Abstract
The alterations in expression and function of circular RNA (circRNA) in human dental pulp cells (hDPCs) during odontogenic differentiation were investigated. To induce odontogenic differentiation, hDPCs (passage 3) were cultured for 14 days in odontogenic induction medium. circRNA high-throughput sequencing was performed using Illumina HiSeqseq™ 2000. Subsequently, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were used to evaluate the bio-functions of the identified circRNAs. To validate the results of circRNA sequencing, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed for two selected differentially expressed circRNAs. The RNA sequencing results revealed that 1,314 and 1,780 circRNAs were upregulated and downregulated, respectively, during odontogenic induction. Their predicted target miRNAs and genes are involved in several biological functions and signaling pathways, including the mitogen-associated protein kinase signaling pathway. The RT-qPCR results of the two selected circRNAs (hsa_circ_0015260 and hsa_circ_0006984) were consistent with the expression trend obtained using high-throughput sequencing. The results of the present study add to the current understanding of the regulatory mechanisms underlying hDPCs differentiation.
Keywords: circular RNA, gene ontology, high-throughput sequencing, human pulp cells, odontogenic differentiation
Introduction
Dental pulp tissue contains precursor cells with regenerative potential, which use their stem cell properties upon pulp injury. They can proliferate and migrate to the damaged location, differentiate into odontoblasts, and form the reparative dentin (1,2). Following isolation from dental pulp and culture in vitro, these cells are termed dental pulp cells (DPCs) (3,4). DPCs are a heterogeneous population that can differentiate into a variety of cell types, including odontoblast-like cells, osteoblast-like cells, chondrocytes, neural-like cells and adipocytes (1,2). The differentiation of DPCs into functional odontoblasts is critical for the production of a mineralized matrix during dentinogenesis and dental pulp regeneration. Although the differentiation process involves various mechanisms (5,6), the potential molecular pathways underlying DPCs differentiation remain incompletely defined.
Circular RNA (circRNA) is a group of endogenous non-coding RNAs first detected in 1991, which was initially thought to be a functionless byproduct of splicing errors (7). With the development of high-throughput RNA sequencing and bioinformatics, it became clear that circRNAs were widespread in animal cells (4,8). The majority of circRNAs are composed of exonic sequences, which are highly conserved across various species and frequently exhibit tissue/developmental-stage-specific expression (9–11). Since circRNAs are not susceptible to digestion by ribonucleases (RNases), they possess higher stability than linear RNA (8,9). This property confers a major advantage in utilizing circRNAs as novel diagnostic markers. Additionally, a number of studies demonstrated that circRNAs serve an important role in the regulation of various cancer pathways (12–15). Additionally, a recent study demonstrated that 19 circRNAs were frequently upregulated and 5 circRNAs were often downregulated at each stage of osteoclast differentiation. These circRNAs profiles suggested that the differentially expressed circRNAs had specific functions during osteoclastogenesis (16). However, to date, little is known about the function of circRNAs during odontogenic differentiation of human dental pulp cells (hDPCs). Specifically, alterations in the expression profiles of circRNAs during odontogenesis have not been reported. Therefore, in the present study, the role of differentially expressed circRNAs during odontogenesis was investigated.
Materials and methods
Cell culture and odontogenic induction
Normal human premolars and third molars were extracted from healthy adults (3 males and 5 females; 12–25 years of age; recruitment date: 6th of July 2016) at the Department of Oral and Maxillofacial Surgery at the Affiliate Stomatology Hospital of Sun Yat-sen University (Guangzhou, China). The patients were clearly informed and their consent was obtained. The protocols were approved by the University Ethics Committee (Sun Yat-sen University; ethics certificate no: ERC-2014-30). hDPCs were isolated and cultured as previously described (17). Briefly, hDPCs from each of the 8 patients were cultuRed Separately in α-modified Eagle medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 15% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 10 U/ml penicillin and 10 mg/ml streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and were incubated at 37°C in an atmosphere of 5% CO2.
For odontogenic differentiation, hDPCs at passage 3 were cultured for 14 days in odontogenic induction medium (10% FBS, 10 mmol/l β-glycerophosphate, 0.2 mmol/l ascorbic acid and 100 nmol/l dexamethasone in α-modified Eagle medium). Control samples were cultured in 10% FBS medium alone.
