Abstract
Osteosarcoma is the most common type of malignant bone cancer, which often affects teenagers and young adults. The present study aimed to screen for critical genes and microRNAs (miRNAs/miRs) involved in osteosarcoma. A total of four microarray datasets (accession numbers GSE32981, GSE21257, GSE14827 and GSE14359) were downloaded from the Gene Expression Omnibus database. Following data preprocessing, module analysis was performed to identify the stable modules using the weighted gene co-expression network analysis (WGCNA) package. The differentially expressed genes (DEGs) between metastatic samples and non-metastatic samples were screened, followed by gene co-expression network construction, and Gene Ontology function and Kyoto Encyclopedia of Genes and Genomes pathway analyses. Subsequently, prognosis-associated genes were screened and a miRNA-target gene regulatory network was constructed. Finally, the data for critical genes were validated. WGCNA analysis identified six modules; blue and yellow modules were significantly positively associated with osteosarcoma metastasis. A total of 1,613 DEGs were screened between primary tissue samples and metastatic samples. Following comparison of the genes in the two (blue and yellow) modules, a total of 166 DEGs were identified (metastatic samples vs. non-metastatic samples). Functional enrichment analysis demonstrated that these DEGs were mainly involved in ‘defense response’, ‘p53 signaling pathway’ and ‘lysosome’. By utilizing the clinical information in GSE21257, 10 critical genes associated with osteosarcoma prognosis were obtained, including CTP synthase 2 (CTPS2), tumor protein p53 inducible protein 3 (TP53I3) and solute carrier family 1 member 1 (SLC1A1). In addition, hsa-miR-422a and hsa-miR-194 were highlighted in the miRNA-target gene network. Finally, matrix metallopeptidase 3 (MMP3) and vascular endothelial growth factor B (VEGFB) were predicted as critical genes in osteosarcoma metastasis. CTPS2, TP53I3 and SLC1A1 may serve major roles in osteosarcoma development, and hsa-miR-422a, hsa-miR-194, MMP3 and VEGFB may be associated with osteosarcoma metastasis.
Keywords: osteosarcoma, differentially expressed genes, metastasis, prognosis
Introduction
Osteosarcoma is the most common type of malignant bone cancer and is prevalent in teenagers and young adults (1). This type of cancer arises from primitive transformed cells of mesenchymal origin that exhibit osteoblastic differentiation and produce malignant osteoids (2). Despite considerable advances in surgery and chemotherapy, the 5-year survival rate remains at 60–70% and patients continue to succumb to osteosarcoma metastasis (3). Therefore, the exploration of novel strategies and noninvasive biomarkers that reflect disease progression is urgently required for the clinical management of patients with osteosarcoma.
With the development of molecular biology techniques, tumor gene therapy for osteosarcoma exhibits a potential clinical strategy (4,5). A number of studies have demonstrated that abnormal gene expression, and alterations in microRNAs (miRNAs/miRs) and molecular signaling pathways contribute to the pathogenesis and development of osteosarcoma (6–8). These affected molecules may be considered potential diagnostic biomarkers and therapeutic targets for patients with osteosarcoma. C-X-C motif chemokine ligand 12 and matrix metallopeptidase 9 (MMP9) serve important roles in the metastasis of osteosarcoma (9). Ren et al (10) revealed that high expression levels of C-X-C motif chemokine receptor 4 and MMP9 are valuable biomarkers for osteosarcoma metastasis and survival rates. A recent study revealed that tumor protein p53 may inhibit cell proliferation and angiogenesis in osteosarcoma cell lines by inhibiting the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT)/mechanistic target of rapamycin pathway; therefore, it may be an effective novel therapeutic candidate against osteosarcoma in the future (11). In addition, Fas cell surface death receptor (Fas) is a death receptor, which has been reported to be involved in osteosarcoma metastasis. An inhibitor of the Fas pathway, c-FLIP, has been developed as a potential treatment for patients with lung metastasis (12).
miRNAs are small non-coding RNA molecules (18–25 nt) and studies have revealed that miRNAs act as critical regulators involved in the pathological process of osteosarcoma (13,14). miR-30a serves as an oncogene, which regulates the proliferation, migration and invasion of human osteosarcoma by targeting runt-related transcription factor 2 (15). In addition, overexpression of miR-21 in the human osteosarcoma cell line MG63 has been reported to significantly increase cell proliferation and invasion (16). Phosphatase and tensin homolog (PTEN) may be a potential target gene of miR-21, and miR-21 may activate the PI3K/Akt pathway by decreasing PTEN expression (16). These previous studies may provide a comprehensive understanding of osteosarcoma development.
Namløs et al (17) explored the potential mechanism underlying osteosarcoma and demonstrated that multiple signaling molecules serve a vital role in promoting metastasis. The present study, according to the gene expression profiles deposited by Namløs et al (17), aimed to identify metastasis-associated genes or miRNAs in osteosarcoma development and to improve the understanding of osteosarcoma metastasis. Firstly, the gene expression in metastatic osteosarcoma samples from four microarray datasets was compared with that in non-metastatic samples; subsequently, a number of differentially expressed genes (DEGs) and miRNAs were screened using the weighted gene co-expression network analysis (WGCNA) algorithm. Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to identify the major signaling pathways involved in osteosarcoma. Subsequently, the gene co-expression network for these DEGs was constructed. Additionally, the miRNA-target gene network was constructed to screen the key miRNAs associated with disease prognosis. Finally, the critical genes and miRNAs were further verified based on validation dataset analysis. The results may provide novel diagnostic biomarkers and therapeutic target molecules in osteosarcoma metastasis.
Materials and methods
Data resources
The microarray datasets associated with osteosarcoma were downloaded from the National Center of Biotechnology Information Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo). The screening standards were as follows: The microarray datasets were gene expression profiles; the datasets were gene expression profiles associated with osteosarcoma tissue samples; osteosarcoma samples were of primary and metastatic origin; gene expression profiling of human osteosarcoma; and, the total number of osteosarcoma samples was >20. Datasets that did not meet any of these criteria were excluded. Eventually, four datasets were screened out for further analysis: GSE32981 (17), GSE21257 (18), GSE14827 (19) and GSE14359 (20) (Table I).
Table I.
Summary of microarray datasets.
| GEO accession no. | Platform | Probe number | Total samples | Metastasis samples | Non-metastasis samples | PMID |
|---|---|---|---|---|---|---|
| GSE32981 | GPL3307-ABI | 14,725 | 23 | 18 | 5 | 22518090 |
| GSE21257 | GPL10295-Illumina | 48,701 | 53 | 34 | 19 | 21372215 |
| GSE14827 | GPL570-Affymetrix | 42,450 | 27 | 9 | 18 | 20159990; 24448647 |
| GSE14359 | GPL96-Affymetrix | 41,059 | 34 | 21 | 13 | 21166698 |
GEO, Gene Expression Omnibus; PMID, PubMed unique identifier.
The GSE32981 dataset was tested based on the GPL3307 ABI Human Genome Survey Microarray v2.0 Array platform (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), including 18 metastatic tissue samples and five non-metastatic samples. The GSE21257 dataset was tested based on the GPL10295 Illumina human-6 v2.0 expression beadchip (using nuIDs as identifiers) platform (Illumina, Inc., San Diego, CA, USA), including 34 metastatic tissue samples and 19 non-metastatic samples. The GSE14827 dataset was tested based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform (Affymetrix; Thermo Fisher Scientific, Inc.), including nine metastatic tissue samples and 18 non-metastatic samples. The GSE14359 dataset was tested based on the GPL96 [HG-U133A] Affymetrix Human Genome U133A Array platform (Affymetrix; Thermo Fisher Scientific, Inc.), including 21 metastatic tissue samples and 13 non-metastatic samples.
Data preprocessing
The GSE14827 and GSE14359 gene expression profiles were preprocessed using oligo software version 1.41.1 (www.bioconductor.org/packages/release/bioc/html/oligo.html) (21) in the R3.4.1 package (22). The original microarray data were converted into gene symbols according to annotation information of the array platform. If several probes corresponded with the same gene, the average scores were calculated as the gene expression value of these probes. Quantile normalization in the preprocessCore package (23) was used to normalize the matrix.
For the GSE32981 and GSE21257 datasets, the limma package (www.bioconductor.org/packages/release/bioc/html/limma.html) in R software 3.1.3 version (24) was used to preprocess the microarray data. The logarithmic value of each microarray data point was calculated and the gene expression data were converted from a skewed distribution to an approximately normal distribution. The median normalization method was used to normalize the microarray data.
