Abstract
Background
Tuberculosis (TB) is a major global health problem and has replaced HIV as the leading cause of death from a single infectious agent.
Methods
Here, we applied high throughput sequencing to study the immune repertoire of nine pulmonary tuberculosis patients and nine healthy control samples.
Results
Tuberculosis patients and healthy controls displayed significantly different high express clones and distinguishable sharing of CDR3 sequences. The TRBV and TRBJ gene usage showed higher expression clones in patients than in controls and we also found specific high express TRBV and TRBJ gene clones in different groups. In addition, six highly expressed TRBV/TRBJ combinations were detected in the CD4 group, 21 in the CD8 group and 32 in the tissue group.
Conclusion
In conclusion, we studied the patients with tuberculosis as well as healthy control individuals in order to understand the characteristics of immune repertoire. Sharing of CDR3 sequences and differential expression of genes was found among the patients with tuberculosis which could be used for the development of potential vaccine and targets treatment.
Keywords: CDR3 sequences, high express clones, high throughput sequencing, TRBJ gene, tuberculosis
1. INTRODUCTION
Tuberculosis (TB) is a major global health problem and has replaced HIV as the leading cause of death from a single infectious agent in 2016. In 2015, an estimated 10.4 million people developed TB and 1.4 million died from the disease. Tuberculosis is a disease caused by mycobacterium tuberculosis infection. The immunology reaction caused by pathogens during the infection and reproduction of mycobacterium tuberculosis has been studied which leads to the development of vaccine, diagnosis, drug resistance (Horwitz, Lee, Dillon, & Harth, 1995; Lindenstrøm et al., 2009; Meintjes et al., 2009; Vanham et al., 1997). Since, the TCR repertoire is a mirror of the human immune response, its characteristics have been widely investigated in infectious and other diseases to study the state of the immune system and the progression of these diseases (Chaudhry, Cairo, Venturi, & Pauza, 2013).
The diversity within the TCR repertoire is ensured through somatic recombination of germline‐encoded variable (V), diversity (D), and junctional (J) gene segments. Nucleotide deletions at the coding ends and nucleotide additions at the V(D)J junctions also contribute substantially to the TCR repertoire diversity (Nikolich‐Žugich, Slifka, & Messaoudi, 2004). The TCR diversity is a function of the third hypervariable complementary‐determining (CDR3) region, which lies at the intersection between the V, D, J and V, J gene segments within the TCR and TCR chains, respectively. The CDR3 region encodes that part of the TCR which predominantly interacts with antigenic peptide/MHC complexes. Thus, even when T cell clones express the same V/J genes rearrangement, they can be identified by the unique combination of their CDR3 sequences (TCR clonotypes) (Toivonen, Arstila, & Hänninen, 2015). Accordingly, the complexity and distribution of TCRs within specific T cell populations will reflect the degree of complexity of the T cell response.
In the present study, we studied the immune repertoire of CD4+, CD8+ T cells of patients and healthy controls and tissue sample of patients to elucidate the effect of tuberculosis on patients’ immune system. The characteristics of diversity and stability, CDR3 length distribution and CDR3 sequences sharing were analyzed. Besides, the usage of TRBJ, TRBV as well as the combination of TRBV/TRBJ were studied. The different repertoire features between tuberculosis patients and controls were then found as future targets for further study.
2. METHODS
2.1. Clinical samples
Tuberculosis tissue samples and blood samples of nine patients and blood samples of nine healthy controls were collected at the Second Clinical Medical College of Jinan University (Shenzhen People's Hospital, Guangdong, China). All patients gave written informed consent and the present study was approved by the Medical Ethics Committee of Shenzhen People's Hospital.
2.2. DNA extraction and mixing
T cell was isolated using superparamagnetic polystyrene beads (Miltenyi) coated with monoclonal antibodies specific for T cells. DNA was prepared from 0.5 to 2 × 106 T cells from each sample (patients and controls), which was sufficient for analyzing the diversity of TCR in the T cell subsets. DNA was extracted from PBMCs using GenFIND DNA (Agencourt, Beckman Coulter, Brea, CA) extraction kits following the manufacturer's instructions.
Ten milligrams of tuberculosis tissue was obtained from each patient sample and DNA was extracted using standard methods. Briefly, dewaxing was done using xylene and followed by over‐night proteinase K digestion for tissues. QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany) was further used for DNA extraction following the manufacturer's instructions. DNA quality was evaluated by loading on a 0.8% agarose gel electrophoresis and DNA concentration was quantified by Qubit fluorometer. DNA from nine patients' peripheral blood samples were mixed together by 1:1:1:1:1:1:1:1:1 according to Qubit value, renamed one blood sample. Meanwhile, DNA from nine patients' tuberculosis tissues and control blood samples were mixed separately in the same way.
2.3. Multiplex‐PCR amplification of TCR‐β CDR3 regions
The human TCR‐β sequences were downloaded from IMGT (http://www.imgt.org/). A relative conserved region in frame region 3, upstream of CDR3, was selected for the puta‐ tive forward primer region. A cluster of primers corresponding to the majority of the V gene family sequence was selected. Similarly, reverse primers corresponding to the J gene family were designed. In total, 30 forward primers and 13 reverse primers were used for multiplex PCR to amplify the rearranged TCR‐β CDR3 regions. The reaction mixtures (50 μl total) comprised 2 μl of pooled TCR‐β variable gene (TRBV; 10 μM), 2 μl of pooled TCR‐β joining gene (TRBJ; 10 μM), 25 μl of 2X Qiagen Multiplex PCR Master Mix, 5 μl of 5X Q‐solution, 500 ng of template DNA (10 μl) and 6 μl of H2O. The PCR conditions comprised 95 °C for 15 min; followed by 25 cycles of 94 °C for 15 s and 60 °C for 3 min; followed by a final extension for 10 min at 72 °C. The PCR products were purified using AMPure XP beads to remove primer sequences (Beckman Coulter, Inc., Brea, CA, USA). A second round of PCR was performed to add a sequencing index to each sample. In this round, each reaction mixture (50 μl total) consisted of 13.5 μl of H2O, 0.5 μl of 2X Q5 DNA polymerase, 10 μl of 5X Q5 buffer, 1 μl of dNTPs (10 mM), 1 μl of P1 (10 μM), 23 μl of DNA, and 1 μl of index (10 μM). The PCR conditions comprised 98 °C for 1 min; followed by 25 cycles of 98 °C for 20 s, 65 °C for 30 s and 72 °C for 30 s; and a final extension for 5 min at 72 °C. The library was separated on an agarose gel, and the target region was isolated and cleaned using QIAquick Gel Extraction kits (Qiagen).
2.4. NGS and data analysis
The library was quantitated using the Agilent 2100 Bioanalyzer instrument (Agilent DNA 1,000 reagents) and real‐time quantitative PCR (TaqMan probes) and sequenced by Illumina MiSeq. Briefly, the adaptor reads and low‐quality reads were filtered from the raw data, the clean data was used in further alignments. Subsequently, the clean data was aligned to the human IGH database and analyzed using the online IMGT/HighV‐QUEST tool. The data included V, J assignment, CDR3 length distribution, clustering and other analyses.
3. RESULT
3.1. Quality control of all sequencing data
Using high‐throughput NGS, we sequenced repertoires CD4+ cells, CD8+ cells and tissue samples of nine pulmonary tuberculosis patients and nine normal controls. All data passed the QC process with an average Q20 >99.99%, Q30 >97.04%. A total sequencing data of total reads number (34094942), immune sequences number (33504380), unknown sequences number (590562), productive sequences number (24345651), nonproductive sequences number (9158729), In‐frame sequences number (27088243), out‐of‐frame sequences number (6326420), total CDR3 sequences number (23670829), Unique CDR3 nucleotide sequences number (1995707) and Unique CDR3 amino acids sequences number (1723256), details including data from each sample are listed in Table 1.
Table 1.