To assess odontogenic differentiation, Alizarin Red S staining and the alkaline phosphatase (ALP) activity test were conducted as previously reported (18). In Alizarin Red S staining, images were captured under a fluorescence microscope (Axio Observer Z1; Zeiss AG, Oberkochen, Germany) and the positively stained nodules appeared orange/red. ALP activity was measured using an ALP assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). To normalize enzymatic activity, the concentrations of protein were measured using a bicinchoninic protein assay kit (Wuhan Boshide Biological Engineering Co., Ltd., Wuhan, China). Furthermore, the expression of odontoblast-associated genes [dentin sialophosphoprotein (DSPP) and dentin matrix acidic phosphoprotein 1 (DMP-1)] in hDPCs was monitored by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) during odontogenic differentiation using the primers listed in Table I. GAPDH was the internal control. The data were analyzed using the ΔCq method (19) and 2−ΔΔCq represented the relative expression level of mRNAs.
Table I.
Primer sequences of odontoblast-associated genes used in reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence | Product size (bp) |
|---|---|---|
| DSPP | F: 5′-TAGCCGAGGAGATGCTTCTTATA-3′ | 180 |
| R: 5′-TTACCTTTGCCACTGTCTGATTT-3′ | ||
| DMP-1 | F: 5′-CAGGAGAGACAGCAAGGGTG-3′ | 178 |
| R: 5′-GGGGTTATCTCCCCTGGACT-3′ |
DSPP, dentin sialophosphoprotein; F, forward; R, reverse; DMP, dentin matrix acidic phosphoprotein 1.
Preparation of RNA samples
Total RNA was extracted from hDPCs (with or without odontogenic induction medium) using TRIzol RNA isolation reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For RNA-sequencing, in order to enrich the circRNAs, ribosomal RNA was removed by Mag-Bind technology and linear RNA was digested by RNases from the samples, according to a previous study (20). Following RNA fragmentation, random hexamer priming, cDNA synthesis [with SuperScript II Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) at 42°C for 50 min followed by 70°C for 15 min for inactivation, First-Strand Buffer, 0.1 M dithiothreitol, deoxyribonucleotides, XhoI primer (Thermo Fisher Scientific, Inc.) and DNA Polymerase I (Promega Corporation, Madison, WI, USA) at 16°C for 2.5 h followed by 70°C for 10 min for inactivation] and adapter ligation, the integrity of purified products was analyzed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) and the concentration and purity of the samples were determined using a NanoDrop™ 2000 UV spectrophotometer (Thermo Fisher Scientific, Inc.) Only RNA samples with absorbance 260/280 ratios between 1.9 and 2.1, and RNA integrity number >8.0 were used for further analysis. One sample from the mixture of three individuals was used for RNA sequencing and the remaining five samples were utilized for RT-qPCR.
circRNA high-throughput sequencing
Sequencing was performed using Illumina HiSeqseq™ 2000 (Illumina, Inc., San Diego, CA, USA). Following raw data filtering, the CIRI software version 1.2 (21) was applied to predict the circRNAs from sequencing samples, that were then labeled and annotated according to their source areas, using circBase (www.circbase.org) (22). P<0.05, or false discovery rate (FDR) ≤0.001 and fold-change ≥2.0 was considered to indicate a statistically significant difference.
RT-qPCR
The high-throughput sequencing demonstrated that the amount of differentially expressed circRNAs was too large to choose from. In view of our previous study investigating decreased miR-135b during odontogenic differentiation of hDPCs (23), and the circRNAs' role as miRNA sponges, a co-expression network of circRNA-miRNA was constructed following a correlation analysis between the differentially expressed circRNAs using Cytoscape (www.cytoscape.org) software version 3.5.1. From this, two differentially expressed circRNAs were selected [both upregulated: Chr1:172520652 |172548407 (ID: hsa_circ_0015260) and chr9:130206308|130207528 (hsa_circ_0006984)] to validate the results obtained with circRNA high-throughput sequencing. Validation was performed via RT-qPCR assay with SYBR-Green (Qiagen, Inc., Valencia, CA, USA) on a StepOnePlus™ Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). RT was performed using M-MLV reverse transcriptase (Promega Corporation), according to the manufacturer's instructions. The PCR cycling conditions were as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. Primers were synthesized by Guangzhou Forevergen Biosciences Co., Ltd. (Guangdong, China) and listed in Table II. The RT-qPCR results were presented as the fold-change of each circRNA in induced cells relative to control cells, as calculated by the 2−ΔΔCq method. These data were produced from three biological replicates consisting of five different cell samples each. The circRNA expression levels were normalized using GAPDH as the reference gene.
