Abstract
Autophagy is a conserved catabolic process by which cytoplasmic components are delivered into lysosomes for degradation. Pigment epithelium-derived factor (PEDF) has been reported to be associated with autophagy and can induce p53 expression; however, the mechanism relating PEDF with autophagy in endothelial cells remains poorly understood. The present study aimed to investigate the association between the PEDF-p53-sestrin pathway and autophagy in human umbilical vein endothelial cells (HUVECs). PEDF-induced autophagy was examined by fluorescence microscopy and western blot analysis. p53 small interfering (si)RNA and sestrin2 siRNA were constructed and transfected into HUVECs prior to PEDF treatment. The protein expression levels of microtubule-associated protein light chain 3 (LC3) I, LC3 II and p62 were evaluated by western blot analysis, and the mRNA expression levels of p53 and sestrin2 were determined using reverse transcription-quantitative polymerase chain reaction analysis. The regulation of mechanistic target of rapamycin (mTOR) was reflected by p70S6 kinase (p70S6K) and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) protein expression levels, as determined by western blot analysis. PEDF could induce HUVEC autophagy by sequentially inducing p53 and sestrin2 expression, as observed by fluorescence microscopy and western blot analysis. Conversely, the induction of sestrin2 by PEDF was eliminated by p53 siRNA. In addition, p53 siRNA and sestrin2 siRNA could attenuate PEDF-induced HUVEC autophagy. Inhibition of mTOR may be the mechanism responsible for PEDF-induced autophagy; as p70S6K and 4E-BP1 phosphorylation levels were significantly upregulated in p53 siRNA-treated and sestrin2 siRNA-treated groups. The findings of the present study indicated that PEDF may trigger autophagy in HUVECs by inducing p53 and sestrin2 expression, and inhibiting mTOR expression; these findings may contribute to the improved understanding of diseases, including cancer and atherosclerosis.
Keywords: pigment epithelium-derived factor, autophagy, p53, sestrin2
Introduction
Neovascularization is vital for the progression of cancer and other diseases, as it provides nutrients and oxygen supply (1). The development, growth and spread of cancer has been proposed to be dependent on the establishment of a vascular network (2). Therefore, anti-angiogenic factors may be potential candidates to treat cancer and other diseases involving neovascularization (3).
Autophagy is a conserved catabolic process by which cytoplasmic components are delivered into lysosomes for degradation (4). While angiogenic factors, such as vascular endothelial growth factor, have been reported to induce endothelial cell autophagy under environmental stress to sustain cell survival (5), anti-angiogenic factors, such as endostatin, can also induce autophagy, in order to promote endothelial cell death (6). Therefore, it has been hypothesized that angiogenic factors induce basal-level autophagy that is protective, whereas anti-angiogenic factors induce overactive autophagy that may cause endothelial cell death.
Pigment epithelium-derived factor (PEDF) is a 50-kDa secreted glycoprotein, considered to be the most potent inhibitor of angiogenesis, which is markedly more potent than angiostatin and endostatin (7). Previous studies have identified reduced PEDF expression in a number of angiogenesis-associated diseases, including diabetic retinopathy (8) and solid tumors (9). Exogenous administration of PEDF has been reported to decrease tumor volume and intratumoral microvessel density (10). PEDF exerts anti-angiogenic activities by targeting multiple pathways that induce vascular endothelial cell apoptosis and inhibit capillary morphogenesis (11). While p53 serves an important role in PEDF-induced endothelial cell apoptosis (12), several earlier studies have reported p53 to be associated with autophagy (13–15). Recently, a number of mechanisms have been identified that connect p53 with autophagy. Sestrin is a novel protein that is reported to regulate autophagy and is itself regulated by p53 (16). Maiuri reported that p53 induces autophagy by targeting sestrin2 in cancer cell lines; conversely, knockdown of sestrin2 reduces autophagy (17). D'Amelio reported that activation of the p53-sestrin2 signaling pathway leads to autophagy in RAW264.7 macrophages (18). However, the connection between the p53-sestrin pathway and endothelial autophagy remains to be elucidated. The present study aimed to investigate the association between the PEDF-p53-sestrin pathway and autophagy in human umbilical vein endothelial cells (HUVECs).
Materials and methods
Cell culture
HUVECs were purchased from AllCells (Alameda, CA, USA). Cells were cultured in Medium 200 with Low Serum Growth Supplement (LSGS) kit (supplement contained 1.9% fetal bovine serum, 3 ng/ml basic fibroblast growth factor, 10 µg/ml heparin, 1 µg/ml hydrocortisone and 10 ng/ml epidermal growth factor; Cascade Biologics, Inc., Portland, OR, USA). Culture plates were pre-coated with 2% gelatin. Cells were grown and maintained at 37°C in a humidified atmosphere containing 5% CO2. Treatment with PEDF (PeproTech EC, Ltd., London, UK) (200 ng/ml) was performed on cells (5×105/ml) seeded in LSGS medium for 12 h at 37°C.