Sequence quality of CD4+, CD8+, tissue sample of tuberculosis patients and normal controls
| M1‐CD4 | M1‐CD8 | M1‐Tissue | M2‐CD4 | M2‐CD8 | M2‐Tissue | M3‐CD4 | M3‐CD8 | M3‐Tissue | M4‐CD4 | M4‐CD8 | M4‐Tissue | M5‐CD4 | M5‐CD8 | M5‐Tissue | M6‐CD4 | M6‐CD8 | M6‐Tissue | M7‐CD4 | M7‐CD8 | M7‐Tissue | M8‐CD4 | M8‐CD8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total reads number | 582217 | 459996 | 677310 | 903177 | 575955 | 335774 | 889156 | 643710 | 573785 | 1065180 | 859602 | 253607 | 889229 | 427415 | 495999 | 971852 | 627698 | 336292 | 776087 | 801797 | 313179 | 1016582 | 892231 |
| Immune sequences number | 550596 | 449546 | 650380 | 901272 | 573975 | 288998 | 887379 | 640989 | 539209 | 1062359 | 857908 | 195758 | 886866 | 424561 | 458664 | 970694 | 625998 | 286683 | 774495 | 799883 | 256256 | 1014869 | 890412 |
| Unknown sequences numebr | 31621 | 10450 | 26930 | 1905 | 1980 | 46776 | 1777 | 2721 | 34576 | 2821 | 1694 | 57849 | 2363 | 2854 | 37335 | 1158 | 1700 | 49609 | 1592 | 1914 | 56923 | 1713 | 1819 |
| Productive sequences number | 353828 | 250365 | 474701 | 699906 | 433221 | 162872 | 685579 | 492654 | 356619 | 841185 | 670586 | 98480 | 691340 | 327224 | 314326 | 764656 | 413577 | 82356 | 611495 | 700658 | 102129 | 810266 | 738949 |
| Nonproductive sequences number | 196768 | 199181 | 175679 | 201366 | 140754 | 126126 | 201800 | 148335 | 182590 | 221174 | 187322 | 97278 | 195526 | 97337 | 144338 | 206038 | 212421 | 204327 | 163000 | 99225 | 154127 | 204603 | 151463 |
| In‐frame sequences number | 377268 | 289584 | 502949 | 746867 | 461590 | 187810 | 731817 | 527307 | 377228 | 900754 | 717545 | 110971 | 738864 | 344971 | 323396 | 816839 | 435942 | 88467 | 654902 | 743398 | 106464 | 861847 | 783627 |
| Out‐of_frame sequences number | 163201 | 158365 | 145359 | 153605 | 111898 | 94557 | 154889 | 113184 | 159463 | 160716 | 139871 | 81018 | 147303 | 78997 | 129220 | 153394 | 189766 | 178497 | 119143 | 55925 | 143195 | 152559 | 106321 |
| Total CDR3 sequences number | 327059 | 233666 | 441542 | 677038 | 420582 | 153780 | 674805 | 484420 | 347379 | 817310 | 622440 | 84721 | 680369 | 321140 | 304849 | 752939 | 407462 | 46143 | 602093 | 691660 | 75485 | 798558 | 728731 |
| Unique CDR3 nt sequences number | 14090 | 12645 | 20153 | 60934 | 26558 | 16661 | 65880 | 32174 | 18011 | 65934 | 51955 | 11956 | 51934 | 18470 | 6677 | 123828 | 18610 | 4807 | 69335 | 22394 | 4851 | 74991 | 48370 |
| Unique CDR3 aa sequences number | 11656 | 10237 | 17021 | 51459 | 21942 | 15302 | 56729 | 27010 | 15661 | 54642 | 43502 | 11351 | 43869 | 15337 | 5265 | 112096 | 15068 | 4416 | 60466 | 18165 | 4371 | 63893 | 40937 |
| Highly expressing clone number all | 41 | 38 | 53 | 2 | 18 | 28 | 5 | 16 | 48 | 0 | 11 | 21 | 4 | 20 | 26 | 2 | 24 | 40 | 1 | 11 | 30 | 0 | 8 |
| Highly expressing clone ratio all | 0.68255575 | 0.52631534 | 0.63128989 | 0.02411238 | 0.31779772 | 0.71275849 | 0.05190685 | 0.34143305 | 0.68128471 | 0 | 0.15724086 | 0.6742012 | 0.05387518 | 0.50208009 | 0.90383436 | 0.05658493 | 0.4683357 | 0.7194374 | 0.00541112 | 0.6621259 | 0.78404981 | 0 | 0.30130597 |
| Shannon entropy all | 0.39719054 | 0.45082821 | 0.41825992 | 0.68833416 | 0.54280623 | 0.43039658 | 0.67979693 | 0.51286718 | 0.40022774 | 0.66933299 | 0.63017905 | 0.41873046 | 0.59746329 | 0.42595744 | 0.26421886 | 0.76166681 | 0.45595582 | 0.45273164 | 0.72728526 | 0.30304497 | 0.37782448 | 0.69998144 | 0.53009339 |
| M8‐Tissue | M9‐CD4 | M9‐CD8 | M9‐Tissue | N1‐CD4 | N1‐CD8 | N2‐CD4 | N2‐CD8 | N3‐CD4 | N3‐CD8 | N4‐CD4 | N4‐CD8 | N5‐CD4 | N5‐CD8 | N6‐CD4 | N6‐CD8 | N7‐CD4 | N7‐CD8 | N8‐CD4 | N8‐CD8 | N9‐CD4 | N9‐CD8 | Sum | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total reads number | 345124 | 920454 | 612229 | 644546 | 734122 | 1015119 | 487365 | 1090624 | 948621 | 1738242 | 609,319 | 790,880 | 691,014 | 675,691 | 528,099 | 811,278 | 1,360,226 | 1,129,827 | 1,023,104 | 569,829 | 907,554 | 1,093,845 | 34,094,942 |
| Immune sequences number | 263278 | 918689 | 609826 | 615559 | 731341 | 1005773 | 486477 | 1089108 | 945180 | 1733207 | 598,136 | 778,657 | 689,656 | 671,243 | 526,754 | 806,631 | 1,355,308 | 1,122,850 | 1,015,052 | 558,523 | 905,671 | 1,089,711 | 33,504,380 |
| Unknown sequences numebr | 81846 | 1765 | 2403 | 28987 | 2781 | 9346 | 888 | 1516 | 3441 | 5035 | 11,183 | 12,223 | 1,358 | 4,448 | 1,345 | 4,647 | 4,918 | 6,977 | 8,052 | 11,306 | 1883 | 4,134 | 590,562 |
| Productive sequences number | 29271 | 740140 | 365631 | 448227 | 572690 | 632787 | 392543 | 748995 | 771280 | 478426 | 461,805 | 655,143 | 543,636 | 540,734 | 421,049 | 459,848 | 1,144,179 | 892,295 | 785,584 | 447,734 | 751,448 | 985,234 | 24,345,651 |
| Nonproductive sequences number | 234007 | 178549 | 244195 | 167332 | 158651 | 372986 | 93934 | 340113 | 173900 | 1254781 | 136,331 | 123,514 | 146,020 | 130,509 | 105,705 | 346,783 | 211,129 | 230,555 | 229,468 | 110,789 | 154,223 | 104,477 | 9,158,729 |
| In‐frame sequences number | 31239 | 791611 | 389714 | 478815 | 611479 | 674818 | 419284 | 803236 | 826442 | 1639631 | 491,438 | 700,926 | 581,739 | 579,260 | 447,370 | 493,172 | 1,214,667 | 940,753 | 837,153 | 476,306 | 801,996 | 1,026,787 | 27,088,243 |
| Out‐of_frame sequences number | 223550 | 126583 | 219578 | 132408 | 119590 | 330324 | 67051 | 285651 | 118315 | 93141 | 105,943 | 76,022 | 107,717 | 91,455 | 79,183 | 312,915 | 140,233 | 181,609 | 177,241 | 81,689 | 103,394 | 62,382 | 6,326,420 |
| Total CDR3 sequences number | 4069 | 729342 | 359173 | 437648 | 562036 | 620952 | 386428 | 737296 | 758769 | 471122 | 452,601 | 637,937 | 526,892 | 524,563 | 414,324 | 451,689 | 1,122,330 | 875,610 | 772,434 | 440,452 | 733,117 | 957,874 | 23,670,829 |
| Unique CDR3 nt sequences number | 2560 | 59322 | 22290 | 23141 | 58045 | 59010 | 47160 | 63725 | 123378 | 36185 | 27,333 | 27,072 | 117,546 | 33,206 | 87,434 | 39,496 | 52,585 | 27,685 | 57,173 | 28,979 | 127,052 | 34,112 | 1,995,707 |
| Unique CDR3 aa sequences number | 2529 | 49316 | 18368 | 20092 | 49690 | 51530 | 41236 | 55749 | 110754 | 30566 | 22,684 | 21,878 | 108,153 | 27,439 | 80,135 | 33,719 | 42,691 | 21,744 | 48,099 | 23,821 | 115,038 | 27,630 | 1,723,256 |
| Highly expressing clone number all | 14 | 0 | 7 | 47 | 0 | 5 | 3 | 10 | 4 | 14 | 15 | 10 | 4 | 12 | 2 | 10 | 7 | 16 | 3 | 12 | 3 | 10 | 655 |
| Highly expressing clone ratio all | 0.16515114 | 0 | 0.24276881 | 0.72208259 | 0 | 0.09839569 | 0.03277972 | 0.33245806 | 0.04359034 | 0.21227835 | 0.24436535 | 0.49173821 | 0.07930088 | 0.31473055 | 0.01548547 | 0.24206478 | 0.41441287 | 0.64089949 | 0.10992525 | 0.21462044 | 0.06475638 | 0.61016585 | 14.5499069 |
| Shannon entropy all | 0.8638929 | 0.66876197 | 0.52750687 | 0.40509952 | 0.67976672 | 0.59714855 | 0.7164873 | 0.55144703 | 0.74832534 | 0.57724095 | 0.49459581 | 0.37677137 | 0.78381492 | 0.52494136 | 0.79603452 | 0.58333764 | 0.44022788 | 0.34205038 | 0.5889533 | 0.56018664 | 0.75464691 | 0.33202264 | 24.7484339 |
3.2. Diversity and stability of repertoire in different groups
The distribution characteristics of the sequences and clone expansion were analyzed firstly. In the current study, the expression level of certain CDR3 clones higher than 0.5% of total clones was defined as high expansion clones (HECs). In tuberculosis patients, the HEC number was higher in the tissue group than that in the CD8 group or the CD4 group, while the comparison between the CD4 and CD8 groups showed no statistical difference. In the control groups, the HEC number in the CD8 group was significantly higher than that of the CD4 group (Figure 1a). In the comparison of HEC ratio, tuberculosis patients’ tissue group showed higher ratio than CD8 or CD4 groups, and that of CD8 group was higher than in CD4 group. In consistent with HEC number, the HEC ratio of CD8 group was higher than in CD4 group in control group (Figure 1b). The Shannon entropy measures multiplex of the immune system. It ranges from 0 to 1, “1” represents the most diversity and “0” represents the least diversity of immune system. In tuberculosis patients, Shannon entropy in CD4 was higher than CD8 or tissues, while tissue group showed the lowest Shannon entropy, although entropy of CD8 was not statistically higher than tissue group. In controls, Shannon entropy in CD4 was also higher than that of CD8 group (Figure 1c).