Table II.
Primer sequences of circRNAs used in reverse transcription-quantitative polymerase chain reaction.
| Genes | Sequence | Size (bp) |
|---|---|---|
| hsa_circ_0015260 | circ F: 5′-CGCCAGGAACTATTTGATGAGG-3′ | 177 |
| circ R: 5′-GTGCGCAGTTGTCATCTTGA-3′ | ||
| linear F: 5′-GTCCACCTGACAGCCTTGTT-3′ | 130 | |
| linear R: 5′-GATTTTGGATCACCACCGCC-3′ | ||
| hsa_circ_0006984 | circ F: 5′-AGCTCTGCCTTCGTTAGACATC-3′ | 133 |
| circ R: 5′-CTTGCTCTGGTGGTGATCCAG-3′ | ||
| linear F: 5′-CCCATGTGTAGAGATGCCCC-3′ | 94 | |
| linear -F: 5′-CCCATGTGTAGAGATGCCCC-3′ |
circ, circular RNA; F, forward; R, reverse.
miRNA prediction and functional analysis
miRanda (issue 2010; www.microrna.org/microrna/) and TargetScan (issue 2011; www.targetscan.org/vert_50/) software were used to predict the binding sites for human miRNA within these differentially expressed circRNAs. To further investigate the biological function and associated pathways of the predicted target genes, the DAVID Database (issue 2016; david-d.ncifcrf.gov/) was used for Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (issue 2016; www.genome.jp/kegg/) was used for pathway analysis of these differentially expressed genes.
Statistical analysis
The experiments were repeated three times and the data are presented as the mean ± standard deviation. SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA) was used to analyze the results. Comparisons between two groups were performed using paiRed Student's t-test and one-way analysis of variance followed by the least significant difference multiple-comparison test was used for multiple group comparisons. P<0.05, or false discovery rate (FDR) ≤0.001 and fold-change ≥2.0 was considered to indicate a statistically significant difference.
Results
Odontogenic differentiation of hDPCs
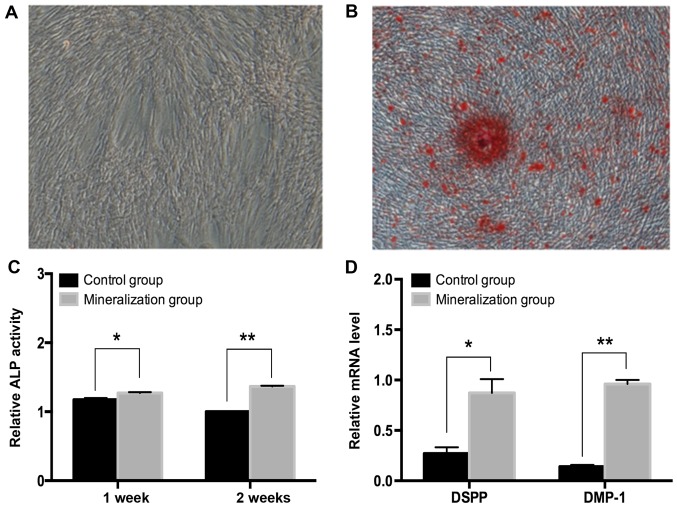
DPCs were incubated in odontogenic induction medium for 14 days (Fig. 1A). Alizarin Red S staining positively demonstrated mineralized matrix deposition in the induced group, which was evident after 14 days (Fig. 1B). Additionally, the ALP activity was measured at 7 and 14 days and demonstrated to be significantly increased (P<0.5; Fig. 1C). The expression of odontoblast marker genes including DSPP and DMP-1 significantly increased following day 14 in the induced group (P<0.05; Fig. 1D). These results were confirmed by RT-qPCR assays.
Figure 1.
Cell culture and odontogenic differentiation of hDPCs. (A) DPCs were incubated in odontogenic medium for 14 days and unstained cells were visualized under an inverted microscope (magnification, ×50). (B) The hDPCs were cultured in odontogenic medium and assayed via Alizarin Red S staining (magnification, ×50) after 14 days. (C) ALP activity results at 7 and 14 days. ‘Relative’ refers to the ALP activity in each group/control group on day 14. (D) The odontogenic-associated genes were assessed by reverse transcription-quantitative polymerase chain reaction and normalized against GAPDH. Data are expressed as the mean ± standard deviation. *P<0.05, **P<0.001. hDPCs, human dental pulp cells; ALP, alkaline phosphatase; DSPP, dentin sialophosphoprotein; DMP-1, dentin matrix acidic phosphoprotein 1.
