Abstract
The prevalence, clinicopathologic correlates, and outcomes of previously untreated chronic lymphocytic leukemia (CLL) patients with IGH-BCL2 and IGH-BCL3 translocations are not well known. Using the Mayo Clinic CLL database, we identified patients seen between March 1, 2002 and September 30, 2016 who had FISH testing performed within 3 years of CLL diagnosis. The prognostic profile, time to first therapy (TTT), and overall survival (OS) of patients with IGH-BCL2 and IGH-BCL3 trans-location were compared to patients without these abnormalities (non-IGH group). Of 1684 patients who met the inclusion criteria, 38 (2.2%) had IGH-BCL2, and 16 (0.9%) had IGH-BCL3 translocation at diagnosis. Patients with IGH-BCL3 translocation were more likely to have high and very-high CLL-International Prognostic Index, compared to patients with IGH-BCL2 translocation and the non-IGH group. The 5-year probability of requiring therapy was significantly higher for IGH-BCL3 compared to IGH-BCL2 and non-IGH groups (84% vs 33% vs 29%, respectively, P < 0.0001). The 5-year OS was significantly shorter for IGH-BCL3 compared to IGH-BCL2 and non-IGH groups (45% vs 89% vs 86%, respectively, P < 0.0001). On multivariable analyses, IGH-BCL3 translocation was associated with a shorter TTT (hazard ratio [HR] = 2.7; P = 0.005) and shorter OS (HR = 5.5; P < 0.0001); IGH-BCL2 translocation did not impact TTT and OS. In conclusion, approximately 3% of all newly diagnosed CLL patients have either an IGH-BCL2 or IGH-BCL3 translocation. Patients with IGH-BCL3 translocations have a distinct prognostic profile and outcome. These results support the inclusion of an IGH probe during the routine evaluation of FISH abnormalities in newly diagnosed CLL.
1 |. INTRODUCTION
Chronic lymphocytic leukemia (CLL) is a common hematological malignancy with an estimated 21 000 new cases diagnosed in the United States in 2018.1 CLL typically represents a clonal neoplasm of small, mature B cells that coexpress CD5 and CD23. CLL has a very heterogeneous clinical profile, with some patients surviving for many years without requiring treatment while others experience a more aggressive clinical course.2 The Rai and Binet clinical staging systems have traditionally been used in evaluating patients with CLL.3–5
Genetic findings play a key role in the prognostication and clinical management of patients with CLL. Since 2000, the Dohner classification has been the gold standard for the genetic characterization and assessment of prognosis of CLL patients.6,7 This hierarchical classification is based predominantly on the findings of 13q14 deletion, 17p13 deletion, 11q22 deletion, and trisomy 12 by fluorescent in situ hybridization (FISH) in these patients. In addition to these well-known genetic prognostic markers in CLL, other less common recurrent abnormalities have also been noted. These latter genomic aberrations include reciprocal translocations involving the immunoglobulin (IG) genes, particularly the immunoglobulin heavy chain (IGH) gene located at the 14q32 chromosomal locus.8–14 IGH-rearrangements typically relocate genes near active regulatory sequences on chromosome 14, leading to activation of a proto-oncogene. There are a number of IGH translocation partners that have been reported in CLL, including IGH-BCL2/t(14;18)(q32;q21), IGH-BCL3/t(14;19)(q32;q13), MYC-IGH/t (8;14)(q24;q32), and BCL11A-IGH/t(2;14)(p16;q32).11 In a report of 35 cases pooled from the published literature, Braekeleer et al reported that the overall frequency of IGH translocations in CLL was approximately 8.3% (range, 1.9% to 26.1% from 18 total studies).11 Other published studies of CLL patients with IGH-BCL2 and IGH-BCL3 translocations included small numbers of patients with relatively short follow-up times and did not report time to first therapy (TTT) and/or overall survival (OS). In addition, these studies did not compare the clinicopathologic characteristics and outcomes of patients with IGH-BCL2 and IGH-BCL3 translocations to CLL patients without these IGH abnormalities.8–10,12–14 In this study, we comprehensively characterized the clinicopathologic features and outcomes of a cohort of newly diagnosed CLL patients with IGH-BCL2 and IGH-BCL3 translocations seen at Mayo Clinic over the past 15 years, and compared these features to CLL patients without IGH abnormalities.