Transient transfection and RNA interference
Specific small interfering (si)RNAs, specific for p53 and sestrin-2, or control scrambled RNA were purchased from Shanghai GeneChem Co., Ltd., (Shanghai, China). In total, 4.8×105/well HUVECs were transfected with siRNAs using 10 mg/ml polybrene and enhanced infection solution (cat. no. REVG0002; Shanghai GeneChem Co., Ltd.). After 24 h, siRNA in the medium was substituted with normal Medium 200 with LSGS. Western blot analysis was performed to confirm the effects of p53 and sestrin-2 gene silencing. The siRNA used were as follows: p53siRNA forward, 5′-CUACUUCCUGAAAACAACGdTdT-3′ and reverse, 5′-CGUUGUUUUCAGGAAGUAGdTdT-3′; sestrin-2siRNA forward, 5′-GCGAGAUCAACAAAUUACUTT-3′ and reverse, 5′-AGUAAUUUGUUGAUCUCGCTT-3′; scramble siRNA forward, 5′-CUAACUAUCUCGAACGCAAdTdT-3′ and reverse, 5′-UUGCGUUCGAGAUAGUUAGdTdT-3′. All siRNAs were transfected at a concentration of nmol/ml.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HUVECs, using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. RNA purity was determined using the 260/280 nm absorbance ratio (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). First-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis kit (cat. no. K1622; Fermentas; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA) according to the manufacturer's protocol. In total, 2 µg total RNA was used as template and 10% of the total cDNA was used for each PCR reaction containing Express SYBR Green (Takara Bio, Inc., Otsu, Japan) and PCR Supermix (Fermentas; Thermo Fisher Scientific, Inc.). The PCR primers used to amplify p53 and sestrin2 are listed in Table I. The PCR mix was subjected to 45-cycle amplification at 95°C for 15 sec, 95°C for 5 sec and 60°C for 30 sec. Relative mRNA expression levels of selected genes were normalized to those of GAPDH and quantified using the 2−ΔΔCq method (19). To investigate the role of p53 in PEDF-induced autophagy, time-course analysis of p53 mRNA expression was performed at 0, 4, 8, 12 and 16 h.
Table I.
Primers for reverse transcription-quantitative polymerase chain reaction.
| Target | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| p53 | ACACGCTTCCCTGGATTG | CAAGAAGCCCAGACGGAAAC |
| Sestrin2 | GCATTACCTGCTGCTGCATA | AAGGCCTGGATATGCTCCTT |
| GAPDH | GCTAGGGACGGCCTGAAG | GCCCAATACGACCAAATCC |
Western blot analysis
Cells were scraped into lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing protease inhibitors (Beyotime Institute of Biotechnology). Subsequently, protein concentrations were determined using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). Blocking was performed using 5% non-fat milk at 4°C overnight. Total proteins (22 µg) were separated by 10% SDSPAGE and transferred onto polyvinylidene difluoride membranes (PVDF; EMD Millipore, Billerica, MA, USA). The PVDF membranes were incubated overnight at 4°C with primary antibodies [anti-p53 (1:200; cat. no. 05-224; EMD Millipore), anti-sestrin2 (1:200; cat. no. sc-101249; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-microtubule-associated protein light chain 3 (LC3; 1:200; cat. no. 4108; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-p62 (1:200; cat. no. 5114S; Cell Signaling Technology, Inc.), anti-p70S6 kinase (p70S6K; 1:200; cat. no. sc-8418; Santa Cruz Biotechnology, Inc.), anti-eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1; 1:200; cat. no. 9452; Cell Signaling Technology, Inc.), anti-phosphorylated (p)-p70S6K (1:200; cat. no. sc-8416; Santa Cruz Biotechnology, Inc.) and anti-p-4E-BP1 (1:200; cat. no. 2855; Cell Signaling Technology, Inc.)]. GAPDH (1:300; cat. no. sc-47724; Santa Cruz Biotechnology, Inc.) or β-actin (1:300; cat. no. sc-sc-517582; Santa Cruz Biotechnology, Inc.) served as a protein loading control. Subsequently, the blots were incubated with an appropriate secondary antibody (horseradish peroxidase-conjugated; 1:500; cat. no. sc-2350; Santa Cruz Biotechnology, Inc.) at 20°C for 2 h. Subsequently, the proteins were detected by enhanced chemiluminescence using BeyoECL Plus (Beyotime Institute of Biotechnology) and scanned using Quantity One analysis software, version 4.6 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). To investigate the involvement of p53 in PEDF-induced autophagy, time-course analysis of p53 protein expression was performed at 0, 4, 8, 12 and 16 h.