Expression profiles of circRNAs during the odontogenic differentiation of hDPCs
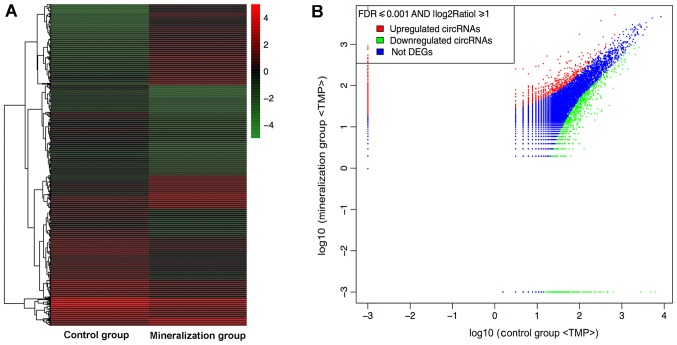
The RNA sequencing results revealed that during odontogenic induction 1,314 circRNAs were upregulated and 1,780 were downregulated (FDR ≤0.001 and fold-change ≥2.0; Fig. 2).
Figure 2.
High-throughput sequencing expression data for circRNAs. (A) Hierarchical clustering reveals significantly different circRNA expression profiles for the induction and control groups. (B) A scatter plot reveals alterations in the induction and control groups. Red indicates upregulation. Green indicates downregulation and blue points indicate unaltered expression of circRNAs, FDR≤0.001 and fold-change ≥2. DEG, differentially expressed gene; TMP, the expression levels following standardized processing; FDR, false discovery rate; circRNA, circular RNA.
Validation of circRNA expression levels by RT-qPCR
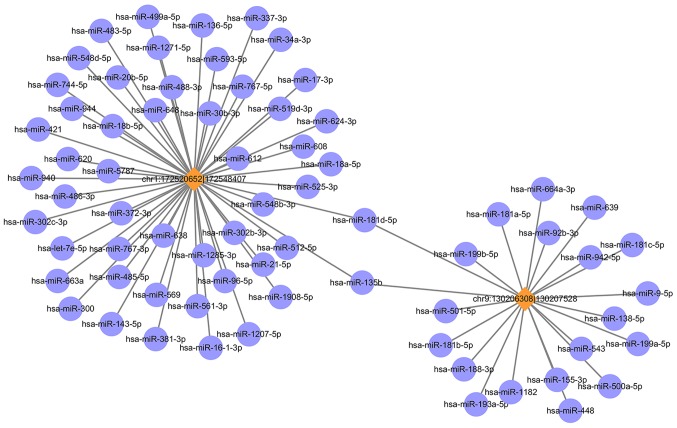
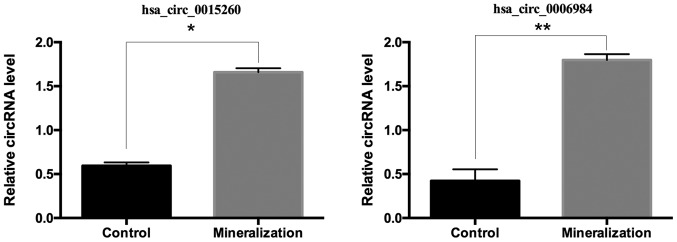
The circRNA-miRNA co-expression network was constructed containing 2 circRNAs and 71 miRNAs (Fig. 3). In the network, the circles represent miRNA and the diamonds represent circRNA. The yellow color represents upregulation. The size of diamond represents the fold-change of circRNAs with the larger size indicating a higher fold-change. In the network, the two circRNA i.e., chr1: 172520652 |172548407 (ID: hsa_circ_0015260) and chr9: 130206308|130207528 (hsa_circ_0006984) have been predicted to contain binding sites for miR-135b. To validate the circRNA results, the RT-qPCR was performed for the above two circRNAs. These results were consistent with the RNA sequencing results, as presented in Fig. 4.