2 |. METHODS
2.1 |. Patient cohort
The Mayo Clinic CLL database includes patients with CLL who have been seen at Mayo Clinic since 1995 and who have granted permission for their records to be used for research purposes.15–17 Previously untreated CLL patients who were seen at Mayo Clinic between March 1, 2002 and September 30, 2016, and who had FISH done within 3 years of diagnosis were identified from the database. Patients with confirmed IGH-BCL2 or IGH-BCL3 translocations were included in this study. Patients with other IGH translocations, such as t(8;14), and those patients with an unidentified partner gene for the rearranged IGH locus were excluded from further analysis. The remaining patients constituted the non-IGH group and were included as the comparator to the IGH-BCL2 and IGH-BCL3 groups. The Dohner hierarchical classification was used to classify patients with non-IGH abnormalities into three groups: low-risk (13q deletion), intermediate risk (normal or trisomy 12), and high risk (11q or 17p deletion). Clinical data obtained from the CLL database included age, gender, Rai stage, serum β2-microglobulin level (β2M), the status of CD38, ZAP70, and CD49d (respectively considered positive when CD38 ≥ 30%, ZAP70 ≥ 20%, and CD49d ≥ 30%), immunoglobulin heavy chain gene (IGHV) mutation status (considered as unmutated when the IGHV mutations were ≤ 2%), genetic abnormalities detected by FISH, therapy administered, and vital status. All diagnostic material, including peripheral blood, bone marrow, and/or tissue biopsy for patients with IGH-BCL2 and IGH-BCL3 translocations were reviewed independently by two hematopathologists (K.K.R. and C.A.H.). Using a combination of morphologic and immunophenotypic features and based on the 2016 WHO classification,2 a diagnosis of CLL was established on all patients included in this study. This study was approved by the Mayo Clinic Institutional Review Board.
2.2 |. Immunophenotypic analysis
Immunophenotypic analysis by flow cytometry was performed on anticoagulated peripheral blood or bone marrow aspirate specimens using previously described methods.18 Samples were examined with antibodies directed against the following antigens: CD3, CD5, CD10, CD11c, CD16, CD19, CD20, CD22, CD23, CD38, CD45, CD103, and kappa and lambda immunoglobulin light chains. The data were analyzed using Kaluza software (Beckman-Coulter, Brea, CA) and/or Diva software (BD Biosciences). Aberrant antigen expression was identified based on comparison with internal positive and negative controls.
2.3 |. Fluorescence in situ hybridization (FISH) panel
FISH was performed on cell suspensions prepared from fresh peripheral blood or bone marrow aspirate using standard techniques.19 FISH analysis was performed primarily using cells harvested directly; rarely, after cell culture stimulated with CpG oligonucleotide or brief cell culture without mitogen stimulation. Two hundred interphase nuclei were examined for each enumeration probe set and 500 interphase nuclei for each dual fusion probe set. FISH for 6q deletion (D6Z1 & MYB), 11q deletion (D11Z1 & ATM), 13q deletion (D13S319 & LAMP1), and 17p deletion (TP53 & D17Z1), trisomy 12 (D12Z3 & MDM2), and IGH rearrangement (CCND1 & IGH) were performed. The level of positive detection sensitivity for the presence of a genomic abnormality was ≥4% for 6q deletion, ≥7.5% for 11q deletion, ≥7.0% for 13q deletion, ≥9.5% for 17p deletion, ≥7% for trisomy 12, and ≥ 0.6% for IGH rearrangement (regardless of partner). Cases showing a rearranged IGH pattern without CCND1 fusion were reflexed to IGH-BCL2 and IGH-BCL3 dual fusion probe sets.
2.4 |. Statistical analysis
Clinical variables were compared across FISH groups using chi-square or Fisher’s Exact (qualitative) and Kruskal-Wallis (quantitative) tests. TTT was measured as time from diagnosis to first treatment date or last known untreated date; TTT was analyzed using the Gray K-sample test accounting for competing risk of death. OS was calculated as time from diagnosis to last known alive date; OS was analyzed using the log-rank test. OS and TTT were compared between the IGH-BCL2, IGH-BCL3, and non-IGH risk groups. Cox regression models were used for univariable and multivariable analyses of TT and OS (models included age, sex, Rai stage, and FISH status); hazard ratios (HR) and 95% confidence intervals (CI) were reported. P ≤ 0.05 were considered statistically significant. Statistical analyses were performed using SAS version 9.4 (SAS Institute, Cary, North Carolina, USA).
3 |. RESULTS
3.1 |. Clinical characteristics
In total, 1684 CLL patients met the inclusion criteria for the study. The median age at diagnosis was 64 years (range: 24–93 years), and 1106 patients (66%) were male. Of 1684 patients, 38 (2.2%) harbored an IGH-BCL2 translocation, 16 (0.9%) had an IGH-BCL3 translocation, and the remaining 1630 (96.7%) did not have either of these abnormalities (non-IGH group). The non-IGH group included 730 low-risk (sole 13q deletion), 692 intermediate-risk (trisomy 12 and negative FISH), and 208 high-risk (11q deletion and 17p deletion) patients according to Dohner classification. Baseline characteristics of all patients are shown in Table 1. There were no significant differences at baseline among patients with IGH-BCL2 compared to the non-IGH group. In contrast, patients with IGH-BCL3 were more likely to have a higher Rai Stage (III or IV, 23% vs 5%; P = 0.02), higher expression of CD38 (100% vs 25%; P < 0.0001) and CD49d (100% vs 33%; P = 0.001), unmutated IGHV genes (100% vs 42%; P = 0.0001), higher serum β2-microglobulin (4.3 vs 2.4 μg/mL; P = 0.002), and a high or very high CLL-IPI score (67% vs 22%; P = 0.002) compared to the non-IGH group.