Figure 1.

CDR3 clones in patients and controls. (a) HEC number comparison of patients and controls. MCD4: CD4+ cells of tuberculosis patients, MCD8: CD8+ cells of patients, MTis: tissue samples of patients. NCD4: CD4+ cells of controls, NCD8: CD8+ cells of controls. (b) HEC ratio analysis of patients and controls. (c) Shannon entropy of patients and controls
The Gini coefficient was then calculated to further understand the stability of tuberculosis patients’ immune system. In patients, Gini coefficient in CD8 was higher than in CD4 group, while other comparisons showed no significant change. In controls, Gini coefficient in CD8 group was higher than that of CD4 group (Figure 2).
Figure 2.
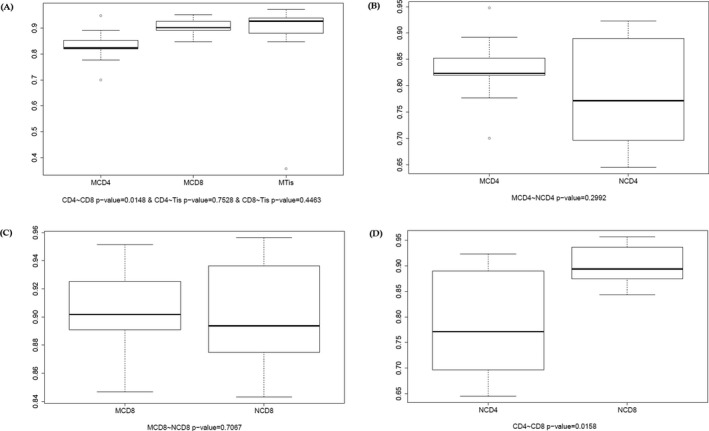
Gini coefficient of patients and controls. (a) Comparison of CD4+ cell group, CD8+ cell group and tissue group of patients. (b) Comparison of CD4+ cell group in patients and CD4+ cell group in controls. (c) Comparison of CD8+ cell in patient group and CD8+ group in control. (d) Comparison of control CD8+ cell group and CD4+ cell group
3.3. CDR3 length distribution mode analysis
In addition, we made further analysis of CDR3 length distribution in all samples and the differences between groups. We first fit the Gaussian distribution of each sample and compared the R2 value between each sample and each group. The R2 value ranged from 0 to 1, suggesting the worst fitted Gaussian distribution to the best fitted distribution. According to the R2 value, the length distribution of all samples was fitted to Gaussian distribution, although no statistical significance was found for comparing between groups, as shown in Figures 3 and 4.
Figure 3.

Gaussian distribution of R 2 value in all samples. The X‐axis depicts each CDR3 length (1–30), and the Y‐axis depicts the total percentage of each length
Figure 4.
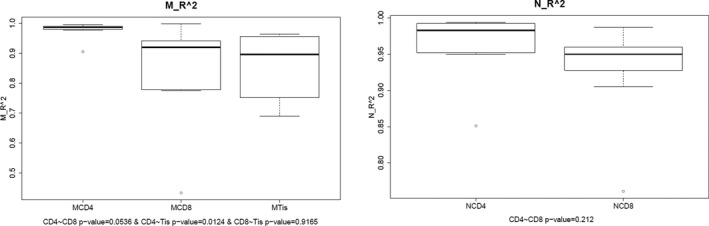
Gaussian distribution of R 2 value between groups. (a) Comparison among CD4+, CD8+ and tissue sample groups in patients. (b) Comparison among CD4+ and CD8+ cell groups in controls
The nucleotides and amino acids length of all samples were analyzed. As shown in Figures 3 and 5, the length distribution of CDR3 sequences ranged from 1–30 nucleotides and followed a Gaussian distribution. Besides, both nucleotides and amino acids length distribution of the high expression clones showed a significant difference between the control and tuberculosis patients. In both tuberculosis patients and controls, the amino acids sequence ranged from 1 to 30 amino acids and the highest percentage for both was 13 amino acid sequences. The CDR3 length of CD4, CD8 and tissue groups was analyzed, and it was observed that all presented with a similar pattern with the whole patient group. However, we found that the amino acid length of 1, 2, 5, 25, 27, 28, 29 were absent in more than seven samples in tissue group (n = 9), which is rare in CD4 and CD8 groups. In healthy controls, the amino acids sequence also ranged from 1 to 30 amino acids and the highest percentage was 13 amino acid sequences. For the distribution of CDR3 length, there were no statistically significant differences as has been found between groups. All samples showed a Gaussian distribution, the highest percentage centralized at 13 amino acids. Besides, CDR3 length of tissue group showed more skewed as length 1, 2, 5, 25, 27, 28, 29 were absent in most of tissue sample (Figure 6).
Figure 5.
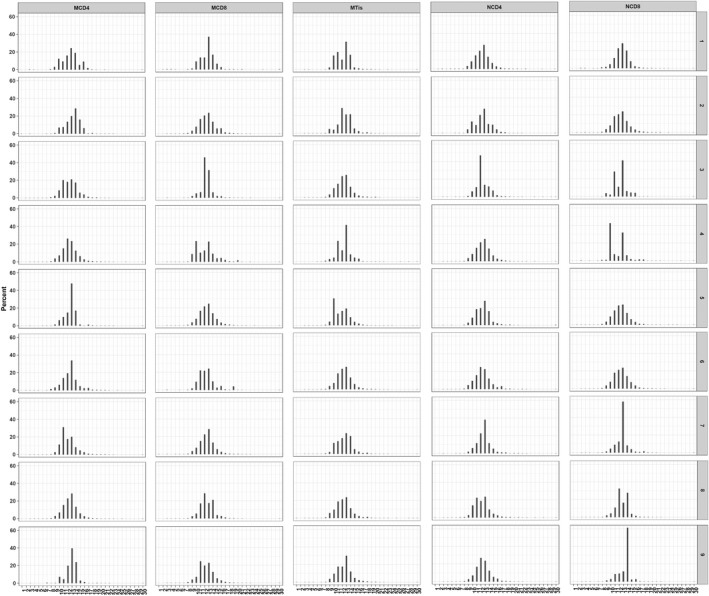
CDR3 length distribution in all samples. X‐axis represents length distribution, Y‐axis depicts the percentage of sequences of the corresponding length
Figure 6.

CDR3 expression status in all samples. X‐axis is sample ID and M1‐M9 represents tuberculosis patient number 1‐9. N1‐N9 represent control sample number 1‐9. CD4, CD8 and Tissue mean different sample types. Y‐axis is the percentage of CDR3 clones in each sample's different sequences. Red dots mean highly expressing clones (≥0.5%) and black dots represent normal expressing clones (<0.5%). (a) CDR3 clones (b) amino acid of CDR3 clones
3.4. CDR3 sequence sharing modes analysis
Different individuals sharing an identical TCR sequence corresponding to the same antigenic epitope, termed public T cell response, were observed in a variety of immune responses, including tumorigenesis, autoimmunity, and viral infections. So, we counted the public T cell clones in each group based on the nucleotide and amino acid sequences of CDR3 (Li, Ye, Ji, & Han, 2012). In order to understand the immunological reaction to the common tuberculosis pathogens, the sharing pattern of CDR3 sequence were analyzed between patients. According to the sequence data, there were 586,248 nucleotide sequences and 504,126 amino acids sequences in CD4 group of patients, and 697,706 nucleotide sequences and 618,480 amino acids sequences in CD4 group of controls. There were 253,466 nucleotide sequences and 210,566 amino acids sequences in CD8 group of patients, 349,470 nucleotide sequences and 294,076 amino acids sequences in CD8 group of control. In addition, in tissue samples of patients, we obtained 108,817 nucleotide sequences and 96,008 amino acids sequences.
To elucidate the characteristics of sharing sequences, we compared the amino acid sequences and nucleotide sequences of highly expressed clones which were expressed in more than 0.5% in either patient group or control group (Figure 7). In patient group, eight amino acid sequences and eight nucleotide sequences were shared in all samples from CD4, CD8 and tissue groups. However, 35 amino acid sequences and 33 nucleotide sequences were shared in CD4 and CD8 samples of patient, while there were 61 amino acid sequences and 61 nucleotide sequences that were shared in CD4 and CD8 samples of control. No amino acid or nucleotide sequences were shared in CD4 and CD8 in both patients and controls. All shared sequences are displayed in Table 2.