Green fluorescent protein (GFP)-LC3 transient transfection and immunofluorescence microscopy
LC3 translocation was detected using a GFP-fused LC3 construct (cat. no. GM-1314P101H) purchased from Genomeditech (Shanghai, China). Briefly, 4.8×105 cells/well cells were plated on glass coverslips in 6-well plates. Following attachment for 24 h, 13 µg/ml GFP-LC3 expression plasmids were transfected using Lipofectamine® LTX reagent (Invitrogen; Thermo Fisher Scientific, Inc.), as shown in Fig. S1. A total of 24 h post-transfection, the cells were treated with PBS, PEDF, PEDF + p53 siRNA, PEDF + sestrin2 siRNA or PEDF + cont siRNA. The working concentration of PEDF was 200 ng/ml, and treatment was performed at 37°C for 12 h. The coverslips containing the attached cells were stained with DAPI (1 µg/ml at 37°C for 15 min) and rinsed three times with PBS. The excess buffer was removed and cells were fixed in 2% paraformaldehyde in PBS for 1 h at 37°C and processed for imaging using a fluorescence microscope (magnification, ×200). In total, six randomly-selected fields of view were analyzed.
Statistical analysis
Data from three independent experiments were analyzed and presented as the means ± standard deviation. Two groups were compared using Student's t-test. Differences between groups were analyzed using oneway analysis of variance and the least significant difference post hoc test. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using SPSS v.11.0 software (SPSS, Inc., Chicago, IL, USA).
Results
PEDF can induce autophagy in HUVECs
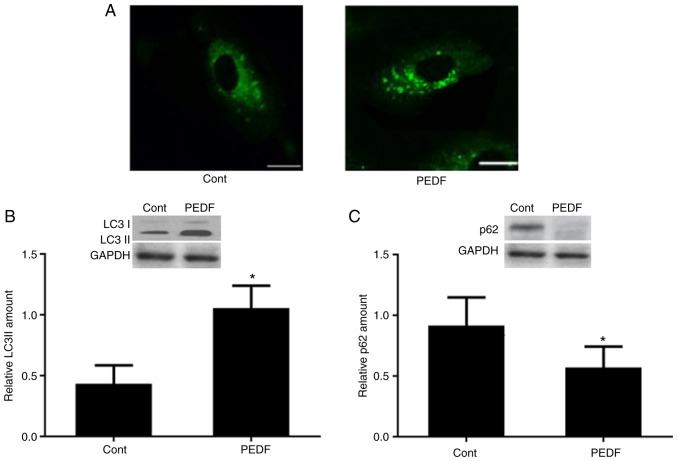
To examine the role of PEDF in autophagy, HUVECs were treated with PEDF for 12 h. A number of GFP-positive autophagosomes were observed in PEDF-treated cells under a fluorescence microscope (Fig. 1A). In addition, compared with control group, PEDF treated group exhibited higher LC3I and LC3II protein expression levels (Fig. 1B) and lower p62 protein expression level (Fig. 1C). These data clearly indicated that PEDF may induce autophagy in HUVECs.
Figure 1.
PEDF can induce autophagy in HUVECs. (A) Fluorescence microscopy of green fluorescent protein-positive autophagosomes in HUVECs treated with or without PEDF for 12 h. Scale bar, 10 µm. Western blotting and corresponding semi-quantitative analysis of (B) LC3 I and LC3 II, and (C) p62 in control groups and PEDF-treated groups. *P<0.05 vs. control. HUVECs, human umbilical vein endothelial cells; LC3, microtubule-associated protein light chain 3; PEDF, pigment epithelium-derived factor.
p53 is critical for PEDF-induced autophagy
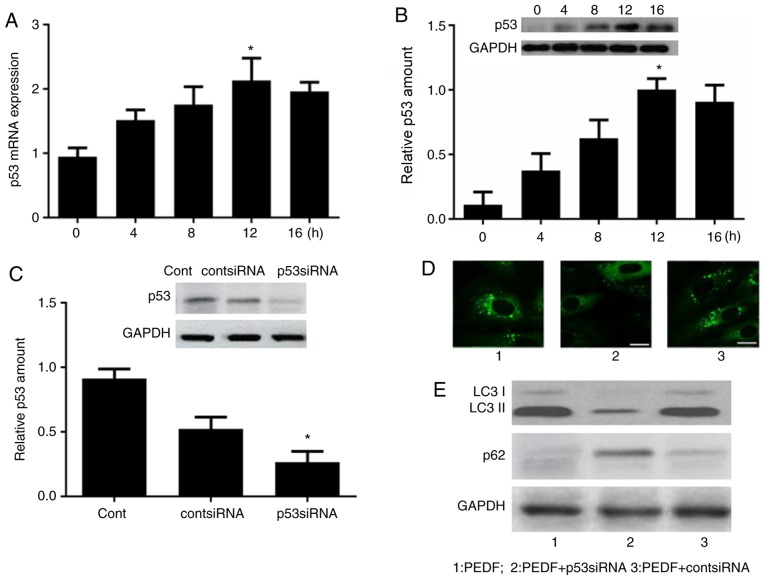
To investigate the involvement of p53 in PEDF-induced autophagy, time-course analysis of p53 mRNA and protein expression was performed by RT-qPCR and western blotting, respectively. As demonstrated in Fig. 2A and B, the mRNA and protein expression levels of p53 were increased at 4 h, peaked at 12 h and were subsequently decreased at 16 h.
