Abstract
Background
20p13 microdeletion syndrome has been reported to be associated with developmental delays, intellectual disability, epilepsy, and unspecific dysmorphic characteristics. However, only a few cases of 20p13 microdeletion have been described, and therefore its typical features and precise pathogenesis remain elusive.
Methods and Results
In this article, we report the case of a 9‐month‐old infant who presented with a large fontanelle, facial dysmorphism, and failure to thrive. Array‐comparative genomic hybridization (aCGH) analysis confirmed a 2.01‐Mb microdeletion in chromosome band 20p13 that involved SOX12 and NRSN2, both of which are considered paramount causative genes in patients with 20p13 microdeletion. To elucidate the typical features of 20p13 microdeletion, we further reviewed these previously reported cases and found that motor delay (90%) was the most common manifestation, followed by language delay (60%), abnormal digits (60%), mental retardation (50%), large fontanelle (50%), electroencephalography abnormalities (50%), and seizure (40%).
Conclusion
This report highlights the potential of aCGH as a practical and powerful tool with which to detect submicroscopic chromosomal abnormalities in individuals presenting with a wide spectrum of phenotypes, ranging from facial dysmorphism to failure to thrive. Additionally, the literature review casts new light on the clinical features of 20p13 microdeletion.
Keywords: 20p13, array‐comparative genomic hybridization, developmental delays, microdeletion
1. INTRODUCTION
Microdeletion syndrome is a genetic disorder characterized by a small (<5 megabases [Mb]) chromosomal deletion that spans several genes but cannot be detected using conventional cytogenetic methods or high‐resolution karyotyping. Microdeletion syndrome has been associated with intellectual disability, multiple congenital anomalies, and autism spectrum disorders (Lupski & Stankiewicz, 2005). Therefore, a high index of suspicion is needed to ensure an early diagnosis and provide appropriate management.
Previously, a microdeletion of the 20p12.2 locus involving JAG1 (OMIM#601920) was found to contribute to the well‐known Alagille syndrome (Saleh, Kamath, & Chitayat, 2016). However, the typical presentations and pathogenesis of its near region in terms of 20p13 microdeletion remain elusive. Although a few reports of 20p13 microdeletion have described developmental delays, mild to moderate intellectual disability, seizure disorders, and dysmorphic features (An et al., 2013; McGill et al., 2010; Moutton et al., 2012; Sebat et al., 2007), the manifestations of this genetic event remain under‐reported because variations in deletion size have led to diverse genotype–phenotype associations. In the absence of specific presentations and consistent genetic defects, individuals affected by 20p13 microdeletion may be at risk of delayed clinical diagnosis.
Here, we report the case of a 9‐month‐old female infant who presented with a large fontanelle, facial dysmorphism, and failure to thrive. Array‐comparative genomic hybridization (aCGH) analysis identified her as a carrier of a 20p13 microdeletion of up to 2.01 Mb in size [chr20:g.(60747_2073671)del]. This microdeletion involved SOX12 (OMIM#601947) and NRSN2 (OMIM#610666), which were previously reported to induce developmental delays in patients with 20p13 microdeletions (An et al., 2013). This case underscores the potential of aCGH as a useful method for elucidating as‐yet unidentified cases involving dysmorphic features and growth disorders and thus ensuring early diagnosis and appropriate therapy.
2. CASE REPORT
A female Taiwanese infant was referred to an outpatient endocrine clinic for the evaluation of a wide anterior fontanelle and failure to thrive. She was the first child born to healthy and nonconsanguineous parents. A review of the prenatal history revealed a normal three‐vessel umbilical cord and normohydramnios. The subject was born to a 34‐year‐old mother by spontaneous vaginal delivery at a gestational age of 39 weeks. At birth, her body weight, body length, and head circumference were 3,300 g (50–75th percentile), 48 cm (10–25th percentile), and 33.5 cm (50th percentile), respectively. A postnatal physical examination revealed a large fontanelle (4 × 4 fingerbreadth), prominent forehead, hypertelorism, flat philtrum, thin upper lip, low set ears (Figure 1), and bilateral postaxial polydactyly. The remaining physical examination findings were unremarkable. She underwent reconstruction of bilateral polydactyly at 2 days of age.