Identification of gene modules associated with osteosarcoma
The WGCNA method was used to identify gene modules associated with osteosarcoma. WGCNA provides the topological properties of co-expression networks, in addition to the correlation of two node genes and relevant other genes (25). The WGCNA package version 1.61 (cran.r-project.org/web/packages/WGCNA/index.html) (26) in R3.4.1 software was used to screen for stable genetic modules. Since the GSE21257 dataset contained the most tumor samples and relatively abundant clinical information, it was used as the training dataset and the other three datasets served as validation datasets. Briefly, for the four datasets, the expression correlation between any two datasets was first calculated, followed by adjacency function definition and module division (the threshold of module screening and division was set as follows: The modules contained at least 150 RNA and cutHeight=0.99). Furthermore, the correlation between each module and clinical information provided by the GSE21257 dataset was analyzed. The clinical information in GSE21257, including age, histological subtype, tumor location and stage is presented in Table II.
Table II.
Clinical information of GSE21257.
| Source name | Age (years) | Group | Histological subtype | Mortality (months) | Tumor location |
|---|---|---|---|---|---|
| GSM530667 | 14.5 | Metastases present at diagnosis | Giant cell rich | Succumbed after 27 | Left distal femur |
| GSM530899 | 13.5 | Metastases at 16 months following diagnosis of the primary tumor | Osteoblastic | Succumbed at 21 following diagnosis of the primary tumor | Left proximal humerus |
| GSM531283 | 14.2 | Metastases present at diagnosis | Osteoblastic | Alive at 46 after diagnosis of the primary tumor | Left distal femur |
| GSM531284 | 11.1 | Metastases at 10 months following diagnosis of the primary tumor | Fibroblastic | Alive at 28 after diagnosis of the primary tumor | Left distal femur |
| GSM531285 | 6.8 | Metastases at 5 months following diagnosis of the primary tumor | Osteoblastic | Succumbed at 11 following diagnosis of the primary tumor | Humerus |
| GSM531286 | 12.0 | Metastases at 30 months following diagnosis of the primary tumor | Chondroblastic | Alive at 37 after diagnosis of the primary tumor | Tibia |
| GSM531287 | 15.0 | Metastases present at diagnosis | Sclerosing | Alive at 45 after diagnosis of the primary tumor | Left distal femur |
| GSM531288 | 15.0 | Metastases present at diagnosis | Chondroblastic | Succumbed at 13 following diagnosis of the primary tumor | Right proximal tibia |
| GSM531289 | 17.0 | Metastases at 9 months following diagnosis of the primary tumor | Osteoblastic | Succumbed at 33 following diagnosis of the primary tumor | Left femur |
| GSM531290 | 15.1 | Metastases at 25 months following diagnosis of the primary tumor | Sclerosing | Alive at 25 after diagnosis of the primary tumor | Right proximal tibia |
| GSM531291 | 79.0 | Metastases at 12 months after diagnosis of the primary tumor | Osteoblastic | Succumbed at 18 following diagnosis of the primary tumor | Humerus |
| GSM531292 | 16.7 | Metastases present at diagnosis | Osteoblastic | Succumbed at 30 following diagnosis of the primary tumor | Right proximal femur |
| GSM531293 | 17.1 | Metastases at 9 months after diagnosis of the primary tumor | Chondroblastic | Succumbed at 35 following diagnosis of the primary tumor | Left distal femur |
| GSM531294 | 10.8 | Metastases present at diagnosis | Chondroblastic | Succumbed at 4 following diagnosis of the primary tumor | Left distal femur |
| GSM531295 | 15.3 | Metastases present at diagnosis | Osteoblastic | Succumbed at 27 following diagnosis of the primary tumor | Distal femur |
| GSM531296 | 18. 3 | Metastases present at diagnosis | Osteoblastic | Alive at 26 after diagnosis of the primary tumor | Right proximal tibia |
| GSM531297 | 32.1 | Metastases at 10 months after diagnosis of the primary tumor | Possibly chondromyxoid fibroma-like | Succumbed at 18 following diagnosis of the primary tumor | Right humerus |
| GSM531298 | 39.0 | Metastases at 36 months after diagnosis of the primary tumor | Fibroblastic | Succumbed at 189 following diagnosis of the primary tumor | Unknown |
| GSM531299 | 22 | Metastases at 10 months after diagnosis of the primary tumor | Osteoblastic | Alive at 36 after diagnosis of the primary tumor | Femur |
| GSM531300 | 19 | Metastases at 24 months after diagnosis of the primary tumor | Osteoblastic | Alive at 123 after diagnosis of the primary tumor | Femur |
| GSM531301 | 12 | Metastases at 44 months after diagnosis of the primary tumor | Osteoblastic | Succumbed at 110 following diagnosis of the primary tumor | Femur |
| GSM531302 | 17 | No metastases | Fibroblastic | Alive at 63 after diagnosis of the primary tumor | Humerus |
| GSM531303 | 22 | No metastases | Fibroblastic | Alive at 60 after diagnosis of the primary tumor | Humerus |
| GSM531304 | 58 | No metastases | Telangiectatic | Alive at 60 after diagnosis of the primary tumor | Tibia |
| GSM531305 | 28 | No metastases | Fibroblastic | Alive at 60 after diagnosis of the primary tumor | Femur |
| GSM531306 | 16.7 | Metastases at 6 months after diagnosis of the primary tumor | Osteoblastic | Succumbed at 10 following diagnosis of the primary tumor | Left proximal tibia |
| GSM531307 | 18.6 | Metastases at 21 months after diagnosis of the primary tumor | Chondroblastic | Succumbed at 39 following diagnosis of the primary tumor | Left proximal tibia |
| GSM531308 | 14.8 | Metastases present at diagnosis | Osteoblastic | Alive at 95 after diagnosis of the primary tumor | Left distal femur |
| GSM531309 | 17. 7 | Metastases at 17 months after diagnosis of the primary tumor | Osteoblastic | Succumbed at 83 following diagnosis of the primary tumor | Right distal femur |
| GSM531310 | 18 | No metastases | Osteoblastic | Alive at 246 after diagnosis of the primary tumor | Left distal femur |
| GSM531311 | 16.7 | Metastases present at diagnosis | Telangiectatic | Succumbed at 25 following diagnosis of the primary tumor | Right distal femur |
| GSM531312 | 13.7 | Metastases at 18 months after diagnosis of the primary tumor | Telangiectatic | Succumbed at 40 following diagnosis of the primary tumor | Right proximal tibia |
| GSM531313 | 14.6 | No metastases | Osteoblastic | Alive at 143 after diagnosis of the primary tumor | Right distal tibia |
| GSM531314 | 16.7 | Metastases present at diagnosis | Osteoblastic | Succumbed at 11 following diagnosis of the primary tumor | Left proximal humerus |
| GSM531319 | 8.4 | No metastases | Osteoblastic | Alive at 105 after diagnosis of the primary tumor | Diaphysis of left femur |
| GSM531320 | 11.3 | No metastases | Osteoblastic | Alive at 78 after diagnosis of the primary tumor | Left proximal tibia |
| GSM531321 | 40.6 | No metastases | Anaplastic | Alive at 97 after diagnosis of the primary tumor | Left proximal femur |
| GSM531322 | 16.5 | Metastases at 10 months after diagnosis of the primary tumor | Osteoblastic | Succumbed at 33 following diagnosis of the primary tumor | Left proximal tibia |
| GSM531323 | 11.4 | No metastases | Osteoblastic | Alive at 77 after diagnosis of the primary tumor | Left proximal tibia |
| GSM531324 | 25.3 | Metastases at 27 months after diagnosis of the primary tumor | Osteoblastic | Succumbed at 47 following diagnosis of the primary tumor | Left proximal tibia |
| GSM531325 | 19.1 | No metastases | Chondroblastic | Alive at 120 after diagnosis of the primary tumor | Right proximal humerus |
| GSM531326 | 20.2 | No metastases | Osteoblastic | Alive at 91 after diagnosis of the primary tumor | Right distal femur |
| GSM531327 | 13.75 | Metastases present at diagnosis | Osteoblastic | Succumbed at 29 following diagnosis of the primary tumor | Left distal femur |
| GSM531328 | 18.2 | Metastases at 24 months after diagnosis of the primary tumor | Osteoblastic | Alive at 32 after diagnosis of the primary tumor | Left proximal fibula |
| GSM531329 | 8 | Metastases present at diagnosis | Osteoblastic | Alive at 31 after diagnosis of the primary tumor | Left distal femur |
| GSM531330 | 10.7 | Metastases present at diagnosis | Osteoblastic | Succumbed at 25 following diagnosis of the primary tumor | Right distal femur |
| GSM531331 | 18.1 | No metastases | Osteoblastic | Alive at 219 after diagnosis of the primary tumor | Left distal femur |
| GSM531332 | 16.0 | No metastases | Osteoblastic | Alive at 193 after diagnosis of the primary tumor | Right proximal tibia |
| GSM531333 | 9.3 | No metastases | Pleomorphic | Alive at 184 after diagnosis of the primary tumor | Right distal femur |
| GSM531334 | 3.1 | No metastases | Osteoblastic | Alive at 194 after diagnosis of the primary tumor | Left proximal tibia |
| GSM531335 | 20.3 | No metastases | Anaplastic | Alive at 94 after diagnosis of the primary tumor | Right distal femur |
| GSM531351 | 18.1 | No metastases | Osteoblastic | Alive at 87 after diagnosis of the primary tumor | Right proximal fibula |
| GSM531352 | 16.0 | No metastases | Osteoblastic | Alive at 60 after diagnosis of the primary tumor | Femur |
Meta-analysis for DEG screening
The MetaDE package (27,28) (cran.r-project.org/web/packages/MetaDE) in R3.4.1 software was used to screen consistent DEGs between metastatic and non-metastatic samples from the four datasets (GSE21257, GSE32981, GSE14827 and GSE14359). τ2=0, Qpval >0.05 and false discovery rate <0.05 were considered as the thresholds. The first two parameters were used for heterogeneity testing and the last parameter was used to evaluate significant differences.