Figure 2.
p53 is critical for PEDF-induced autophagy. Human umbilical vein endothelial cells were treated with PEDF for 0, 4, 8, 12 and 16 h. (A) p53 mRNA and (B) protein expression was detected by reverse transcription-quantitative polymerase chain reaction and western blot analysis. *P<0.05 vs. 0 h. (C) p53 siRNA was established and western blot analysis was performed to confirm the effect of p53 gene silencing. *P<0.05 vs. control group. (D) Fluorescence microscopy of green fluorescent protein-positive autophagosomes in the PEDF, PEDF + p53 siRNA and PEDF + cont siRNA groups. Scale bar, 10 µm. (E) Western blot analysis in the PEDF, PEDF + p53 siRNA and PEDF + cont siRNA groups. LC3, microtubule-associated protein light chain 3; PEDF, pigment epithelium-derived factor; si, small interfering.
To validate the hypothesis that p53 is involved in mediating PEDF-induced autophagy, p53 siRNA was established and western blot analysis was performed to confirm the effect of p53 gene silencing (Fig. 2C). HUVECs were transfected with p53 siRNA, prior to PEDF treatment, to inhibit p53 expression. Fluorescence microscopy revealed a punctuate pattern of autophagosomes in PEDF-treated cells and a diffuse pattern in the PEDF + p53 siRNA-treated cells (Fig. 2D). Furthermore, western blot analysis of LC3 and p62 demonstrated attenuation of PEDF-induced LC3 accumulation and p62 downregulation in cells transfected with p53 siRNA prior to PEDF treatment (Fig. 2E).
PEDF induces sestrin2 gene expression and activates sestrin2 transcription via p53
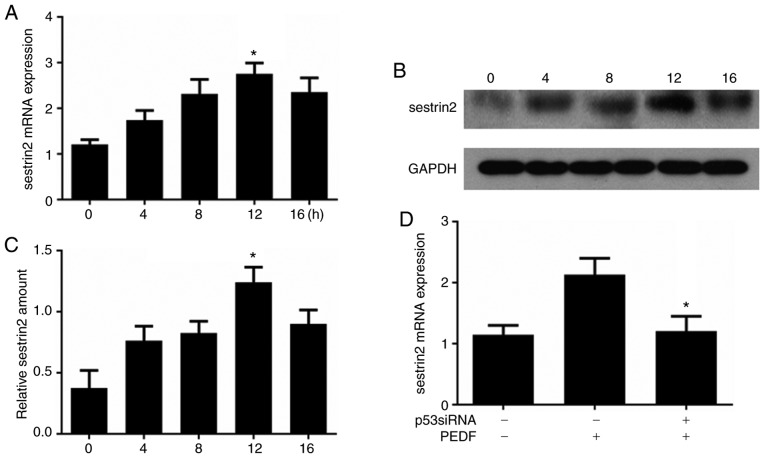
To examine the influence of PEDF on sestrin2 expression, time-course analysis of sestrin2 mRNA expression was performed by RT-qPCR; the results demonstrated that the mRNA expression levels of sestrin2 were increased at 4 h, peaked at 12 h and were subsequently decreased at 16 h (Fig. 3A). Similar results were also observed for sestrin2 protein expression in HUVECs following PEDF treatment (Fig. 3B). Semi-quantitative analysis demonstrated that sestrin2 protein expression increased at 4 h, peaked at 12 h and was eventually decreased at 16 h (Fig. 3C). The effects of p53 on PEDF-induced sestrin2 overexpression were further investigated; the results revealed that transfection of HUVECs with p53 siRNA attenuated PEDF-induced sestrin2 overexpression (Fig. 3D).
Figure 3.