Figure 1.
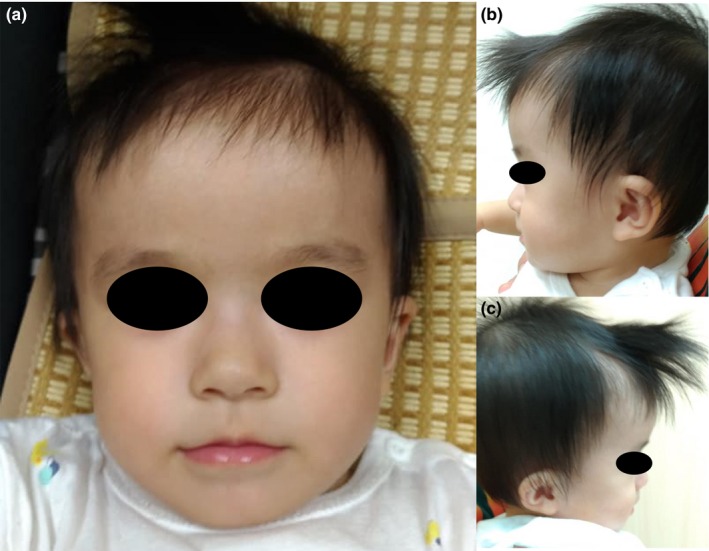
Image of facial dysmorphism in a 9‐month‐old female infant with 20p13 microdeletion syndrome. Note the flat philtrum, thin upper lip, hypertelorism (a), prominent forehead (b), and bilateral low‐set ears (c)
The subject was brought to our pediatric endocrine clinic at the age of 7 months because of poor weight gain and a persistently wide fontanelle. At that time, her body weight, body length, and head circumference were 5,800 g (<third percentile), 62 cm (<third percentile), and 42.5 cm (25–50th percentile), respectively. She was unable to roll over or sit unassisted, and a backward head position was observed when she was pulled up from her bed. Her short stature and failure to thrive warranted an analysis of thyroid function, which revealed a free T4 level of 1.10 ng/dl (normal: 0.89–1.78 ng/dl), T4 level of 5.49 µg/dl (5.38–12.40 µg/dl), and thyroid‐stimulating hormone level of 1.90 µIU/ml (0.25–5.00 µIU/ml). Venous blood gas analysis yielded a pH of 7.405 (7.32–7.45), PCO2 of 28.9 mmHg (41–51 mmHg), PO2 of 63.8 mmHg (20–49 mmHg), HCO3 of 17.7 mmol/L (24–28 mmol/L), and base excess of −5.3 mmol/L ([−10]–[−2] mmol/L). Metabolic profile analysis revealed an NH3 level of 110 µg/dl (68–136 µg/dl) and a lactate level of 2.1 mmol/L (0.6–3.2 mmol/L). Urine organic acid profile and plasma ammonic acid studies yielded negative results.
Chromosome analysis revealed no chromosomal aberrations, despite the observed facial dysmorphism. In addition, brain magnetic resonance imaging showed no prominent Virchow–Robin spaces, abnormal basal ganglia, or cerebral atrophy. Accordingly, oligonucleotide aCGH analysis was conducted, which detected a microdeletion of up to 2.01 Mb at chromosome 20p13 [chr20:g.(60747_2073671)del]. This microdeleted chromosomal region contains the following OMIM genes: SOX12 (OMIM#601947), NRSN2 (OMIM#610666), TRIB3 (OMIM#607898), RBCK1 (OMIM#610924), TBC1D20 (OMIM#611663), CSNK2A1 (OMIM#115440), TCF15 (OMIM#601010), SLC52A3 (OMIM#613350), FAM110A (OMIM#611393), ANGPT4 (OMIM#603705), RSPO4 (OMIM#610573), SNPH (OMIM#604942), FKBP1A (OMIM#186945), SIRPG (OMIM#605466), PDYN (OMIM#131340), ect (Figure 2). The above microdeletion was not detected in her parents, as it was considered de novo.