TABLE 1.
Baseline characteristics of CLL patients with IGH-BCL2, IGH-BCL3, and the non-IGH group at diagnosis
| IGH-BCL2 (n = 38) | IGH-BCL3 (n = 16) | Non-IGH (n = 1630) |
P-value IGH-BCL2 vs non-IGH |
P-value IGH-BCL3 vs non-IGH |
|
|---|---|---|---|---|---|
| Age (median [range], years) | 66 [46–86] | 60 [36–77] | 64 [24–93] | 0.24 | 0.10 |
| Gender (%male) | 26 (68%) | 8 (50%) | 1072 (66%) | 0.73 | 0.19 |
| Rai stage | 0.33 | 0.02 | |||
| 0 | 23 (61%) | 5 (39%) | 1083 (67%) | ||
| I or II | 11 (29%) | 5 (39%) | 457 (28%) | ||
| III or IV | 4(11%) | 3 (23%) | 84 (5%) | ||
| Data not available | 0 | 3 | 6 | ||
| CD38 (%positive; ≥30%) | 7 (20%) | 11 (100%) | 391 (25%) | 0.54 | <0.0001 |
| Data not available | 3 | 5 | 35 | ||
| ZAP70 (%positive; ≥20%) | 7 (24%) | 5 (63%) | 499 (34%) | 0.26 | 0.13 |
| Data not available | 9 | 8 | 171 | ||
| IGHV (%unmutated; ≤2%) | 8 (30%) | 11 (100%) | 563 (42%) | 0.19 | 0.0001 |
| Data not available | 11 | 5 | 293 | ||
| CD49d (%positive; ≥30%) | 12 (43%) | 6 (100%) | 446 (33%) | 0.25 | 0.001 |
| Data not available | 10 | 10 | 258 | ||
| β2M (median [range], ug/mL) | 2.5 [1.5–8.4] | 4.3 [2.0–7.8] | 2.4 [0.2–32.4] | 0.71 | 0.002 |
| Data not available | 6 | 6 | 145 | ||
| CLL-IPI risk category | 0.63 | 0.002 | |||
| Low | 14 (54%) | 0 (0%) | 603 (46%) | ||
| Intermediate | 6 (23%) | 3 (33%) | 429 (33%) | ||
| High | 6 (23%) | 5 (56%) | 247 (19%) | ||
| Very high | 0 (0%) | 1 (11%) | 42 (3%) | ||
| Data not available | 12 | 7 | 309 | ||
| Dohner FISH risk categorya | 0.09 | 0.002 | |||
| Low | 16 (42%) | 1(6%) | 730 (45%) | ||
| Intermediate | 21 (55%) | 10 (63%) | 692 (43%) | ||
| High | 1 (3%) | 5 (31%) | 208 (13%) | ||
| Any FISH result | |||||
| 13q deletion | 19 (50%) | 2 (13%) | 914 (56%) | 0.67 | 0.005 |
| Trisomy 12 | 11 (29%) | 11 (69%) | 282 (17%) | 0.02 | <0.0001 |
| 11q deletion | 0 (0%) | 2 (13%) | 156 (10%) | 0.04 | 0.66 |
| 17p deletion | 1 (3%) | 3 (19%) | 61 (4%) | >0.99 | 0.02 |
CLL-IPI, chronic lymphocytic leukemia-international prognostic index; IGHV, immunoglobulin heavy chain variable region; β2M, β2-microglobulin.
Dohner FISH risk category: Low = 13q deletion; Intermediate = normal or trisomy 12; High = 11q deletion or 17p deletion.
3.2 |. Concomitant FISH abnormalities
Patients with IGH translocations had other concomitant FISH abnormalities as shown in Table 1. In our cohort, 50% (19/38) of patients with IGH-BCL2 had concurrent 13q deletion and 29% (11/38) had a concurrent trisomy 12. In the IGH-BCL3 group, 69% (11/16) of patients had coincident trisomy 12, and 19% (3/16) had coincident 17p deletion. The detailed FISH findings for the 38 IGH-BCL2 and 16 IGH-BCL3 cases are presented in Supporting Information Tables S1 and S2.
3.3 |. Morphologic and immunophenotypic findings
In patients with IGH-BCL2 translocation, the overwhelming majority (95%, 36/38) presented with typical CLL morphologic characteristics. These characteristics included small-sized lymphocytes, with clumped chromatin, inconspicuous nucleoli, and scant cytoplasm. These typical cases expressed positive CD5, positive CD23, dim CD20, and dim surface immunoglobulin expression. The two cases with atypical morphology showed predominantly small-sized neoplastic lymphocytes with scant cytoplasm and occasional intermediate size cells with eccentric nuclear localization. The concomitant FISH abnormality for these two patients was trisomy 12 and 13q deletion, respectively.