Figure 7.

Top 60 expressing clones in all samples. Red rectangle: expression percentage ≥0.5%, green rectangle: expression percentage >0, gray rectangle = 0. (a) Nucleotide sequences of top 60 expressed clones in CD4+ and CD8+ group of patients. (b) Nucleotide sequences of top 60 expressed clones in CD4+, CD8+ and tissue group of patients. (c) Amino acids sequences of top 60 expressed clones in CD4+ and CD8+ group of patients. (d) Nucleotide sequences of top 60 expressed clones in CD4+ and CD8+ group of controls.
Table 2.
Shared sequence in all patient samples, in CD4+ and CD8+ cells of patients, in CD4+ and CD8+ cells of controls
| Group | Shared sequences (NT) | Shared sequences (AA) |
|---|---|---|
| Shared sequences in all patients | GCCTGGAGCTTCGGGAGAACTGAAGCTTTC | AWSFGRTEAF |
| GCCAGCATGGGTAACACCGGGGAGCTGTTT | ASMGNTGELF | |
| GCCAGCAGTTACTCTGGGACAGGGGGCGAGCAGTAC | ASSYSGTGGEQY | |
| GCCAGCAGTGAGAGCGGGGACTCCTCCTACGAGCAGTAC | ASSESGDSSYEQY | |
| GCCTGGGTAAGGGACTACCCGTCGGACGAGCAGTAC | AWVRDYPSDEQY | |
| GCCTGGAGTCCCCGTACGAAGTTAGCTTTC | AWSPRTKLAF | |
| GCCTGGGTTAGCGGGAGCACGGACACCGGGGAGCTGTTT | AWVSGSTDTGELF | |
| GCCAGCAGTGTCGGGACTCTCATCAATGAGCAGTTC | ASSVGTLINEQF | |
| CD4 and CD8 of patient | GCCTGGAGCTTCGGGAGAACTGAAGCTTTC | AWSFGRTEAF |
| GCCAGCATGGGTAACACCGGGGAGCTGTTT | ASMGNTGELF | |
| GCCAGCAGTTACTCTGGGACAGGGGGCGAGCAGTAC | ASSYSGTGGEQY | |
| GCCAGCAGAGATATTGACAGGGAAGACAATGAGCAGTTC | ASRDIDREDNEQF | |
| GCCAGCAGTGAGAGCGGGGACTCCTCCTACGAGCAGTAC | ASSESGDSSYEQY | |
| GCCTGGAGTGATGTGGGGGAGACCCAGTAC | AWSDVGETQY | |
| GCCAGCAGCCAAGAGGGTAGCGGGAGTCAGGAGACCCAGTAC | ASSQEGSGSQETQY | |
| GCCAGCAGTTACTCGGACAGGAGCTCCTACGAGCAGTAC | ASSYSDRSSYEQY | |
| GCCAGCAGATTTGACAGGGACCATTCACCCCTCCAC | ASRFDRDHSPLH | |
| GCCAGCAGTTACAGGCCGAACACCGGGGAGCTGTTT | ASSYRPNTGELF | |
| GCCAGCAGTTGGGGGGAGACCCAGTAC | ASSWGETQY | |
| GCCTGGGTTAGCGGGAGCACGGACACCGGGGAGCTGTTT | AWVSGSTDTGELF | |
| GCCAGCAGTTACAGGTCAGGATCCTACGAGCAGTAC | ASSYRSGSYEQY | |
| GCCAGCGCAACCGGGACAGGGGTTCAAGAGACCCAGTAC | ASATGTGVQETQY | |
| GCCTGGGTAAGGGACTACCCGTCGGACGAGCAGTAC | AWVRDYPSDEQY | |
| GCCTGGAGAACTGGGAACTATGGCTACACC | AWRTGNYGYT | |
| GCCAGCAGGCCAAGGGGCGGGGGAGGTTTCGGGGAGCTGTTT | ASRPRGGGGFGELF | |
| GCCTGGAGTGGCAGGGTCTTGTGGGACACCGGGGAGCTGTTT | ASSSEQAVREKLF | |
| GCCAGCAGCTCGGAACAGGCAGTACGGGAAAAACTGTTT | AWSGRVLWDTGELF | |
| GCCAGCAGTTTATCGTGGGGAGAGACCCAGTAC | ASSLSWGETQY | |
| GCCAGCAGTGTGGGGAGGAACACTGAAGCTTTC | ASSVGRNTEAF | |
| GCCAGCAGCTTGGAGCAGACGGCACGCAGCAATGAGCAGTTC | ASSLGWGTGSTNEKLF | |
| GCCTGGAGTTTGGGGGTGTCTAACTATGGCTACACC | ASSLEQTARSNEQF | |
| GCCTGGAGCGGACAGGGCTACGAGCAGTAC | AWSLGVSNYGYT | |
| GCCAGCAGGCGGGGGTGGGACACCGGGGAGCTGTTT | AWSGQGYEQY | |
| GCCTGGAACCGGGACACTGAAGCTTTC | ASRRGWDTGELF | |
| GCCTGGAGTCCCCGTACGAAGTTAGCTTTC | AWNRDTEAF | |
| GCCAGCAGCTTAGAGGGGGGGAGTTTTAATCATGAAAAACTGTTT | AWSPRTKLAF | |
| GCCTGGACTGAGTTCGGAGGCACCGGGGAGCTGTTT | ASSLEGGSFNHEKLF | |
| GCCAGCAGTTTACGTCCCGTTGAGGTCAATGAGCAGTTC | AWTEFGGTGELF | |
| GCCTGGAGTCTCCGGACAGGGTTCTGGGGGCAGGGCGCGGGTACGGGAGAGACCCAGTAC | ASSLEQGARSDEQF | |
| GCCAGCAGTTACGGGCGGGAGAAGTCCGGGGAGCTGTTT | ASSLRPVEVNEQF | |
| GCCAGCAGTTTCTCTCTAAACACCGGGGAGCTGTTT | AWSLRTGFWGQGAGTGETQY | |
| ASSYGREKSGELF | ||
| ASSFSLNTGELF | ||
| CD4 and CD8 of controls | GCCTGGAGTGTAGGGACAGGGGACTCCTACGAGCAGTAC | AWSVGTGDSYEQY |
| AGTGCTAGAGATGCGAGGGTTCAAGACACCGGGGAGCTGTTT | SARDARVQDTGELF | |
| GCCTGGAGTCGAGTGATGAACACTGAAGCTTTC | AWSRVMNTEAF | |
| GCCTGGAGGATAGGGAGTCTAGTAGGCGAGCAGTAC | AWRIGSLVGEQY | |
| AGTGCTCATGCCGGACAGGGAGAAACAGATACGCAGTAT | SAHAGQGETDTQY | |
| AGTGCTAGACCTTACGACAGGGGGACCACCGGGGAGCTGTTT | SARPYDRGTTGELF | |
| AGTGCTAGTCGGGATTGGGACGATACTAATGAAAAACTGTTT | SASRDWDDTNEKLF | |
| GCCAGCAGCTTGGAACAGGGGGCTCGCACAGATACGCAGTAT | ASSLEQGARTDTQY | |
| GCCTGGAGGGGAAAGGGTTTCACTGAAGCTTTC | AWSVLAGETGELF | |
| GCCTGGAGCGTCCTAGCGGGAGAAACCGGGGAGCTGTTT | AWRGKGFTEAF | |
| GCCAGCAGTTACAGGGGCCTATTGACTGAAGCTTTC | ASSYRGLLTEAF | |
| GCCAGCACTCCGGACAGGGGACGAGGCTACACC | ASTPDRGRGYT | |
| GCCAGCAGTTTAAAGGGACCTTACACTGAAGCTTTC | ASSLKGPYTEAF | |
| GCCAGCAGTTACTCAGGCGAGCGGCCCTACAATGAGCAGTTC | ASSYSGERPYNEQF | |
| GCCTGGAGTGTGACAGCCTACGAGCAGTAC | AWSVTAYEQY | |
| GCCTGGAGTGTACAGGTCGGGTTTGGAGAGACCCAGTAC | AWSVQVGFGETQY | |
| GCCAGCAGTTACAAGGGAAACAACTATGGCTACACC | ASSYKGNNYGYT | |
| GCCTGGGGGGCGGAGACCTACGAGCAGTAC | AWGAETYEQY | |
| GCTAGTGCCACAGACTCCTACAATGAGCAGTTC | ASATDSYNEQF | |
| GCCAGCAGTTACGGGGCTACTGAAGCTTTC | ASSYGATEAF | |
| GCCAGCAGTTTAGCACCCACCTCCTACAATGAGCAGTTC | ASSLAPTSYNEQF | |
| GCCAGCAGTTGGAGGGTAAGGAAGCCTGGAAACACCATATAT | ASSWRVRKPGNTIY | |
| GCCAGCAGCTTGGTGATCGGGGATCGCCCCTACGAGCAGTAC | ASSLVIGDRPYEQY | |
| GCCAGCAGCCAAGGAAGACAGGGCTACGAGCAGTAC | ASSQGRQGYEQY | |
| GCCAGCAGTTACAGTTTTAGCAATCAGCCCCAGCAT | ASSYSFSNQPQH | |
| GCCAGCAGTTCCCCCACCTACGAGCAGTAC | ASSSPTYEQY | |
| AGTGCCGAGGACAGTTCGTTGGGGAGCAATCAGCCCCAGCAT | SAEDSSLGSNQPQH | |
