Abstract
Background
Recently, accumulating evidence have revealed that circular RNA (circRNA) was deregulated in multiple types of cancer, suggesting that circRNA might serve as a novel candidate biomarker of cancer diagnosis. However, inconsistent results have become an obstacle in applying circRNAs to clinical practice. The aim of this study is to evaluate diagnostic value of circRNAs among cancers.
Methods
A literature search was systematically performed among PubMed, Sciencedirect, Cochrane Library, Web of Science, Wanfang, and Chinese National Knowledge Infrastructure databases up to February 15, 2019. The pooled sensitivity (SEN), specificity (SPE), positive likelihood ratios, negative likelihood ratios, diagnostic odds ratio, and area under the SROC curve (AUC) were applied to evaluate diagnostic performance of circRNAs.
Results
In total, the study included 64 studies with single circRNA and 13 studies with combined circRNAs. Overall, the study presented that a single circRNA had moderate diagnostic value, with a SEN of 0.75, a SPE of 0.76, and an AUC of 0.82. The plasma circRNAs had higher diagnostic accuracy than tissue (AUC: 0.87, 95% confidence interval [CI]: 0.83–0.89 for plasma/serum subgroup; AUC: 0.79, 95% CI: 0.75–0.82 for tissue subgroup). Furthermore, the combined circRNAs had good diagnostic efficacy for GC, with a SEN of 0.89, a SPE of 0.94, and an AUC of 0.97, respectively.
Conclusion
This study confirmed that circRNAs may be candidate biomarkers for cancer diagnosis. In particular, diagnosis of combined circRNAs will be a new alternative applied to clinical research and practice for cancer.
Keywords: biomarker, cancer, CircRNA, diagnosis
1. INTRODUCTION
The incidence and mortality of cancer has been of great public concern worldwide (Chen, Sun, et al., 2018; Dal Maso et al., 2018; Kimura & Egawa, 2018). One of the important reasons for the rapid increase of mortality and morbidity is the lack of sensitive biomarkers that can be used for early detection and diagnosis among various types of cancer. As it is a most convenient and common clinical detection method, the serum or plasma marker detection has been widely used in clinical practice involving applying auxiliary diagnosis, predicting prognostic outcome, evaluating therapeutic efficacy, and monitoring the severity of the condition (Shimada, Noie, Ohashi, Oba, & Takahashi, 2014). There are many traditional biomarkers such as CEA (carcinoembryonic antigen), CA‐15.3, and CA‐125, failing to achieve utility of cancer diagnosis (Baskic et al., 2007; Stieber et al., 2015; Zhang et al., 2013). Therefore, in the recent decade, several studies have been trying to find new breakthroughs from miroRNAs, lncRNAs, and circular RNAs (circRNAs) to explore more effective tumor biomarkers.
Circular RNAs, a special class of endogenous noncoding RNAs, are characterized by closed loop structures formed by covalent bonds of the head and tail, which are universally generated from exons of precursor mRNAs via backsplicing (Hsu & Coca‐Prados, 1979; Meng et al., 2017). Consequently, circRNAs are more stable and conserved than linear RNAs, so large numbers of circRNAs can extensively exist in peripheral blood or tissue, and even in exosomes (Jeck et al., 2013; Li, Zheng, et al., 2015; Meng et al., 2017; Rybak‐Wolf et al., 2015). CircRNAs might regulate carcinogenesis of different cancer through their complex biological functions, involving serving as ceRNAs or miRNA sponge, regulation of gene transcription, or splicing and interaction with other proteins as well as translation into proteins (Barrett & Salzman, 2016; Chen, Li, Tan, & Liu, 2016; Cortes‐Lopez & Miura, 2016; Hansen, Kjems, & Damgaard, 2013; Kulcheski, Christoff, & Margis, 2016; Li, Yang, et al., 2015; Qu et al., 2015). For example, circRNAs containing shared MREs (miRNA response elements) could competitively absorb miRNAs like sponges, resulting in functional changes of the relevant miRNAs. The biological mechanism suggested that lessening the inhibitory effect on their target genes might cause overexpression of the genes (Hansen et al., 2013; Memczak et al., 2013; Qiu et al., 2018). CircRNAs interact with other genes to promote carcinogenesis via oncogenic signal pathways in multiple types of cancer (Xu et al., 2017, 2018; Yao et al., 2017). Moreover, circRNAs might also inhibit or promote proliferation, migration, metastasis, and invasion of tumor cell (Chen, Li, Zheng, et al., 2017; Guo et al., 2016; Ma, Yao, Yu, Chen, & Li, 2018; Yu et al., 2018; Zhang, Xu, et al., 2018; Zhong et al., 2018). Additionally, differentially expressed circRNAs may be novel biomarkers with higher accuracy and efficacy for cancer diagnosis due to their stable structure and intricate gene regulatory functions.
Specific circRNAs could serve as potential candidate biomarkers for detection of different kinds of cancers, including colorectal cancer, breast cancer, gastric cancer, lung cancer, and hepatocellular carcinoma, through the discovery of circRNAs and their different expression profiles. The purpose of the study was to reveal the potential value of circRNAs for cancer diagnosis. First, this study aimed to evaluate the diagnostic accuracy of a single circRNA detected in plasma or serum and tissue. Furthermore, we also explored whether diagnosis of combined circRNAs is more effective than a single circRNA.
2. MATERIALS AND METHODS
2.1. Literature search
The meta‐analysis was strictly performed in accordance with the statement of the PRISMA (Preferred Reporting Items for Systematic Review and Meta‐Analyses) (Moher, Liberati, Tetzlaff, & Altman, 2009). To obtain relevant literatures, we searched six databases including PubMed, Sciencedirect, Cochrane Library, Web of Science, Wanfang, and Chinese National Knowledge Infrastructure databases up to February 15, 2019. Four English databases adopted the search strategy (“circular RNA” OR circRNA OR “circularRNA”) and (diagnosis OR specificity [SPE] OR sensitivity [SEN] OR “receiver operating characteristics” OR ROC) and (cancer OR carcinoma OR tumor OR malignancy OR neoplasm OR lymphoma OR leukemia). Two Chinese databases use the following search terms (“circular RNA” OR circRNA OR “circularRNA”) and (cancer OR carcinoma OR tumor OR malignancy OR neoplasm OR lymphoma OR leukemia). The references of the retrieved articles were also retrieved manually to guarantee that each eligible study was not missed. All included literatures were independently assessed by three reviewers (L.J., G.M., and L.X.). Any disagreement opinions were resolved by fully discussing to consensus with a fourth reviewer (Y.Z.).
2.2. Literature selection
All eligible studies consistently met the following included criteria: (a) expression of circRNA was detected in tissue or plasma/serum from peripheral blood; (b) studies focused on the association between circRNA expression and diagnosis for multiple kinds of cancers; (c) the all included studies provided available data covering the sample size of case and control, SEN, SPE, and area under curve (AUC). In addition, excluded criteria of unqualified studies: (a) reviews, meta‐analyses, and letters; (b) duplicated publications; (c) unqualified or not available data; (d) non‐English literatures.