Figure 2.

Schematic view of chromosome 20. A zoom of the 20p13 region shows the deleted genes. The observed microdeletion in 20p13 was up to 2.01 Mb in size (genomic location: chr20:60,747‐2,073,671)
Electroencephalography (EEG) was performed to survey potential associated clinical defects. This exam revealed a bilateral occipital background with high‐voltage synchronous spikes and waves in F3 and F4. Rapid bilateral frontal poly spikes and C3 spikes were also detected. An abdominal sonography and echocardiogram showed no significant abnormalities. Although a normal fundus was observed by an ophthalmologist, bilateral sensorineural hearing loss was confirmed by an otolaryngologist.
3. DISCUSSION
We report the case of a female infant who presented with a large fontanelle, facial dysmorphism, and failure to thrive, which were attributed to a subtelomeric deletion of 20p13. This deletion included SOX12 and NRSN2, both of which have been identified as pivotal genes associated with developmental delays (An et al., 2013). Notably, the pure microdeletion of 20p13 without other genetic defects is rare, and such events involve a wide range of chromosomal deletion sizes. Therefore, it is difficult to establish a genotype–phenotype correlation in affected patients (Jezela‐Stanek, Kucharczyk, Pelc, Gutkowska, & Krajewska‐Walasek, 2013). Although 20p13 deletions may be associated with developmental delays in motor and speech, mental retardation and epilepsy at various levels of severity, and facial dysmorphism (Moutton et al., 2012), the typical characteristics of such cases remain enigmatic, leading to diagnostic delays. Interestingly, previous studies showed that patients who simultaneously harbored a duplication and subtelomeric deletion of 20p exhibited facial dysmorphism, psychomotor retardation, delayed development, and speech difficulties (Leclercq et al., 2009; Trachoo, Assanatham, Jinawath, & Nongnuch, 2013). However, the precise mechanisms underlying duplications and/or deletions of the 20p13 chromosome band remain unclear and require further study.
To date, only a few cases of 20p13 microdeletion have been reported, and most involved de novo events like our patient (An et al., 2013). To elucidate the typical features of 20p13 microdeletion, we further reviewed these previously reported cases and found that motor delay (90%) was the most common manifestation, followed by language delay (60%), abnormal digits (60%), mental retardation (50%), large fontanelle (50%), EEG abnormalities (50%), and seizure (40%) (Table 1) (An et al., 2013; Baker et al., 2002; Jezela‐Stanek et al., 2013; McGill et al., 2010; Moutton et al., 2012). Last but not least, facial dysmorphism was identified as a crucial presentation even though its occurrence varied from 30% to 70% and depended on the involved facial sites. As all reported patients with 20p13 microdeletion exhibited at least one facial abnormality, we emphasize that microdeletion syndrome should be considered in any patient who presents with facial dysmorphism to facilitate an early diagnosis and appropriate management. To be closer to the real‐world clinical practice, 52 additional cases exhibited pathogenic condition were also reviewed through ClinVar database. Similar to Table 1, the most common phenotype of these informally reported cases was developmental delays (13 cases, 25%) followed by facial dysmorphism (five cases, 10%). Interestingly, we found that most of the affected patients with developmental delays were caused by the defects of multiple genes which were usually associated with TBC1D20, SOX12, and NRSN2 genes. Therefore, further functional study is required to elucidate the mechanisms behind those causative genes and developmental process.
Table 1.