PEDF induces sestrin2 gene expression and activates sestrin2 transcription through p53. (A) HUVECs were treated with PEDF for 0, 4, 8, 12 and 16 h, and sestrin2 mRNA expression was detected by RT-qPCR. *P<0.05 vs. 0 h. (B) Western blot analysis of sestrin2 protein expression in PEDF-treated HUVECs at different time points. (C) Semi-quantitative analysis of blot in (B). *P<0.05 vs. 0 h. (D) p53 siRNA-transfected HUVECs were treated with PEDF for 12 h and sestrin2 was detected by RT-qPCR. *P<0.05 vs. PEDF-treated group. HUVECs, human umbilical vein endothelial cells; PEDF, pigment epithelium-derived factor; si, small interfering; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
PEDF triggers sestrin2-dependent autophagy
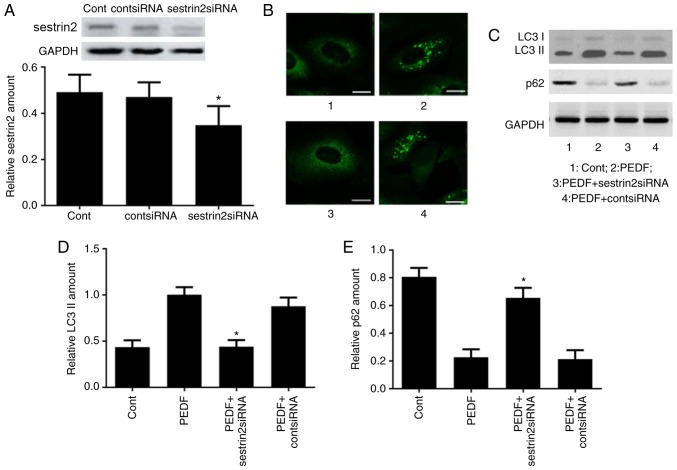
To verify that sestrin2 is critical in PEDF-induced autophagy, HUVECs were transfected with sestrin2 siRNA to inhibit sestrin2 expression. Western blot analysis confirmed the effect of sestrin2 gene silencing (Fig. 4A). Fluorescence microscopy demonstrated fewer GFP-positive autophagosomes in HUVECs that were transfected with sestrin2 siRNA prior to PEDF treatment (Fig. 4B). Western blotting was performed to evaluate LC3 and p62 expression in HUVECs from different groups (Fig. 4C); semi-quantitative analysis revealed that LC3 II expression was decreased (Fig. 4D), whereas p62 expression was increased (Fig. 4E) in the PEDF + sestrin2 siRNA-treated group compared with in the PEDF-treated group, which indicated that sestrin2 expression may regulate PEDF-induced autophagy.
Figure 4.
PEDF trigger sestrin2-dependent autophagy. (A) Western blot analysis was performed to confirm the effect of sestrin2 gene silencing. *P<0.05 vs. control group. (B) Fluorescence microscopy of green fluorescent protein-positive autophagosomes in the control, PEDF, PEDF + cont siRNA and PEDF + sestrin2 siRNA groups. Scale bar, 10 µm. (C) Western blot analysis of LC3 I, LC3 II and p62 in the control, PEDF, PEDF + cont siRNA and PEDF + sestrin2 siRNA groups. (D and E) Semi-quantitative analysis of LC3II and p62 blots in (C). *P<0.05 vs. PEDF-treated group. LC3, microtubule-associated protein light chain 3; PEDF, pigment epithelium-derived factor; si, small interfering.
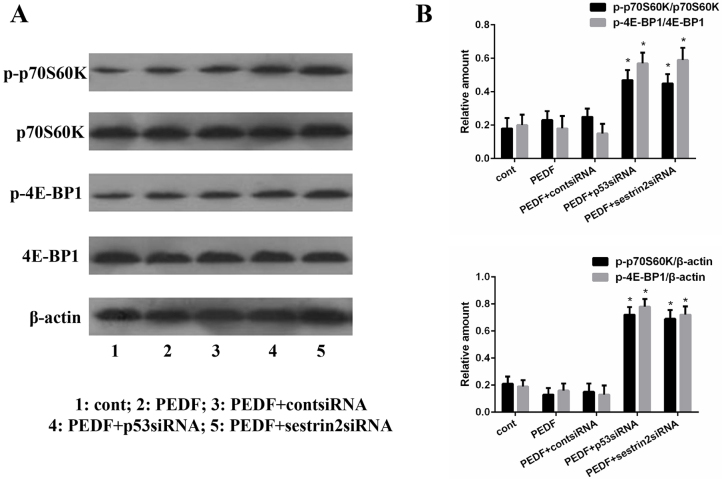
PEDF-induced HUVEC autophagy is mediated by inhibition of mechanistic target of rapamycin (mTOR)
To explore whether PEDF induces autophagy by regulating mTOR, p70S6K and 4E-BP1, which are targets of mTOR and are phosphorylated at a number of sites, were analyzed. Western blot analysis was conducted to examine alterations in the phosphorylation of p70S6K and 4E-BP1 in HUVECs following transfection with p53 siRNA and sestrin2 siRNA (Fig. 5A). The results revealed that p-p70S6K and p-4E-BP1 expression levels were significantly upregulated in p53 siRNA- and sestrin2 siRNA-treated groups (Fig. 5B), thus indicating that PEDF may regulate mTOR by activating the p53-sestrin2 signaling pathway.