2.3. Data extraction and quality assessment
The following information was collected for each included literature: first author, year and country of publication, sources of control, cancer type, specimen type, detection method, sample size of case and control, downregulated or upregulated expression of circRNAs, and other pivotal data involving the SEN, SPE, AUC, and the cutoff value. Two reviewers repeatedly appraised the methodological quality assessment of each diagnostic test according to the Quality Assessment of Diagnostic Accuracy Studies‐2 (QUADAS‐2) criteria (Wade, Corbett, & Eastwood, 2013).
2.4. Statistical methods
Meta‐analysis was conducted using STATA 11.0, Meta‐DiSc 1.4, and Review Manager 5.3 software. Review Manager 5.3 software was applied to assess the quality of the original literature. The random effect model was adopted to calculate the pooled SEN, SPE, positive likelihood ratios (PLR), negative likelihood ratios (NLR), and diagnostic odds ratio (DOR) with corresponding 95% confidence intervals (CIs) to evaluate diagnostic value of circRNAs. The summary receiver operator characteristic (SROC) curve was used to calculate the corresponding area under the SROC curve (AUC) for the quantitative assessment of diagnostic accuracy. All diagnostic parameters were evaluated by the rates of true positive, false positive, false negative, and true negative from original studies. Furthermore, the Cochran's‐Q and I‐squared (I 2) statistics index was used to evaluate the heterogeneity across studies. A p value less than 0.05 from Q test and an I‐square of more than 50% from I‐squared test indicated the existence of significant heterogeneity among eligible studies. The ROC plane/Spearman's correlation coefficient was conducted to examine the heterogeneity caused by the threshold effect. Meta‐regression and subgroup analysis were used to explore other possible sources of heterogeneity not related with threshold effect. A bivariate box plot was used to detect the heterogeneity of each study. Fagan's nomogram was constructed to calculate the posttest probabilities for the clinical practicality. Deeks’ funnel plot asymmetry test was applied to examine the probability of publication bias.
3. RESULTS
3.1. Study characteristics
Initially, a total of 1,414 articles were retrieved. Finally, a total of 64 studies about a single circRNA and 13 studies about combined circRNAs from 43 publications were included in the meta‐analysis after removing unqualified literature. The flow diagram of the literature selection was provided in Figure 1. Among the included publications, a total of 43 articles on 64 studies were analyzed involving 10,123 subjects in Table S1. Of the 64 studies, five studies focused on colorectal cancer (Ji et al., 2018; Wang, Li, et al., 2018; Wang et al., 2015; Zhang, Yang, et al., 2018; Zhuo, Lin, Chen, Huang, & Hu, 2017), 10 on breast cancer (Lu et al., 2017; Yin et al., 2018), 22 on gastric cancer (Chen, Li, Zhao, Xiao, & Guo, 2017; Huang, He, Liang, Huang, & Zhu, 2017; Li, et al., 2017, 2015; Li, Shao, et al., 2018; Li, Song, et al., 2017; Lu et al., 2018; Lu, Shao, Ye, Xiao, & Guo, 2017; Shao, Chen, et al., 2017; Shao, Li, et al., 2017; Shao, Yang, et al., 2017; Sun, Tang, et al., 2018; Tian, Chen, Li, & Xiao, 2018; Wei et al., 2019; Xie et al., 2018; Zhao, Chen, Li, Xiao, & Zhang, 2018), four on lung cancer (Li, Wang, Chen, Chen, & Jin, 2018; Zhu et al., 2017; Zong et al., 2018), 16 on hepatocellular carcinoma (Chen, Zhang, Lin, Song, & Wang, 2018; Fu et al., 2018, 2017; Li et al., 2019; Qin et al., 2016; Shang et al., 2016; Yao et al., 2017; Yao et al., 2018; Zhang, Zhang, Lin, & Wang, 2018; Zhang, Zhou, et al., 2018; Zhang, Xu, et al., 2018) and seven on other cancers (Fan, Cao, Liu, Zhang, & Li, 2019; Lan et al., 2018; Li et al., 2019; Sun, Li, et al., 2018; Teng et al., 2017; Wang, Xue, et al., 2018; Yang et al., 2017, 2018). Moreover, three articles with 13 studies explored the diagnostic value of the combined circRNAs (Chen, Li, Zhao, et al., 2017; Fan et al., 2019; Li, Shao, et al., 2018) in Table S2. Finally, detailed results of the QUADAS‐2 quality assessment are provided in Figure S1 and Figure S2. Each study showed risk of bias from four primary domains: patient selection, index test, reference standard, and, flow and timing. Also, the first three domains are applied to evaluate clinical applicability. The existence of the risk of bias is caused by many reasons. For example, index test studies with design deficiencies, predefinition of thresholds of index test positivity, and the inclusion of healthy controls or volunteers. About 20 studies included healthy participants, and several original literatures did provided explicit selection of subjects. The quality of all included studies ranged from medium to high, indicating that the study is relatively reliable.
Figure 1.
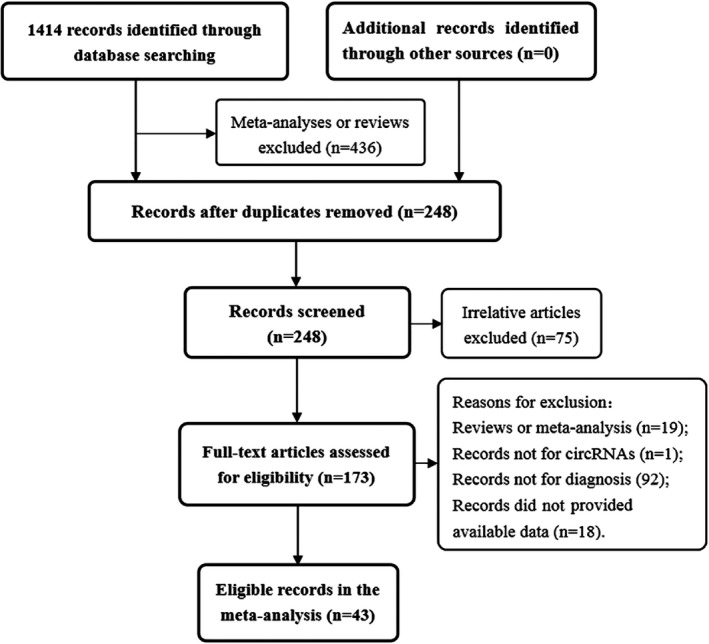
The flow diagram of this meta‐analysis
3.2. Diagnostic performance of a single circRNA
All 64 studies covering 5,373 cases and 4,750 controls were available for evaluating diagnostic accuracy of single circRNA (presented in Table 1). As presented in the forest plots (Figure 2a,b) and SROC curve (Figure 3d), the overall pooled results were 0.75 (95% CI: 0.71–0.78) for SEN, 0.76 (95% CI: 0.71, 0.79) for SPE, and 0.82 (95% CI: 0.78–0.85) for AUC. Compared with breast cancer subgroup, the subgroups of gastric cancer, colorectal cancer, hepatocellular carcinoma, and lung cancer presented higher diagnostic value with AUC values of 0.79 (95% CI: 0.75–0.83), 0.85 (95% CI: 0.82–0.88), 0.86 (95% CI: 0.83–0.89), and 0.78 (95% CI: 0.74–0.81) respectively. The results of specimen subgroup implied that plasma & serum circRNAs had a relatively high diagnostic accuracy compared with tissue subgroup (SEN: 0.78, SPE: 0.81, and AUC: 0.87 for plasma/serum subgroup). In addition, plasma/serum circRNAs existed with higher diagnostic accuracy in BC and GC (BC: AUC = 0.78; GC: AUC = 0.83; HCC = 0.90) (Figure 4a–c). Similar results were observed in tissue circRNA (CRC: AUC = 0.85; GC: AUC = 0.77; HCC: AUC = 0.82; BC: AUC = 0.66) (Shown in Figure 4d–f). The Fagan's diagram was regarded as a useful tool for predicting the value of circRNA applied to clinical diagnosis. When 20% value was selected as the pretest probability, the positive posttest probability of circRNAs would rise to 43% with a PLR of 3, while the negative posttest probability would drop to 8% with a NLR of 0.33, as presented in the Fagan's nomogram of Figure 3a.