Figure 3.
circRNA-miR co-expression network analysis. Circles represent miR and diamonds represent circRNA. The yellow color represents upregulation. The size of the diamond represents the circRNA fold-change with a larger size indicating a higher fold-change. The two circRNA chr1:172520652 |172548407 (ID: hsa_circ_0015260) and chr9:130206308|130207528 (hsa_circ_0006984) have been both predicted to contain binding sites for miR-135b. circRNA, circular RNA; miR, microRNA; chr, chromosome.
Figure 4.
Two differentially expressed circRNAs during odontogenic differentiation of hDPCs assessed by reverse transcription-quantitative polymerase chain reaction. The expression of hsa_circ_0015260 and hsa_circ_0006984 were significantly increased in the mineralization groups. Data are expressed as the mean ± standard deviation. *P<0.05, **P<0.001. circRNA, circular RNA; hDPCs, human dental pulp cells.
miRNA prediction and function analysis
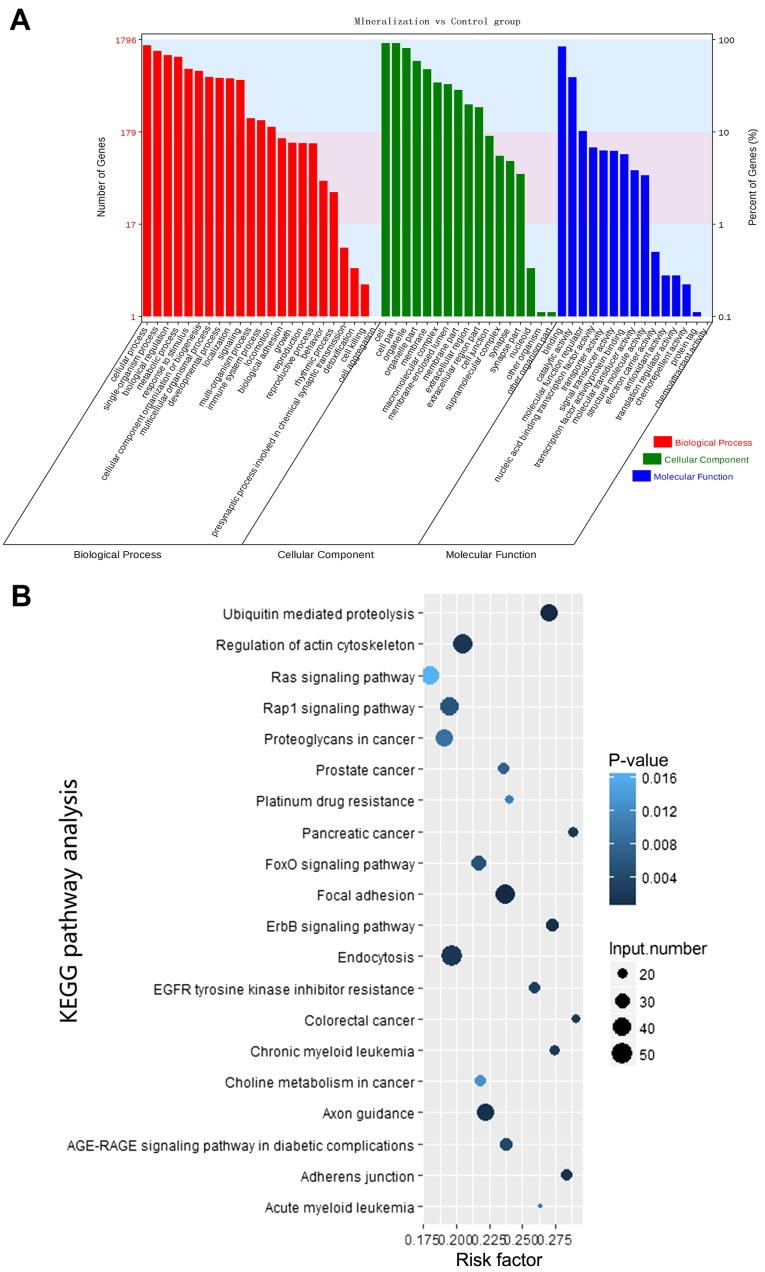
A total of 2,588 miRNAs were identified as differentially expressed circRNAs. Through GO analysis, it was demonstrated that a great quantity of the differentially expressed circRNA parental genes were associated with important biological processes, including binding and catalytic activity, molecular transducer activity, biological regulation, cellular components organization or biogenesis, cellular process, immune system process, metabolic processes and response to stimulus (Fig. 5A). Additionally, KEGG pathway analysis identified several enriched pathways, including adherens junction, regulation of actin cytoskeleton, forkhead box protein O1 signaling pathway and mitogen activated protein kinase (MAPK) signaling pathway, associated with odontogenic differentiation of hDPCs (Table III). Certain identified KEGG pathways were consistent with the biological processes identified by the GO analysis. The top 20 enriched signaling pathways are shown in Fig. 5B; however, in the present study there were a total of 37 significantly enriched signaling pathways, which included the MAPK signaling pathway. Therefore, the results from the functional analysis of the identified circRNAs target genes, using different bioinformatics approaches, suggest that circRNAs are essential regulators of the odontogenic differentiation of hDPCs through their role in key signaling pathways.