Figure 5.
PEDF-induced human umbilical vein endothelial cell autophagy is mediated through inhibition of mechanistic target of rapamycin. (A) Western blot analysis was conducted to examine the alterations in p70S6K and 4E-BP1 phosphorylation in the control, PEDF, PEDF + cont siRNA, PEDF + p53 siRNA and PEDF + sestrin2 siRNA groups. (B) Semi-quantitative analysis of blots in (A). *P<0.05 vs. PEDF-treated group. 4E-BP1, eukaryotic translation initiation factor 4E-binding protein 1; p, phosphorylated; p70S6K, p70S6 kinase; PEDF, pigment epithelium-derived factor; si, small interfering.
Discussion
PEDF is an important endogenous anti-angiogenesis factor that induces apoptosis of endothelial cells and causes endothelial cell death (20). Recently, studies have demonstrated that controlled autophagy is a cytoprotective response (21–23); however, unchecked autophagy may trigger cell death by activating p53 (24). While PEDF has been reported to be associated with autophagy and can induce p53 expression, the mechanism underlying PEDF and autophagy in endothelial cells is poorly understood. The present study identified that PEDF can induce autophagy in HUVECs by activating the p53-sestrin2 signaling pathway.
The current study demonstrated the participation of p53 in PEDF-induced autophagy in HUVECs. It was identified that PEDF increased the expression of p53 in HUVECs. To investigate the role of p53 in PEDF-induced autophagy, p53 siRNA was used to inhibit p53 signaling; the results demonstrated that p53 siRNA prevented autophagy, thus indicating that p53 mediated autophagy. Similarly, other studies have demonstrated the capability of p53 to induce autophagy; activation of p53 can cause excess autophagy in cancer cells (25–27). However, inhibition of p53 has also been reported to enhance autophagy under environmental stress conditions (28). Therefore, the present study hypothesized that PEDF may induce excess autophagy via p53 to induce HUVEC death, which may prevent the progression of atherosclerotic plaque formation and tumor growth. The association between PEDF and p53 in HUVEC autophagy requires further exploration.
The present study also explored the role of a downstream factor of p53 in autophagy and the results revealed that PEDF may upregulate sestrin2 expression via p53, thus suggesting sestrin2 as a major target in p53-mediated autophagy. The present study also revealed that sestrin2 siRNA was able to reduce the number of autophagosomes, and attenuate LC3 conversion and p62 degradation, thus suggesting that sestrin2 may serve a critical role in PEDF-induced autophagy. Furthermore, disruption of the sestrin2 gene in HUVECs attenuated the inhibition of p70S6K and 4E-BP1 activities. Notably, PEDF treatment alone was not able to inhibit such effect, possibly due to the influence of PEDF on p70S6K and 4E-BP1 via other signaling pathways and further studies are required to understand this effect. Previous studies have revealed the critical role of sestrin2 in p53 and mTOR signaling (29–31). An association between p53 and autophagy, including sestrin2, 5′AMP-activated protein kinase and DNA damage-regulated autophagy modulator 1, has previously been reported (17). In addition, sestrin2 interacts with tuberous sclerosis (TSC)1:TSC2 complex to regulate mTOR activity in a TSC2-dependent manner (29).
mTOR is a serine/threonine kinase, which acts as a master regulator of cellular metabolism (32). mTOR regulates cell growth and proliferation in response to a wide range of cues, and its signaling pathway is deregulated in a number of human diseases (33). p53 deficiency and mTOR signaling activation are hallmarks of human cancer. Several mechanisms account for mTOR activation in cancer, including the activation of Ras, phosphatidylinositol 3-kinase and protein kinase B, and inactivation of tumor suppressors that negatively regulate these molecules: Phosphatase and tensin homolog, TSC1, TSC2 and liver kinase B1. mTOR has also been reported to serve a key role in regulating autophagy (34). In this study, p70S6K and 4E-BP1, which are targets of mTOR and phosphorylated at a number of sites, were used to detect mTOR regulation. It was revealed that p53 siRNA and sestrin2 siRNA increased p70S6K and 4E-BP1 phosphorylation levels, thus demonstrating that PEDF inhibits mTOR signaling by activating p53 and sestrin2.