Table 1.
Assessment of diagnostic accuracy of circRNA for cancer diagnosis
| Subgroup | N | SEN (95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) |
|---|---|---|---|---|---|---|---|
| Overall | 64 | 0.75 [0.71, 0.78] | 0.76 [0.71, 0.79] | 3.1 [2.6, 3.6] | 0.33 [0.29, 0.38] | 9 [7, 12] | 0.82 [0.78, 0.85] |
| Specimen | |||||||
| Tissue | 40 | 0.73 [0.70, 0.77] | 0.72 [0.68, 0.76] | 2.6 [2.3, 3.0] | 0.37 [0.33, 0.42] | 7 [6, 9] | 0.79 [0.75, 0.82] |
| Plasma/Serum | 24 | 0.78 [0.70, 0.84] | 0.81 [0.73, 0.87] | 4.2 [2.9, 6.0] | 0.27 [0.20, 0.36] | 16 [9, 26] | 0.87 [0.83, 0.89] |
| Source of control | |||||||
| Healthy | 20 | 0.79 [0.70, 0.86] | 0.83 [0.75, 0.89] | 4.7 [3.1, 7.1] | 0.25 [0.18, 0.36] | 19 [11, 33] | 0.88 [0.85, 0.91] |
| ANT, PNT | 40 | 0.73 [0.70, 0.77] | 0.72 [0.68, 0.76] | 2.6 [2.3, 3.0] | 0.37 [0.33, 0.42] | 7 [6, 9] | 0.79 [0.75, 0.82] |
| Other | 4 | 0.74 [0.70, 0.78]* | 0.72 [0.56, 0.84] | 2.7 [1.6, 4.4] | 0.36 [0.28, 0.46] | 7 [4, 15] | 0.75 [0.71, 0.78] |
| Sample size | |||||||
| ≥160 | 31 | 0.75 [0.69, 0.80] | 0.78 [0.72, 0.84] | 3.5 [2.7, 4.4] | 0.32 [0.27, 0.40] | 11 [7, 15] | 0.83 [0.80, 0.86] |
| <160 | 33 | 0.75 [0.71, 0.79] | 0.72 [0.67, 0.77] | 2.7 [2.3, 3.3] | 0.34 [0.29, 0.41] | 8 [6, 11] | 0.80 [0.77, 0.84] |
| Cancer | |||||||
| GC | 22 | 0.70 [0.63, 0.76] | 0.77 [0.68, 0.84] | 3.0 [2.3, 4.0] | 0.39 [0.33, 0.46] | 8 [6, 10] | 0.79 [0.75, 0.83] |
| HCC | 16 | 0.80 [0.73, 0.86] | 0.78 [0.69, 0.84] | 3.6 [2.5, 5.0] | 0.26 [0.18, 0.36] | 14 [8, 26] | 0.86 [0.83, 0.89] |
| CRC | 5 | 0.83 [0.76, 0.88] | 0.72 [0.63, 0.80] | 3.0 [2.3, 4.0]* | 0.23 [0.17, 0.32]* | 13 [9, 19] | 0.85 [0.82, 0.88] |
| BC | 10 | 0.67 [0.61, 0.72]* | 0.64 [0.58, 0.69]* | 1.8 [1.5, 2.2]* | 0.52 [0.43, 0.64]* | 4 [2, 5] | 0.69 [0.65, 0.73] |
| LC | 4 | 0.76 [0.70, 0.81]* | 0.84 [0.73, 0.91] | 4.6 [2.7, 8.0]* | 0.29 [0.22, 0.37]* | 16 [8, 32] | 0.78 [0.74, 0.81] |
| Other | 7 | 0.79 [0.68, 0.87] | 0.78 [0.65, 0.88] | 3.7 [2.1, 6.3] | 0.27 [0.17, 0.43] | 14 [6, 34] | 0.86 [0.82, 0.88] |
Abbreviations: ANT, adjacent noncancerous/normal tissue; AUC, area under the curve; BC, breast cancer; CI, confidence interval; CRC, colorectal cancer; DOR, diagnostic odds ratio; GC, gastric cancer; HCC, hepatocellular carcinoma; LC, lung cancer; NLR, negative likelihood ratio; PLR, positive likelihood ratio; PNT, para‐cancerous normal tissues; SEN, sensitivity; SPE, specificity.
No statistically significant heterogeneity.
Figure 2.
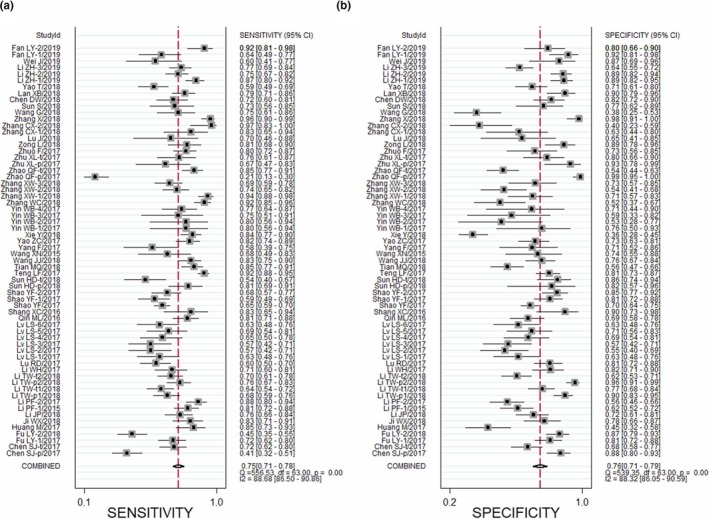
The diagnostic accuracy index of circRNA. (a) Sensitivity, (b) Specificity. circRNA, circular RNA
Figure 3.