Comparison of phenotypic features of the patients with a 20p13 subtelomeric microdeletion
| Yu An et al. | McGill et al. | Baker et al. | Moutton et al. | Jezela‐Stanek et al. | Our case | Affected/Total case (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | Case 4 | Case 1 | Case 2 | Case 1 | Case 1 | Case 1 | |||
| Size of deletion | 1.4 Mb | 1.1 Mb | 799 kb | 274 kb | 1.7 Mb | 1.2 Mb | NA | 2.08 Mb | 1.15 Mb | 2.01 Mb | |
| Gender | Male | Male | Female | Male | Female | Female | Male | Female | Female | Female | |
| Diagnostic age | 16 years 9 months | 11 years 6 months | 3 years 4 months | 16 years | 11 months | 15 years | 10 years | 19 years | 9 years | 9months | |
| Large fontanelles | − | + | − | NA | + | + | NA | − | + | + |
5/10 (50%) |
| Forehead | Normal | Frontal bossing | Frontal bossing | Normal | Normal | NA | NA | Normal | Normal | Frontal bossing |
3/10 (30%) |
| Head | Normal | Macrocephaly | Macrocephaly | Normal | Normal | Microcephaly | NA | Microcephaly | Normal | Normal |
4/10 (40%) |
| Hearing | NA | NA | Normal | Normal | NA | NA | NA | NA | NA |
Bilateral sensorineural hearing loss |
1/10 (10%) |
| Vision | NA | NA | NA | Normal | NA | Poor visual acuity | NA | Peripheral retinopathy | Normal | Normal |
2/10 (20%) |
| Facial feature | |||||||||||
| Prominent nasal root and ridge | − | − | A short nasal root with broad tip and anteverted nares | − | − | + | NA | + | − | − |
3/10 (30%) |
| Eyes | Narrow palpebral fissures | Deep‐set eyes | Downslanting palpebral fissures, slightly deep set eyes | Slightly hypertelorism | Microcorneas, marked edema of the lids, reversed epicanthal, synophrys, telecanthus | NA | NA | Normal | Normal | Hypertelorism |
6/10 (60%) |
| Ears | Hypoplastic helix | − | − | − | Thicked, posteriorly rotated | Low‐set and posteriorly rotated ears with thickened overfolded helices | NA | Low‐set, dysplastic, thick overfolded helices | − | Low‐set ears |
5/10 (50%) |
| Flat philtrum | NA | Everted | NA | NA | + | − | NA | − | + | + |
4/10 (40%) |
| philtrum | |||||||||||
| Thin upper lip | Normal | Inverted upper lip | Normal | Bowed upper lip | + | + | NA | + | + | + |
7/10 (70%) |
| High palate | + | − | − | − | − | + | NA | + | + | − |
4/10 (40%) |
| Others | |||||||||||
| Hypoplastic nails | + | − | NA | − | − | + | NA | Broad finger tips | + | − |
4/10 (40%) |
| Digits | Normal | Syndactyly | NA | long toes | NA | Short, broad thumbs | NA | Brachydactyly | discrete fifth finger clinodactyly | Polydactyly |
6/10 (60%) |
| Seizures | − | − | NA | − | Neonatal seizure | Generalized seizure | Generalized seizure | Absence‐epilepsy | − | − |
4/10 (40%) |
| EEG abnormalities | + | + | NA | − | NA | + | NA | + | NA | + |
5/10 (50%) |
| Weight deficiency | − | − | − | − | − | + | NA | − | + | Growth retardation |
3/10 (30%) |
| Neonatal complication | Jaundice | − | Gastroesophageal reflux | NA | Respiratory stress | Mild jaundice | NA | Jaundice | NA | NA |
5/10 (50%) |
| Developmental delay | |||||||||||
| Motor delay | + | + | + | + | + | + | NA | + | + | + |
9/10 (90%) |
| Language delay | + | + | + | + | NA | + | NA | + | − | na |
6/10 (60%) |
| Mental retardation | − | Mild | − | NA | NA | Mild | Moderate | Moderate | + | na |
5/10 (50%) |
NA, no available; +, present; −, absent; na, no assessment.