| GCCAGCAGTCCGACAGGGGGCGGGGAGTATGGCTACACC | ASSPTGGGEYGYT | |
| GCCTGGGAACCCCCGACTAGCGGGGGGTACGAGCAGTAC | AWEPPTSGGYEQY | |
| GCCAGTAGCGGGAGTAACACCGGGGAGCTGTTT | ASSGSNTGELF | |
| GCCTGGAGGAGTTTACGGGGCGTAAGGTCCTACGAGCAGTAC | AWRSLRGVRSYEQY | |
| GCCAGCAGGACTAGCTCCACGAACACCGGGGAGCTGTTT | ASRTSSTNTGELF | |
| GCCAGCAGTTTAGATGAGGGGGGGCCGAACACTGAAGCTTTC | ASSLDEGGPNTEAF | |
| GCCTGGAGCTCGGGAGGCCTCAATCAGCCCCAGCAT | AWSSGGLNQPQH | |
| GCCAGCGGGAGGACGGGACAGGGCTACGAGCAGTAC | ASGRTGQGYEQY | |
| GCCTGGACCGAGGGACCGAACACCGGGGAGCTGTTT | AWTEGPNTGELF | |
| AGTGCTAGAGTTTCTTCGGGTGGAGGGATGAACACTGAAGCTTTC | SARVSSGGGMNTEAF | |
| GCCAGCAGTTCTGGGACTAGCAGTTACAATGAGCAGTTC | ASSSGTSSYNEQF | |
| GCCTGGAGACAGGCGAACACTGAAGCTTTC | ASSLGTDRSTEAF | |
| GCCAGCAGCTTGGGGACAGATCGCTCGACTGAAGCTTTC | AWRQANTEAF | |
| GCCAGCAGCCTCGTGGGCGACAGACACTACTCTGGAAACACCATATAT | ASSLVGDRHYSGNTIY | |
| GCCAGCAGCTCGCCCTACAGGGGCGGCCACTCTGGGGCCAACGTCCTGACT | ASSSPYRGGHSGANVLT | |
| GCCAGCAGTGCAGGGTCCACTGAAGCTTTC | ASSAGSTEAF | |
| GCCTGGAGTGTTGGAGGGAGGTTTGAAGATACGCAGTAT | AWSVGGRFEDTQY | |
| GCCAGCAGCCAAGATCTCGGGGTTTCGTCAGGGGGAGGGGTGGGGGAGCAGTAC | ASSQDLGVSSGGGVGEQY | |
| GCCTGGAGTTTAAGCGGGAGGGCCGGCGAGCAGTAC | AWSLSGRAGEQY | |
| GCCTGGAGTGTACCAGGGGAGGACACCGGGGAGCTGTTT | AWSVPGEDTGELF | |
| GCCAGCAGTTTAGTCCCAGGGGGAAATGGCTACACC | ASSLVPGGNGYT | |
| GCCTGGAGTGGTCCCCCAACTAATGAAAAACTGTTT | AWSGPPTNEKLF | |
| GCCAGCAGTTGGGGGGGGACTCCCTACGAGCAGTAC | ASSWGGTPYEQY | |
| GCCAGCAGTCGCGGGGCAGGGGGTTACAATGAGCAGTTC | ASSRGAGGYNEQF | |
| GCCTGGAGTGAGGGGGTCGGAAACACCATATAT | AWSEGVGNTIY | |
| GCCTGGAGTTCGACAGGGTCTAGCACAGATACGCAGTAT | AWSSTGSSTDTQY | |
| GCCAGCAGTTATGGACAGTTCTCCATTCAGTAC | ASSYGQFSIQY | |
| GCCAGCAGCTCCTACTTCCGACAGGGCCCCCAAGAGACCCAGTAC | ASSSYFRQGPQETQY | |
| GCCAGCAGCAGTCATGGGGGGCGAGAGGACGAGCAGTAC | ASSSHGGREDEQY | |
| GCCTGGGCCGACAGCTCTGGAAACACCATATAT | AWADSSGNTIY | |
| GCCTGGAGGGACGGGGGGCGGGACACCGGGGAGCTGTTT | AWRDGGRDTGELF | |
| GCCAGCAGCTCGGGGACAGGGAGAGGGGATGGCTACACC | ASSSGTGRGDGYT | |
| GCCAGCAGCTTGGGCCCATTAGGGGCGGCTAACTATGGCTACACC | ASSLGPLGAANYGYT | |
| GCCAGCAGCTTGGGCCTACTAGCAGATACGCAGTAT | ASSLGLLADTQY |
3.5. Significance of TRBV and TRBJ usage in patients and controls
In addition, to elucidate the potential specific immune reaction to tuberculosis, usage of TRBV and TRBJ was analyzed in all groups.
In patients, TRBV13 and TRBV6‐4 were significantly highly expressed in CD8 group than in CD4 group, on the contrary TRBV10‐3, TRBV20‐1, TRBV3‐1, TRBV5‐1, TRBV5‐4, TRBV6‐6 showed significantly lower expression in CD8 group than in CD4 group. The expression of TRBV10‐3, TRBV18, TRBV24‐1, RBV24/OR9‐2, TRBV28, TRBV3‐1, TRBV5‐5, TRBV5‐8, TRBV6‐1, TRBV6‐6, TRBV6‐7, TRBV6‐9, TRBV7‐6, TRBV7‐7 was also higher in CD4 than in tissue group. TRBV10‐3, TRBV13, TRBV27, TRBV28, TRBV6‐2, TRBV6‐3, TRBV6‐4, TRBV6‐7, TRBV6‐9 were highly expressed in CD8 samples than in tissues while TRBV20‐1, TRBV5‐1 showed lower expression in CD8 samples than in tissue samples. In controls, TRBV11‐1, TRBV14, TRBV3‐1, TRBV6‐6, TRBV7‐2 and TRBV7‐7 exhibited significant higher expression in CD4 group than in CD8 group, while TRBV27, TRBV7‐6, TRBV9 all showed lower expression in CD4 group than in CD8 group. Moreover, TRBV3‐1 and TRBV4‐1, TRBV5‐8 and TRBV7‐3 in controls’ CD4 group showed statistically lower expression than that of in patients. TRBV11‐1 and TRBV7‐2 exhibited higher expression in patients’ CD8 group than that of control's (Figure 8).
Figure 8.
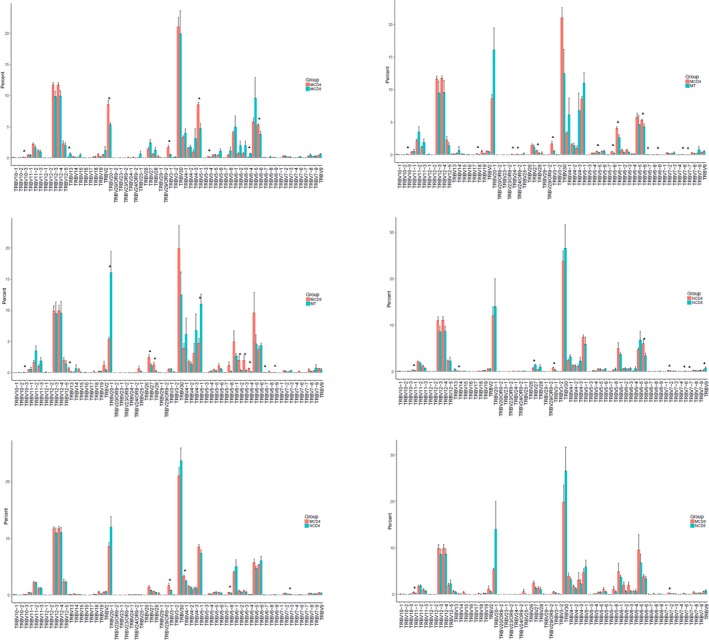
Comparison of TRBV gene usage in groups. X‐axis represents the genes of TRBV. Y‐axis represents the expressing percentage of corresponding gene: (a) CD4+ cell and CD8+ cell groups in patients. (b) CD4+ cell and tissue groups in patients. (c) CD8+ cell and tissue groups in patients. (d) CD4+ cell and CD8+ cell groups in controls. (e) CD4+ cell group of patients and CD4+ cell group of controls. (f) CD8+ cell group of patients and CD8+ cell group of controls
In patients, TRBJ1‐3, TRBJ2‐6 were significantly highly expressed in CD8 group than in CD4 group, on the contrary TRBJ1‐5 showed lower expression in CD8 group than in CD4 group. The expression of TRBJ1‐3, TRBJ1‐5, TRBJ2‐4, TRBJ2‐5 was also higher in CD4 than in tissue group, and TRBJ2‐7 exhibited lower expression in CD4 than in tissue group. TRBJ1‐2 showed lower expression and TRBJ2‐5 presented higher expression in CD8 group than in tissue group respectively. In controls, TRBJ1‐2, TRBJ1‐6, TRBJ2‐4, TRBJ2‐5 exhibited significant higher expression in CD4 group than in CD8 group. Besides, TRBJ1‐6 and TRBJ2‐7 in controls’ CD4 group showed statistical higher expression than that of patients. While TRBJ1‐3 in controls’ CD4 group showed lower expression than patients’ expression. TRBJ2‐5 exhibited higher expression in patients’ CD8 group than that of control's (Figure 9). Top 20 genes in each group are shown in Figure 10.