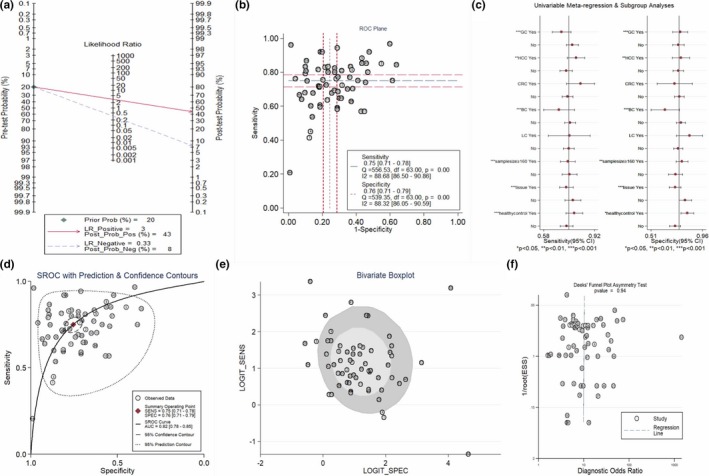
Evaluation of the diagnostic accuracy of circRNA. (a) Fagan's nomogram, (b) ROC Plane, (c) Meta‐regression analysis, d SROC curve, (e) Bivariate boxplot, f Deeks’ funnel plot. circRNA, circular RNA; ROC, receiver operating characteristics; SROC, summary receiver operator characteristic
Figure 4.
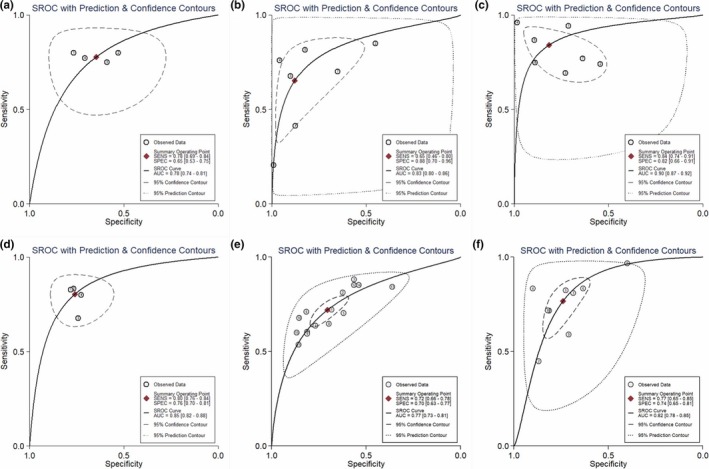
The SROC curve of circRNA in different cancers. (a) BC, (b) GC, (c) HCC in plasma/serum group. (d) CRC, (e) GC, (f) HCC in tissue group. BC, breast cancer; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; SROC, summary receiver operator characteristic
The heterogeneity were observed by I‐squared (SEN: I 2 = 88.68% and SPE: I 2 = 88.32%), which indicated existence of the significant heterogeneity among the all included studies (Figure 2a,b). Hence, we conducted meta‐regression analysis, bivariate boxplot, and ROC plane/Spearman's correlation coefficient to identify the potential sources of heterogeneity across the studies. ROC plane/Spearman's correlation coefficient was applied to examine whether the heterogeneity was caused by the diagnostic threshold effect. The ROC plane presented the absence of typical shoulder arm, proving nil threshold effect (Figure 3b). Spearman's correlation coefficient was 0.228 (p = 0.07), suggesting that the threshold effect did not exist. The presence of heterogeneity may result from sample size, specimens, control sources, breast cancer, gastric cancer, and hepatocellular carcinoma according to univariable meta‐regression analysis (Figure 3c). As shown in bivariate boxplot of Figure 3e, there are 13 studies not located in the boxplot. Of the 13 studies, six studies (study 4, 17, 18, 35, 52, and 54) belong to tissue group and other studies belong to plasma group (study 1, 5, 12, 10, 41, 44, and 53), implying that specimen was the major causes of heterogeneity. After excluding the 13 studies, the pooled SEN, SPE, and AUC were 0.75 (95% CI: 0.72–0.77), 0.76 (95% CI: 0.72–0.79), and 0.82(95% CI: 0.78–0.85) respectively, which had only minimal changes compared with previous results. Additionally, the heterogeneity across studies was also reduced (SEN: I 2 = 75.72% and SPE: I 2 = 78.36%). No significant publication bias was detected by Deeks’ funnel plot asymmetry test, with a p value of 0.94 as shown in Figure 3f.
3.3. Diagnostic performance of combined circRNAs
Thirteen studies from three publications evaluated the diagnostic practicality of the combined circRNAs. The pooled SEN and SPE were 0.89 (95% CI: 0.83, 0.93) and 0.94 (95% CI: 0.90, 0.97), respectively (Figure 5a,b). Additionally, the corresponding AUC was 0.97 (95% CI: 0.95–0.98) (shown in Figure 5e), demonstrating that the combined circRNA improved the diagnostic accuracy. The AUC of GC subgroup was also 0.97 (95% CI: 0.95–0.98), thereby also indicating well diagnostic performance of circRNAs (SEN: 0.89 (95% CI: 0.83, 0.93); SPE: 0.94 (95% CI: 0.89, 0.96)) (The data is not presented). Fagan's nomogram was adopted to examine the posttest probabilities for the clinical value of the combined circRNAs diagnosis. Fagan's diagram presented that the posttest probability of accurately diagnosing cancer would increase to 79%, while the negative probability would drop to 3%, with a pretest probability of 20% (Figure 5d). These results suggested that the combined circRNAs serve as the best potential biomarkers for cancer diagnosis. The ROC plane of nontypical shoulder arm (Figure 5c) and Spearman's correlation coefficient (−0.055, p = 0.857) mean the absence of the threshold effect. The p value of Deek's test was 0.93, indicating that the significant publication bias did not exist (Figure 5g).
Figure 5.
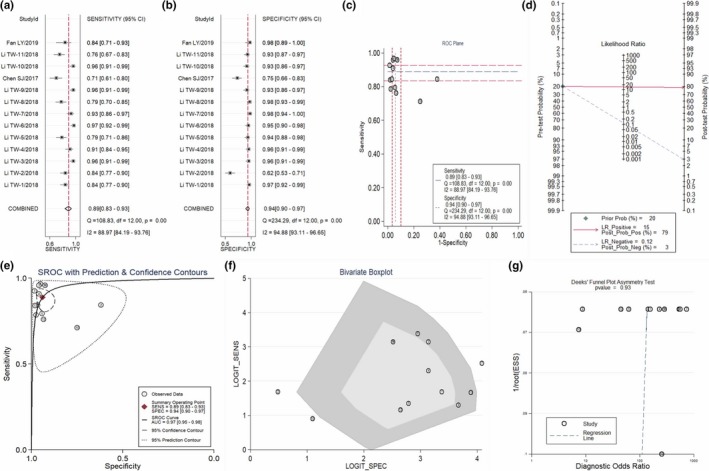
Evaluation of the diagnostic accuracy of the combined circRNA. (a) Sensitivity, (b) Specificity, (c) ROC Plane, (d) Fagan's nomogram, (e) SROC curve, (f) Bivariate boxplot, (g) Deeks’ funnel plot. circRNA, circular RNA; ROC, receiver operating characteristics; SROC, summary receiver operator characteristic
4. DISCUSSION
In the past decade, because of their special advantages as biomarkers, circRNAs have attracted attention in the detection, diagnosis, and prognosis of cancers (Chen, Li, Zheng, et al., 2017; Jiang et al., 2018; Shang et al., 2016; Zhang et al., 2017; Zhu et al., 2017). First, dysregulation of circRNAs was identified in tissues, plasma, and even exosomes from patients with many types of cancer (Li, Song, et al., 2017; Lu et al., 2017; Shao, Chen, et al., 2017). Second, circRNAs possess conserved sequences and close stable loop structures (Jeck et al., 2013; Qu et al., 2015). Third, circRNAs closely correlated with carcinogenesis as they have complex biological functions (Barrett & Salzman, 2016; Cortes‐Lopez & Miura, 2016). Therefore, circRNAs might be deemed to be promising candidate biomarkers for early diagnosis of carcinoma.