PEDF is involved in in the progression of a number of diseases. To the best of our knowledge, this is the first study to reveal that HUVEC autophagy is induced by PEDF via p53-sestrin2 signaling. The findings may contribute to improved understanding of various diseases, including cancer and arthrosclerosis. p53 has been revealed to serve an important role in PEDF-induced endothelial cell apoptosis (20); therefore, the association between p53 induced-autophagy and p53 induced-apoptosis will be investigated in a future study. The present study is a preliminary examination that established a link between the p53 signaling pathway and PEDF-induced autophagy in HUVECs; in vivo experiments and further in vitro studies are required to detect the detailed mechanism underlying PEDF, p53/sestrin2 signaling and autophagy.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The study was supported by funding from the National Natural Science Foundation of China (grant no. 81473502).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
TC and TL conceived and designed the present study. TC and TL performed the experiments. TL analyzed the data. JW analyzed the data and provided reagents and materials. TC wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kozin SV, Duda DG, Munn LL, Jain RK. Neovascularization after irradiation: What is the source of newly formed vessels in recurring tumors? J Natl Cancer Inst. 2012;104:899–905. doi: 10.1093/jnci/djs239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Small DM, Burden RE, Jaworski J, Hegarty SM, Spence S, Burrows JF, McFarlane C, Kissenpfennig A, McCarthy HO, Johnston JA, et al. Cathepsin S from both tumor and tumor-associated cells promote cancer growth and neovascularization. Int J Cancer. 2013;133:2102–2112. doi: 10.1002/ijc.28238. [DOI] [PubMed] [Google Scholar]
- 3.Lee S, Wurtzel JG, Singhal SS, Awasthi S, Goldfinger LE. RALBP1/RLIP76 depletion in mice suppresses tumor growth by inhibiting tumor neovascularization. Cancer Res. 2012;72:5165–5173. doi: 10.1158/0008-5472.CAN-12-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida GJ. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: From pathophysiology to treatment. J Hematol Oncol. 2017;10:67. doi: 10.1186/s13045-017-0436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanton MJ, Dutta S, Zhang H, Polavaram NS, Leontovich AA, Hönscheid P, Sinicrope FA, Tindall DJ, Muders MH, Datta K. Autophagy control by the VEGF-c/NRP-2 axis in cancer and its implication for treatment resistance. Cancer Res. 2013;73:160. doi: 10.1158/0008-5472.CAN-11-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen TM, Subramanian IV, Xiao X, Ghosh G, Nguyen P, Kelekar A, Ramakrishnan S. Endostatin induces autophagy in endothelial cells by modulating Beclin 1 and beta-catenin levels. J Cell Mol Med. 2009;13:3687–3698. doi: 10.1111/j.1582-4934.2009.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becerra SP, Notario V. The effects of PEDF on cancer biology: Mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–271. doi: 10.1038/nrc3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elahy M, Baindur-Hudson S, Cruzat VF, Newsholme P, Dass CR. Mechanisms of PEDF-mediated protection against reactive oxygen species damage in diabetic retinopathy and neuropathy. J Endocrinol. 2014;222:R129–R139. doi: 10.1530/JOE-14-0065. [DOI] [PubMed] [Google Scholar]
- 9.Nwani NG, Deguiz ML, Jimenez B, Vinokour E, Dubrovskyi O, Ugolkov A, Mazar AP, Volpert OV. Melanoma cells block PEDF production in fibroblasts to induce the tumor-promoting phenotype of cancer-associated fibroblasts. Cancer Res. 2016;76:2265–2276. doi: 10.1158/0008-5472.CAN-15-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimura T, Yamagishi S, Ueda S, Fukami K, Shibata R, Matsumoto Y, Kaida Y, Hayashida A, Koike K, Matsui T, et al. Administration of pigment epithelium-derived factor, (PEDF) reduces proteinuria by suppressing decreased nephrin and increased VEGF expression in the glomeruli of adriamycin-injected rats. Nephrol Dial Transplant. 2008;24:1397–1406. doi: 10.1093/ndt/gfn659. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Wei T, Jiang X, Li Z, Cui H, Pan J, Zhuang W, Sun T, Liu Z, Zhang Z, Dong H. PEDF and 34-mer inhibit angiogenesis in the heart by inducing tip cells apoptosis via up-regulating PPAR-γ to increase surface FasL. Apoptosis. 2016;21:60–68. doi: 10.1007/s10495-015-1186-1. [DOI] [PubMed] [Google Scholar]
