. doi: 10.1111/pce.12069. [DOI] [PubMed] [Google Scholar]
- 31.Xiao Y, Thatcher S, Wang M, Wang T, Beatty M, Zastrow G, et al. Transcriptome analysis of near-isogenic lines provides molecular insights into starch biosynthesis in maize kernel. J Integr Plant Biol. 2016;58:713. doi: 10.1111/jipb.12455. [DOI] [PubMed] [Google Scholar]
- 32.Fu F, Xue H. Coexpression analysis identifies Rice starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010;154:927–938. doi: 10.1104/pp.110.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Vendramin S, Shi L, McGinnis KM. Construction and optimization of a large gene co-expression network in maize using RNA-Seq data. Plant Physiol. 2017;175:568–583. doi: 10.1104/pp.17.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai J, Dey N, Kim C, Bharti AK, Rudd S, Mayer KFX, et al. Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Res. 2004;14:1932–1937. doi: 10.1101/gr.2780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Wang D, Yang R, Logan K, Chen H, Zhang S, et al. Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. P Natl Acad Sci USA. 2014;111:7582–7587. doi: 10.1073/pnas.1406383111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Wang J, Ye J, Zheng X, Xiang X, Li C, et al. The maize imprinted gene floury3 encodes a PLATZ protein required for tRNA and 5S rRNA transcription through interaction with RNA polymerase III. Plant Cell. 2017;29:2661–2675. doi: 10.1105/tpc.17.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang B, Hennen-Bierwagen TA, Myers AM. Functions of multiple genes encoding ADP-glucose pyrophosphorylase subunits in maize endosperm, embryo, and leaf. Plant Physiol. 2014;164:596–611. doi: 10.1104/pp.113.231605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie G, Li Z, Ran Q, Wang H, Zhang J. Over-expression of mutated ZmDA1 or ZmDAR1 gene improves maize kernel yield by enhancing starch synthesis. Plant Biotechnol J. 2018;16:234–244. doi: 10.1111/pbi.12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohdan T, Jr FP, Sawada T, Hirose T, Terao T, Satoh H, et al. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- 40.Hannah LC, Jr OEN. Characterization of ADP-glucose pyrophosphorylase from shrunken-2 and brittle-2 mutants of maize. Biochem Genet. 1976;14:547–560. doi: 10.1007/BF00485834. [DOI] [PubMed] [Google Scholar]
- 41.Nelson OE, Rines HW. The enzymatic deficiency in the waxy mutant of maize. Biochem Biophys Res Commun. 1962;9:297–300. doi: 10.1016/0006-291X(62)90043-8. [DOI] [PubMed] [Google Scholar]
- 42.Fisher DK, Boyer CD, Hannah LC. Starch branching enzyme II from maize endosperm. Plant Physiol. 1993;102:1045. doi: 10.1104/pp.102.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 44.Chuck GS, Brown PJ, Meeley R, Hake S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. P Natl Acad Sci USA. 2014;111:18775–18780. doi: 10.1073/pnas.1407401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Z, Cao Y, Yang R, Qi T, Hang Y, Lin H, et al. Switchgrass SBP-box transcription factors PvSPL1 and 2 function redundantly to initiate side tillers and affect biomass yield of energy crop. Biotechnol Biofuels. 2016;9:101. doi: 10.1186/s13068-016-0516-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bian H, Xie Y, Guo F, Han N, Ma S, Zeng Z, et al. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa) New Phytol. 2012;196:149–161. doi: 10.1111/j.1469-8137.2012.04248.x. [DOI] [PubMed] [Google Scholar]
- 47.Xia K, Ren W, Ou X, Fang Z, Tian C, Duan J, et al. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One. 2012;7:e30039. doi: 10.1371/journal.pone.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol Plant. 2015;8:998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 50.Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. P Natl Acad Sci USA. 2004;101:13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 52.Vercruyssen L, Gonzalez N, Werner T, Schmulling T, Inze D. Combining enhanced root and shoot growth reveals cross talk between pathways that control plant organ size in Arabidopsis. Plant Physiol. 2011;155:1339–1352. doi: 10.1104/pp.110.167049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hewezi T, Maier TR, Nettleton D, Baum TJ. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012;159:321–335. doi: 10.1104/pp.112.193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bazin J, Khan GA, Combier JP, Bustos-Sanmamed P, Debernardi JM, Rodriguez R, et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013;74:920–934. doi: 10.1111/tpj.12178. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, et al. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell. 2015;27:3354–3366. doi: 10.1105/tpc.15.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Hua W, Yang H, Zhan G, Li R, Deng L, et al. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J Exp Bot. 2012;63:3727–3740. doi: 10.1093/jxb/ers066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 58.Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol J. 2016;14:2134–2146. doi: 10.1111/pbi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 61.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet. 2006;38(Suppl):S31. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 62.Saminathan T, Bodunrin A, Singh NV, Devarajan R, Nimmakayala P, Jeff M, et al. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 2016;16:122. doi: 10.1186/s12870-016-0807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ge F, Luo X, Huang X, Zhang Y, He X, Liu M, et al. Genome-wide analysis of transcription factors involved in maize embryonic callus formation. Plant Physiol. 2016;158:452–462. doi: 10.1111/ppl.12470. [DOI] [PubMed] [Google Scholar]