In previous meta‐analysis, the diagnostic implication of tissue‐based circRNAs was only appraised in hepatocellular carcinoma and gastric cancer (Chen, Zhang, Han, et al., 2018; Wang, Yang, et al., 2018). However, this study was the first to quantitatively analyze the diagnostic value of circRNA in breast cancer and lung cancer. Additionally, diagnostic significance of blood‐based and combined circRNAs was also first evaluated in the meta‐analysis. All the parameters of the meta‐analyses indicated moderate test performance of circRNAs for the diagnosis of lung cancer, gastric cancer, hepatocellular carcinoma, and colorectal cancer, with AUC values of 0.78, 0.79, 0.86, and 0.85 respectively. The blood‐based circRNAs showed a diagnostic superiority compared to tissue circRNAs by the pooled AUC (blooded circRNA: 0.87; tissue circRNA: 0.79). The combined circRNAs serve as good candidate biomarkers for GC diagnosis(SEN: 89%; SPE: 94%; AUC: 0.97). Therefore, results of the study suggested that specific circRNAs may be potential biomarkers to discriminate cancer patients from nontumorous individuals.
Overall, significant heterogeneity existed in this meta‐analysis (I 2 = 88.68% for SEN). The value of the spearman correlation coefficient was 0.228, which demonstrated that the heterogeneity of interstudy did not result from the threshold effect. The regression analyses suggested that cancer, type of control, and specimen were probably the sources of heterogeneity. In addition, abnormal expression of circRNAs in tissue is different from plasma/serum. Therefore, we speculated that the heterogeneity of interstudy may be caused by the specimen. Subsequently, we respectively analyzed the diagnostic accuracy of circRNA in tissue and blood among different types of cancer. For tissue group, the study presented the following strengths compared with the previous studies which only assessed the diagnostic role of tissue circRNAs in GC and HCC (Chen, Zhang, Han, et al., 2018; Wang, Yang, et al., 2018). First, it was first time to comprehensively evaluate the diagnostic role of tissue circRNAs in CRC and BC. Also, the circRNAs for CRC diagnosis have moderate diagnosis role with the corresponding AUC of 0.85 (95% CI: 0.82, 0.88). However, the AUC of BC subgroup was lower than that of HCC, GC, and CRC groups, which implied that the circRNAs may possess lower diagnostic performance for BC patients. Second, we avoided the potential bias derived from the specimen to reveal the diagnostic performance of tissue‐based circRNAs compared with the study of Chen et al. (Chen, Zhang, Han, et al., 2018).
The study first analyzed circRNA expression in plasma/serum to differentiate patients with cancer from healthy people and conducted a preliminary assessment of clinical utility for plasma/serum circRNAs. The pooled AUC was 0.87 (95% CI: 0.83, 0.89), implying that the blood‐based circRNAs had higher diagnostic performance than tissue circRNAs. Subsequently, we conducted subgroup analyses by sample size and cancer type. CircRNAs of GC group (AUC: 0.83) presented better diagnostic value compared with BC group (AUC: 0.78). Therefore, blood circRNAs play different diagnostic roles among multiple types of cancer.
Multiple studies provided that the detection of a single biomarker shows lower sensitivity and poor specificity. Hence, the detection of combined markers is necessary for clinical diagnosis as combined markers can remarkably improve the sensitivity and specificity of screen test. Therefore, it is essential to assess the diagnostic practicality of the combined circRNAs in the pooled meta‐analysis (Chen, Li, Zheng, et al., 2017; Li, Shao, et al., 2018). We identified that the combined circRNAs was rather served as better diagnostic biomarkers for GC diagnosis, with a pooled SEN of 0.89, a SPE of 0.94, and an AUC of 0.97. Additionally, Glas et al. provided that the higher DOR index signifies better diagnostic performance as it combined SEN and SPE (Glas, Lijmer, Prins, Bonsel, & Bossuyt, 2003). In the study, the DOR value was equal to 124, indicating that the combined circRNAs might act as relatively perfect biomarkers for cancer diagnosis. Fagan's plot demonstrated that detection of the combined circRNAs possess good clinical utility. Therefore, we can explore the combined currently valid biomarkers and novel markers with higher diagnostic accuracy for clinical diagnosis in the future.
We found that the diagnosis of combined circRNAs apparently possess better diagnostic value for clinical practice compared with that of a single circRNA. Multiple studies found that single circRNAs yield good diagnostic performance in tissue or plasma (Shang et al., 2016; Sun, Tang, et al., 2018; Wang, Li, et al., 2018; Zong et al., 2018). Moreover, several studies provided that single circRNAs in combination with other plasma protein biomarkers such as CEA, CA‐15.3, CA‐125 and AFP possess better diagnostic performance (Huang et al., 2017; Prefumo, 2016; Yang et al., 2017; Yin et al., 2018; Zhang, Yang, et al., 2018). Additionally, a single circRNA combined with other circRNAs obtained also had better diagnostic accuracy (Li, Chen, et al., 2017; Li, Shao, et al., 2018; Ren & Xin, 2017). For example, Li et al. identified that has_circ_0000096 and hsa_circ_002059 have moderate diagnosis value for gastric cancer, with an AUC of 0.82 and 0.73 respectively, while the AUC value of hsa_circ_0000096 combined with hsa_circ_002509 increased to 0.91 (Li, et al., 2017, 2015). Besides, when hsa_circ_0001017 in tissue combined with plasma, diagnostic accuracy was greatly improved for gastric cancer (AUC: 0.952) (Li, Shao, et al., 2018). In addition, the combination of hsa_circ_0001017 and hsa_circ_0061276 in tissue and plasma gained better efficacy for gastric cancer (AUC: 0.966). Consequently, we should continue to explore novel biomarkers, such as circRNAs, lncRNAs, and miRNAs with good diagnostic performance, combined with other effective biomarkers to yield better performance for cancer diagnosis, and ultimately for clinical practice.
The meta‐analysis had several limitations. First, there were no studies in non‐Asian population, which reduced the clinical applicability of the pooled results across different ethnicities. Second, the sample sizes of included studies were limited. Third, some confounding information including stage of cancer, age distributions, and gender proportions were unknown, so further subgroup analysis could not be conducted. Finally, a small number of studies included in healthy subjects would lead to overestimation of the diagnostic value of circRNA in this study. Hence, more large‐scale studies are indispensable to evaluate the diagnostic accuracy of plasma or tissue circRNAs among different malignant cancers due to the existence of the drawbacks.