Similar to other reports in the literature, a subset of our patients with IGH-BCL3 translocation exhibited some atypical morphologic and/or immunophenotypic features. These included larger neoplastic cells, nuclear indentations, modest to abundant basophilic cytoplasm, and/or plasmacytoid appearance seen in 71% (10/14; 2 cases without slides for re-review) of cases (Figure 1; Table 2), and negative CD23, brighter CD20, and/or brighter surface immunoglobulin expression seen in 31% (5/16; Table 2). Definitive exclusion of an alternative malignant lymphoma diagnosis (especially mantle cell lymphoma or marginal zone lymphoma) was made by combining morphologic and immunophenotypic pattern of bone marrow findings in all patients (lymph node biopsy samples were also reviewed in 5 patients and a spleen specimen in 1 patient) as well as confirming the absence of t(11;14) by FISH.
FIGURE 1.
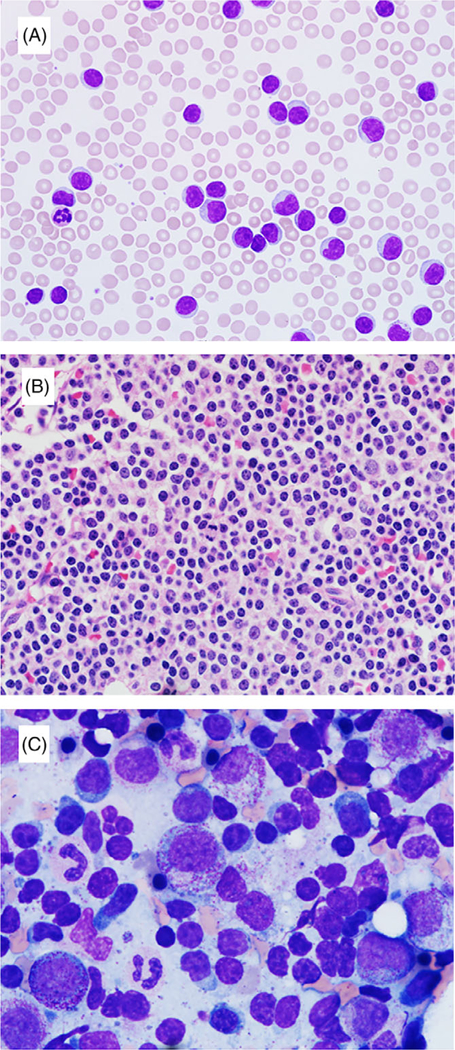
Morphologic findings in CLL patients with IGH-BCL3 translocation: A peripheral blood (A; 600×) and a core biopsy (B; 600×) from a same case show mostly small-sized lymphocytes with nuclear irregularity, modest cytoplasm, and some with single, small nucleolus. A bone marrow aspirate smear (C; 1000×) from another case shows a subset of cells with slightly irregular nuclear contours, moderate basophilic cytoplasm, and plasmacytoid appearance
TABLE 2.
Morphologic and immunophenotypic features in patients with IGH-BCL3 translocation (n = 16)
| ID | Cytomorphology | Immunophenotype |
|---|---|---|
| 1 | No slides for re-review | Atypical: CD5+, CD19+, CD20+, CD23– |
| 2 | Atypical; occasional atypical cells with nuclear indentations and modest to abundant cytoplasm | CD5+, CD19+, CD20+(dim), CD23+ |
| 3 | Typical; rare larger cells | CD5+, CD19+, CD20+, CD23+(partial) |
| 4 | Atypical; predominance of larger cells with basophilic cytoplasm | CD5+, CD19+, CD20+, CD23+(partial) |
| 5 | Atypical; cells with more abundant cytoplasm and occasional single nucleolus | CD5+, CD19+, CD20+(dim), CD23+(partial) |
| 6 | Atypical; occasional larger cells with slightly more basophilic cytoplasm and single small nucleolus | CD5+, CD19+, CD20+, CD23+ |
| 7 | Atypical; cells with moderately abundant pale cytoplasm | CD5+, CD19+, CD20+(moderate), CD23+ |
| 8 | Atypical; occasional larger cells with deeply basophilic cytoplasm | Atypical: CD5+, CD19+, CD20+, CD23– |
| 9 | Typical; rare larger cells | Atypical: CD5+, CD19+, CD20+(bright), CD23+(partial) |
| 10 | Atypical; subset of cells with irregular nuclear contours, moderate basophilic cytoplasm, plasmacytoid appearance, and scattered larger cells | CD5+, CD19+, CD20–, CD23+ |
| 11 | No slides for re-review | CD5+(dim), CD19+, CD20+, CD23+ |
| 12 | Atypical; subset of cells with irregular nuclear contours, modest cytoplasm, and some with single, small nucleolus | CD5+, CD19+, CD20+(dim), CD23+ |
| 13 | Typical | Atypical: CD5+(dim), CD19+, CD20+, CD23– |
| 14 | Atypical; population of intermediate and rare larger cells with irregular nuclear contours and relatively abundant cytoplasm | CD5+(partial), CD19+, CD20+, CD23+(partial) |
| 15 | Atypical; small cells with irregular nuclear contours and moderate pale cytoplasm | Atypical: CD5+(partial), CD19+, CD20+(dim), CD23– |
| 16 | Typical | CD5+, CD19+, CD20+, CD23+ |
3.4 |. Outcomes
The median follow-up for all patients was 5.9 years. Among the 38 patients with IGH-BCL2 translocation, 25 patients had not required treatment with current follow-up, 8 had received chemoimmunotherapy, 4 had been treated with anti-CD20 monoclonal antibody alone, and 1 patient received zanubrutinib (a novel Bruton tyrosine kinase inhibitor). Among the 16 patients with IGH-BCL3 translocation, 3 patients had not required treatment with current follow-up, 10 had received chemoimmunotherapy, 1 patient had received a combination of an alkylating agent and anti-CD20 monoclonal antibody, 1 patient had received ibrutinib, and 1 patient had been treated with anthracycline-based therapy.