Among the affected cases on ClinVar database, 16 patients with 20p13 microdeletion exhibited TBC1D20 gene deletion (16/52, 30%), and 10 out of them (63%) were reported to have developmental delays, suggesting the importance of TBC1D20 gene for neurological development. Furthermore, Warburg Micro syndrome‐4 (WARBM4) (OMIM#615663) caused by homozygous mutation of the TBC1D20 gene has been associated with corpus callosum hypoplasia and severe mental retardation (Liegel et al., 2013; Martsolf, Hunter, & Haworth, 1978; Warburg, Sjo, Fledelius, & Pedersen, 1993). On the other hand, An et al. retrospectively analyzed 32 affected cases and reported that two genes expressed in the nervous system, SOX12 and NRSN2, are candidate genes involved in developmental deficits (An et al., 2013). However, an animal model study identified the transcription factors Sox4 and Sox11 as important regulators of diverse developmental processes in various organ systems, including the nervous system, heart, lung, spleen, and pancreas, as well as β‐cell development (Hoser et al., 2008). Although Sox4, Sox11, and Sox12 are jointly referred to as SoxC proteins, Sox12 is a weaker transactivator than the others (Hoser et al., 2008). Sox12‐deficient mice of both sexes were reported to exhibit normal fertility with no gross phenotypic abnormalities, suggesting that Sox4 and Sox11 compensate for the loss of function of Sox12 during mouse organogenesis (Hoser et al., 2008). This discrepant effect of a defect in Sox12 on embryonic development may be related to the presence or absence of functionally compensatory genes (Hoser et al., 2008), as well as to differences in the genetic backgrounds of humans and mice. Further studies are warranted to elucidate the precise roles of SOX12 and NRSN2 in developmental processes.
The patient in this case was diagnosed at a younger age than those in previously reported cases. Accordingly, we were unable to assess her developmental function. However, she harbored a large‐scale genetic defect comprising a microdeletion of up to 2.01 Mb in 20p13. This microdeletion, which was confirmed by oligonucleotide aCGH analysis, showed that the involved region contained TBC1D20, SOX12, and NRSN2. These findings suggest that a long‐term follow‐up of the subject's cognitive ability is needed to provide a sufficiently early intervention. Even though our review of previously reported patients failed to clarify a specific genotype–phenotype association in 20p13 microdeletion syndrome, we still demonstrated that the most common presentation was motor and language delay, followed by abnormal digits, mental retardation, large fontanelle, EEG abnormalities, and seizure. Our findings suggest that aCGH should be considered as a screening tool for any patients with the above presentations.
ETHICAL COMPLIANCE
Signed informed consent was obtained from the patient's parents in accordance with Institutional Review Board of Tri‐Service General Hospital. The ethics committee approved this study.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
The authors would like to thank the patient and her family to participate in this study.
Fang H‐H, Liu S‐Y, Wang Y‐F, Chiang C‐M, Liu C‐C, Lin C‐M. Phenotypic features of a microdeletion in chromosome band 20p13: A case report and review of the literature. Mol Genet Genomic Med. 2019;7:e739 10.1002/mgg3.739
REFERENCES
- An, Y. , Amr, S. S. , Torres, A. , Weissman, L. , Raffalli, P. , Cox, G. , … Shen, Y. (2013). SOX12 and NRSN2 are candidate genes for 20p13 subtelomeric deletions associated with developmental delay. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 162B(8), 832–840. 10.1002/ajmg.b.32187 [DOI] [PubMed] [Google Scholar]