After screening the DEGs based on the MetaDE method, these genes were compared with those from screening stable gene modules according to the WGCNA method. The common genes in these two sets of DEGs were deemed the common critical genes. In addition, based on these gene interactions, the gene co-expression network was constructed and visualized using Cytoscape 3.3 (29) (www.cytoscape.org). GO function (biological process, molecular function and cellular component) and KEGG pathway enrichment analyses were performed using the online search annotation software tool Database for Annotation, Visualization and Integrated Discovery (30) (version 6.8, david.ncifcrf.gov). P<0.05 was considered to indicate a statistically significant difference.
Critical genes screening associated with osteosarcoma prognosis
Based on the key node sets in the gene co-expression network, combined with the clinical prognostic information of samples, the critical genes associated with osteosarcoma prognosis were identified using the univariate Cox regression analysis in Survival package (31) (version 2.4, cran.r-project.org/web/packages/survival/index.html) in R3.4.1 software. Survival data were plotted using Kaplan-Meier analysis and Log-Rank test was used to compare the statistical significance. P<0.05 was considered to indicate a statistically significant difference.
Construction of miRNA-target gene regulatory network
The miRNAs directly associated with osteosarcoma were searched from the miR2Disease database (32) (http://watson.compbio.iupui.edu:8080/miR2Disease/index.jsp). Each entry in miR2Disease contains information about a miRNA and its association with disease, in addition to the ID of the miRNA, disease name and a brief description of the miRNA-disease association, references and detection methods of miRNA expression. ‘Osteosarcoma’ was used as the disease name to screen key miRNAs associated with osteosarcoma in this database.
Furthermore, the target genes of miRNAs were searched using miRanda database (33) (www.microrna.org/microrna/home.do). Finally, a miRNA-target gene regulatory network was constructed. Cytoscape 3.3 software was used to visualize the interactions among miRNAs and related target genes.
Validation of critical genes
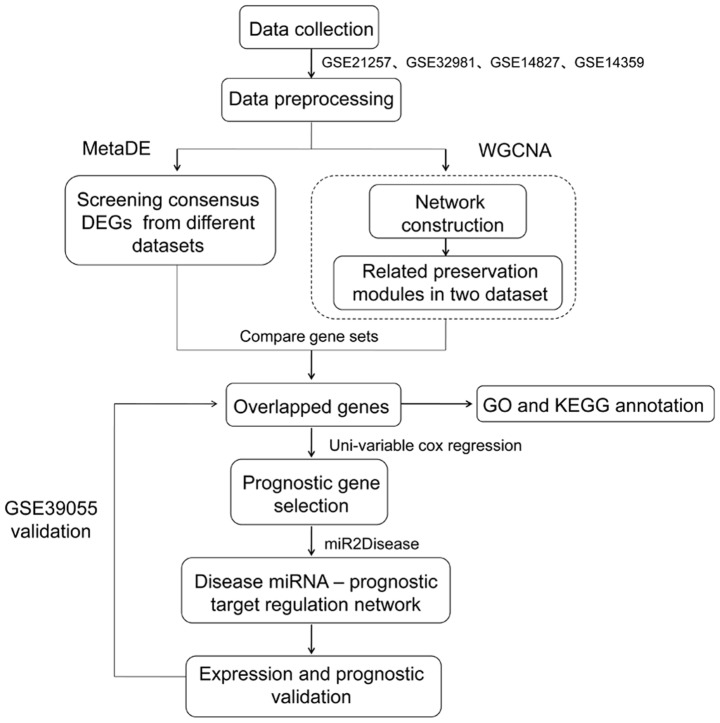
To validate the universality of critical genes, the GSE39055 (34) expression profile (platform GPL14951 Illumina HumanHT-12 WG-DASL V4.0 R2 expression beadchip) was downloaded from the GEO database as a novel validation dataset. This dataset included 37 osteosarcoma samples that possessed associated survival rate information. This dataset was used to validate the associations between key genes and survival outcomes. Additionally, the expression levels of these key genes between metastatic and non-metastatic samples in GSE21257, GSE32981, GSE14827 and GSE14359 were analyzed. The analysis flow chart is presented in Fig. 1.
Figure 1.
Analysis process for the four microarray datasets. DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA/miR, microRNA; WGCNA, weighted gene co-expression network analysis.
Results
Identification of stable modules associated with osteosarcoma
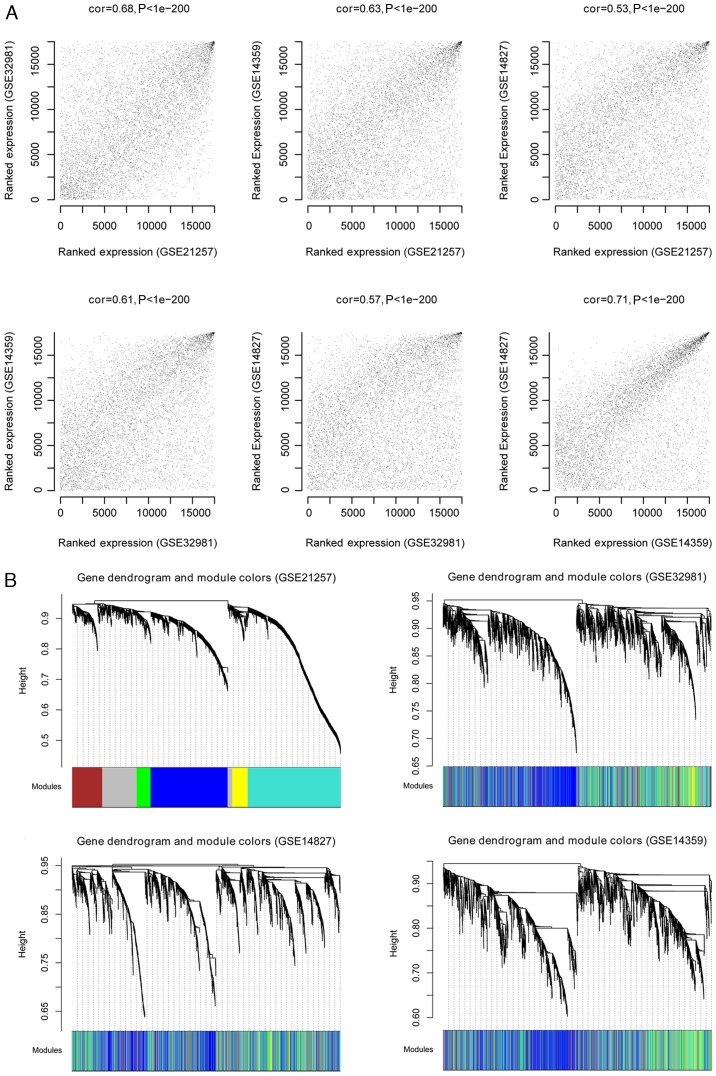
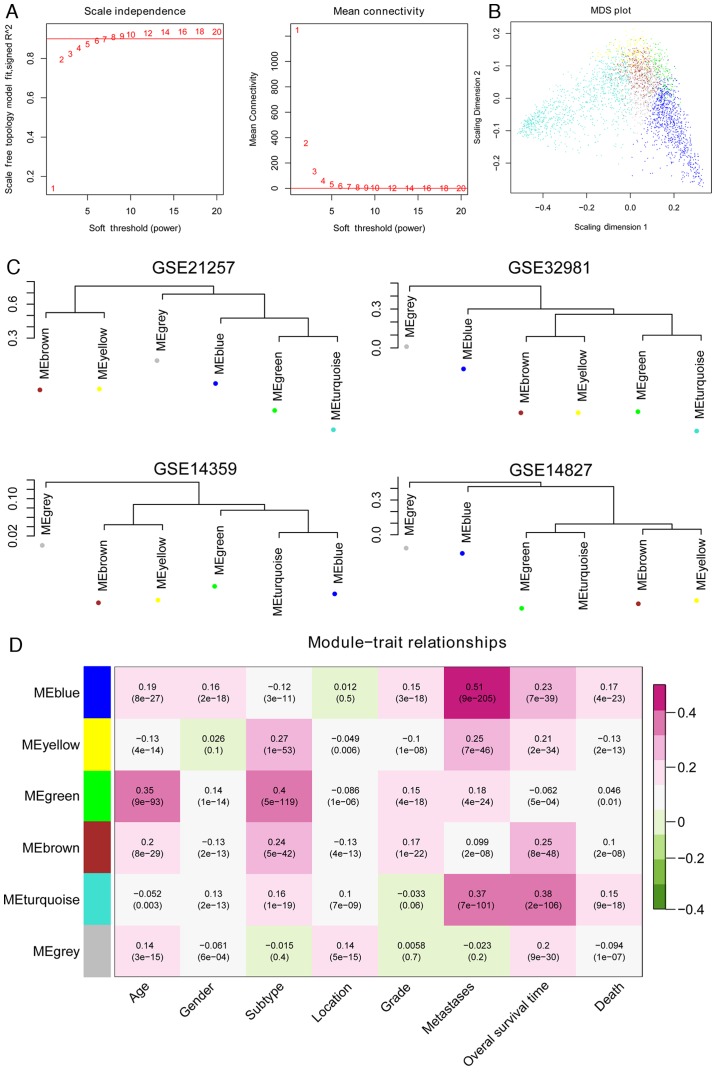
Following the normalization of the four datasets, GSE21257 was used as the training set and the other three were considered validation datasets. The WGCNA algorithm was used to screen for significant modules associated with osteosarcoma. The processes were as follows: First, consensus analysis was performed for the overlapping genes in the four datasets. The results revealed that the correlation values between either of two datasets were >0.5 and all P-values were <1×10−200, which indicated that the data were comparable (Fig. 2A). The cluster dendrogram based on the all modules is presented in Fig. 2B. Second, in order to satisfy the precondition of scale-free network distribution, the weight parameter (power) of the adjacency matrix was fixed by setting the selection range of network parameter and calculating the scale-free topological matrix. The scale-free distribution of the topological matrix was calculated based on the GSE21257 dataset and the results are presented in Fig. 3A (‘scale independence’). The horizontal axis represents weight parameters of the power, while the vertical axis represents the square values of correlation coefficient between log (k) and log [p (k)]. A higher square value meant the scale-free distribution of these data. Once the square value reached 0.9 for the first time, the power value (power=7) was selected and the mean connectivity of genes calculated. As presented in Fig. 3A (‘mean connectivity’), the mean connectivity of genes was only 1, which is in line with the connectivity feature of nodes in a scale-free small network.