Figure 2A shows the TTT among patients in the IGH-BCL2, IGH-BCL3, and non-IGH groups (5-year risk of therapy was 33%, 84%, and 29%, respectively) (P < 0.0001). Figure 2B shows the TTT for patients in IGH-BCL2, IGH-BCL3, and the non-IGH cohort categorized into low-risk, intermediate-risk, and high-risk FISH groups according to the Dohner hierarchical risk categorization (P < 0.0001). TTT was similar between patients with IGH-BCL2 translocation and the low risk FISH non-IGH group (P = 0.15; Figure 2C). In contrast, the patients with IGH-BCL3 translocation showed a shorter TTT than patients in the high-risk FISH non-IGH group (median 1.0 vs 5.3 year, P = 0.007; Figure 2D). The TTT was also significantly shorter in the IGH-BCL3 group compared to the IGH-BCL2 group (P = 0.002; Figure 2E).
FIGURE 2.
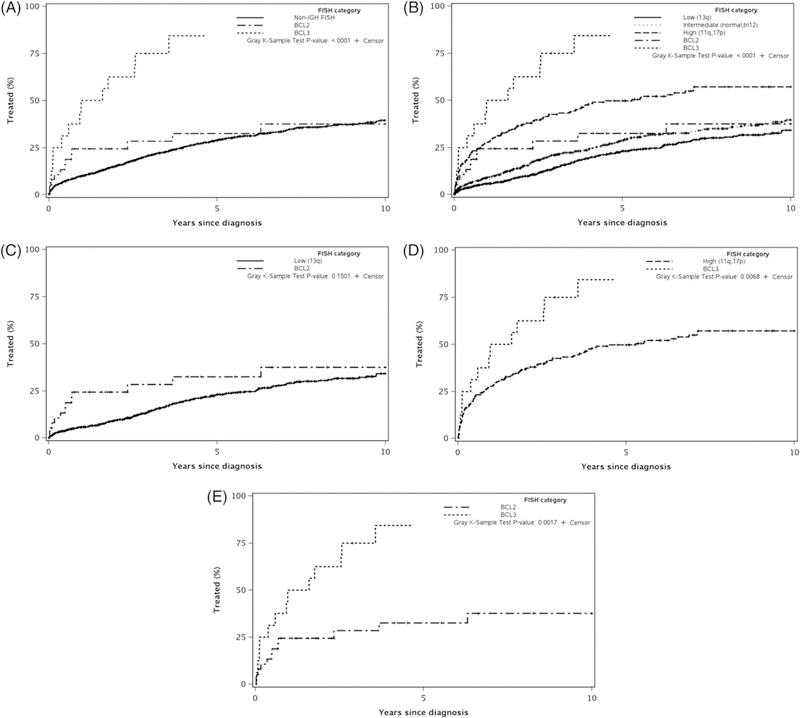
Time to first treatment in the IGH-BCL2, IGH-BCL3, and the non-IGH groups
The 5-year OS for all patients in our study was 85% (95% CI: 83%−87%). The 5-year OS for the IGH-BCL2, IGH-BCL3, and non-IGH groups was 89%, 45%, and 86%, respectively (P < 0.0001; Figure 3A). Figure 3B shows the OS for the IGH-BCL2 and IGH-BCL3 compared to the non-IGH groups categorized according to the Dohner hierarchical risk into low, intermediate, and high-risk FISH (P < 0.0001; Figure 3B). There was no difference in the OS of patients in the IGH-BCL2 group compared to the Dohner low risk non-IGH group (P = 0.69; Figure 3C). In contrast, the OS of patients in the IGH-BCL3 group was significantly shorter compared to the Dohner high risk non-IGH group (P = 0.01; Figure 3D). Finally, the OS of patients in the IGH-BCL3 was significantly shorter than those in the IGH-BCL2 group (P < 0.0001; Figure 3E).
FIGURE 3.