Figure 5.
Bioinformatics analysis. (A) Gene ontology analysis. (B) Pathway enrichment. The risk factor represents the ratio of enriched differentiated genes to all genes enriched for a certain pathway term. Spot size represents the number of significantly differentiated genes; larger spots indicate more differentiated genes. Q values result from adjustments of P-values. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table III.
Enriched KEGG pathways involving target genes of the differentially expressed circular RNAs during the odontogenic differentiation of human dental pulp cells.
| Term | KEGG ID | N | P-value |
|---|---|---|---|
| Adherence junction | hsa04520 | 21 | 0.00115035 |
| Regulation of actin cytoskeleton | hsa04810 | 44 | 0.00186187 |
| FoxO signaling pathway | hsa04068 | 29 | 0.00529183 |
| MAPK signaling pathway | hsa04010 | 38 | 0.04811483 |
N, number of input genes in the pathway; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; FoxO, forkhead box protein O1.
Discussion
In previous years, increasing evidence has indicated that circular RNAs (circRNAs) participate in the regulation of gene expression and can be used as ideal diagnostic biomarkers for certain diseases (24–26). A recent study focusing on osteogenesis identified circ19142 and circ5846 as being associated with osteoblast differentiation (27). It also reported that BMP2 may induce osteogenic differentiation through the circ19142/circ5846-targeted miRNA-mRNA axis. In an investigation of the circRNA landscape of periodontal ligament stem cells (PDLSCs) during osteogenesis, a temporal dynamic alteration in the expression was observed during the osteoblast differentiation and biomineralization of PDLSCs. This suggested that circRNAs served a role in osteoblast differentiation and bone formation (28). However, the role of circRNAs in odontogenic differentiation has never been reported. In the present study, RNA-Seq was used to screen and compare the 3,094 differentially expressed circRNAs during odontogenesis ofhuman dental pulp cells (hDPCs). In this analysis, 1,314 circRNAs were upregulated, while 1,780 circRNAs were downregulated in the induction group as compared with the control group. Two circRNAs (hsa_circ_0015260 and hsa_circ_0006984) were selected for RT-qPCR, which produced results that were consistent with those obtained via the circRNA high-throughput sequencing. Subsequently, whether the differentially expressed circRNAs had a regulatory function during odontogenic differentiation was investigated.
Recently, circRNAs have been proposed to act as miRNA sponges to regulate gene expression and consequently regulate linear RNA transcription and protein synthesis (29,30). The most well-known circRNA is CDR1 antisense RNA, which contains 70 common binding sites and functions as a sponge for miR-7.11 (24). The expression levels of circRNAs are higher by comparison to their linear isomers and circRNAs are enriched with miRNA binding sites (31). The results suggested that specific circRNAs may function as competing endogenous RNAs to promote PDLSC osteogenic differentiation and periodontal regeneration (32). In addition, circRNAs can chelate miRNAs more efficiently. Therefore, the roles of circRNAs in osteoblast differentiation may be associated with their miRNA-mediated effects. A previous study has demonstrated that miR-135b was decreased prominently during odontogenic differentiation of hDPCs (23). Through target prediction and functional analysis, it was also hypothesized that the downregulation of miR-135b increased the expression of APC, myocyte enhancer factor 2C, and collagen type V α1 chain, subsequently mediating the MAPK and the Wnt signaling pathways and therefore promoting hDPC odontogenic differentiation (23). In the present study, the two RT-qPCR validated circRNAs (hsa_circ_0015260 and hsa_circ_0006984) increased. In addition, both circRNAs were enriched with miRNA binding sites for miR-135b, which was consistent with the hypothesis of the circRNAs acting as miRNA sponges. Based on the previous studies (23,29,32), it is reasonable to hypothesize that the upregulation of the circRNAs hsa_circ_0015260 and hsa_circ_0006984 may competitively release the inhibition of miR-135b on its target genes, and consequently mediate the associated signaling pathways, which ultimately facilitate hDPC odontogenic differentiation. The specific interaction between the two validated circRNAs and miR-135b requires further investigation.