Figure 2.
Identification of stable gene modules associated with osteosarcoma as determined by weighted gene co-expression network analysis. (A) Correlation values between any two datasets from GSE21257, GSE32981, GSE14827 and GSE14359. The charts represent correlations between GSE21257-GSE32981, GSE21257-GSE14359, GSE21257-GSE14827, GSE32981-GSE14359, GSE32981-GSE14827 and GSE14359-GSE14827. (B) Cluster dendrogram based on the dynamic tree (GSE21257, GSE32981, GSE14827 and GSE14359). Different dendrogram colors represent various modules.
Figure 3.
Assessment of the stability of the modules. (A) Adjacency function definition for the genes. The left chart represents the power selection diagram of adjacency matrix weight parameter. The horizontal axis represents weight parameters of the power, while the vertical axis represents the square values of correlation coefficient between log (k) and log [p (k)]. A higher square value indicates the scale-free distribution of these data. The red line represents the standard line while square value reached 0.9. The right chart represents the mean connectivity of genes under different adjacency matrix weight parameters. (B) Multidimensional scaling plot of genes in each module. The X- and Y-axes represent the first and second principal components, respectively. (C) Cluster dendrogram of modules in the four datasets, GSE21257, GSE32981, GSE14359 and GSE14827. (D) Heat map for the correlation between each module and clinical factors. The horizontal axis represents clinical factors and the vertical axis represents different colored modules; the color changes from green to pink indicate changes from negative to positive, the numbers in the grid indicate the correlation coefficient and the numbers in parentheses indicate the significance of the correlation (P-value).
Subsequently, the gene dendrogram and modules were identified based on WGCNA. The GSE21257 dataset was used as the training set to screen the modules associated with osteosarcoma. The dissimilarity in easements of different nodes was calculated and a hierarchical clustering tree was generated. Based on the dynamic tree, the minimum number of genes for each network was set as 100 and the cut height was set to 0.99. A total of six modules were obtained, namely M1-blue, M2-brown, M3-green, M4-grey, M5-turquoise and M6-yellow (Table III).
Table III.
Preservation of modules associated with microarray datasets.
| Module | Color | Module size | Preservation Z-score |
|---|---|---|---|
| 1 | Blue | 908 | 29.979 |
| 2 | Brown | 354 | 2.198 |
| 3 | Green | 164 | 7.264 |
| 4 | Grey | 461 | 5.492 |
| 5 | Turquoise | 1,092 | 2.576 |
| 6 | Yellow | 183 | 18.147 |
Finally, the stability of gene modules was evaluated. The other three datasets (GSE32981, GSE14827 and GSE14359) were also subjected to module partition and the stability of modules obtained from the GSE21257 dataset was evaluated. The results of module partition in GSE32981, GSE14827 and GSE14359 datasets are presented in Fig. 2B. The correlation of modules in each dataset is presented in Fig. 3B and C. The genes in the same module (same color) were inclined to cluster together, indicating that these genes had similar expression levels. The overall expression of modules on the same dendrogram branch was more similar, including brown and yellow modules, in addition to blue, green and turquoise modules.
Following analysis of the correlation of gene expression for the same-colored modules, three modules (M1-blue, M3-green and M6-yellow) with preservation Z scores >5 were identified, which were considered significantly stable modules. The three stable modules may be major functional modules associated with osteosarcoma. According to the clinical information provided by the GSE21257 training dataset, the correlation between each module and clinical factors was analyzed. As presented in Fig. 3D, genes in blue and yellow modules were significantly positively correlated with osteosarcoma metastasis (correlation coefficient values, 0.51 and 0.25; P-values, 9×10−205 and 7×10−46, respectively). Finally, the 1,091 genes in these two modules were selected for further analysis.
DEG screening and gene co-expression network analysis
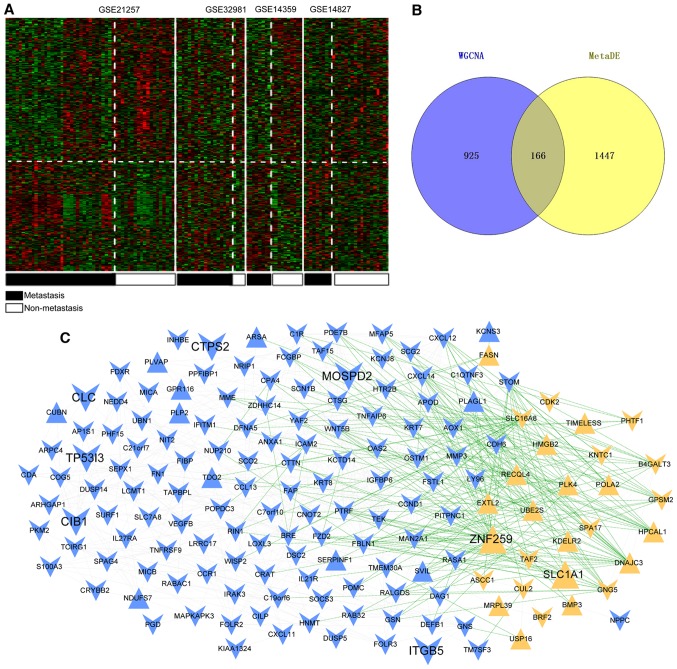
Based on the thresholds, a total of 1,613 consistent DEGs were screened out between the osteosarcoma primary tissue samples and metastatic samples. These DEGs were subjected to hierarchical clustering analysis using the MetaDE package (Fig. 4A). The cluster analysis revealed that DEGs screened from four datasets could accurately distinguish primary osteosarcoma samples from metastatic samples. Subsequently, these genes were compared with the 1,091 genes obtained from blue and yellow modules, and 166 common genes were obtained (Fig. 4B).
Figure 4.
DEG screening and gene co-expression network analysis. (A) Heat map for the significant DEGs. Black bars represent metastatic osteosarcoma samples and white bars represent non-metastatic osteosarcoma samples. (B) Venn diagram of key genes screened according to the WGCNA method and using the MetaDE package. (C) Co-expression network of overlapping genes. Blue and yellow represent the genes screened from blue and yellow modules, respectively. The equilateral and inverted triangles represent upregulated genes and downregulated genes; the green and gray lines represent negative and positive correlations, respectively. DEGs, differentially expressed genes; WGCNA, weighted gene co-expression network analysis.
Based on the expression correlation among the 166 genes, a gene co-expression network was constructed that consisted of 166 nodes (28 from yellow module and 138 from blue module) and 1,344 edges (434 negative correlation and 910 positive correlation). Among these nodes, 28 were upregulated genes and 138 were downregulated (Fig. 4C).