In conclusion, the study proved that specific circRNAs may be used as candidate biomarkers for diagnosis of cancers including colorectal cancer, breast cancer, gastric cancer, lung cancer, and hepatocellular carcinoma. The combined circRNAs may possess well diagnostic efficacy for gastric cancer. Nonetheless, further large‐scale studies, involving novel and effective circRNAs as well as their in combination with other circRNAs or clinical biomarkers, are needed to contribute to the application of clinical diagnosis in the future.
CONFLICTS OF INTEREST
No conflicts of interest proclaimed.
Supporting information
ACKNOWLEDGMENTS
This study was supported by Program for Innovation Talents in Universities of Liaoning Province (No. LR2016026).
Li J, Li H, Lv X, et al. Diagnostic performance of circular RNAs in human cancers: A systematic review and meta‐analysis. Mol Genet Genomic Med. 2019;7:e749 10.1002/mgg3.749
REFERENCES
- Barrett, S. P. , & Salzman, J. (2016). Circular RNAs: Analysis, expression and potential functions. Development, 143(11), 1838–1847. 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskic, D. , Ristic, P. , Matic, S. , Bankovic, D. , Popovic, S. , & Arsenijevic, N. (2007). Clinical evaluation of the simultaneous determination of CA 15–3, CA 125 and sHER2 in breast cancer. Biomarkers, 12(6), 657–667. 10.1080/13547500701520563 [DOI] [PubMed] [Google Scholar]
- Chen, D. , Zhang, C. , Lin, J. , Song, X. , & Wang, H. (2018). Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Management and Research, 10, 1275–1283. 10.2147/cmar.s166740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Li, Y. , Zheng, Q. , Bao, C. , He, J. , Chen, B. , … Huang, S. (2017). Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Letters, 388, 208–219. 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Chen, S. , Li, T. , Zhao, Q. , Xiao, B. , & Guo, J. (2017). Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clinica Chimica Acta, 466, 167–171. 10.1016/j.cca.2017.01.025 [DOI] [PubMed] [Google Scholar]
- Chen, W. , Sun, K. , Zheng, R. , Zeng, H. , Zhang, S. , Xia, C. , … He, J. (2018). Cancer incidence and mortality in China, 2014. Chinese Journal of Cancer Research, 30(1), 1–12. 10.21147/j.issn.1000-9604.2018.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Li, C. , Tan, C. , & Liu, X. (2016). Circular RNAs: A new frontier in the study of human diseases. Journal of Medical Genetics, 53(6), 359–365. 10.1136/jmedgenet-2016-103758 [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Zhang, L. , Han, G. , Zuo, X. , Zhang, Y. , Zhu, Q. , … Wang, X. (2018). A meta‐analysis of the diagnostic accuracy of circular RNAs in digestive system malignancy. Cellular Physiology and Biochemistry, 45(3), 962–972. 10.1159/000487291 [DOI] [PubMed] [Google Scholar]
- Cortes‐Lopez, M. , & Miura, P. (2016). Emerging functions of circular RNAs. The Yale Journal of Biology and Medicine, 89(4), 527–537. [PMC free article] [PubMed] [Google Scholar]
- Dal Maso, L. , Panato, C. , Franceschi, S. , Serraino, D. , Buzzoni, C. , Busco, S. , … Stoppa, G. (2018). The impact of overdiagnosis on thyroid cancer epidemic in Italy, 1998–2012. European Journal of Cancer, 94, 6–15. 10.1016/j.ejca.2018.01.083 [DOI] [PubMed] [Google Scholar]
- Fan, L. , Cao, Q. , Liu, J. , Zhang, J. , & Li, B. (2019). Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Molecular Cancer, 18(1), 16 10.1186/s12943-018-0936-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, L. , Wu, S. , Yao, T. , Chen, Q. , Xie, Y. I. , Ying, S. , … Hu, Y. (2018). Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. Journal of Clinical Laboratory Analysis, 32(2). 10.1002/jcla.22239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, L. , Yao, T. , Chen, Q. , Mo, X. , Hu, Y. , & Guo, J. (2017). Screening differential circular RNA expression profiles reveals hsa_ circ_ 0004018 is associated with hepatocellular carcinoma. Oncotarget, 8(35), 58405–58416. 10.18632/oncotarget.16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas, A. S. , Lijmer, J. G. , Prins, M. H. , Bonsel, G. J. , & Bossuyt, P. M. (2003). The diagnostic odds ratio: A single indicator of test performance. Journal of Clinical Epidemiology, 56(11), 1129–1135. 10.1016/S0895-4356(03)00177-X [DOI] [PubMed] [Google Scholar]
- Guo, J. , Li, J. , Zhu, C. , Feng, W. , Shao, J. , Wan, L., … He, J. (2016). Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. OncoTargets and Therapy, 9, 7451–7458. 10.2147/ott.s123220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, T. B. , Kjems, J. , & Damgaard, C. K. (2013). Circular RNA and miR‐7 in cancer. Cancer Research, 73(18), 5609–5612. 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- Hsu, M. T. , & Coca‐Prados, M. (1979). Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature, 280(5720), 339–340. 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- Huang, M. , He, Y. , Liang, L. , Huang, Q. , & Zhu, Z. (2017). Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World Journal of Gastroenterology, 23(34), 6330–6338. 10.3748/wjg.v23.i34.6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck, W. R. , Sorrentino, J. A. , Wang, K. , Slevin, M. K. , Burd, C. E. , Liu, J. , … Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19(2), 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, W. , Qiu, C. , Wang, M. , Mao, N. , Wu, S. , & Dai, Y. (2018). Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochemical and Biophysical Research Communications, 497(1), 122–126. 10.1016/j.bbrc.2018.02.036 [DOI] [PubMed] [Google Scholar]
- Jiang, X. M. , Li, Z. L. , Li, J. L. , Xu, Y. , Leng, K. M. , Cui, Y. F. , & Sun, D. J. (2018). A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. European Review for Medical and Pharmacological Sciences, 22(2), 365–371. 10.26355/eurrev_201801_14182 [DOI] [PubMed] [Google Scholar]
- Kimura, T. , & Egawa, S. (2018). Epidemiology of prostate cancer in Asian countries. International Journal of Urology, 25, 524–531. 10.1111/iju.13593 [DOI] [PubMed] [Google Scholar]
- Kulcheski, F. R. , Christoff, A. P. , & Margis, R. (2016). Circular RNAs are miRNA sponges and can be used as a new class of biomarker. Journal of Biotechnology, 238, 42–51. 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Lan, X. , Cao, J. , Xu, J. , Chen, C. , Zheng, C. , Wang, J. , … Ge, M. (2018). Decreased expression of hsa_circ_0137287 predicts aggressive clinicopathologic characteristics in papillary thyroid carcinoma. Journal of Clinical Laboratory Analysis, 32(8), e22573 10.1002/jcla.22573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Wang, J. , Chen, Z. , Chen, Y. , & Jin, M. (2018). Hsa_circ_0079530 promotes cell proliferation and invasion in non‐small cell lung cancer. Gene. 10.1016/j.gene.2018.04.059 [DOI] [PubMed] [Google Scholar]