Figure 9.
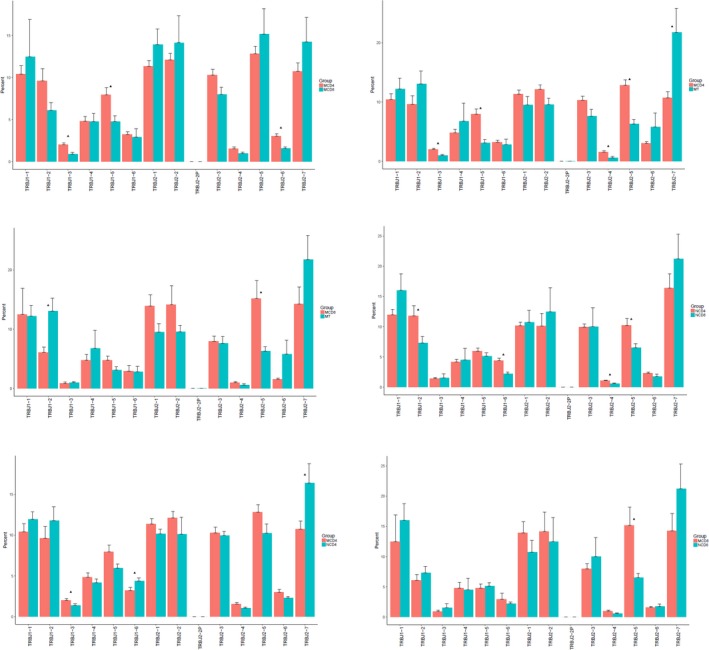
Comparison of TRBJ gene usage in groups. X‐axis represents the genes of TRBJ. Y‐axis depicts expressing percentage of corresponding gene. (a) CD4+ cell and CD8+ cell groups in patients. (b) CD4+ cell and tissue groups in patients. (c) CD8+ cell and tissue groups in patients. (d) CD4+ cell and CD8+ cell groups in controls. (e) CD4+ cell group of patients and CD4+ cell group of controls. (f) CD8+ cell group of patients and CD8+ cell group of controls
Figure 10.

Top 20 used TRBV genes in each group. X‐axis is sample ID, N1‐N9 represent control sample number 1–9. CD4, CD8 and Tissue mean different sample types. Y‐axis represents the frequencies of corresponding reads. (a) Top 20 used TRBV genes in CD4+ cell group of patients. (b) Top 20 used TRBV genes in CD8+ cell group of patients. (c) Top 20 used TRBV genes in tissue group of patients. (d) Top 20 used TRBV genes in CD4+ cell group of controls. (e) Top 20 used TRBV genes in CD8+ cell group of controls. (f) Top 20 used TRBV genes in all sample groups
Additionally, we combined the expression data of all samples on TRBV or TRBJ to understand the correlation between the expression in the samples. The heatmaps are shown in Figure 11.
Figure 11.

Heatmap of TRBV and TRBJ usage in all samples. Heatmap of gene usage in comparison of all samples for (a) TRBV (b) TRBJ
3.6. Combination of usage of TRBV and TRBJ in tuberculosis patients and control samples
TRBV/TRBJ combination was an important source of CDR3 sequence diversification. Within all TRBV/TRBJ combinations, we first counted the highly expressed which represent more than 0.5% of all combinations in each group. For CD4 group in controls, there were six TRBV/TRBJ combinations which were used more than 0.5%, and the number is 22 in CD8 group in controls. In the tuberculosis patient group, there were also six TRBV/TRBJ combinations which were used more than 0.5% in CD4 group, and 21 high expression TRBV/TRBJ in CD8 group, and 32 high expression TRBV/TRBJ in tissue group (Table 3).
Table 3.
Combination of usage of TRBV and TRBJ in tuberculosis patients and control samples
| V_type | J_type | count | individual | Percent |
|---|---|---|---|---|
| TRBV30 | TRBJ2‐7 | 243,097 | N7‐CD4 | 21.6600287 |
| TRBV20‐1 | TRBJ2‐2 | 233,941 | N7‐CD4 | 20.8442259 |
| TRBV30 | TRBJ2‐7 | 77,626 | N4‐CD4 | 17.1510889 |
| TRBV30 | TRBJ2‐5 | 51,206 | N5‐CD4 | 9.71850019 |
| TRBV6‐1 | TRBJ1‐2 | 66,181 | N8‐CD4 | 8.56785175 |
| TRBV6‐6 | TRBJ1‐2 | 49,082 | N9‐CD4 | 6.69497502 |
| TRBV30 | TRBJ2‐7 | 285,440 | N4‐CD8 | 44.7442302 |
| TRBV20‐1 | TRBJ2‐2 | 361,501 | N9‐CD8 | 37.7399324 |
| TRBV30 | TRBJ1‐1 | 164,581 | N2‐CD8 | 22.3222424 |
| TRBV20‐1 | TRBJ2‐3 | 190,384 | N7‐CD8 | 21.7430134 |
| TRBV20‐1 | TRBJ1‐4 | 178,703 | N9‐CD8 | 18.6562116 |
| TRBV30 | TRBJ1‐1 | 62,806 | N6‐CD8 | 13.9046999 |
| TRBV30 | TRBJ2‐2 | 108,417 | N7‐CD8 | 12.3818823 |
| TRBV30 | TRBJ2‐7 | 80,921 | N2‐CD8 | 10.9753749 |
| TRBV5‐1 | TRBJ2‐3 | 87,949 | N7‐CD8 | 10.044312 |
| TRBV30 | TRBJ2‐7 | 48,860 | N5‐CD8 | 9.31441981 |
| TRBV6‐5 | TRBJ1‐1 | 48,079 | N5‐CD8 | 9.16553398 |
| TRBV12‐3 | TRBJ1‐1 | 73,219 | N7‐CD8 | 8.36205617 |
| TRBV6‐5 | TRBJ2‐1 | 41,462 | N5‐CD8 | 7.90410303 |
| TRBV12‐3 | TRBJ2‐1 | 32,958 | N8‐CD8 | 7.4827677 |
| TRBV30 | TRBJ2‐7 | 63,002 | N7‐CD8 | 7.19521248 |
| TRBV30 | TRBJ1‐1 | 42,289 | N1‐CD8 | 6.81034927 |
| TRBV30 | TRBJ2‐7 | 29,944 | N6‐CD8 | 6.6293401 |
| TRBV4‐3 | TRBJ2‐7 | 36,285 | N1‐CD8 | 5.84344684 |
| TRBV12‐3 | TRBJ1‐2 | 25,505 | N8‐CD8 | 5.79064234 |
| TRBV6‐5 | TRBJ1‐1 | 25,641 | N6‐CD8 | 5.67669348 |
| TRBV12‐5 | TRBJ2‐1 | 28,982 | N5‐CD8 | 5.52497984 |
| TRBV12‐3 | TRBJ1‐1 | 24,064 | N3‐CD8 | 5.10780647 |
| TRBV30 | TRBJ1‐2 | 45,654 | M1‐CD4 | 13.9589493 |
| TRBV12‐3 | TRBJ2‐2 | 33,833 | M1‐CD4 | 10.3446167 |
| TRBV12‐3 | TRBJ1‐1 | 24,993 | M1‐CD4 | 7.64174048 |
| TRBV6‐5 | TRBJ2‐7 | 44,913 | M6‐CD4 | 5.96502506 |
| TRBV20‐1 | TRBJ2‐1 | 39,180 | M3‐CD4 | 5.80612177 |
| TRBV30 | TRBJ1‐1 | 17,997 | M1‐CD4 | 5.50267689 |
| TRBV30 | TRBJ1‐1 | 281,888 | M7‐CD8 | 40.7552844 |
| TRBV6‐5 | TRBJ2‐2 | 177,317 | M8‐CD8 | 24.3322982 |
| TRBV6‐5 | TRBJ2‐7 | 68,199 | M5‐CD8 | 21.2365324 |
| TRBV12‐3 | TRBJ2‐1 | 132,063 | M7‐CD8 | 19.0936298 |
| TRBV30 | TRBJ2‐5 | 73,658 | M6‐CD8 | 18.0772686 |
| TRBV6‐1 | TRBJ2‐7 | 55,800 | M9‐CD8 | 15.5356889 |
| TRBV4‐3 | TRBJ2‐5 | 39,717 | M3‐CD8 | 8.19887701 |
| TRBV30 | TRBJ2‐7 | 39,449 | M3‐CD8 | 8.14355312 |
| TRBV12‐3 | TRBJ1‐6 | 38,230 | M3‐CD8 | 7.89191198 |
| TRBV6‐2 | TRBJ2‐7 | 31,203 | M2‐CD8 | 7.41900509 |
| TRBV12‐3 | TRBJ2‐5 | 43,944 | M4‐CD8 | 7.05995759 |
| TRBV6‐2 | TRBJ2‐2 | 22,112 | M5‐CD8 | 6.88547051 |
| TRBV30 | TRBJ2‐2 | 15,645 | M1‐CD8 | 6.6954542 |