From the bioinformatics analyses, the GO analysis revealed several differentially expressed circRNA parental genes, which were associated with many important biological processes. The results suggested that the regulation of the affected genes during cellular processes was vital during odontogenic differentiation. Furthermore, the KEGG pathway analysis indicated that the majority of the identified genes were enriched in several signaling pathways. Certain pathways, including focal adhesion, regulation of the actin cytoskeleton and MAPK signaling were associated with odontogenic differentiation. The results of the present study, in accordance with previous studies (7–10), infer that circRNA has a potential role as a regulator of various biological processes. Among the enriched signaling pathways identified in this study, MAPK signaling is widely associated with the differentiation of dental pulp cells and the process of tooth development (33,34). Woo et al (35) reported that 17-estradiol induced odontoblastic differentiation via activation of the c-Src/MAPK signaling pathway in hDPCs. Insulin-like growth factor 1 has been revealed to promote the proliferation and osteogenic/odontogenic differentiation of human dental pulp stem cells through MAPK signaling pathways (36). Additionally, the magnetic nanofiber scaffold could induce the stimulation of odontogenesis and pro-angiogenesis of hDPCs through the Wnt/MAPK/nuclear factor-κB signaling pathway (37). Furthermore, a previous study observed that the elasticity of the hDPCs cytoskeleton increased over time and the focal adhesion was enhanced by the odontogenic medium (38).
In conclusion, in the present study, high-throughput sequencing was used to screen for differentially expressed circRNAs during the odontogenic differentiation of hDPCs. In addition, the potential functions of these circRNAs using GO and KEGG analyses were examined. The obtained results suggest that the enrichment of target genes involved in the MAPK signaling pathway is of particular importance. This study adds to the current understanding of the regulatory mechanisms underlying the differentiation of hDPCs.
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science Foundation of Guangdong Province (grant no. 2017A030313713).
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to confidentiality of another study in our group, but are available from the corresponding author on reasonable request.
Authors' contributions
HJ conceived the study. CL designed and performed the experiments, analyzed the data, and wrote the manuscript.
Ethics approval and consent to participate
The patients were clearly informed and their consent was obtained. The protocols were approved by the University Ethics Committee (ethic certificate no: ERC-2014-30).
Patient consent for publication
The patients were clearly informed and their consent was obtained.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang J, Ma H, Jin X, Hu J, Liu X, Ni L, Ma PX. The effect of scaffold architecture on odontogenic differentiation of human dental pulp stem cells. Biomaterials. 2011;32:7822–7830. doi: 10.1016/j.biomaterials.2011.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambrichts I, Driesen RB, Dillen Y, Gervois P, Ratajczak J, Vangansewinkel T, Wolfs E, Bronckaers A, Hilkens P. Dental pulp stem cells: Their potential in reinnervation and angiogenesis by using scaffolds. J Endod. 2017;43(Suppl 9):S12–S16. doi: 10.1016/j.joen.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Wei X, Ling J, Huang Y, Gong Q, Huo Y. Identification and characterization of side population cells from adult human dental pulp after ischemic culture. J Endod. 2012;38:1489–1497. doi: 10.1016/j.joen.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Hilkens P, Gervois P, Fanton Y, Vanormelingen J, Martens W, Struys T, Politis C, Lambrichts I, Bronckaers A. Effect of isolation methodology on stem cell properties and multilineage differentiation potential of human dental pulp stem cells. Cell Tissue Res. 2013;353:65–78. doi: 10.1007/s00441-013-1630-x. [DOI] [PubMed] [Google Scholar]
- 5.Qin W, Yang F, Deng R, Li D, Song Z, Tian Y, Wang R, Ling J, Lin Z. Smad 1/5 is involved in bone morphogenetic protein-2-induced odontoblastic differentiation in human dental pulp cells. J Endod. 2012;38:66–71. doi: 10.1016/j.joen.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Geng YW, Zhang Z, Liu MY, Hu WP. Differentiation of human dental pulp stem cells into neuronal by resveratrol. Cell Biol Int. 2017;41:1391–1398. doi: 10.1002/cbin.10835. [DOI] [PubMed] [Google Scholar]