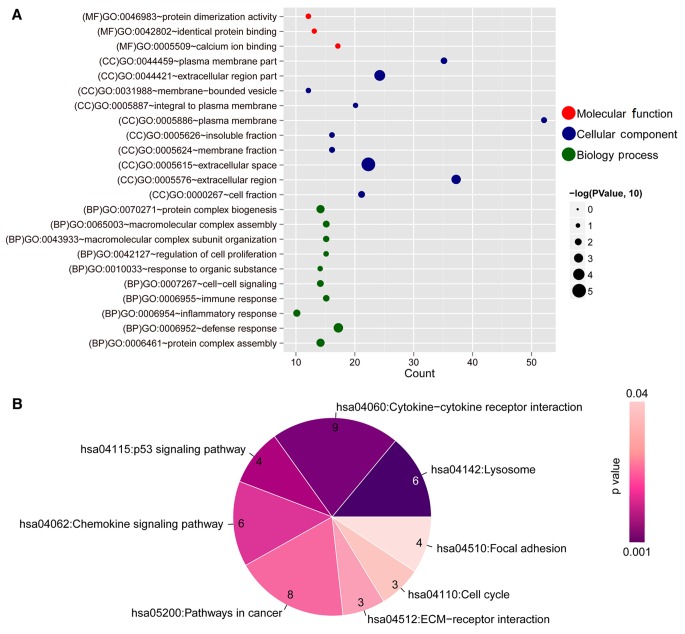
To further explore the function of these 166 DEGs, GO function and KEGG pathway analyses were performed. The results demonstrated that these DEGs were enriched in several functional terms and pathways (Fig. 5 and Table IV). The DEGs were mainly involved in the following GO terms: ‘Defense response’, ‘extracellular region’, ‘calcium ion binding’, etc. The major pathways the DEGs were involved in were ‘lysosome’, ‘cytokine-cytokine receptor interaction’, ‘chemokine signaling pathway’, ‘p53 signaling pathway,’ ‘ECM-receptor interaction’, ‘cell cycle’ and ‘focal adhesion’.
Figure 5.
Functional annotation of the key overlapping genes in the co-expression network. (A) GO annotation. The horizontal axis represents the number of genes and the vertical axis represents the name of the GO terms. The size of the dot represents a significant P-value; larger dots and lower P-values indicate a higher significance. (B) Kyoto Encyclopedia of Genes and Genomes pathway analysis for genes in the network. The color changes from purple to light pink represent changes in significance from high to low. The numbers in each component represent the number of genes involved in a pathway. GO, Gene Ontology.
Table IV.
GO terms and KEGG pathways for the critical DEGs in the gene co-expression network.
| Term | Count | P-value |
|---|---|---|
| Biological process | ||
| GO:0006952-defense response | 17 | 6.710×10−4 |
| GO:0070271-protein complex biogenesis | 14 | 2.379×10−3 |
| GO:0006461-protein complex assembly | 14 | 2.379×10−3 |
| GO:0006954-inflammatory response | 10 | 6.995×10−3 |
| GO:0065003-macromolecular complex assembly | 15 | 9.548×10−3 |
| GO:0007267-cell-cell signaling | 14 | 9.895×10−3 |
| GO:0006955-immune response | 15 | 1.289×10−2 |
| GO:0043933-macromolecular complex subunit organization | 15 | 1.618×10−2 |
| GO:0042127-regulation of cell proliferation | 15 | 3.508×10−2 |
| GO:0010033-response to organic substance | 14 | 3.794×10−2 |
| Cellular component | ||
| GO:0005615-extracellular space | 22 | 9.670×10−6 |
| GO:0044421-extracellular region part | 24 | 1.680×10−4 |
| GO:0005576-extracellular region | 37 | 7.410×10−4 |
| GO:0000267-cell fraction | 21 | 9.733×10−3 |
| GO:0044459-plasma membrane part | 35 | 1.210×10−2 |
| GO:0005624-membrane fraction | 16 | 2.361×10−2 |
| GO:0005886-plasma membrane | 52 | 2.390×10−2 |
| GO:0005626-insoluble fraction | 16 | 3.132×10−2 |
| GO:0031988-membrane-bounded vesicle | 12 | 3.805×10−2 |
| GO:0005887-integral to plasma membrane | 20 | 4.399×10−2 |
| GO:0046983-protein dimerization activity | 12 | 3.398×10−2 |
| GO:0005509-calcium ion binding | 17 | 4.100×10−2 |
| GO:0042802-identical protein binding | 13 | 4.578×10−2 |
| KEGG pathway | ||
| hsa04142: Lysosome | 6 | 1.733×10−3 |
| hsa04060: Cytokine-cytokine receptor interaction | 9 | 1.914×10−3 |
| hsa04115: p53 signaling pathway | 4 | 5.762×10−3 |
| hsa04062: Chemokine signaling pathway | 6 | 9.418×10−3 |
| hsa05200: Pathways in cancer | 8 | 1.309×10−2 |
| hsa04512: ECM-receptor interaction | 3 | 2.978×10−2 |
| hsa04110: Cell cycle | 3 | 4.860×10−2 |
| hsa04510: Focal adhesion | 4 | 4.878×10−2 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Screening critical genes associated with osteosarcoma metastasis
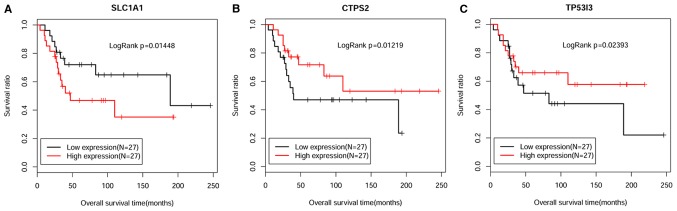
By combining the clinical information of the GSE21257 dataset, the critical genes associated with osteosarcoma metastasis were identified using a Cox regression model. Eventually, 10 genes associated with osteosarcoma prognosis were obtained (Table V). For the top three genes with higher P-values compared with the other DEGs, a Kaplan-Meier survival curve analysis was performed. All samples were divided into high expression and low expression groups in terms of their median numerical boundary (Fig. 6). The results demonstrated that tumor samples with high CTP synthase 2 (CTPS2) and tumor protein p53 inducible protein 3 (TP53I3) expression were associated with improved survival outcome. The expression levels of these genes were downregulated in metastatic tumor samples, indicating that these patients had a worse prognosis. Furthermore, the hazard ratio values were <1, meaning that these genes may be major factors for promoting osteosarcoma metastasis. Conversely, high expression of solute carrier family 1 member 1 (SLC1A1) was associated with a worse prognostic effect. In terms of expression, SLC1A1 was increased in metastatic tumor samples, which indicated that these patients had a worse prognosis. In addition, its hazard ratio value was >1 (Table V), thus suggesting that SLC1A1 expression may be a risk factor for osteosarcoma metastasis.
Table V.
Critical genes associated with the prognosis of osteosarcoma.
| Gene | P-value | Hazard ratio | 95% CI |
|---|---|---|---|
| SLC1A1 | 0.013 | 3.497 | 1.299–9.414 |
| CTPS2 | 0.034 | 0.479 | 0.204–1.126 |
| TP53I3 | 0.034 | 0.505 | 0.247–1.031 |
| CIB1 | 0.041 | 0.596 | 0.265–1.339 |
| VEGFB | 0.041 | 0.555 | 0.224–1.373 |
| ITGB5 | 0.045 | 0.519 | 0.233–1.159 |
| MMP3 | 0.045 | 0.518 | 0.127–2.112 |
| MOSPD2 | 0.046 | 0.820 | 0.320–2.099 |
| FAP | 0.047 | 0.760 | 0.538–1.075 |
| CLC | 0.047 | 0.726 | 0.246–2.137 |
CI, confidence interval.
Figure 6.
Kaplan-Meier survival curve analysis for the top three genes, (A) SLC1A1, (B) CTPS2 and (C) TP53I3, associated with the prognosis of osteosarcoma. The black and red curves represent low expression and high expression sample groups, respectively. CTPS2, CTP synthase 2; SLC1A1, solute carrier family 1 member 1; TP53I3, tumor protein p53 inducible protein 3.