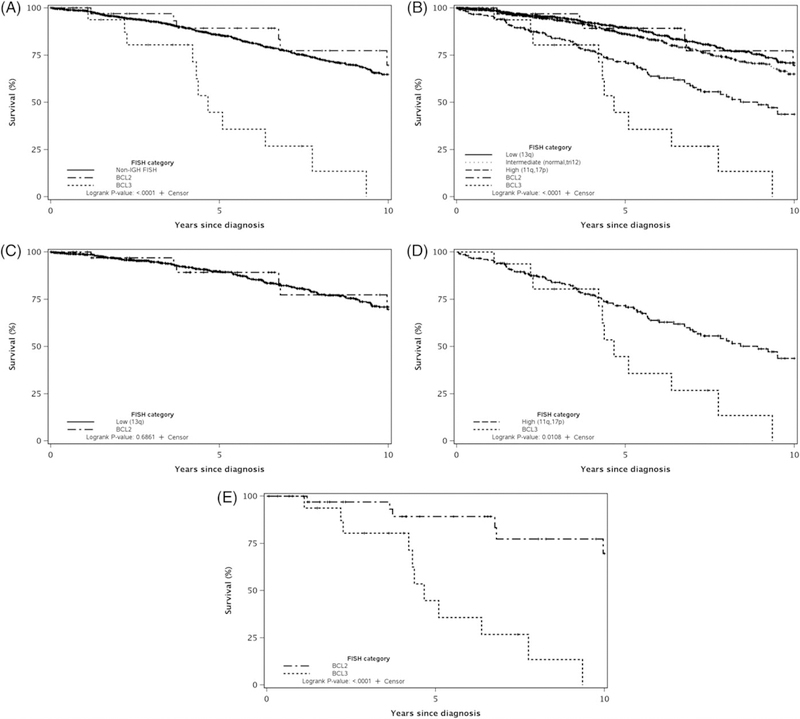
Overall survival in the IGH-BCL2, IGH-BCL3, and the non-IGH groups
In univariable analyses, the presence of IGH-BCL2 was not associated with TTT or OS. In univariable analyses, the presence of IGH-BCL3 was associated with both shorter TTT (HR = 6.3; 95% CI = 3.6–11.0; P < 0.0001) and shorter OS (HR = 4.8; 95% CI = 2.6–8.7; P < 0.0001). In multivariable analyses (including age, gender, Rai stage, and FISH), IGH-BCL3 remained statistically significantly associated with both shorter TTT (HR = 2.7; 95% CI = 1.3–5.4; P = 0.005) and shorter OS (HR = 5.5; 95% CI = 2.6–11.6; P < 0.0001). IGHV mutation status was not included in the multivariable model due to multicollinearity with IGH-BCL3 translocations as all patients with IGH-BCL3 translocations had unmutated IGHV genes.
4 |. DISCUSSION
The results in the present study including >1600 patients show that approximately 3% of newly diagnosed CLL patients have either an IGH-BCL2 or IGH-BCL3 translocation at the time of diagnosis. Although the prognostic profile of patients with IGH-BCL2 does not appear to differ substantially from non-IGH CLL patients, patients with an IGH-BCL3 translocation have a more aggressive clinical presentation at baseline, including a high and very-high CLL-IPI score. Consistent with this adverse prognostic profile, patients with an IGH-BCL3 translocation have a shorter TTT and OS compared to patients with IGH-BCL2 translocation and those without IGH abnormalities, even after adjusting for other variables known to be associated with adverse outcomes.
In a prior study from our group, of 1032 patients with a B-cell lymphoproliferative disorder evaluated in the Mayo Clinic Cytogenetic laboratory, 7% had an IGH translocation identified by interphase FISH.20 The partner gene present in 45 patients was cyclin D1, consistent with a diagnosis of mantle cell lymphoma. The partner gene present in the remaining patients was BCL2 in 24%, BCL3 in 8%, MYC and BCL-11a in <3% patients, and unknown in the remaining 17%. The results of our current study extend our understanding of the prevalence of these abnormalities in a large cohort of newly diagnosed CLL patients evaluated at Mayo Clinic where a FISH panel was consistently obtained at the time of diagnosis. At our institution, the standard CLL FISH panel includes probes to detect 6q23 deletion, 13q14 deletion, 17p13 deletion, 11q22 deletion, trisomy 12, and IGH-CCND1. The inclusion of the IGH-CCND1 probe is essential in ruling out mantle cell lymphoma, which has a very different therapeutic approach. Cases showing a rearranged IGH pattern without CCND1 fusion are reflexed to look for additional IGH abnormalities, including IGH-BCL2 and IGH-BCL3 dual fusion probe sets. In some laboratories, it may not be routine to include an IGH probe in the CLL FISH prognostic panel. The results of this study suggest that it may be important to employ an IGH probe in the FISH panel test at diagnosis for all CLL patients.