| TRBV12‐3 | TRBJ2‐5 | 27,212 | M6‐CD8 | 6.67841418 |
| TRBV6‐2 | TRBJ2‐5 | 27,203 | M6‐CD8 | 6.67620539 |
| TRBV30 | TRBJ1‐4 | 15,233 | M1‐CD8 | 6.51913415 |
| TRBV12‐3 | TRBJ2‐2 | 30,744 | M3‐CD8 | 6.34655877 |
| TRBV12‐3 | TRBJ2‐2 | 38,148 | M4‐CD8 | 6.1287835 |
| TRBV5‐1 | TRBJ2‐1 | 24,851 | M2‐CD8 | 5.90871697 |
| TRBV2 | TRBJ2‐5 | 12,950 | M1‐CD8 | 5.54209855 |
| TRBV5‐8 | TRBJ1‐6 | 16,347 | M5‐CD8 | 5.09030329 |
| TRBV12‐3 | TRBJ2‐7 | 27,534 | M4‐Tissue | 32.4996164 |
| TRBV4‐3 | TRBJ1‐2 | 73,434 | M5‐Tissue | 24.0886472 |
| TRBV4‐1 | TRBJ2‐6 | 54,498 | M5‐Tissue | 17.8770473 |
| TRBV20‐1 | TRBJ1‐4 | 27,013 | M2‐Tissue | 17.5660034 |
| TRBV5‐1 | TRBJ2‐7 | 13,014 | M7‐Tissue | 17.2405114 |
| TRBV30 | TRBJ1‐1 | 70,700 | M1‐Tissue | 16.0120668 |
| TRBV4‐3 | TRBJ1‐2 | 6,465 | M6‐Tissue | 14.0107925 |
| TRBV20‐1 | TRBJ2‐6 | 9,564 | M7‐Tissue | 12.6700669 |
| TRBV20‐1 | TRBJ2‐7 | 9,212 | M7‐Tissue | 12.2037491 |
| TRBV5‐1 | TRBJ1‐2 | 39,469 | M3‐Tissue | 11.3619419 |
| TRBV6‐5 | TRBJ1‐1 | 16,242 | M2‐Tissue | 10.5618416 |
| TRBV4‐1 | TRBJ2‐6 | 4,634 | M6‐Tissue | 10.0426934 |
| TRBV5‐1 | TRBJ1‐2 | 8,463 | M4‐Tissue | 9.98925886 |
| TRBV12‐3 | TRBJ1‐1 | 38,671 | M9‐Tissue | 8.83609659 |
| TRBV30 | TRBJ2‐7 | 29,333 | M3‐Tissue | 8.44409132 |
| TRBV12‐3 | TRBJ2‐2 | 22,954 | M5‐Tissue | 7.52962942 |
| TRBV30 | TRBJ2‐1 | 25,885 | M3‐Tissue | 7.45151549 |
| TRBV12‐3 | TRBJ2‐2 | 25,845 | M3‐Tissue | 7.44000069 |
| TRBV5‐1 | TRBJ2‐1 | 11,003 | M2‐Tissue | 7.15502666 |
| TRBV30 | TRBJ1‐6 | 30,986 | M1‐Tissue | 7.01767895 |
| TRBV30 | TRBJ1‐1 | 285 | M8‐Tissue | 7.00417793 |
| TRBV30 | TRBJ2‐7 | 285 | M8‐Tissue | 7.00417793 |
| TRBV12‐3 | TRBJ2‐7 | 29,615 | M9‐Tissue | 6.76685373 |
| TRBV12‐3 | TRBJ2‐7 | 19,908 | M5‐Tissue | 6.53044622 |
| TRBV12‐3 | TRBJ2‐7 | 4,895 | M7‐Tissue | 6.48473207 |
| TRBV30 | TRBJ2‐7 | 27,923 | M9‐Tissue | 6.38024166 |
| TRBV30 | TRBJ1‐4 | 26,309 | M9‐Tissue | 6.01145213 |
| TRBV5‐1 | TRBJ2‐7 | 2,719 | M6‐Tissue | 5.89255142 |
| TRBV14 | TRBJ2‐3 | 8,461 | M2‐Tissue | 5.50201587 |
| TRBV20‐1 | TRBJ1‐1 | 2,425 | M6‐Tissue | 5.25540169 |
| TRBV5‐1 | TRBJ2‐1 | 4,310 | M4‐Tissue | 5.08728651 |
| TRBV11‐3 | TRBJ2‐2 | 22,117 | M1‐Tissue | 5.00903651 |
In order to examine the potential contribution of specific TRBV/TRBJ combinations to disease progress, comparison of the relative frequencies of TRBV/TRBJ combinations between patients and controls was performed. There were 46 up‐regulated and 10 down‐regulated TRBV/TRBJ combinations as has been found after comparison between CD4 group of patients and controls. There were 12 up‐regulated combinations and 12 down‐regulated combinations as has been found after comparison between CD8 group of patients and controls (Figure 12). We then compared different TRBV/TRBJ combinations in CD4, CD8, tissue group of patients as shown in Figure 13.
Figure 12.

TRBV/TRBJ combination in groups. (a) Comparison between CD4+ cell group in patients and CD4+ cell group in controls. (b) Comparison between CD8+ cell group in patients and CD8+ cell group in controls
Figure 13.
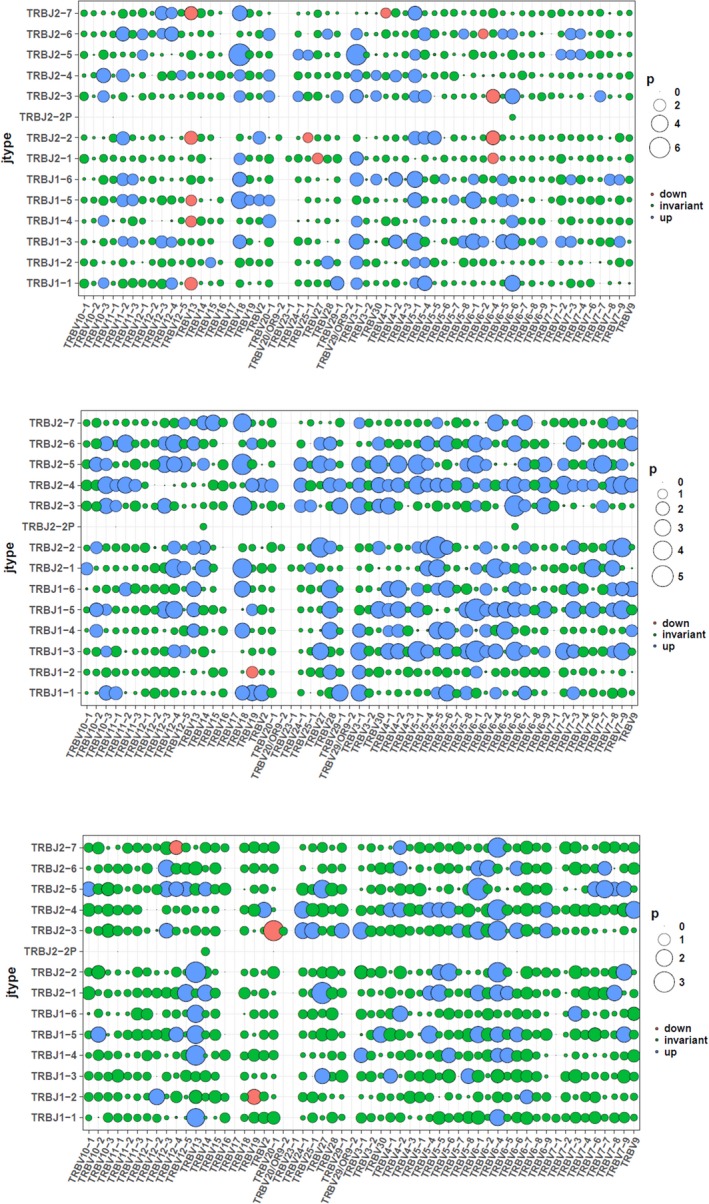
TRBV/TRBJ combination in groups. (a) Comparison between CD8+ cell group in patients and CD4+ cell group in patients. (b) Comparison between tissue group in patients and CD4+ cell group in patients. (c) Comparison between CD8+ cell group in patients and tissue group in patients
4. DISCUSSION
Tuberculosis, a well‐known infectious disease, is closely related to immune reaction in its development, diagnosis and treatment process (Janis, Kaufmann, Schwartz, & Pardoll, 1989; Cooper, 2009; Andersen et al., 2000; MacMicking, Taylor, & McKinney, 2003). The immune repertoire is characterized by a complex and dynamic organization, a highly organized, dynamic and coherent structure to assist in the understanding of the generation and selection of immune TCRs. Thus, fully measuring the diversity of the T cell repertoire, which determines the flexibility and specificity in the cellular immune response, could provide new insights into the underlying disease process (Burgos, 1996; Liu et al., 2017). In 1996, Li, et al., investigated disease‐specific change in gamma‐delta T cell repertoire of pulmonary tuberculosis patients by flow cytometric analysis of blood and bronchoalveolar lavage gamma‐delta T cells (Li et al., 1996). They demonstrated the hypothesis that gamma‐delta T cells play a role in the protective immune response to Mtb infection (Li et al., 1996). In 2018, Chaofei Cheng, et al., found that the CDR3δ tended to be more polyclonal and CDR3γ tended to be longer in TB patients; the γδ T cells expressing CDR3 sequences using a Vγ9‐JγP rearrangement expanded significantly during Mtb infection by NGS study of repertoire (Cheng et al., 2018).