- 64.Huang V, Place R, Portnoy V, Wang J, Qi Z, Jia Z, et al. Upregulation of cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Majid S, Dar A, Saini S, Yamamura S, Hirata H, Tanaka Y, et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Place R, Li L, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. P Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, et al. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol. 2009;10:29. doi: 10.1186/1471-2199-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ni Z, Hu Z, Jiang Q, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol. 2013;82:113–129. doi: 10.1007/s11103-013-0040-5. [DOI] [PubMed] [Google Scholar]
- 69.Hanemian M, Barlet X, Sorin C, Yadeta KA, Keller H, Favery B, et al. Arabidopsis CLAVATA1 and CLAVATA2 receptors contribute to Ralstonia solanacearum pathogenicity through a miR169-dependent pathway. New Phytol. 2016;211:502–515. doi: 10.1111/nph.13913. [DOI] [PubMed] [Google Scholar]
- 70.Mu J, Tan H, Hong S, Liang Y, Zuo J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant. 2013;6:188–201. doi: 10.1093/mp/sss061. [DOI] [PubMed] [Google Scholar]
- 71.Richter R, Behringer C, Muller IK, Schwechheimer C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 2010;24:2093–2104. doi: 10.1101/gad.594910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell. 2006;18:3252–3274. doi: 10.1105/tpc.106.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Hirsch CN, Sekhon RS, De Leon N, Kaeppler SM. Evidence for maternal control of seed size in maize from phenotypic and transcriptional analysis. J Exp Bot. 2016;67:1907–1917. doi: 10.1093/jxb/erw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie S, Zhang X, Zhou Z, Li X, Huang Y, Zhang J, et al. Identification of genes alternatively spliced in developing maize endosperm. Plant Biol. 2018;20:59–66. doi: 10.1111/plb.12631. [DOI] [PubMed] [Google Scholar]
- 75.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 76.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aparicio G, Götz S, Conesa A, Segrelles D, Blanquer I, García JM, et al. Blast2GO goes grid: developing a grid-enabled prototype for functional genomics analysis. Stud Health Technol Inform. 2006;120:194. [PubMed] [Google Scholar]
- 78.Walley JW, Shen Z, Sartor R, Wu KJ, Osborn J, Smith LG, et al. Reconstruction of protein networks from an atlas of maize seed proteotypes. P Natl Acad Sci USA. 2013;110:E4808. doi: 10.1073/pnas.1319113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Analysis of GO term and KEGG pathway enrichment among genes differentially expressed in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP. a-b GO terms of differentially expressed genes in the endosperm of Mo17 and Ji419 at 15 DAP (a) and 25 DAP (b); c-d KEGG pathways of differentially expressed genes in the endosperm of Mo17 and Ji419 at 15 DAP (c) and 25 DAP(d). (TIF 1660 kb)
Table S1. The expression of 40 starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S2. GO term and KEGG pathway enrichment of 40 starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S3. Differentially expressed miRNAs in the endosperm of maize inbred lines Mo17 and Ji419 at 15 DAP; Table S4. Differentially expressed miRNAs in the endosperm of maize inbred lines Mo17 and Ji419 at 25 DAP; Table S5. Differentially expressed miRNAs and their target genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 DAP; Table S6. Differentially expressed miRNAs and their target genes in the endosperm of maize inbred lines Mo17 and Ji419 at 25 DAP; Table S7. Predicted target genes of miRNAs in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S8. Pearson’s correlation coefficients between the expression of 35 target genes and eight key starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP; Table S9. Primers used in this study. (XLSX 508 kb)
Figure S2. Starch content in the mature endosperm of maize inbred lines Mo17 and Ji419. All data are means ± SD (n = 3). *, ** significant at p ≤ 0.05 and p ≤ 0.01 by the student’s t test. (TIF 840 kb)
Figure S3. Size distribution of unique miRNAs (a) and Venn diagram of miRNAs differentially expressed (b) in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP. Red represents miRNAs down-regulated in Ji419 compared with Mo17 at 15 DAP; blue represents miRNAs up-regulated in Ji419 at 15 DAP; yellow represents miRNAs down-regulated in Ji419 at 25 DAP; green represents miRNAs up-regulated in Ji419 at 25 DAP. (TIF 358 kb)
Figure S4. Heat map depicting co-expression analysis based on Pearson’s correlation coefficients between the expression of 35 target genes and eight key starch biosynthetic genes in the endosperm of maize inbred lines Mo17 and Ji419 at 15 and 25 DAP. (TIF 401 kb)
Figure S5. Scanning electron micrograph of starchy endosperm cells in seeds of maize inbred lines Mo17 and Ji419 at different developmental stages. a Endosperm of Mo17 at 15 DAP; b Endosperm of Mo17 at 25 DAP; c Endosperm of Mo17 at 35 DAP; d Mature endosperm of Mo17; e Endosperm of Ji419 at 15 DAP; f Endosperm of Ji419 at 25 DAP; g Endosperm of Ji419 at 35 DAP; h Mature endosperm of Ji419. Scale bar, 50 μm. (TIF 3589 kb)
Data Availability Statement
Raw sequence data for small RNAs, degradome, and transcriptome in this study are available at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under accession number SRP099290.