Prior studies of IGH abnormalities in CLL have reported divergent results, with some suggesting that the presence of IGH translocations is associated with similar or better outcomes,11,21–23 whereas others have reported that they portend inferior outcomes.24 This is likely due to the fact that these studies had a small sample size, included patients with mantle cell lymphoma, and/or combined patients with IGH-BCL2 and IGH-BCL3 together into one analyses. Results from our study show that patients with IGH-BCL2 translocations have comparable outcomes to low-risk FISH (13q deletion as the sole abnormality) CLL patients, with TTT and OS being similar in these two groups of patients. Other prognostic factors in patients with IGH-BCL2 are also very favorable (70% have mutated IGHV genes, 77% have either low or intermediate-risk CLL-IPI score, and only 3% have high-risk FISH (1 patient with concomitant 17p deletion). In contrast, the median TTT in patients with IGH-BCL3 translocation was 18 months (and >75% of these patients were treated within 3 years of diagnosis), suggesting that these patients had biologically aggressive disease at presentation. To further determine if this was associated with other adverse features at the time of diagnosis (such as 17p or 11q deletion), we conducted a multivariable analyses of other known adverse prognostic factors in CLL (that included age, sex, Rai stage, and CLL FISH) and found that the presence of IGH-BCL3 was independently associated with shorter TTT and OS.
IGH-BCL3 translocation can appear at diagnosis in many B-lymphoproliferative disorders, similar to what we observed in our CLL case series. This translocation is associated with BCL3 overexpression, which activates an inhibitor of kappa B (IκB)-like protein, contributes to the regulation of the NF-κB signaling pathway, and therefore modulates cell proliferation.25,26 Prior studies have suggested that BCL3 gene is a proto-oncogene and may contribute to the malignant development of B-lymphocytes following the chromosome translocation.27–29 This evidence might partly indicate the role of BCL3 in the disease pathogenesis of CLL and its inferior outcomes.
Our study has several limitations. First, this is a single center retrospective study, which may not be generalizable to the entire CLL population. Second, too few patients had other less commonly characterized IGH partners such as MYC, BCL11A, BCL11B, BCL7A, IKZF3, and NFKBIE to be included in the analysis. Finally, we do not have information about next generation sequencing results in this group of patients to correlate if specific recurrent mutations associate preferentially with IGH-BCL2 and IGH-BCL3 translocations and if they impact outcomes of these patients.
In summary, our study is a comprehensive analysis of the clinico-pathologic features and clinical outcome of previously untreated CLL patients with IGH-BCL2 and IGH-BCL3 translocations identified by FISH. CLL patients with IGH-BCL2 have a favorable prognosis, which is similar to those in the Dohner hierarchical FISH low-risk group. In contrast, CLL patients with IGH-BCL3 have a shorter TTT and shorter OS than patients in the Dohner hierarchical FISH high-risk group. These results indicate that the use of an IGH probe in CLL FISH panel to identify this subset of patients may have important prognostic implications.
Supplementary Material
ACKNOWLEDGMENTS
Sameer A. Parikh and Saad S. Kenderian are recipients of the K12 CA090628 grant from the National Cancer Institute (Paul Calabresi Career Development Award for Clinical Oncology). Tait D. Shanafelt and Neil E. Kay are supported by R01CA197120. Daniel Van Dyke is supported by UO1 CA 81534 (NIH, CRC - CLL Research Consortium - Core C).
Funding information
National Cancer Institute, Grant/Award Numbers: K12 CA090628, R01CA197120; NIH Clinical Center, Grant/Award Number: UO1 CA 81534
Research funding has been provided to the institution from Pharma-cyclics, MorphoSys, Janssen, AbbVie, Genentech, GlaxoSmithKline, Celgene, and AstraZeneca for clinical studies in which Sameer A. Parikh is a principal investigator.
Sameer A. Parikh has also participated in Advisory Board meetings of Pharmacyclics, AstraZeneca, Gilead, Genentech and AbbVie (he was not personally compensated for his participation).