However, for further understanding of immune reaction, repertoire diversity and stability within tuberculosis patients and controls still needs more comprehensive studies. Here, we present a study of enormous characterization data of tuberculosis patients and comparable controls. HEC number, HEC ratio, Shannon entropy and Gini coefficient were applied to evaluate the general characteristics of the repertoires. In both patients and normal controls, the HEC number and HEC ratio showed higher frequency in tissue samples than in CD8 or CD4 samples. Besides, HEC number and HEC ratio showed higher frequency in CD8 than CD4 samples in both patients and normal controls. This suggested that a more centralized and stronger immune reaction in tissue samples than in the CD4+ or CD8+ cell samples provides potential evidence for further elucidation for the understanding of the mechanism in depth. The Shannon entropy which was previously used as an economy parameter, was introduced in this study to illustrate the multiplex of the immune system. According to the criteria, we found that the tissue group's repertoire showed lowest complexity in patients which also suggests the strong immune reaction in tuberculosis tissue than other sample type groups.
The length distribution of CDR3 in each sample was fitted to Gaussian distribution, which provides an evenly distributed data set. Consistent with the previous study, we found that there were significant differences between patients and control groups. Since, the CDR3 sequence which was commonly expressed in patient samples could provide a solid clue for disease‐specific immune reaction research, we evaluated all amino acids shared in all samples. The CD4, CD8 groups showed similar sharing of sequences which were quite different to that of tissue groups. In the analysis of TRBV and TRBJ gene usage, we found significant difference between CD4, CD8 and tissue groups of patients. These differently expressed genes showed a disease‐specific gene expression profile which provides further information for tuberculosis and control study. To find further disease‐specific CDR3 sequences, we also investigated the TRBV/TRBJ combination in patients and controls. Within same group analysis, we found highly expressed combination sequences in each certain group. And we also found differential expression level of recombination sequences in the comparison between patients’ samples and normal control samples, which revealed the important function of TRBV/TRBJ combination in the immune of tuberculosis and provide presupposition for further study of diagnosis or treatment application.
In conclusion, our study first elucidated the immune repertoire characteristics of tuberculosis patients using NGS based methods. We found that the CDR3 sequences were extremely highly expressed in tuberculosis patients’ tissue samples than other type of samples, which suggested a specific and strong immune reaction during the development of tuberculosis. We then analyzed the CDR3 sequence sharing in all samples and each group. Later, we elucidated the TRBV, TRBJ usage and TRBV/TRBJ combination in all samples. This study provides a whole spectrum and profile of tuberculosis patients and studied the specific recombination CRD3 sequences which differ between the patients and controls. Although the sample number is relatively small, we still provide a useful resource of further study on the diagnosis, prognosis and prevention of tuberculosis by understanding candidates’ immunology repertoire features.
CONFLICT OF INTEREST
The authors have no conflict of interests to declare regarding this manuscript.
AUTHOR'S CONTRIBUTIONS
YF, BL, SL and YD designed the study and drafted the manuscript. YL, MW, YY, LX, ShL and QH acquired and interpreted the data. SL, YF and YD revised the manuscript for important content. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All patients gave written informed consent and the present study was approved by the Medical Ethics Committee of Shenzhen People's Hospital.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGMENT
Not applicable.
Fu Y, Li B, Li Y, et al. A comprehensive immune repertoire study for patients with pulmonary tuberculosis. Mol Genet Genomic Med. 2019;7:e792 10.1002/mgg3.792
Yingyun Fu and Bo Li contributed equally to this work.
Funding information
The present study was supported by funds from the Science and Technology Commission of Shenzhen (HTP study of sarcoidosis TCRß chain CDR3 immune repertoire, JCYJ20160422142121988), Science and Technology Department of Guangdong province, (Kartagener syndrome pathogenic gene diagnosis and microarray diagnosis kit, 2017A020214016s), Science and Technology Commission of Shenzhen (identification of gene mutation sites of genetic pathogenic genes in pulmonary embolism and diagnosis panel design, JCYJ20170413093032806), Key Laboratory of respiratory diseases in Shenzhen (ZDSYS201504301616234).
Data Availability Statement: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- Andersen, P. , Munk, M. E. , Pollock, J. M. , & Doherty, T. M. (2000). Specific immune‐based diagnosis of tuberculosis. Lancet, 356, 1099–1104. 10.1016/S0140-6736(00)02742-2 [DOI] [PubMed] [Google Scholar]
- Burgos, J. D. (1996). Fractal representation of the immune B cell repertoire. BioSystems, 39, 19–24. 10.1016/0303-2647(95)01574-4 [DOI] [PubMed] [Google Scholar]
- Chaudhry, S. , Cairo, C. , Venturi, V. , & Pauza, C. D. (2013). The gammadelta T‐cell receptor repertoire is reconstituted in HIV patients after prolonged antiretroviral therapy. AIDS, 27, 1557–1562. 10.1097/qad.0b013e3283611888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C. , Wang, B. , Gao, L. , Liu, J. , Chen, X. , Huang, H. , & Zhao, Z. (2018). Next generation sequencing reveals changes of the γδ T cell receptor repertoires in patients with pulmonary tuberculosis. Scientific Reports, 8, 3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A. M. (2009). Cell‐mediated immune responses in tuberculosis. Annual Review of Immunology, 27, 393–422. 10.1146/annurev.immunol.021908.132703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz, M. A. , Lee, B. W. , Dillon, B. J. , & Harth, G. (1995). Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences, 92, 1530–1534. 10.1073/pnas.92.5.1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janis, E. M. , Kaufmann, S. H. , Schwartz, R. H. , & Pardoll, D. M. (1989). Activation of gamma delta T cells in the primary immune response to Mycobacterium tuberculosis. Science, 244, 713–716. 10.1126/science.2524098 [DOI] [PubMed] [Google Scholar]
- Li, B. , Rossman, M. D. , Imir, T. , Oner‐Eyuboglu, A. F. , Lee, C. W. , Biancaniello, R. , & Carding, S. R. (1996). Disease‐specific changes in gammadelta T cell repertoire and function in patients with pulmonary tuberculosis. The Journal of Immunology, 157, 4222–4229. [PubMed] [Google Scholar]
- Li, H. , Ye, C. , Ji, G. , & Han, J. (2012). Determinants of public T cell responses. Cell Research, 22, 33–42. 10.1038/cr.2012.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenstrom, T. , Agger, E. M. , Korsholm, K. S. , Darrah, P. A. , Aagaard, C. , Seder, R. A. , … Andersen, P. (2009). Tuberculosis subunit vaccination provides long‐term protective immunity characterized by multifunctional CD4 memory T cells. The Journal of Immunology, 182, 8047–8055. 10.4049/jimmunol.0801592 [DOI] [PubMed] [Google Scholar]
- Liu, S. , Zhang, Q. , Huang, D. , Zhang, W. , Zhong, F. , Feng, J. , … Zhang, H. (2017). Comprehensive assessment of peripheral blood TCRbeta repertoire in infectious mononucleosis and chronic active EBV infection patients. Annals of Hematology, 96, 665–680. 10.1007/s00277-016-2911-8 [DOI] [PubMed] [Google Scholar]
- MacMicking, J. D. , Taylor, G. A. , & McKinney, J. D. (2003). Immune control of tuberculosis by IFN‐gamma‐inducible LRG‐47. Science, 302, 654–659. [DOI] [PubMed] [Google Scholar]
- Meintjes, G. , Rangaka, M. X. , Maartens, G. , Rebe, K. , Morroni, C. , Pepper, D. J. , … Wilkinson, R. J. (2009). Novel relationship between tuberculosis immune reconstitution inflammatory syndrome and antitubercular drug resistance. Clinical Infectious Diseases, 48, 667–676. 10.1086/596764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolich‐Žugich, J. , Slifka, M. K. , & Messaoudi, I. (2004). The many important facets of T‐cell repertoire diversity. Nature Reviews Immunology, 4, 123–132. 10.1038/nri1292 [DOI] [PubMed] [Google Scholar]
- Toivonen, R. , Arstila, T. P. , & Hänninen, A. (2015). Islet‐associated T‐cell receptor‐β CDR sequence repertoire in prediabetic NOD mice reveals antigen‐driven T‐cell expansion and shared usage of VβJβ TCR chains. Molecular Immunology, 64, 127–135. 10.1016/j.molimm.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Vanham, G. , Toossi, Z. , Hirsch, C. S. , Wallis, R. S. , Schwander, S. K. , … Ellner, J. J. (1997). Examining a paradox in the pathogenesis of human pulmonary tuberculosis: Immune activation and suppression/anergy. Tuberculosis (Edinb), 78, 145–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