- Li, J. , Yang, J. , Zhou, P. , Le, Y. , Zhou, C. , & Wang, S., …Gong, Z. (2015). Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. American Journal of Cancer Research, 5(2), 472–480. [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Chen, H. , Chen, S. , Mo, X. , Li, T. , Xiao, B. , … Guo, J. (2017). Circular RNA 0000096 affects cell growth and migration in gastric cancer. British Journal of Cancer, 116(5), 626–633. 10.1038/bjc.2016.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Chen, S. , Chen, H. , Mo, X. , Li, T. , Shao, Y. , … Guo, J. (2015). Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clinica Chimica Acta, 444, 132–136. 10.1016/j.cca.2015.02.018 [DOI] [PubMed] [Google Scholar]
- Li, T. , Shao, Y. , Fu, L. , Xie, Y. I. , Zhu, L. , Sun, W. , … Guo, J. (2018). Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT‐PCR detection. Journal of Molecular Medicine, 96(1), 85–96. 10.1007/s00109-017-1600-y [DOI] [PubMed] [Google Scholar]
- Li, W. H. , Song, Y. C. , Zhang, H. , Zhou, Z. J. , Xie, X. , Zeng, Q. N. , … Chang, D. M. (2017). Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Disease Markers. 10.1155/2017/4587698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zheng, Q. , Bao, C. , Li, S. , Guo, W. , Zhao, J. , … Huang, S. (2015). Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Research, 25(8), 981–984. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Zhou, Y. E. , Yang, G. , He, S. , Qiu, X. , Zhang, L. , … Zheng, F. (2019). Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clinica Chimica Acta, 492, 37–44. 10.1016/j.cca.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Lu, J. , Zhang, P. Y. , Xie, J. W. , Wang, J. B. , Lin, J. X. , Chen, Q. Y. , …Zheng, C. H. (2018). Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. Journal of Clinical Laboratory Analysis, 33, e22726 10.1002/jcla.22726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü, L. , Sun, J. , Shi, P. , Kong, W. , Xu, K. , He, B. , … Wang, J. (2017). Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget, 8(27), 44096–44107. 10.18632/oncotarget.17307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Shao, Y. , Ye, G. , Xiao, B. , & Guo, J. (2017). Low expression of hsa_circ_0006633 in human gastric cancer and its clinical significances. Tumor Biology, 39(6), 1–7. 10.1177/1010428317704175 [DOI] [PubMed] [Google Scholar]
- Ma, H. B. , Yao, Y. N. , Yu, J. J. , Chen, X. X. , & Li, H. F. (2018). Extensive profiling of circular RNAs and the potential regulatory role of circRNA‐000284 in cell proliferation and invasion of cervical cancer via sponging miR‐506. American Journal of Translational Research, 10(2), 592–604. [PMC free article] [PubMed] [Google Scholar]
- Memczak, S. , Jens, M. , Elefsinioti, A. , Torti, F. , Krueger, J. , Rybak, A., …Rajewsky, N. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Meng, S. , Zhou, H. , Feng, Z. , Xu, Z. , Tang, Y. , Li, P. , & Wu, M. (2017). CircRNA: Functions and properties of a novel potential biomarker for cancer. Molecular Cancer, 16(1), 94 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher, D. , Liberati, A. , Tetzlaff, J. , & Altman, D. G. (2009). Reprint–preferred reporting items for systematic reviews and meta‐analyses: The PRISMA statement. Physical Therapy, 89(9), 873–880. 10.1093/ptj/89.9.873 [DOI] [PubMed] [Google Scholar]
- Prefumo, F. (2016). Circular RNA and pre‐eclampsia: On the long road from the laboratory to the bedside. BJOG: an International Journal of Obstetrics & Gynaecology, 123(13), 2119 10.1111/1471-0528.13965 [DOI] [PubMed] [Google Scholar]
- Qin, M. , Liu, G. , Huo, X. , Tao, X. , Sun, X. , Ge, Z. , … Qin, W. (2016). Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers, 16(1), 161–169. 10.3233/CBM-150552 [DOI] [PubMed] [Google Scholar]
- Qiu, L. P. , Wu, Y. H. , Yu, X. F. , Tang, Q. , Chen, L. , & Chen, K. P. (2018). The emerging role of circular RNAs in hepatocellular carcinoma. Journal of Cancer, 9(9), 1548–1559. 10.7150/jca.24566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, S. , Yang, X. , Li, X. , Wang, J. , Gao, Y. , Shang, R. , … Li, H. (2015). Circular RNA: A new star of noncoding RNAs. Cancer Letters, 365(2), 141–148. 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Ren, S. , Xin, Z. , Xu, Y. , Xu, J. , & Wang, G. (2017). Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle, 16( 22), 2204 – 2211. 10.1080/15384101.2017.1346754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak‐Wolf, A. , Stottmeister, C. , Glažar, P. , Jens, M. , Pino, N. , Giusti, S. , … Rajewsky, N. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell, 58(5), 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Shang, X. , Li, G. , Liu, H. , Li, T. , Liu, J. , Zhao, Q. , & Wang, C. (2016). Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore), 95(22). 10.1097/MD.0000000000003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Y. , Chen, L. , Lu, R. , Zhang, X. , Xiao, B. , Ye, G. , & Guo, J. (2017). Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumor Biology, 39(4). 10.1177/1010428317699125 [DOI] [PubMed] [Google Scholar]
- Shao, Y. , Li, J. , Lu, R. , Li, T. , Yang, Y. , Xiao, B. , & Guo, J. (2017). Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Medicine, 6(6), 1173–1180. 10.1002/cam4.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Y. , Yang, Y. , Lu, R. , Xiao, B. , Ye, G. , & Guo, J. (2017). Identification of tissue‐specific circRNA hsa_circ_0000705 as an indicator for human gastric cancer. International Journal of Clinical and Experimental Pathology, 10(3), 3151–3156. [Google Scholar]
- Shimada, H. , Noie, T. , Ohashi, M. , Oba, K. , & Takahashi, Y. (2014). Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer, 17(1), 26–33. 10.1007/s10120-013-0259-5 [DOI] [PubMed] [Google Scholar]
- Stieber, P. , Nagel, D. , Blankenburg, I. , Heinemann, V. , Untch, M. , Bauerfeind, I. , & Di Gioia, D. (2015). Diagnostic efficacy of CA 15–3 and CEA in the early detection of metastatic breast cancer‐A retrospective analysis of kinetics on 743 breast cancer patients. Clinica Chimica Acta, 448, 228–231. 10.1016/j.cca.2015.06.022 [DOI] [PubMed] [Google Scholar]
- Sun, H. , Tang, W. , Rong, D. , Jin, H. , Fu, K. , Zhang, W. , … Cao, X. (2018). Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomarkers, 21(2), 299–306. 10.3233/CBM-170379 [DOI] [PubMed] [Google Scholar]