Footnotes
CONFLICT OF INTEREST
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Rev 4th ed Lyon: IARC; 2017. [Google Scholar]
- 3.Binet JL, Auquier A, Dighiero G, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48:198–206. [DOI] [PubMed] [Google Scholar]
- 4.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–234. [DOI] [PubMed] [Google Scholar]
- 5.International CLLIPIwg. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17:779–790. [DOI] [PubMed] [Google Scholar]
- 6.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343: 1910–1916. [DOI] [PubMed] [Google Scholar]
- 7.Van Dyke DL, Werner L, Rassenti LZ, et al. The Dohner fluorescence in situ hybridization prognostic classification of chronic lymphocytic leukaemia (CLL): the CLL research consortium experience. Br J Haematol. 2016;173:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapiro E, Radford-Weiss I, Bastard C, et al. The most frequent t(14; 19)(q32;q13)-positive B-cell malignancy corresponds to an aggressive subgroup of atypical chronic lymphocytic leukemia. Leukemia. 2008; 22:2123–2127. [DOI] [PubMed] [Google Scholar]
- 9.Huh YO, Abruzzo LV, Rassidakis GZ, et al. The t(14;19)(q32; q13)-positive small B-cell leukaemia: a clinicopathologic and cytogenetic study of seven cases. Br J Haematol. 2007;136: 220–228. [DOI] [PubMed] [Google Scholar]
- 10.Huh YO, Schweighofer CD, Ketterling RP, et al. Chronic lymphocytic leukemia with t(14;19)(q32;q13) is characterized by atypical morphologic and immunophenotypic features and distinctive genetic features. Am J Clin Pathol. 2011;135:686–696. [DOI] [PubMed] [Google Scholar]
- 11.De Braekeleer M, Tous C, Gueganic N, et al. Immunoglobulin gene translocations in chronic lymphocytic leukemia: a report of 35 patients and review of the literature. Mol Clin Oncol. 2016;4: 682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen-Khac F, Chapiro E, Lesty C, et al. Specific chromosomal IG translocations have different prognoses in chronic lymphocytic leukemia. Am J Blood Res. 2011;1:13–21. [PMC free article] [PubMed] [Google Scholar]
- 13.Schweighofer CD, Huh YO, Luthra R, et al. The B cell antigen receptor in atypical chronic lymphocytic leukemia with t(14;19)(q32;q13) demonstrates remarkable stereotypy. Int J Cancer. 2011;128:2759–2764. [DOI] [PubMed] [Google Scholar]
- 14.Shin SY, Park CJ, Lee KH, Huh J, Chi HS, Seo EJ. An illustrative case of t(14;19)/BCL3 rearrangement as a karyotypic evolution of chronic lymphocytic leukemia. Ann Hematol. 2013;92:1717–1719. [DOI] [PubMed] [Google Scholar]
- 15.Hampel PJ, Chaffee KG, King RL, et al. Liver dysfunction in chronic lymphocytic leukemia: prevalence, outcomes, and pathological findings. Am J Hematol. 2017;92:1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanafelt TD, Geyer SM, Bone ND, et al. CD49d expression is an independent predictor of overall survival in patients with chronic lymphocytic leukaemia: a prognostic parameter with therapeutic potential. BrJ Haematol. 2008;140:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanafelt TD, Parikh SA, Noseworthy PA, et al. Atrial fibrillation in patients with chronic lymphocytic leukemia (CLL). Leuk Lymphoma. 2017;58:1630–1639. [DOI] [PubMed] [Google Scholar]
- 18.Lim KH, Tefferi A, Lasho TL, et al. Systemic mastocytosis in 342 consecutive adults: survival studies and prognostic factors. Blood. 2009; 113:5727–5736. [DOI] [PubMed] [Google Scholar]
- 19.Smoley SA, Van Dyke DL, Kay NE, et al. Standardization of fluorescence in situ hybridization studies on chronic lymphocytic leukemia (CLL) blood and marrow cells by the CLL Research onsortium. Cancer Genet Cytogenet. 2010;203:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowakowski GS, Dewald GW, Hoyer JD, et al. Interphase fluorescence in situ hybridization with an IGH probe is important in the evaluation of patients with a clinical diagnosis of chronic lymphocytic leukaemia. BrJ Haematol. 2005;130:36–42. [DOI] [PubMed] [Google Scholar]
- 21.Baseggio L, Geay MO, Gazzo S, et al. In non-follicular lymphoproliferative disorders, IGH/BCL2-fusion is not restricted to chronic lymphocytic leukaemia. BrJ Haematol. 2012;158:489–498. [DOI] [PubMed] [Google Scholar]
- 22.Put N, Meeus P, Chatelain B, et al. Translocation t(14;18) is not associated with inferior outcome in chronic lymphocytic leukemia. Leukemia. 2009;23:1201–1204. [DOI] [PubMed] [Google Scholar]
- 23.Davids MS, Vartanov A, Werner L, Neuberg D, Dal Cin P, Brown JR. Controversial fluorescence in situ hybridization cytogenetic abnormalities in chronic lymphocytic leukaemia: new insights from a large cohort. BrJ Haematol. 2015;170:694–703. [DOI] [PubMed] [Google Scholar]
- 24.Tang G, Banks HE, Sargent RL, Medeiros LJ, Abruzzo LV. Chronic lymphocytic leukemia with t(14;18)(q32;q21). Hum Pathol. 2013;44: 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dechend R, Hirano F, Lehmann K, et al. The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators. Oncogene. 1999;18:3316–3323. [DOI] [PubMed] [Google Scholar]
- 26.Michaux L, Dierlamm J, Wlodarska I, Bours V, Van den Berghe H, Hagemeijer A. T(14;19)/BCL3 rearrangements in lymphoproliferative disorders: a review of 23 cases. Cancer Genet Cytogenet. 1997;94:36–43. [DOI] [PubMed] [Google Scholar]
- 27.Bours V, Franzoso G, Azarenko V, et al. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. [DOI] [PubMed] [Google Scholar]
- 28.McKeithan TW, Ohno H, Diaz MO. Identification of a transcriptional unit adjacent to the breakpoint in the 14;19 translocation of chronic lymphocytic leukemia. Genes Chromosomes Cancer. 1990;1:247–255. [DOI] [PubMed] [Google Scholar]
- 29.Ohno H, Takimoto G, McKeithan TW. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990;60:991–997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