- 7.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: Possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 13.Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, Zhang H, Li H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385–387. doi: 10.1016/j.gdata.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16:2469–2476. doi: 10.3892/mmr.2017.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 16.Dou C, Cao Z, Yang B, Ding N, Hou T, Luo F, Kang F, Li J, Yang X, Jiang H, et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep. 2016;6:21499. doi: 10.1038/srep21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, Ling J, Wu L, Liu L, Xiao Y. Expression of mineralization markers in dental pulp cells. J Endod. 2007;33:703–708. doi: 10.1016/j.joen.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, He H, Tang C, Zhang G, Li Y, Wang R, Shi J, Jin Y. Differentiation potential of STRO-1+ dental pulp stem cells changes during cell passaging. BMC Cell Biol. 2010;11:32. doi: 10.1186/1471-2121-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Yin QF, Chen LL, Yang L. Fractionation of non-polyadenylated and ribosomal-free RNAs from mammalian cells. Methods Mol Biol. 2015;1206:69–80. doi: 10.1007/978-1-4939-1369-5_6. [DOI] [PubMed] [Google Scholar]
- 21.Gao Y, Wang J, Zhao F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glažar P, Papavasileiou P, Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Q, Wang R, Jiang H, Lin Z, Ling J. Alteration of microRNA expression of human dental pulp cells during odontogenic differentiation. J Endod. 2012;38:1348–1354. doi: 10.1016/j.joen.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 25.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 27.Qian DY, Yan GB, Bai B, Chen Y, Zhang SJ, Yao YC, Xia H. Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed Pharmacother. 2017;90:492–499. doi: 10.1016/j.biopha.2017.03.051. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Li X, Huang Y, Jia L, Li W. The circular RNA landscape of periodontal ligament stem cells during osteogenesis. J Periodontol. 2017;88:906–914. doi: 10.1902/jop.2017.170078. [DOI] [PubMed] [Google Scholar]
- 29.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular cacinoma development. Medicine. 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu X, Li M, Jin Y, Liu D, Wei F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18:100. doi: 10.1186/s12863-017-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyata H, Genma T, Ohshima M, Yamaguchi Y, Hayashi M, Takeichi O, Ogiso B, Otsuka K. Mitogen-activated protein kinase/extracellular signal-regulated protein kinase activation of cultured human dental pulp cells by low-power gallium-aluminium-arsenic laser irradiation. Int Endod J. 2006;39:238–244. doi: 10.1111/j.1365-2591.2006.01080.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao JW, Gao ZL, Mei H, Li YL, Wang Y. Differentiation of human mesenchymal stem cells: The potential mechanism for estrogen-induced preferential osteoblast versus adipocyte differentiation. Am J Med Sci. 2011;341:460–468. doi: 10.1097/MAJ.0b013e31820865d5. [DOI] [PubMed] [Google Scholar]
- 35.Woo SM, Seong KJ, Oh SJ, Park HJ, Kim SH, Kim WJ, Jung JY. 17β-estradiol induces odontoblastic differentiation via activation of the c-Src/MAPK pathway in human dental pulp cells. Biochem Cell Biol. 2015;93:587–595. doi: 10.1139/bcb-2015-0036. [DOI] [PubMed] [Google Scholar]
- 36.Lv T, Wu Y, Mu C, Liu G, Yan M, Xu X, Wu H, Du J, Yu J, Mu J. Insulin-like growth factor 1 promotes the proliferation and committed differentiation of human dental pulp stem cells through MAPK pathways. Arch Oral Biol. 2016;72:116–123. doi: 10.1016/j.archoralbio.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 37.Yun HM, Kang SK, Singh RK, Lee JH, Lee HH, Park KR, Yi JK, Lee DW, Kim HW, Kim EC. Magnetic nanofiber scaffold-induced stimulation of odontogenesis and pro-angiogenesis of human dental pulp cells through Wnt/MAPK/NF-κB pathways. Dent Mater. 2016;32:1301–1311. doi: 10.1016/j.dental.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Jones TD, Naimipour H, Sun S, Cho M, Alapati SB. Mechanical changes in human dental pulp stem cells during early odontogenic differentiation. J Endod. 2015;41:50–55. doi: 10.1016/j.joen.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to confidentiality of another study in our group, but are available from the corresponding author on reasonable request.