- Sun, S. , Li, B. , Wang, Y. , Li, X. , Wang, P. , Wang, F. , … Yang, H. (2018). Clinical significance of the decreased expression of hsa_circ_001242 in oral squamous cell carcinoma. Disease Markers, 2018, 6514795 10.1155/2018/6514795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, L. , Chen, Y. U. , Chen, H. , He, X. , Wang, J. , Peng, Y. , … Shao, B. O. (2017). Circular RNA hsa_circ_0021001 in peripheral blood: A potential novel biomarker in the screening of intracranial aneurysm. Oncotarget, 8(63), 107125–107133. 10.18632/oncotarget.22349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M. , Chen, R. , Li, T. , & Xiao, B. (2018). Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. Journal of Clinical Laboratory Analysis, 32(3). 10.1002/jcla.22281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, R. , Corbett, M. , & Eastwood, A. (2013). Quality assessment of comparative diagnostic accuracy studies: Our experience using a modified version of the QUADAS‐2 tool. Research Synthesis Methods, 4(3), 280–286. 10.1002/jrsm.1080 [DOI] [PubMed] [Google Scholar]
- Wang, G. , Xue, W. , Jian, W. , Liu, P. , Wang, Z. , Wang, C. , … Zhang, C. (2018). The effect of Hsa_circ_0001451 in clear cell renal cell carcinoma cells and its relationship with clinicopathological features. Journal of Cancer, 9(18), 3269–3277. 10.7150/jca.25902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Li, X. , Lu, L. , He, L. , Hu, H. , & Xu, Z. (2018). Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. Journal of Clinical Laboratory Analysis, 32(5), e22379– 10.1002/jcla.22379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Yang, Y. , Xu, J. , Bai, W. , Ren, X. , & Wu, H. (2018). CircRNAs as biomarkers of cancer: A meta‐analysis. BMC Cancer, 18(1), 10.1186/s12885-018-4213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zhang, Y. , Huang, L. , Zhang, J. , Pan, F. , Li, B., …Zheng, W. (2015). Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. International Journal of Clinical and Experimental Pathology, 8(12), 16020–16025. [PMC free article] [PubMed] [Google Scholar]
- Wei, J. , Wei, W. , Xu, H. , Wang, Z. , Gao, W. , Wang, T. , … De, W. (2019). Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomarkers, 1–7. 10.3233/CBM-182029 [DOI] [PubMed] [Google Scholar]
- Xie, Y. , Shao, Y. , Sun, W. , Ye, G. , Zhang, X. , Xiao, B. , & Guo, J. (2018). Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomarkers in Medicine, 12(1), 11–20. 10.2217/bmm-2017-0114 [DOI] [PubMed] [Google Scholar]
- Xu, L. , Zhang, M. , Zheng, X. , Yi, P. , Lan, C. , & Xu, M. (2017). The circular RNA ciRS‐7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. Journal of Cancer Research and Clinical Oncology, 143(1), 17–27. 10.1007/s00432-016-2256-7 [DOI] [PubMed] [Google Scholar]
- Xu, X. , Jia, S. , Dai, Y. , Zhang, J. , Li, X. , Shi, J. , … Lang, J. (2018). Identification of circular RNAs as a novel biomarker for ovarian endometriosis. Chinese Medical Journal, 131( 5), 559 – 566. 10.4103/0366-6999.226070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. , Liu, D. Y. , Guo, J. T. , Ge, N. , Zhu, P. , Liu, X. , … Sun, S. Y. (2017). Circular RNA circ‐LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World Journal of Gastroenterology, 23( 47), 8345 – 8354. 10.3748/wjg.v23.i47.8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, T. , Chen, Q. , Shao, Z. , Song, Z. , Fu, L. , & Xiao, B. (2018). Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. Journal of Clinical Laboratory Analysis, 32(8), e22572 10.1002/jcla.22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Z. , Luo, J. , Hu, K. , Lin, J. , Huang, H. , Wang, Q., …Yang, Y. (2017). ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Molecular Oncology, 11(4), 422–437. 10.1002/1878-0261.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W. B. , Yan, M. G. , Fang, X. , Guo, J. J. , Xiong, W. , & Zhang, R. P. (2018). Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clinica Chimica Acta. 487, 363 – 368. 10.1016/j.cca.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Yu, J. , Xu, Q. G. , Wang, Z. G. , Yang, Y. , Zhang, L. , Ma, J.Z. , … Zhou, W.P. (2018). Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. Journal of Hepatology, 68( 6), 1214 – 1227. 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Zhang, C. , Lin, J. , & Wang, H. (2018). Circular RNA Hsa_Circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma. Cellular Physiology and Biochemistry, 51(1), 290–300. 10.1159/000495230 [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Liu, H. , Hou, L. , Wang, G. E. , Zhang, R. , Huang, Y. , … Zhu, J. (2017). Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR‐424‐5p and regulating LATS1 expression. Molecular Cancer, 16(1), 151 10.1186/s12943-017-0719-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. J. , Hu, Y. , Qian, H. L. , Jiao, S. C. , Liu, Z. F. , Tao, H. T. , & Han, L. (2013). Expression and significance of ER, PR, VEGF, CA15‐3, CA125 and CEA in judging the prognosis of breast cancer. Asian Pacific Journal of Cancer Prevention, 14(6), 3937–3940. 10.7314/APJCP.2013.14.6.3937 [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Yang, S. , Liu, Y. , Wang, Y. , Lin, T. , Li, Y. , & Zhang, R. (2018). Hsa_circ_0007534 as a blood‐based marker for the diagnosis of colorectal cancer and its prognostic value. International Journal of Clinical and Experimental Pathology, 11(3), 1399–1406. [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Xu, Y. , Qian, Z. , Zheng, W. , Wu, Q. I. , Chen, Y. , … Yu, Y. (2018). circRNA_104075 stimulates YAP‐dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death & Disease, 9(11), 1091 10.1038/s41419-018-1132-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhou, H. U. , Jing, W. , Luo, P. , Qiu, S. , Liu, X. , … Tu, J. (2018). The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Disease Markers, 2018, 1–9. 10.1155/2018/3073467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q. , Chen, S. , Li, T. , Xiao, B. , & Zhang, X. (2018). Clinical values of circular RNA 0000181 in the screening of gastric cancer. Journal of Clinical Laboratory Analysis, 32(4), e22333– 10.1002/jcla.22333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, L. , Wang, Y. , Cheng, Y. , Wang, W. , Lu, B. , Zhu, L. , & Ma, Y. (2018). Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR‐4641/PCK1 pathway. Biochemical and Biophysical Research Communications, 499(4), 1044–1049. 10.1016/j.bbrc.2018.03.221 [DOI] [PubMed] [Google Scholar]
- Zhu, X. , Wang, X. , Wei, S. , Chen, Y. , Chen, Y. , Fan, X. , … Wu, G. (2017). hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. The FEBS Journal, 284(14), 2170–2182. 10.1111/febs.14132 [DOI] [PubMed] [Google Scholar]
- Zhuo, F. , Lin, H. , Chen, Z. , Huang, Z. , & Hu, J. (2017). The expression profile and clinical significance of circRNA0003906 in colorectal cancer. OncoTargets and Therapy, 10, 5187–5193. 10.2147/ott.s147378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, L. , Sun, Q. , Zhang, H. , Chen, Z. , Deng, Y. , Li, D. , & Zhang, L. (2018). Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomedicine & Pharmacotherapy, 102, 639–644. 10.1016/j.biopha.2018.03.084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