- 12.Gao Q, Zhu X, Chen J, Mao C, Zhang L, Xu Z. Upregulation of P53 promoted G1 arrest and apoptosis in human umbilical cord vein endothelial cells from preeclampsia. J Hypertens. 2016;34:1380–1388. doi: 10.1097/HJH.0000000000000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. Kimmelman, Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mrakovcic M, Fröhlich LF. p53-mediated molecular control of autophagy in tumor cells. Biomolecules. 2018;8:E14. doi: 10.3390/biom8020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, Xiao H, Li J, Zhang J, Li Y, Zhang J, Wang X, Bai X, Tao K, Hu D, Guan H. Wild-type p53-modulated autophagy and autophagic fibroblast apoptosis inhibit hypertrophic scar formation. Lab Invest. 2018;98:1423–1437. doi: 10.1038/s41374-018-0099-3. [DOI] [PubMed] [Google Scholar]
- 16.Wang N, Pan W, Zhu M, Zhang M, Hao X, Liang G, Feng Y. Fangchinoline induces autophagic cell death via p53/sestrin2/AMPK signalling in human hepatocellular carcinoma cells. Br J Pharmacol. 2011;164:731–742. doi: 10.1111/j.1476-5381.2011.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 18.D'Amelio M, Cecconi F. A novel player in the p53-mediated autophagy: Sestrin2. Cell Cycle. 2009;8:1467. [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Ho TC, Chen SL, Yang YC, Liao CL, Cheng HC, Tsao YP. PEDF induces p53-mediated apoptosis through PPAR gamma signaling in human umbilical vein endothelial cells. Cardiovasc Res. 2007;76:213–223. doi: 10.1016/j.cardiores.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 21.Sumitomo Y, Koya J, Nakazaki K, Kataoka K, Tsuruta-Kishino T, Morita K, Sato T, Kurokawa M. Cytoprotective autophagy maintains leukemia-initiating cells in murine myeloid leukemia. Blood. 2016;128:1614–1624. doi: 10.1182/blood-2015-12-684696. [DOI] [PubMed] [Google Scholar]
- 22.Miao L, Dong Y, Zhou FB, Chang YL, Suo ZG, Ding HQ. Protective effect of tauroursodeoxycholic acid on the autophagy of nerve cells in rats with acute spinal cord injury. Eur Rev Med Pharmacol Sci. 2018;22:1133–1141. doi: 10.26355/eurrev_201802_14402. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Zhu Y, Zhang W, Peng X, Zhou J, Li F, Han B, Liu X, Ou Y, Yu X. Delphinidin induced protective autophagy via mTOR pathway suppression and AMPK pathway activation in HER-2 positive breast cancer cells. BMC Cancer. 2018;18:342. doi: 10.1186/s12885-018-4231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis M, Stern O, Ashur-Fabian O. The double benefit of Spalax p53: Surviving underground hypoxia while defying lung cancer cells in vitro via autophagy and caspase-dependent cell death. Oncotarget. 2016;7:63242–63251. doi: 10.18632/oncotarget.11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakhar R, Paul S, Bhardwaj M, Kang SC. Astemizole-Histamine induces Beclin-1-independent autophagy by targeting p53-dependent crosstalk between autophagy and apoptosis. Cancer Lett. 2015;372:89–100. doi: 10.1016/j.canlet.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 26.Haque E, Kamil M, Irfan S, Sheikh S, Hasan A, Nazir A, Mir SS. Blocking mutation independent p53 aggregation by emodin modulates autophagic cell death pathway in lung cancer. Int J Biochem Cell Biol. 2018;96:90–95. doi: 10.1016/j.biocel.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, Li R, Guo X, Pang L, Zang Y, Liu K, Chen D. Chen, DNA damage regulated autophagy modulator 1 recovers the function of apoptosis-stimulating of p53 protein 2 on inducing apoptotic cell death in Huh7.5 cells. Oncol Lett. 2018;15:9333–9328. doi: 10.3892/ol.2018.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang QY, Jin R, Zhang X, Sheng JP, Yu F, Tan RX, Pan Y, Huang JJ, Kong LD. The putative oncotarget CSN5 controls a transcription-uncorrelated p53-mediated autophagy implicated in cancer cell survival under curcumin treatment. Oncotarget. 2016;7:69688–69702. doi: 10.18632/oncotarget.11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayaraj P, Sen S, Rangarajan S, Ray N, Vasu K, Singh VK, Phartyal R, Yadav S, Verma A. Immunohistochemical evaluation of stress-responsive protein sestrin2 and its correlation with p53 mutational status in eyelid sebaceous gland carcinoma. Br J Ophthalmol. 2018;102:848–854. doi: 10.1136/bjophthalmol-2017-311283. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Zhang Q, Luo L, Ning B, Fang Y. β-asarone inhibited cell growth and promoted autophagy via P53/Bcl-2/Bclin-1 and P53/AMPK/mTOR pathways in Human Glioma U251 cells. J Cell Physiol. 2018;233:2434–2443. doi: 10.1002/jcp.26118. [DOI] [PubMed] [Google Scholar]
- 32.Kassem L, Abdel-Rahman O. Targeting mTOR pathway in gynecological malignancies: Biological rationale and systematic review of published data. Crit Rev Oncol Hematol. 2016;108:1–12. doi: 10.1016/j.critrevonc.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munson MJ, Ganley IG. MTOR, PIK3C3 and autophagy: Signaling the beginning from the end. Autophagy. 2015;11:2375–2376. doi: 10.1080/15548627.2015.1106668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.