- Baker, E. , Hinton, L. , Callen, D. F. , Altree, M. , Dobbie, A. , Eyre, H. J. , … Haan, E. (2002). Study of 250 children with idiopathic mental retardation reveals nine cryptic and diverse subtelomeric chromosome anomalies. American Journal of Medical Genetics, 107(4), 285–293. 10.1002/ajmg.10159 [DOI] [PubMed] [Google Scholar]
- Hoser, M. , Potzner, M. R. , Koch, J. M. , Bosl, M. R. , Wegner, M. , & Sock, E. (2008). Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Molecular and Cellular Biology, 28(15), 10.1128/MCB.00338-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezela‐Stanek, A. , Kucharczyk, M. , Pelc, M. , Gutkowska, A. , & Krajewska‐Walasek, M. (2013). 1.15 Mb microdeletion in chromosome band 20p13 associated with moderate developmental delay‐additional case and data's review. American Journal of Medical Genetics. Part A, 161A(1), 172–178. 10.1002/ajmg.a.35654 [DOI] [PubMed] [Google Scholar]
- Leclercq, S. , Maincent, K. , Baverel, F. , Tessier, D. L. , Letourneur, F. , Lebbar, A. , & Dupont, J. M. (2009). Molecular cytogenetic characterization of the first reported case of inv dup del 20p compatible with a U‐type exchange model. American Journal of Medical Genetics. Part A, 149A(3), 437–445. 10.1002/ajmg.a.32640 [DOI] [PubMed] [Google Scholar]
- Liegel, R. P. , Handley, M. T. , Ronchetti, A. , Brown, S. , Langemeyer, L. , Linford, A. , … Sidjanin, D. J. (2013). Loss‐of‐function mutations in TBC1D20 cause cataracts and male infertility in blind sterile mice and Warburg micro syndrome in humans. American Journal of Human Genetics, 93(6), 10.1016/j.ajhg.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski, J. R. , & Stankiewicz, P. (2005). Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genetics, 1(6), 10.1371/journal.pgen.0010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martsolf, J. T. , Hunter, A. G. W. , Haworth, J. C. , & Herrmann, J. (1978). Severe mental retardation, cataracts, short stature, and primary hypogonadism in two brothers. American Journal of Medical Genetics, 1(3), 291–299. 10.1002/ajmg.1320010305 [DOI] [PubMed] [Google Scholar]
- McGill, A. K. , Pastore, M. T. , Herman, G. E. , Alliman, S. , Rosenfeld, J. A. , & Weaver, D. D. (2010). A tale of two deletions: A report of two novel 20p13 –> pter deletions. American Journal of Medical Genetics. Part A, 152A(4), 10.1002/ajmg.a.33339 [DOI] [PubMed] [Google Scholar]
- Moutton, S. , Rooryck, C. , Toutain, J. , Cailley, D. , Bouron, J. , Villega, F. , … Goizet, C. (2012). Dysmorphic features in subtelomeric 20p13 deletion excluding JAG1: A recognizable microdeletion phenotype? European Journal of Medical Genetics, 55(2), 151–155. 10.1016/j.ejmg.2011.12.009 [DOI] [PubMed] [Google Scholar]
- Saleh, M. , Kamath, B. M. , & Chitayat, D. (2016). Alagille syndrome: Clinical perspectives. The Application of Clinical Genetics, 9, 75–82. 10.2147/TACG.S86420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat, J. , Lakshmi, B. , Malhotra, D. , Troge, J. , Lese‐Martin, C. , Walsh, T. , … Wigler, M. (2007). Strong association of de novo copy number mutations with autism. Science, 316(5823), 445–449. 10.1126/science.1138659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachoo, O. , Assanatham, M. , Jinawath, N. , & Nongnuch, A. (2013). Chromosome 20p inverted duplication deletion identified in a Thai female adult with mental retardation, obesity, chronic kidney disease and characteristic facial features. European Journal of Medical Genetics, 56(6), 319–324. 10.1016/j.ejmg.2013.03.011 [DOI] [PubMed] [Google Scholar]
- Warburg, M. , Sjo, O. , Fledelius, H. C. , & Pedersen, S. A. (1993). Autosomal recessive microcephaly, microcornea, congenital cataract, mental retardation, optic atrophy, and hypogenitalism. Micro syndrome. American Journal of Diseases of Children, 147(12), 10.1001/archpedi.1993.02160360051017 [DOI] [PubMed] [Google Scholar]