Analysis of miRNA-target gene network
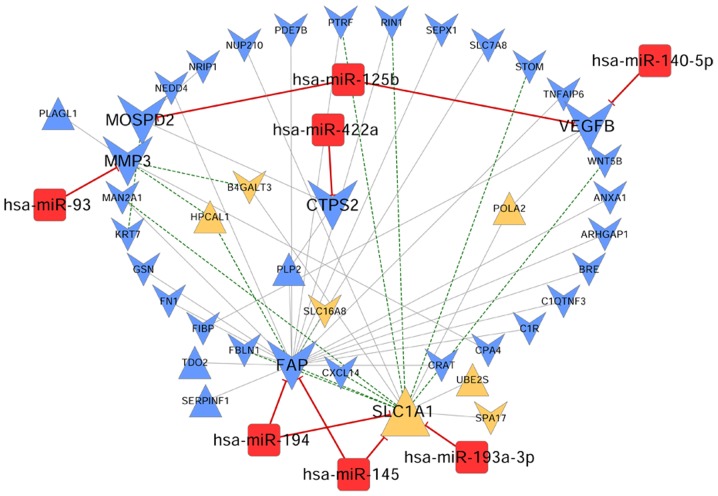
Following searches in the miR2Disease database, seven miRNAs and multiple target genes associated with osteosarcoma were obtained (Table VI). The interactions between miRNAs and their target genes were visualized by a biological network (Fig. 7). The miRNA-target gene network consisted of 48 nodes, including seven miRNAs (including hsa-miR-422a, hsa-miR-145 and hsa-miR-194), six disease prognosis-associated DEGs [CTPS2, fibroblast activation protein α (FAP), SLC1A1, MMP3, motile sperm domain containing 2 (MOSPD2) and vascular endothelial growth factor B (VEGFB)] and 35 DEGs co-expressed with critical genes or miRNAs.
Table VI.
Critical miRNAs related to the prognosis of OS.
| Disease miRNA | OS DEGs | PMID |
|---|---|---|
| hsa-miR-422a | CTPS2 | 20949564 |
| hsa-miR-145 | FAP, SLC1A1 | 26339404 |
| hsa-miR-194 | FAP, SLC1A1 | 26339404 |
| hsa-miR-93 | MMP3 | 28260111 |
| hsa-miR-125b | MOSPD2, VEGFB | 25661090 |
| hsa-miR-193a-3p | SLC1A1 | 20949564 |
| hsa-miR-140-5p | VEGFB | PMC2783211 |
DEGs, differentially expressed genes; miRNA/miR, microRNA; OS, osteosarcoma; PMID, PubMed unique identifier.
Figure 7.
miRNA-target gene network associated with osteosarcoma. Blue and yellow colors represent the genes screened from blue and yellow modules, respectively. The equilateral and inverted triangles represent upregulated and downregulated genes, respectively. The red squares refer to miRNAs associated with osteosarcoma. The green and gray lines represent the negative and positive connections. The red lines represent the interactions between miRNAs and target genes. miRNA/miR, microRNA.
KEGG analysis was performed to identify the major pathways for the miRNAs and target genes. Eventually, nine pathways were identified, including ‘pathways in cancer’, ‘ubiquitin-mediated proteolysis’ and the ‘Ras signaling pathway’ (Table VII). Among the genes, VEGFB was identified to be involved in several signaling pathways (‘pathways in cancer’, ‘focal adhesion’, ‘Ras signaling pathway’ and ‘cytokine-cytokine receptor interaction’), meaning that it may serve a vital role in the development of osteosarcoma.
Table VII.
Kyoto Encyclopedia of Genes and Genomes pathway analysis for the target genes in the regulatory network.
| Term | Count | P-value | Genes |
|---|---|---|---|
| hsa05200: Pathways in cancer | 3 | 0.029 | VEGFB, WNT5B, FN1 |
| hsa04120: Ubiquitin mediated proteolysis | 2 | 0.032 | NEDD4, UBE2S |
| hsa00230: Purine metabolism | 2 | 0.039 | PDE7B, POLA2 |
| hsa05205: Proteoglycans in cancer | 2 | 0.043 | WNT5B, FN1 |
| hsa04510: Focal adhesion | 2 | 0.044 | VEGFB, FN1 |
| hsa01100: Metabolic pathways | 5 | 0.044 | MAN2A1, TDO2, B4GALT3, CTPS2, POLA2 |
| hsa04810: Regulation of actin cytoskeleton | 2 | 0.045 | GSN, FN1 |
| hsa04014: Ras signaling pathway | 2 | 0.047 | VEGFB, RIN1 |
| hsa04060: Cytokine-cytokine receptor interaction | 2 | 0.047 | VEGFB, CXCL14 |
Expression and prognostic validation
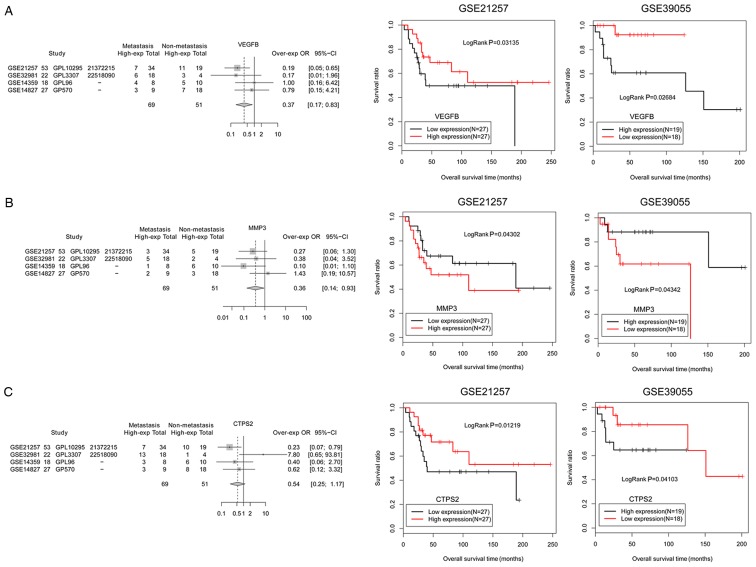
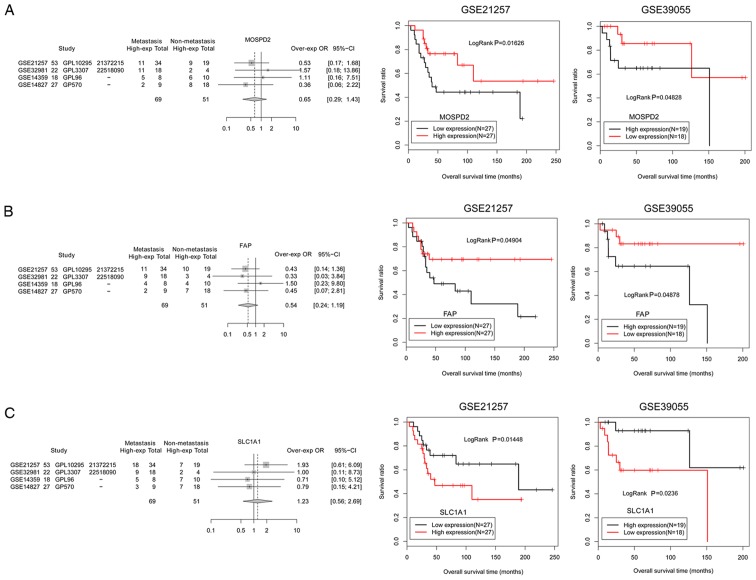
To validate the general characteristics of the six prognosis-associated DEGs involved in the miRNA-target network, a different dataset, GSE39055 (including 37 osteosarcoma samples and related survival outcome information), was applied as a validation dataset to verify the association between the genes and survival outcome. The results demonstrated that the prognostic analysis of six genes in the GSE39055 validation dataset was consistent with that in the GSE21257 dataset (Figs. 8 and 9). Among these genes, MMP3 (Fig. 8B) and SLC1A1 (Fig. 9C) had improved survival outcomes in low expression groups of osteosarcoma samples. In addition, the expression levels of SLC1A1 were upregulated in metastatic osteosarcoma samples and patients with high expression of this gene exhibited worse survival outcomes, indicating that SLC1A1 expression may be a risk factor for osteosarcoma metastasis. In addition, VEGFB, CTPS2, MOSPD2 and FAP had improved survival outcomes in high expression groups of osteosarcoma samples (Figs. 8 and 9). The expression levels of these genes were downregulated in metastatic osteosarcoma samples and patients exhibited worse survival outcomes, indicating that decreased expressions of these four genes (VEGFB, CTPS2, MOSPD2 and FAP) may be risk factors for osteosarcoma metastasis.
Figure 8.
Expression and prognostic validation for critical genes, including (A) VEGFB, (B) MMP3 and (C) CTPS2. The black and red curves represent low expression and high expression osteosarcoma sample groups, respectively. CTPS2, CTP synthase 2; MMP3, matrix metallopeptidase 3; VEGFB, vascular endothelial growth factor B.
Figure 9.
Expression and prognostic validation for critical genes, including (A) MOSPD2, (B) FAP and (C) SLC1A1. The black and red curves represent low expression and high expression osteosarcoma sample groups, respectively. FAP, fibroblast activation protein α; MOSPD2, motile sperm domain containing 2; SLC1A1, solute carrier family 1 member 1.
Discussion
In the present study, gene expression profile analysis (accession numbers: GSE32981, GSE21257, GSE14827 and GSE14359) was performed and a total of 166 critical DEGs were identified in metastatic osteosarcoma tissue samples compared with non-metastatic samples, including 28 upregulated genes and 138 downregulated genes. Functional enrichment analysis results demonstrated that these DEGs were mainly enriched in ‘defense response’, ‘lysosome’ and ‘p53 signaling pathway’. In the gene co-expression network, CTPS2, TP53I3 and SLC1A1 were key nodes and may be considered risk factors for osteosarcoma metastasis. In addition, hsa-miR-422a and hsa-miR-194 were highlighted in the miRNA-target gene network. Finally, MMP3 and VEGFB were predicted as critical genes in osteosarcoma metastasis.
CTPS2 is a critical enzyme that controls the synthesis of cytosine nucleotides, and CTPS2 serves a vital role in numerous metabolic processes (35). Cancer cells that exhibit increased cell proliferation also exhibit increased activity of CTPS2. Patients with colorectal cancer with low CTPS2 expression did not receive a survival benefit from 5-fluorouracil treatment (P=0.072), whereas those with high expression did (P=0.003); therefore, low CTPS2 expression may be a major determinant for chemoresistance (36). TP53I3 encodes the putative quinone oxidoreductase, an enzyme that is involved in cellular responses to oxidative stress and irradiation in humans (37). TP53I3 is involved in p53-mediated cell death and can be induced by the tumor suppressor p53 (38). A recent study indicated that p53 is able to directly regulate target genes, including TP53I3, associated with several drug treatments in an osteosarcoma cell line (39). SLC1A1, also known as excitatory amino-acid transporter 3, is a high-affinity glutamate transporter (40). This protein serves an essential role in glutamate transport from plasma membranes to neurons. However, studies on SLC1A1 in osteosarcoma are few. In the present study, CTPS2, TP53I3 and SLC1A1 were abnormally expressed in metastatic osteosarcoma tissue samples, thus suggesting that these three genes may be considered as prognostic biomarkers of osteosarcoma.
The miRNA-target gene network demonstrated that several miRNAs were involved in osteosarcoma prognosis. Downregulation of miR-422a has been reported to be associated with poor prognosis in human osteosarcoma (41). Increased expression levels of miR-422a can inhibit cell proliferation and invasion, and can enhance chemosensitivity in osteosarcoma cells (42). Zhang et al (43) demonstrated that miR-422a may serve as a tumor inhibitor in osteosarcoma via suppression of BCL2 like 2 (BCL2L2) and KRAS proto-oncogene, GTPase (KRAS) translation. Therefore, miR-442a may be involved in the progression of osteosarcoma via targeting BCL2L2 and KRAS. In addition, hsa-miR-194 has been reported to be a major factor involved in tumor progression and metastasis (44). Han et al (45) indicated that miR-194 may suppress osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting cadherin 2 and insulin-like growth factor 1 receptor. Therefore, these findings suggested that hsa-miR-422a and hsa-miR-194 may be critical molecules in osteosarcoma metastasis.
MMP3 or stromelysin-1 is an enzyme that regulates the breakdown of extracellular matrix proteins in normal physiological processes, in addition to in disease processes, including tumor metastasis or arthritis (46). The MMP family, including MMP3, has been confirmed to interact with collagen type I α 2 chain and serve an important role in osteosarcoma tumorigenesis (47). A previous study revealed that upregulation of MMP13 can result in suppression of osteosarcoma metastasis in a mouse model (48). As a member of the MMP family, MMP3 may be an important diagnostic marker for osteosarcoma metastasis. VEGFs are signaling proteins produced by cells that stimulate the formation of blood vessels, which is an important process for tumorigenesis and metastasis (49). It has also reported the abnormal expression of VEGF in osteosarcoma and explored the prognostic value in cancer patients (50). VEGFB belongs to the VEGF family and serves a role in maintaining newly formed blood vessels during pathological conditions (51). It has been reported that VEGF correlates with a poor histologic response to chemotherapy in osteogenic sarcoma (52). In the present study, the expression levels of MMP3 and VEGFB were downregulated in metastatic osteosarcoma samples and these patients exhibited worse survival outcomes. The survival analysis of MMP3 for GSE21257 training set is consistent with that of the GSE39055 validation set in the present study. Together with the present findings, it was suggested that these two genes may be risk factors for promoting osteosarcoma metastasis.
However, certain limitations existed in the present study. One important factor is the lack of experimental verification. The number of osteosarcoma samples in metastatic or non-metastatic groups was small; the clinical information in microarray datasets was also scarce. Therefore, more studies are required to explore the diagnostic roles of critical genes and miRNAs in osteosarcoma metastasis; for example, the expression levels of the critical genes and miRNAs in osteosarcoma samples should be validated. Besides, the roles of these genes and miRNAs in predicting the prognosis of osteosarcoma should be analyzed in a large sample size in future studies.
In conclusion, the present study identified 28 upregulated and 138 downregulated genes in metastatic osteosarcoma samples. The DEGs were associated with ‘defense response’, ‘p53 signaling pathway’ and ‘lysosome’. In addition, it was revealed that CTPS2, TP53I3 and SLC1A1 may serve a major role in osteosarcoma metastasis, and hsa-miR-422a, hsa-miR-194, MMP3 and VEGFB may also be associated with the metastasis of osteosarcoma. The present study provided a reliable strategy to discover non-invasive biomarkers for osteosarcoma prognosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SZ and HF made substantive intellectual contributions to the study design. HF, SL, SW and SZ searched and downloaded the information about microarray datasets from the Gene Expression Omnibus. HF, SL, SW and SZ made substantial contributions to analysis and interpretation of microarray datasets. HF, SL, SW and SZ were involved in the revising the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment-where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Ferrari S, Smeland S, Mercuri M, Bertoni F, Longhi A, Ruggieri P, Alvegard TA, Picci P, Capanna R, Bernini G, et al. Neoadjuvant chemotherapy with high-dose ifosfamide, high-dose methotrexate, cisplatin, and doxorubicin for patients with localized osteosarcoma of the extremity: A joint study by the Italian and Scandinavian Sarcoma Groups. J Clin Oncol. 2005;23:8845–8852. doi: 10.1200/JCO.2004.00.5785. [DOI] [PubMed] [Google Scholar]
- 4.Ragland BD, Bell WC, Lopez RR, Siegal GP. Cytogenetics and molecular biology of osteosarcoma. Lab Invest. 2002;82:365–373. doi: 10.1038/labinvest.3780431. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Zeng X, Oliver P, Le LP, Chen J, Chen L, Zhou W, Agrawal S, Zhang R. MDM2 oncogene as a target for cancer therapy: An antisense approach. Int J Oncol. 1999;15:653–660. [PubMed] [Google Scholar]
- 6.Kushlinskii NE, Fridman MV, Braga EA. Molecular mechanisms and microRNAs in osteosarcoma pathogenesis. Biochemistry (Mosc) 2016;81:315–328. doi: 10.1134/S0006297916040027. [DOI] [PubMed] [Google Scholar]
- 7.Li H, He Y, Hao P, Liu P. Identification of characteristic gene modules of osteosarcoma using bioinformatics analysis indicates the possible molecular pathogenesis. Mol Med Rep. 2017;15:2113–2119. doi: 10.3892/mmr.2017.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shabani P, Izadpanah S, Aghebati-Maleki A, Baghbani E, Baghbanzadeh A, Fotouhi A, Bakhshinejad B, Aghebati-Maleki L, Baradaran B. Role of miR-142 in the pathogenesis of osteosarcoma and its potential as therapeutic approach. J Cell Biochem. 2019;120:4783–4793. doi: 10.1002/jcb.27857. [DOI] [PubMed] [Google Scholar]
- 9.Lv Z, Yang D, Li J, Hu M, Luo M, Zhan X, Song P, Liu C, Bai H, Li B, et al. Bone morphogenetic protein 9 overexpression reduces osteosarcoma cell migration and invasion. Mol Cells. 2013;36:119–126. doi: 10.1007/s10059-013-0043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren Z, Liang S, Yang J, Han X, Shan L, Wang B, Mu T, Zhang Y, Yang X, Xiong S, Wang G. Coexpression of CXCR4 and MMP9 predicts lung metastasis and poor prognosis in resected osteosarcoma. Tumour Biol. 2016;37:5089–5096. doi: 10.1007/s13277-015-4352-8. [DOI] [PubMed] [Google Scholar]
- 11.Song R, Tian K, Wang W, Wang L. P53 suppresses cell proliferation, metastasis, and angiogenesis of osteosarcoma through inhibition of the PI3K/AKT/mTOR pathway. Int J Surg. 2015;20:80–87. doi: 10.1016/j.ijsu.2015.04.050. [DOI] [PubMed] [Google Scholar]
- 12.Rao-Bindal K, Rao CK, Yu L, Kleinerman ES. Expression of c-FLIP in pulmonary metastases in osteosarcoma patients and human xenografts. Pediatr Blood Cancer. 2013;60:575–579. doi: 10.1002/pbc.24412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen GB, Knudsen A, Hager H, Hansen LL, Tost J. miRNA profiling identifies deregulated miRNAs associated with osteosarcoma development and time to metastasis in two large cohorts. Mol Oncol. 2018;12:114–131. doi: 10.1002/1878-0261.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilbao-Aldaiturriaga N, Gutierrez-Camino A, Martin-Guerrero I, Pombar-Gomez M, Zalacain-Diez M, Patiño-Garcia A, Lopez-Lopez E, Garcia-Orad A. Polymorphisms in miRNA processing genes and their role in osteosarcoma risk. Pediatr Blood Cancer. 2015;62:766–769. doi: 10.1002/pbc.25416. [DOI] [PubMed] [Google Scholar]
- 15.Zhang R, Yan S, Wang J, Deng F, Guo Y, Li Y, Fan M, Song Q, Liu H, Weng Y, Shi Q. MiR-30a regulates the proliferation, migration, and invasion of human osteosarcoma by targeting Runx2. Tumour Biol. 2016;37:3479–3488. doi: 10.1007/s13277-015-4086-7. [DOI] [PubMed] [Google Scholar]
- 16.Lv C, Hao Y, Tu G. MicroRNA-21 promotes proliferation, invasion and suppresses apoptosis in human osteosarcoma line MG63 through PTEN/Akt pathway. Tumour Biol. 2016;37:9333–9342. doi: 10.1007/s13277-016-4807-6. [DOI] [PubMed] [Google Scholar]
- 17.Namløs HM, Kresse SH, Müller CR, Henriksen J, Holdhus R, Sæter G, Bruland OS, Bjerkehagen B, Steen VM, Myklebost O. Global gene expression profiling of human osteosarcomas reveals metastasis-associated chemokine pattern. Sarcoma. 2012;2012:639038. doi: 10.1155/2012/639038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buddingh EP, Kuijjer ML, Duim RA, Bürger H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PC, Lankester AC, Cleton-Jansen AM. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: A rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17:2110–2119. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535–544. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 20.Fritsche-Guenther R, Noske A, Ungethüm U, Kuban RJ, Schlag PM, Tunn PU, Karle J, Krenn V, Dietel M, Sers C. De novo expression of EphA2 in osteosarcoma modulates activation of the mitogenic signalling pathway. Histopathology. 2011;57:836–850. doi: 10.1111/j.1365-2559.2010.03713.x. [DOI] [PubMed] [Google Scholar]
- 21.Parrish RS, Spencer HJ., III Effect of normalization on significance testing for oligonucleotide microarrays. J Biopharm Stat. 2004;14:575–589. doi: 10.1081/BIP-200025650. [DOI] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frigola R, Rasmussen CE. Integrated pre-processing for bayesian nonlinear system identification with gaussian processes. Ieee Decis Contr P. 2013:5371–5376. doi: 10.1109/CDC.2013.6760734. [DOI] [Google Scholar]
- 24.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai X, Xue Q, Liu Q, Guo Y, Chen Z. Colon cancer recurrenceassociated genes revealed by WGCNA coexpression network analysis. Mol Med Rep. 2017;16:6499–6505. doi: 10.3892/mmr.2017.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi C, Hong L, Cheng Z, Yin Q. Identification of metastasis-associated genes in colorectal cancer using metaDE and survival analysis. Oncol Lett. 2016;11:568–574. doi: 10.3892/ol.2015.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Kang DD, Shen K, Song C, Lu S, Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al. An R package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics. 2012;28:2534–2536. doi: 10.1093/bioinformatics/bts485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P, Wang Y, Hang B, Zou X, Mao JH. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 2016;7:55343–55351. doi: 10.18632/oncotarget.10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res 36 (Database Issue) 2008:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly AD, Haibe-Kains B, Janeway KA, Hill KE, Howe E, Goldsmith J, Kurek K, Perez-Atayde AR, Francoeur N, Fan JB, et al. MicroRNA paraffin-based studies in osteosarcoma reveal reproducible;independent prognostic profiles at 14q32. Genome Med. 2013;5:2. doi: 10.1186/gm406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Identification of cDNA encoding an isoform of human CTP synthetase. Biochim Biophys Acta. 2000;1492:548. doi: 10.1016/S0167-4781(00)00141-X. [DOI] [PubMed] [Google Scholar]
- 36.Tan WL, Bhattacharya B, Loh M, Balasubramanian I, Akram M, Dong D, Wong L, Thakkar B, Salto-Tellez M, Soo RA, et al. Low cytosine triphosphate synthase 2 expression renders resistance to 5-fluorouracil in colorectal cancer. Cancer Biol Ther. 2011;11:599–608. doi: 10.4161/cbt.11.6.14670. [DOI] [PubMed] [Google Scholar]
- 37.Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 38.Voltan R, Secchiero P, Corallini F, Zauli G. Selective induction of TP53I3/p53-inducible gene 3 (PIG3) in myeloid leukemic cells, but not in normal cells, by Nutlin-3. Mol Carcinog. 2014;53:498–504. doi: 10.1002/mc.21985. [DOI] [PubMed] [Google Scholar]
- 39.Zhou H, Cui X, Yuan H, Zhang B, Meng C, Zhao D. Effects of distinct drugs on gene transcription in an osteosarcoma cell line. Oncol Lett. 2017;14:4694–4700. doi: 10.3892/ol.2017.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith CP, Weremowicz S, Kanai Y, Stelzner M, Morton CC, Hediger MA. Assignment of the gene coding for the human high-affinity glutamate transporter EAAC1 to 9p24: Potential role in dicarboxylic aminoaciduria and neurodegenerative disorders. Genomics. 1994;20:335–336. doi: 10.1006/geno.1994.1183. [DOI] [PubMed] [Google Scholar]
- 41.Bahador R, Taheriazam A, Mirghasemi A, Torkaman A, Shakeri M, Yahaghi E, Goudarzi PK. Tissue expression levels of miR-29b and miR-422a in children, adolescents, and young adults' age groups and their association with prediction of poor prognosis in human osteosarcoma. Tumor Biol. 2016;37:3091–3095. doi: 10.1007/s13277-015-4140-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu M, Xiusheng H, Xiao X, Wang Y. Overexpression of miR-422a inhibits cell proliferation and invasion, and enhances chemosensitivity in osteosarcoma cells. Oncol Rep. 2016;36:3371–3378. doi: 10.3892/or.2016.5182. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, He QY, Wang GC, Tong DK, Wang RK, Ding WB, Li C, Wei Q, Ding C, Liu PZ, et al. miR-422a inhibits osteosarcoma proliferation by targeting BCL2L2 and KRAS. Biosci Rep. 2018;38:BSR20170339. doi: 10.1042/BSR20170339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khella WZ, Bakhet M, Allo G, Jewett MA, Girgis A, Bjarnason GA, Yousef GM. Supression of tumor progression and metastasis in renal cell carcinoma by miR-192, miR-194, and miR-215. J Clin Oncol. 2013;31:385–385. doi: 10.1200/jco.2013.31.6_suppl.385. [DOI] [PubMed] [Google Scholar]
- 45.Han K, Zhao T, Chen X, Bian N, Yang T, Ma Q, Cai C, Fan Q, Zhou Y, Ma B. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol. 2014;45:1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendes O, Kim HT, Stoica G. Expression of MMP2, MMP9 and MMP3 in breast cancer brain metastasis in a rat model. Clin Exp Metastasis. 2005;22:237–246. doi: 10.1007/s10585-005-8115-6. [DOI] [PubMed] [Google Scholar]
- 47.Wu D, Chen K, Bai Y, Zhu X, Chen Z, Wang C, Zhao Y, Li M. Screening of diagnostic markers for osteosarcoma. Mol Med Rep. 2014;10:2415–2420. doi: 10.3892/mmr.2014.2546. [DOI] [PubMed] [Google Scholar]
- 48.Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T, et al. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther. 2011;19:1123–1130. doi: 10.1038/mt.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senger DR, Galli SJ, Dvorak AM, Perruzzi CA, Harvey VS, Dvorak HF. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science. 1983;219:983–985. doi: 10.1126/science.6823562. [DOI] [PubMed] [Google Scholar]
- 50.Qu JT, Wang M, He HL, Tang Y, Ye XJ. The prognostic value of elevated vascular endothelial growth factor in patients with osteosarcoma: A meta-analysis and systemic review. J Cancer Res Clin Oncol. 2012;138:819–825. doi: 10.1007/s00432-012-1149-7. [DOI] [PubMed] [Google Scholar]
- 51.Zhang F, Tang Z, Hou X, Lennartsson J, Li Y, Koch AW, Scotney P, Lee C, Arjunan P, Dong L, et al. VEGF-B is dispensable for blood vessel growth but critical for their survival, and VEGF-B targeting inhibits pathological angiogenesis. Proc Natl Acad Sci USA. 2009;106:6152–6157. doi: 10.1073/pnas.0813061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdeen A, Chou AJ, Healey JH, Khanna C, Osborne TS, Hewitt SM, Kim M, Wang D, Moody K, Gorlick R. Correlation between clinical outcome and growth factor pathway expression in osteogenic sarcoma. Cancer. 2009;115:5243–5250. doi: 10.1002/cncr.24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.